Abstract
Previous studies have demonstrated leukemic complications in mice after high-copy retroviral gene transfer of the multidrug resistance 1 (MDR1) cDNA, encoding a membrane-located efflux pump expressed in hematopoietic stem cells. In contrast, no such complications or MDR1-associated alterations of hematopoiesis were observed in numerous other studies exploring MDR1 gene transfer into cell lines, mice, dogs, nonhuman primates, and human subjects. Here, we show that leukemias associated with retroviral expression of MDR1 depend on high vector dose, and involve the selection of clones with combinatorial insertional mutagenesis of proto-oncogenes or other signaling genes. Compared with insertion patterns in normal long-term repopulating hematopoietic cells, such hits were overrepresented in leukemic clones, pointing to a causal role. A similar constellation of insertion sites was also observed in a leukemia arising after high-copy retroviral gene transfer of a fluorescent protein. Spectral karyotyping demonstrated additional chromosomal translocations in a subset of cases, indicative of secondary genetic instability. We also show that insertional mutants can be amplified in vitro prior to transplantation. On the basis of these findings, we suggest the use of preclinical dose-escalation studies to define a therapeutic index for retroviral transgene delivery.
Introduction
Multidrug resistance 1 (MDR1) encodes P-glycoprotein,1 a member of the superfamily of adenosine triphosphate (ATP)–binding cassette (ABC) transporters that is involved in the rhodamine-dull phenotype of primitive hematopoietic stem cells.2 This gene is subject to a tight developmental control, and silenced in most differentiating blood cell lineages except some T-cell subsets.2 Besides conferring resistance to a wide spectrum of clinically important anticancer drugs,1 MDR1 expression may also affect some endogenous substrates involved in apoptosis induction.3 Accordingly, an antiapoptotic function has also been associated with MDR1.2,3 Since transgenic mice overexpressing MDR1 in hematopoietic cells were shown to acquire cancer drug resistance without obvious alterations of multilineage hematopoiesis,4 numerous studies including clinical trials have been conducted to explore the utility of this gene as a selectable marker expressed from retroviral vectors. This work has shown the ability to select hematopoietic cells retrovirally overexpressing MDR1 by systemic treatment with myelotoxic doses of anthracyclines, vinca alkaloids, or taxanes, in cell lines, mice, and dogs5-7 (and references therein). Studies in nonhuman primates and clinical trials were less successful, providing little evidence of a selective advantage despite detectable gene transfer of MDR1.8-11 In part, this could be explained by insufficient levels of gene transfer and expression available at that time in large animals and humans.
The use of MDR1 in human gene therapy was re-evaluated when a leukemia-like disorder was repeatedly observed after retroviral gene transfer into murine hematopoietic cells.12-14 These studies showed a tight correlation of cellular transformation with functional overexpression of MDR1, whereas cells transduced with control vectors lacking a functional MDR1 gene behaved normally. Experiments were performed with HaMDR1, a retroviral vector encoding a “splice-corrected” version of MDR1 under control of murine leukemia virus (MLV) long terminal repeats (LTR). Using identical vectors but less efficient gene transfer conditions, leukemic complications could not be reproduced in nonhuman primates, despite systemic challenge with growth factors.10
As MDR1-associated leukemias in mice were shown12,13 to be associated with high transgene copy numbers (typically > 10 per clone), we hypothesized that insertional activation of protooncogenes might be required for their manifestation. This may well explain the discrepancies noted between the different studies performed to date.
Our hypothesis follows the concept that retroviral vector insertions near a proto-oncogene or another locus involved in regulating cell growth and proliferation may contribute to leukemogenesis, especially when collaborating with signal alterations evoked by the retrovirally encoded transgene product.15,16 Only 2 examples for such combinatorial side effects have been published so far. First, we reported a murine monocytic leukemia occurring after a single insertion of a retroviral vector expressing the cytoplasmically deleted p75 neurotrophin receptor had activated the Evi1 proto-oncogene.15 Shortly thereafter, 2 cases of malignant lymphoproliferation were observed in an otherwise successful clinical trial performed to correct an inborn deficiency of the interleukin 2 receptor gamma chain (IL2RG).17 In both patients, the proto-oncogene LMO2 was upregulated in the leukemic clone following IL2RG vector insertion into this locus.17 Suggesting a predictive value of the mouse model for human insertional leukemogenesis, insertions in both Il2rg and Lmo2 were recently observed in murine lymphoid malignancies induced by replication-competent retrovirus.18 Thus, 2 collaborating signal alterations may be required for leukemia initiation, neither of which would be sufficient to elicit complications.
To date, combinatorial insertional events in crucial growth-regulatory genes have not been reported when using replication-deficient retroviral vectors. Such constellations were considered highly unlikely, based on hypotheses that proto-oncogene activation by a single retroviral insertion may occur in the range of 10-6 or less. However, recent insights have revealed that the probability of insertion in a gene of interest not only depends on the accessibility (transcriptional activity) of the locus,19 but also on the type of gene transfer vector used.20 MLV-based vectors not only show a preference for insertions in the 5-kb window surrounding the transcriptional start site of actively expressed genes,20 but may also affect target gene regulation from larger distances (> 100 kb). Considering that “cancer genes” comprise at least 1% of the genome of higher mammals,21 “dangerous” retroviral insertions may in fact occur with a similar frequency.16 In hematopoietic stem cells, which were found to express about 1000 different signaling genes (∼ 50% of their transcriptome),22 the risk may even be higher. As predicted by Poisson distribution and recently demonstrated in human hematopoietic cells, clones with multiple vector insertions may occur.23-25 With an average transgene copy number of only 2 per cell and unrestricted transduction conditions, 5% of exposed cells may contain 5 or more insertions.25 Considering the treatment of more than 108 cells per patient in clinical gene therapy, this may indeed lead to combinatorial proto-oncogene hits. Predictive preclinical models are thus urgently required, both for risk assessment and prevention. The present study reveals that dose-escalation studies are an important step in this direction, potentially allowing to establish a therapeutic index for retroviral transgene delivery.
Materials and methods
Animals
C57BL/6J-CD45.1 mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and represented the recipient population. C57BL/6J mice with the wild-type CD45 (CD45.2) were used as donor animals. All mice were bred and kept in the pathogen-free animal facilities of the Heinrich-Pette-Institute, Hamburg, Germany. Animal experiments were approved by the local ethical committee and performed according to their guidelines.
Retroviral vectors and vector production
The retroviral vector HaMDR1 and the HaMDR1 GP+E86 producer cells were generated as described.13 The HaMDR1 vector backbone was derived from the Harvey murine sarcoma retrovirus. The MDR1 cDNA in the HaMDR1 vector encodes an MDR-1 protein with the wild-type glycine as amino acid 185 and silent mutations in cryptic splice sites at positions + 336 and + 2319. The SF91m3 vector has been described.26 It contains a partially splice corrected human MDR-1 cDNA (correction at position + 2319). The SF91 backbone contains the spleen focus-forming virus LTR and modifications of RNA processing elements as described.27 SF91PdsRED2 expresses the dsRED2 fluorescent protein in this backbone. SF11tCD34 encoding human truncated CD34 has been described.28 Cell-free supernatants were generated by transient transfection of Phoenix gp packaging cells by using a packaging construct coding for the ecotropic envelope protein. Virus titrations were performed on SC1 fibroblasts (SFa11tCD34, SF91PdsRED2) or NIH 3T3 fibroblasts (SF91m3, HaMDR1).
Bone marrow transduction and transplantation
Lineage-negative bone marrow (BM) cells were transduced as previously described.29 Briefly, lineage-negative cells were isolated from complete BM by magnetic sorting using lineage-specific antibodies (Gr1, CD11b, CD45R/B220, CD3e, TER-119; Pharmingen, Hamburg, Germany). The lineage-negative cell population was then cultured for 3 days in StemSpan HS2000 medium (CellSystems, St Katharinen, Germany) containing 50 ng/mL murine stem cell factor (mSCF), 100 ng/mL human Flt-3 ligand, 100 ng/mL human interleukin-11 (IL-11), 20 ng/mL murine IL-3, and 1% penicillin/streptomycin, 2 mM glutamine. Lineage-negative cells were infected on the 2 following days (days 4 and 5) using plates preloaded with Retronectin (TaKaRa, Otsu, Japan) and vector. For preparation of plates, the bottom surface of the wells was coated with 10 μg/cm2 Retronectin and virus loaded by centrifugation of viral supernatants at 900g to 1000g for 30 minutes at 4°C. Virus preloading was repeated up to 4 times. Transgene expression was measured on day 6 just prior to transplantation.
For retrovirus infection by coculture, unseparated BM cells were seeded on irradiated (15 Gy) HaMDR1 virus producing GP+E86 cells as described,13 using Dulbecco modified essential medium containing 15% fetal calf serum (FCS), 10 ng/mL murine IL-3, 5 ng/mL human IL-6, 50 ng/mL mSCF, 1% penicillin/streptomycin, 2 mM glutamine, 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), and 4 μg/mL protamine sulfate. In vitro–expanded BM cells were cultured in the same medium without protamine sulfate with medium changes every 3 days. In the case of lineage-negative BM cells, 5 × 105 to 8 × 105 cells were transplanted into lethally (9 Gy) irradiated mice. Mice that underwent transplantation with in vitro–expanded BM cells received 5 × 106 expanded cells plus 1 × 105 fresh marrow cells.
Analysis of MDR1 transgene expression
MDR1 transgene expression was measured by rhodamine 123 efflux activity.26 Cells were incubated in medium containing 100 ng/mL rhodamine at 37°C for 30 minutes. After 2 washes, cells were further incubated for 1 hour to allow rhodamine efflux. Rhodamine-positive and -negative cells were detected by flow cytometry using a FACSCalibur (Becton Dickinson, Heidelberg, Germany). In mice, efflux activity in donor cells was compared with efflux activity in recipient cells as could be distinguished by the CD45 chimerism.
Analysis of leukemias
Mice were killed when symptomatic, and were macroscopically examined for pathologic abnormalities. Enlarged organs were weighed. A panel of tissues was collected, fixed in 4% formalin, and paraffin-embedded for histologic examinations. Cells from BM, spleen, thymus (if enlarged), and blood were subjected to flow cytometry. After red blood cell lysis, cells were stained with lineage-specific antibodies against Gr1, CD11b (myeloid cells), CD19 (B cells), Ter119 (erythroid cells), CD3, CD4, CD8 (T cells), Sca1, and c-Kit (Pharmingen, Hamburg, Germany). Antibodies were usually directly linked to fluorescein isothiocyanate (FITC), phycoerythrin (PE), or allophycocyanin (APC) fluorochromes. Dead cells were excluded by propidium iodide staining. Leukocyte morphology was also evaluated in Giemsa-stained blood smears and cytospins of BM and spleen cells. Blood cell counts were collected by microscopic counting or by using an automatic analyzer (MAXM; Coulter, Miami, FL). Mice that did not develop leukemia were analyzed according to this protocol after an observation period of 30 to 35 weeks. Secondary transplantations were performed using selected mice of the high-dose gene transfer groups as donors. BM cells of 1 animal (2 × 106 each) were transplanted into 2 secondary C57BL/6J-CD45.1 recipients.
Spleen colony-forming unit (CFU-S) assay
Mononuclear BM cells (5 × 104 to 5 × 105) harvested from primary recipients were transplanted by tail vein injection into lethally irradiated (9 Gy) recipients. After 12 days, well-isolated spleen colonies were dissected from spleens. Cells were suspended by passing through a G18 needle for preparation of genomic DNA.
Southern blot
Genomic DNA for Southern blot analysis was isolated from frozen spleen tissues. Tissues were homogenized and digested with 0.8 mg/mL Proteinase K at 50°C overnight. Samples were treated with 10 μg/mL RNAse A and subsequently purified by phenol/chlorophorm extraction. DNA was precipitated and resolved in Tris/EDTA (ethylenediaminetetraacetic acid) buffer. Genomic DNA (10 μg) was digested with appropriate enzymes and Southern blot performed according to standard protocols. DNA of MDR1 transgenic cells was hybridized with a 1200-bp probe of the human MDR1 cDNA (108-1294, GenBank accession no. AF016 535). The dsRED2 transgene was detected using the full-length 700-bp cDNA.
Real-time quantitative PCR
DNA of peripheral blood leukocytes was purified using QIAmp Blood DNA Preparation Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. TaqMan real-time polymerase chain reaction (PCR) was performed using the ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems, Weiterstadt, Germany). Primers mdr-f (5′-AGAAAGCGAAGCAGTGGTTCA-3′) and mdr-r (5′-CGAACTGTAGACAAACGATGAGCTA-3′) amplified a 90-bp fragment of the MDR1 cDNA, which was detected by the FAM-labeled TaqMan probe mdr-p (5′-TGGTCCGACCTTTCTGGCCTTATCCA-3′). Primers epo-f (5′-TGCATCGACCTTTCTGGCCTTATCCA-3′) and epo-r (5′-GGTGCGCTCCTGGACGTAG-3′) amplified a 78-bp control fragment of the murine erythropoietin receptor gene, which was detected by the VIC-labeled TaqMan probe epo-p (5′-CCCGACCACATGAAGCAGCACGAC-3′). Quantitative PCR for dsRED2 was performed on an i-Cycler (BioRad) using QuantiTect SYBR Green (Qiagen). The dsRED2-specific primers (forward 5′-CAAGTGGGAGCGCGTGATGAA-3′ and reverse 5′-GGGCCGTCGGAGGGGAAGTT-3′) amplified a 130-bp fragment. The dsRED2-specific signal was normalized by the signal of a housekeeping gene, flk-1 intron enhancer (GenBank accession no. AF061 804; bases 352-459; forward 5′-GTGAATTGCAGAGCTGTTGTGTTG-3′ and reverse 5′-ATTCATTGTATAAAGGTGGGATTG-3′). Quantification was calculated using the comparative CT method.
LAM-PCR and LM-PCR
Linear amplification-mediated (LAM)–PCR and ligation-mediated (LM)–PCR were performed as described.30,31 For LM-PCR of HaMDR1-transduced cells, DNA was digested with 2.5 U of HpyCH4V (New England BioLabs, Frankfurt a. Main, Germany). For primer extension, 0.25 pmol biotinylated retroviral primer A1HARV (5′-AGCTGTTCTATCTGTTCTTGGCCCT-3′) was used. The first PCR (95°C for 5 minutes; 95°C for 1 minute, 55°C for 30 seconds, 68°C for 2 minutes for 30 cycles; 68°C for 10 minutes) was performed using Extensor Hi-Fidelity PCR Master Mix (ABGENE, Hamburg, Germany), primer A2HARV (5′-ATCTGAACTTCTCTATTCTC-3′) and linker-specific primer OCI.31 The nested PCR was performed under identical conditions, but using primer A3HARV (5′-CCATGCCTTGTAAAATGGC-3′) and linker-specific primer OCII.31 PCR products were isolated after gel electrophoresis using QIA quick Gel Extraction Kit (Qiagen) and sequenced directly using the primer RAseq (5′-CTTGCAAAATGGCGTTAC-3′). Recovered sequences were screened using the National Center for Biotechnology Information (NCBI) mouse genome database. In case of unclear results (such as hits in bacterial artifical chromosome [BAC] clones), the University of California Santa Cruz (UCSC) database was used. Gene classification followed database records and PubMed literature.
For LM-PCR of dsRED2-transduced BM cells, DNA was digested with Tsp509 I (New England BioLabs). For primer extension reaction, first and nested PCRs biotinylated retroviral primer A1RV (5′-CTGGGGACCATCTGTTCTTGGCCTC-3′), A2RV (5′-GCCCTTGATCTGAACTTCTC-3′), or A3RV (5′-CCATGCCTTGCAAAATGGC-3′) were used, respectively.
Chromosome preparation and spectral karyotyping
Chromosomes were prepared according to standard protocols.32 Spectral karyotyping (SKY) analyses were performed as described.32 Briefly, after separate denaturation of mouse chromosomes and mouse SKY probe mixture (Applied Spectral Imaging, Migdal Haemek, Israel), hybridization took place for 72 hours at 37°C. Signal detection and image acquisition were carried out according to the manufacturer's instructions (Applied Spectral Imaging). Using the SKYView software package (Applied Spectral Imaging), 15 metaphases of each cell line were analyzed.
Results
Reproducible leukemia induction after dose-escalated gene transfer of retroviral vectors into murine hematopoietic cells
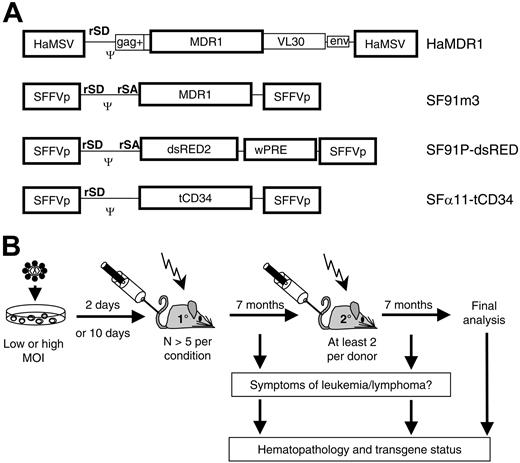
To address the role of vector copy number in MDR1-associated leukemogenesis, we transplanted C57Bl/6 mice with bone marrow (BM) cells that were transduced with a high dose (HD) of HaMDR1 (Figure 1A). Due to the relatively low titer of this vector (< 5 × 105 infectious units/mL), high gene transfer rates could only be achieved by coculture, as in previous studies.12-14 One cohort of recipients received HaMDR1-transduced cells that were expanded for 10 days in vitro after HD gene transfer, because this procedure has been shown to increase the incidence of HaMDR1-associated leukemias.13 Control animals received cells after supernatant transduction with low doses (LDs) of the vectors HaMDR1 or SF91m3 (Figure 1A), another retroviral vector expressing relatively high levels of MDR1.26 Of note, HaMDR1 and SF91m3 are the most potent retroviral MDR1 expression vectors known to date. Further control animals underwent transplantation with cells transduced using an LD of a vector encoding a signal-deficient human CD34 gene (SF11tCD34, titer 1 × 106/mL; Figure 1A), or an HD of a vector expressing the dsRED2 fluorescent protein (SF91PdsRED2, titer 5 × 106/mL; Figure 1A, Table 1).
Gene transfer efficiency and leukemia development
Group . | Mouse no. . | Transgene expression before BMT, % . | Frequency of transgene expression after BMT, frequency (% expression in positive mice) . | Mean transfer efficiency, by PCR, % . | Outcome . |
|---|---|---|---|---|---|
| Experiment I | |||||
| SF91m3 supernatants (LD) | 17-25, n = 9 | 16 | 0/9 mice | 5 | Healthy |
| HaMDR1 supernatants (LD) | 8-16, n = 8 | 15 | 2/8 mice (8) | 14 | Healthy |
| SF91PdsRED2 supernatants (HD) | 1-7, n = 7 | 75 | 7/7 mice (80-90) | 420 | 1 leukemia (L5) |
| HaMDR1 coculture (HD) | 26-31, n = 6 | 30 | 3/6 mice (6) | 147 | 2 leukemias (L29, L31); 1 recipient-type leukemia after secondary BMT (L28.2) |
| Experiment II | |||||
| SF91m3 supernatants (LD) | 54-61, n = 8 | 10 | 1/8 mice (8) | < 1 | Healthy |
| HaMDR1 supernatants (LD) | 64-69, n = 6 | 30 | 1/6 mice (9) | 10 | Healthy |
| SF11tCD34 supernatants (LD) | 48-53, n = 6 | 30 | 6/6 mice (8) | ND | Healthy |
| Experiment III | |||||
| HaMDR1 coculture + in vitro expansion (HD) | 40-47, n = 8 | 65 | 6/8 mice (10) | 1100 | 5 leukemias (L40, L43, L44, L45, L46) |
| Mock-transduced + in vitro expansion | 32-39, n = 8 | — | 0/8 mice | 0 | Healthy |
Group . | Mouse no. . | Transgene expression before BMT, % . | Frequency of transgene expression after BMT, frequency (% expression in positive mice) . | Mean transfer efficiency, by PCR, % . | Outcome . |
|---|---|---|---|---|---|
| Experiment I | |||||
| SF91m3 supernatants (LD) | 17-25, n = 9 | 16 | 0/9 mice | 5 | Healthy |
| HaMDR1 supernatants (LD) | 8-16, n = 8 | 15 | 2/8 mice (8) | 14 | Healthy |
| SF91PdsRED2 supernatants (HD) | 1-7, n = 7 | 75 | 7/7 mice (80-90) | 420 | 1 leukemia (L5) |
| HaMDR1 coculture (HD) | 26-31, n = 6 | 30 | 3/6 mice (6) | 147 | 2 leukemias (L29, L31); 1 recipient-type leukemia after secondary BMT (L28.2) |
| Experiment II | |||||
| SF91m3 supernatants (LD) | 54-61, n = 8 | 10 | 1/8 mice (8) | < 1 | Healthy |
| HaMDR1 supernatants (LD) | 64-69, n = 6 | 30 | 1/6 mice (9) | 10 | Healthy |
| SF11tCD34 supernatants (LD) | 48-53, n = 6 | 30 | 6/6 mice (8) | ND | Healthy |
| Experiment III | |||||
| HaMDR1 coculture + in vitro expansion (HD) | 40-47, n = 8 | 65 | 6/8 mice (10) | 1100 | 5 leukemias (L40, L43, L44, L45, L46) |
| Mock-transduced + in vitro expansion | 32-39, n = 8 | — | 0/8 mice | 0 | Healthy |
— indicates not applicable.
Overview of the experimental groups. Three independent experiments were performed (experiments I-III), containing 4, 3, and 2 groups of mice that underwent transplantation with bone marrow cells that were transduced by different retroviral vectors. Note that the efflux assays used to detect MDR1 transgene expression may not be as sensitive as detection of dsRED2 or tCD34 by flow cytometry. Data in the fourth column show the frequency of mice expressing the transgene and the average expression level in the positive animals. In an independent experiment involving 20 primary recipients observed for one year and serial BMT into secondary animals observed for another 7 months, no evidence was obtained for a leukemogenic capacity of SF91PdsRED2 under LD conditions (data not shown).
BMT indicates bone marrow transplantation; LD, low-dose gene transfer; HD, high-dose gene transfer according to in vivo marking rates (based on PCR) determined 9 weeks after BMT.
Study design. (A) Retroviral vectors, expressing MDR1, dsRED2, or human truncated CD34 (htCD34). SFFVp (spleen focus forming virus LTR), HaMSV (Harvey murine sarcoma virus LTR), gag (residual retroviral gag sequences), env (residual retroviral envelope sequences), VL30 (residual sequences of virus-like 30 element), Ψ (packaging signal), wPRE (woodchuck hepatitis virus posttranscriptional regulatory element). rSD, retroviral splice donor; rSA, retroviral splice acceptor. (B) Experimental setup; bone marrow cells were transduced and transplanted 2 or 10 days later in lethally irradiated C57BL/6J recipient mice. After a 7-month observation, secondary recipients underwent transplantation. Symptomatic animals were killed for detailed analysis. All other mice were analyzed at the end of the experiment. MOI indicates multiplicity of infection.
Study design. (A) Retroviral vectors, expressing MDR1, dsRED2, or human truncated CD34 (htCD34). SFFVp (spleen focus forming virus LTR), HaMSV (Harvey murine sarcoma virus LTR), gag (residual retroviral gag sequences), env (residual retroviral envelope sequences), VL30 (residual sequences of virus-like 30 element), Ψ (packaging signal), wPRE (woodchuck hepatitis virus posttranscriptional regulatory element). rSD, retroviral splice donor; rSA, retroviral splice acceptor. (B) Experimental setup; bone marrow cells were transduced and transplanted 2 or 10 days later in lethally irradiated C57BL/6J recipient mice. After a 7-month observation, secondary recipients underwent transplantation. Symptomatic animals were killed for detailed analysis. All other mice were analyzed at the end of the experiment. MOI indicates multiplicity of infection.
For at least 7 months, we prospectively screened mice for symptoms of hematopoietic malignancies, following consensus criteria.33,34 We also performed secondary BM transplantation (BMT) to promote potential leukemic progression or to confirm the diagnosis of leukemia (Figure 1B).15,16 Prior to BMT, MDR1 expression ranged from 10% to 65%, depending on the vector dose. Thus, multiple cells with MDR1 overexpression were infused (> 105/animal) (Table 1).
All animals (n = 31) that underwent transplantation with cells that received retroviral MDR1 vectors under LD conditions remained healthy (Table 1). Similarly, no leukemia was observed after LD transduction with control vectors SF11tCD34 (n = 6) (Table 1). Using identical transplantation conditions, we also observed no leukemia in more than 50 animals that underwent transplantation with a “normal dose” of retroviral vectors encoding fluorescent proteins (data not shown). In line with previous reports,12,13 we reproduced the high leukemia incidence (5/8) in recipients of expanded BM cells after HD gene transfer with HaMDR1 (Tables 1 and 2). In contrast, expansion of mock-transduced cells was not leukemogenic (n = 8, Table 1). Without ex vivo expansion, leukemia was diagnosed in 2 of 6 recipients of HD HaMDR1 cells (Tables 1 and 2). Of note, one leukemic condition was also observed among the 7 recipients of SF91PdsRED2.
Leukemias diagnosed in this study
Vector and mouse no. . | Leukocyte counts (× 106/mL) . | Latency of manifestation (wks) . | Flow cytometry . | Spleen weight, mg . | Copy number after 9 weeks‡ . | Copy number in leukemia§ . | Diagnosis . |
|---|---|---|---|---|---|---|---|
| SF91PDsRED2 | |||||||
| 5* | 12.6 | 26 | CD11b+, GR1+ | 200 | 14 | 8 | MPD-like myeloid leukemia |
| 5.1* (secondary of 5) | 250 | 26 + 12 | CD11b+ c-Kit+ | 850 | ND | 8 | Myeloid leukemia with maturation |
| 5.2* (secondary of 5) | 430 | 26 + 20 | CD11b+ c-Kit+ | 380 | ND | 8 | Myeloid leukemia with maturation |
| HaMDR1 | |||||||
| 29* | 5.7 | 34 | BM: all marker low; spleen:CD3+, CD4+, CD8+ | 170, thymus enlarged | 0.8 | 7 | Precursor T-lymphoblastic lymphoma/leukemia |
| 31* | 3.9 | 16 | Ter119 low, c-Kit+ | 300 | 3 | 6 | Erythroid leukemia |
| 31.2* (secondary of 31) | 10 | 16 + 10 | Ter119 low, c-Kit+ | 580 | ND | 6 | Erythroid leukemkia |
| 40† | 78 | 26 | CD11b+, Gr1+/- | 510 | 20 | 10 | Myeloid leukemia with maturation |
| 43† | 14 | 33 | CD11b+, Gr1+/- | 780 | 7.6 | ND | Myeloid leukemia with maturation |
| 44† | 1.5 | 33 | CD11b+, c-Kit+ | 510 | 5.5 | 10 | Myeloid leukemia without maturation |
| 45† | NA | 24 | NA | Thymus/spleen enlarged | 12.8 | NA | (Most likely) T-cell leukemia |
| 46† | 1 | 32 | CD11b+, GR1+ | 635 | 38.8 | 12 | Myeloid leukemia with maturation |
| 28.2* (secondary of 28) | 40 | 26 + 21 | CD3+ recipient type | 860 | Mouse 28 = 0.82 | 0 | Precursor T-lymphoblastic lymphoma/leukemia |
Vector and mouse no. . | Leukocyte counts (× 106/mL) . | Latency of manifestation (wks) . | Flow cytometry . | Spleen weight, mg . | Copy number after 9 weeks‡ . | Copy number in leukemia§ . | Diagnosis . |
|---|---|---|---|---|---|---|---|
| SF91PDsRED2 | |||||||
| 5* | 12.6 | 26 | CD11b+, GR1+ | 200 | 14 | 8 | MPD-like myeloid leukemia |
| 5.1* (secondary of 5) | 250 | 26 + 12 | CD11b+ c-Kit+ | 850 | ND | 8 | Myeloid leukemia with maturation |
| 5.2* (secondary of 5) | 430 | 26 + 20 | CD11b+ c-Kit+ | 380 | ND | 8 | Myeloid leukemia with maturation |
| HaMDR1 | |||||||
| 29* | 5.7 | 34 | BM: all marker low; spleen:CD3+, CD4+, CD8+ | 170, thymus enlarged | 0.8 | 7 | Precursor T-lymphoblastic lymphoma/leukemia |
| 31* | 3.9 | 16 | Ter119 low, c-Kit+ | 300 | 3 | 6 | Erythroid leukemia |
| 31.2* (secondary of 31) | 10 | 16 + 10 | Ter119 low, c-Kit+ | 580 | ND | 6 | Erythroid leukemkia |
| 40† | 78 | 26 | CD11b+, Gr1+/- | 510 | 20 | 10 | Myeloid leukemia with maturation |
| 43† | 14 | 33 | CD11b+, Gr1+/- | 780 | 7.6 | ND | Myeloid leukemia with maturation |
| 44† | 1.5 | 33 | CD11b+, c-Kit+ | 510 | 5.5 | 10 | Myeloid leukemia without maturation |
| 45† | NA | 24 | NA | Thymus/spleen enlarged | 12.8 | NA | (Most likely) T-cell leukemia |
| 46† | 1 | 32 | CD11b+, GR1+ | 635 | 38.8 | 12 | Myeloid leukemia with maturation |
| 28.2* (secondary of 28) | 40 | 26 + 21 | CD3+ recipient type | 860 | Mouse 28 = 0.82 | 0 | Precursor T-lymphoblastic lymphoma/leukemia |
ND indicates not determined; NA, not applicable (mouse found dead).
Bone marrow transplantation (BMT) was performed with cells that were expanded for 2 days after the first exposure to retroviral particles
BMT was performed with cells that were expanded for 10 days after the first exposure to retroviral particles
Transgene copy number was determined in peripheral blood cells by real-time PCR
Transgene copy number in leukemias was determined by Southern blot
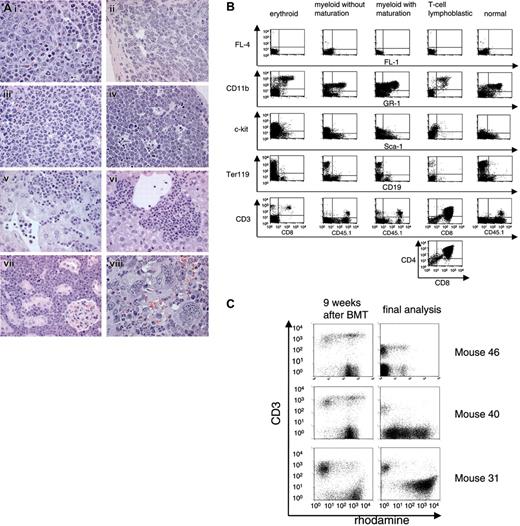
Two donor cell–derived leukemias (L5; L31) required secondary BMT for clear diagnosis (mice 31.2, 5.1, and 5.2, Table 2). Underlining the leukemia-promoting effect of secondary BMT, we also observed one case of a host-derived (CD45.1+), thus probably irradiation-induced, T-lymphoblastic leukemia in a secondary recipient (Table 2). The 7 leukemias associated with MDR1 vectors differed strongly with respect to both latency (16 to 34 weeks in primary recipients) and phenotype (Table 2), the latter determined by histopathology (Figure 2A) and flow cytometry (Figure 2B). Leukemia phenotypes included myeloid (with different stages of maturation), erythroid, and T-lymphoblastic forms (Table 2). Moreover, the expression levels of MDR1 differed strongly among these leukemias. In one case, infiltrating T cells had higher MDR1 expression than the leukemic clone (as determined by the efflux assay; Figure 2C).
Importantly, producer cells as well as leukemic clones and tissues showed no evidence of ecotropic, amphotropic, or polytropic replication-competent retroviruses, as determined by a validated PCR, Southern blot, and sensitive marker rescue assays (data not shown).
Leukemias arise from clones with multiple hits in cellular growth-regulatory genes
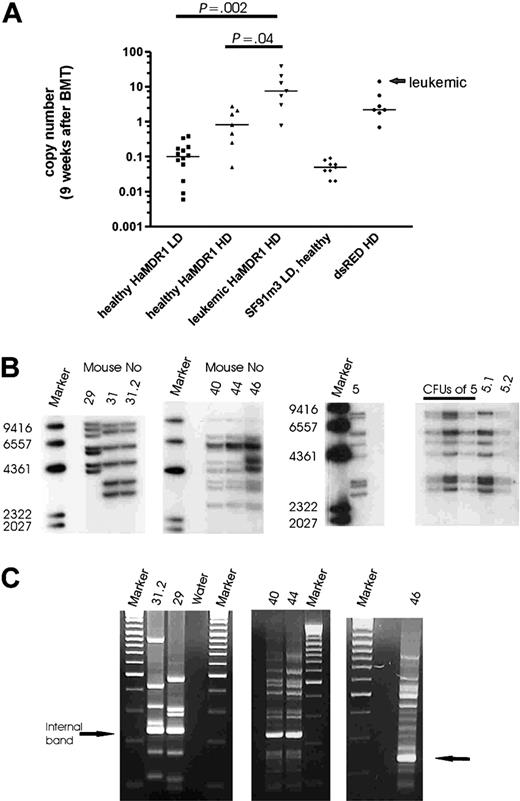
To determine in vivo gene marking rates following donor engraftment, we used quantitative PCR (qPCR) with DNA of peripheral blood leukocytes harvested 9 weeks after BMT. This confirmed that HD conditions resulted in repopulation with clones showing multiple hits per cell, as opposed to an average transgene copy number less than 1 in LD conditions (Figure 3A). Moreover, we found a correlation of disease disposition and transgene copy number determined by qPCR 9 weeks after primary BMT (Figure 3A). In recipients of HaMDR1-transduced cells, each mouse with an average transgene copy number more than 5 per peripheral blood leukocyte subsequently developed leukemia, resulting in a highly significant difference in average transgene copy number between pre-leukemic and healthy animals (Figure 3A). In addition, 2 animals presenting with a lower marking 9 weeks after transplantation (0.8 and 3 average transgene copies per cell) developed leukemic clones containing 7 and 6 copies, respectively.
One of the leukemic mice was found dead (L45). From the 7 remaining leukemias, we obtained DNA of sufficient quality for molecular studies in 6 cases. By Southern blot analysis of secondary recipients and of CFU-S, we revealed that the leukemias were clonal, and had acquired 6 to 12 vector insertions per clone (Figure 3B). This clonal origin explains the significant latency of manifestation (Table 2). Thus we also found 2 primary recipients developing the same clonal disease following transplantation of ex vivo–expanded cells (L40; L44). In 3 cases (L5, L40, and L46), the copy number in the leukemias was below the marking levels measured 9 weeks after BMT (Table 2), suggesting that at earlier time points other clones with even higher copy numbers dominated hematopoiesis. These data clearly revealed clonal selection as the basis of MDR1 vector–associated leukemias.
For detailed molecular studies of insertion sites, we used LM-PCR (Figure 3C) and LAM-PCR.30,31 To avoid potential contaminations by insertions present in healthy cells, we used the less sensitive LM-PCR method for all leukemias. The results were highly reproducible (eg, see LM-PCR gels of identical leukemias L40 and L44 in Figure 3C). We excised only the major bands and sequenced the PCR products either directly or after subcloning. Overall, 81% (35/43) of the insertions predicted by Southern blot in leukemic mice were identified (Table 3). As controls, we sequenced 72 insertions from healthy recipients: 60 insertions were recovered from 8 healthy primary recipients of HaMDR1 cells, including 3 animals that underwent transplantation with ex vivo–expanded cells, and 12 insertions from 2 healthy primary recipients of SF91PdsRED2 cells. We selected animals with high marking levels (Table 4).
Vector insertion sites recovered from leukemic clones
No. of the leukemic clone (recovered loci): locus hit, nearest gene* . | Gene ID . | Chromosome . | Distance to transcriptional start . | Forward (F) or reverse (R) orientation . | (Predicted) gene function . | Class (localization) . | RTCGD . | CGD . |
|---|---|---|---|---|---|---|---|---|
| 5.1 (6/8) | ||||||||
| Hivep1 | 110521 | 13A4 | 40111 | R | transcription factor | 1(B) | 2 hits | - |
| LOC236667 | 236667 | XA1 + 1 | 337984 | F | (unknown) | 3 | - | - |
| Csfr3 | 12986 | 4D2 + 2 | -625 | R | GPR for G-CSF | 1(A) | - | - |
| Mllt3 | 70122 | 4C4 | -1243 | R | transcription factor | 1(B) | - | + |
| Slcl2a3 | 20497 | 8C5 | 33950 | R | solute carrier | 3 | - | - |
| Klrb1c | 17059 | 6F3 | -9039 | R | NK cell receptor | 2(A) | - | - |
| 31 (5/6) | ||||||||
| Sos1 | 20662 | 17E3 | -4055 | R | Ras activating | 2(A) | - | - |
| Gpr43 | 233079 | 7A3 | -6455 | R | GPR, myeloid differentiation | 2(A) | - | - |
| Hrh2 | 15466 | 13B1 | 43483 | R | GPR, myeloid differentiation | 2(A) | - | - |
| Fli1 | 14247 | 9A4 | -14094 | F | transcription factor | 1(B) | 7 hits | + |
| Foxo3 | 56484 | 10B2 | 3990 | F | transcription factor | 1(B) | + | |
| 29 (4/7) | ||||||||
| Plcg2 | 234779 | 8E1 | 36215 | R | survival signaling | 2(A) | - | - |
| F2r | 14062 | 13D1 | -38130 | R | GPR for thrombin | 2(A) | - | - |
| Rab11a | 53869 | 9C | -49969 | R | Ras binding protein | 2(A) | - | (RAB5EP) |
| Arkadia | 93836 | 9D | -413 | R | transcription modulator | 2(B) | - | - |
| 46 (10/12) | ||||||||
| HoxA7 | 15404 | 6B2 + 3 | -138 | F | transcription factor | 1(B) | 19 hits | (HoxA9) |
| Brd2 | 14312 | 17B1 | -2135 | F | transcription factor | 1(B) | - | (BRD4) |
| Rab31-like | 383272 | 17E1 + 1 | 48640 | F | (GTP binding protein) | 2(A) | - | (RAB5EP) |
| Myo18a | 360013 | 11B2 | 7940 | F | myosin | 3 | - | - |
| Lsm10pending | 116748 | 4D2 + 2 | 12041 | F | RNA protein | 3 | - | - |
| Itm2b | 16432 | 14D1 | -14564 | F | (apoptosis regulation) | 2(A) | - | - |
| 1700060C2 | 73399 | 2H2 | -14847 | F | unknown | 3 | - | - |
| 0Rik | ||||||||
| Asb5 | 76294 | 8D1+1 | -94219 | F | SOCS box signaling | 2(A) | - | - |
| Bcl11a | 14025 | 11A3+2 | 149048 | F | transcription factor | 1(B) | 6 hits | + |
| Tpst2 | 22022 | 5F | 9467 | F | sulfotransferase | 3 | - | - |
| 40 (10/10) | ||||||||
| Sema4b | 20352 | 7D1 | -41046 | R | receptor activity | 1(A) | 4 hits | - |
| Ski | 20481 | 4E2 | -12065 | F | transcription factor | 1(B) | - | - |
| Rab31-like | 383272 | 17E1+1 | 48640 | F | (GTP binding protein) | 2(A) | - | (RAB5EP) |
| Evil | 14013 | 3A3 | -119998 | R | transcription factor | 1(B) | 20 hits | + |
| Gpr43 | 233079 | 7A3 | 13904 | R | GPR, myeloid differentiation | 2(A) | - | - |
| Tpk1 | 29807 | 6B2.1 | -27024 | F | thiamin pyrophosphokinase | 3 | - | - |
| LOC383539 | 383539 | 1C5 | -571 | F | (ribosomal protein) | 3 | - | - |
| Sip1 | 66603 | 12C2 | 10227 | F | spliceosome | 3 | - | - |
| Zfp3612 | 12193 | 17 E4 | 72206 | R | Zinc finger protein | 2(B) | - | - |
| Znrf1 | 170737 | 8E1 | 58558 | R | Zinc and ring finger protein | 2(B) | - | - |
No. of the leukemic clone (recovered loci): locus hit, nearest gene* . | Gene ID . | Chromosome . | Distance to transcriptional start . | Forward (F) or reverse (R) orientation . | (Predicted) gene function . | Class (localization) . | RTCGD . | CGD . |
|---|---|---|---|---|---|---|---|---|
| 5.1 (6/8) | ||||||||
| Hivep1 | 110521 | 13A4 | 40111 | R | transcription factor | 1(B) | 2 hits | - |
| LOC236667 | 236667 | XA1 + 1 | 337984 | F | (unknown) | 3 | - | - |
| Csfr3 | 12986 | 4D2 + 2 | -625 | R | GPR for G-CSF | 1(A) | - | - |
| Mllt3 | 70122 | 4C4 | -1243 | R | transcription factor | 1(B) | - | + |
| Slcl2a3 | 20497 | 8C5 | 33950 | R | solute carrier | 3 | - | - |
| Klrb1c | 17059 | 6F3 | -9039 | R | NK cell receptor | 2(A) | - | - |
| 31 (5/6) | ||||||||
| Sos1 | 20662 | 17E3 | -4055 | R | Ras activating | 2(A) | - | - |
| Gpr43 | 233079 | 7A3 | -6455 | R | GPR, myeloid differentiation | 2(A) | - | - |
| Hrh2 | 15466 | 13B1 | 43483 | R | GPR, myeloid differentiation | 2(A) | - | - |
| Fli1 | 14247 | 9A4 | -14094 | F | transcription factor | 1(B) | 7 hits | + |
| Foxo3 | 56484 | 10B2 | 3990 | F | transcription factor | 1(B) | + | |
| 29 (4/7) | ||||||||
| Plcg2 | 234779 | 8E1 | 36215 | R | survival signaling | 2(A) | - | - |
| F2r | 14062 | 13D1 | -38130 | R | GPR for thrombin | 2(A) | - | - |
| Rab11a | 53869 | 9C | -49969 | R | Ras binding protein | 2(A) | - | (RAB5EP) |
| Arkadia | 93836 | 9D | -413 | R | transcription modulator | 2(B) | - | - |
| 46 (10/12) | ||||||||
| HoxA7 | 15404 | 6B2 + 3 | -138 | F | transcription factor | 1(B) | 19 hits | (HoxA9) |
| Brd2 | 14312 | 17B1 | -2135 | F | transcription factor | 1(B) | - | (BRD4) |
| Rab31-like | 383272 | 17E1 + 1 | 48640 | F | (GTP binding protein) | 2(A) | - | (RAB5EP) |
| Myo18a | 360013 | 11B2 | 7940 | F | myosin | 3 | - | - |
| Lsm10pending | 116748 | 4D2 + 2 | 12041 | F | RNA protein | 3 | - | - |
| Itm2b | 16432 | 14D1 | -14564 | F | (apoptosis regulation) | 2(A) | - | - |
| 1700060C2 | 73399 | 2H2 | -14847 | F | unknown | 3 | - | - |
| 0Rik | ||||||||
| Asb5 | 76294 | 8D1+1 | -94219 | F | SOCS box signaling | 2(A) | - | - |
| Bcl11a | 14025 | 11A3+2 | 149048 | F | transcription factor | 1(B) | 6 hits | + |
| Tpst2 | 22022 | 5F | 9467 | F | sulfotransferase | 3 | - | - |
| 40 (10/10) | ||||||||
| Sema4b | 20352 | 7D1 | -41046 | R | receptor activity | 1(A) | 4 hits | - |
| Ski | 20481 | 4E2 | -12065 | F | transcription factor | 1(B) | - | - |
| Rab31-like | 383272 | 17E1+1 | 48640 | F | (GTP binding protein) | 2(A) | - | (RAB5EP) |
| Evil | 14013 | 3A3 | -119998 | R | transcription factor | 1(B) | 20 hits | + |
| Gpr43 | 233079 | 7A3 | 13904 | R | GPR, myeloid differentiation | 2(A) | - | - |
| Tpk1 | 29807 | 6B2.1 | -27024 | F | thiamin pyrophosphokinase | 3 | - | - |
| LOC383539 | 383539 | 1C5 | -571 | F | (ribosomal protein) | 3 | - | - |
| Sip1 | 66603 | 12C2 | 10227 | F | spliceosome | 3 | - | - |
| Zfp3612 | 12193 | 17 E4 | 72206 | R | Zinc finger protein | 2(B) | - | - |
| Znrf1 | 170737 | 8E1 | 58558 | R | Zinc and ring finger protein | 2(B) | - | - |
— indicates no hit in the given database.
Gene hits were determined according to the NCBI database of the mouse genome, January 2005. UCSC database results are given if NCBI showed unclear results. Genes listed in parentheses represent close relatives of hits found in leukemias. Class 1, protooncogene; class 2, signaling gene; class 3, other gene. Class 1 classification required entry as a common insertion site (CIS) in RTCGD, representation in CGD, or additional literature providing evidence for transforming potential.
RTCGD indicates retrovirally tagged cancer gene database35 ; CGD, cancer gene database of human cancer genes11 ; NA, not applicable; A, transmembrane receptor or cytoplasmic protein; B, nuclear protein; GPR, G-protein-coupled receptor.
The total number of locus hits was 35. There were 11 (31.4%) class 1 genes; 15 (42.9%) class 2 genes, and 9 (25.7%) class 3 genes
Gene hits in healthy animals
No. of the leukemic clone: locus hit, nearest gene* . | Gene ID . | Chromosome . | Distance to transcriptional start . | Forward (F) or reverse (R) orientation . | (Predicted) gene function . | Class (localization) . | RTCGD . | CGD . |
|---|---|---|---|---|---|---|---|---|
| HaMDR1 26 | ||||||||
| Siat4c | 20443 | 9A4 | -1848 | R | sialyltransferase, hemostasis | 3 | - | - |
| Evil | 14013 | 3A3 | -16655 | R | transcription factor | 1(B) | 20 hits | + |
| Nif311 | 65102 | 1C1.3 | 944 | F | transcriptional corepressor | 2(B) | - | - |
| Plcg2 | 234779 | 8E1 | -2770 | R | survival signaling | 2(A) | - | - |
| Gng12 | 14701 | 6C1 | 59426 | F | G protein | 2(B) | - | - |
| Arrb1 | 109689 | 7E1 | -3800 | F | arrestin, IGF-1 pathway, antiapoptotic | 2(A) | - | - |
| HaMDR1 27 | ||||||||
| AW558560 | 106821 | 17C | 8584 | F | (unknown function) | 3 | - | - |
| LOC381313 | 381313 | 1H5 | 610 | F | (similar to Rab3 GTPase activating protein) | 2(A) | - | - |
| LOC381410 | 381410 | 2E1 | -1108 | R | (similar to Zn finger protein) | 2(B) | - | - |
| Neil1 | 72774 | 9B | -854 | F | DNA repair | 3 | - | - |
| RP24-179M18 | NA | 19 | NA | NA | unknown | 3 | - | - |
| LOC381079 | 381079 | 17A3.3 | -27788 | R | (similar to Bcl2/E1B interacting protein) | 2(A) | - | - |
| Dsc2 | 13506 | 18A2 | 39460 | F | cadherin-like, cell adhesion | 2(A) | - | - |
| D630010B17 Rik | 328594 | 15E3 | 7228 | R | (unknown function) | 3 | - | - |
| Xylt1 | 233781 | 7F1 | -93391 | F | xylosyltransferase | 3 | - | - |
| LOC215999 (Gm64) | 215999 | 10B4 | 37233 | R | acetylglucosaminyltransferase | 3 | - | - |
| Mgat4a | 269181 | 1(B) | 48026 | F | metabolic enzyme | 3 | - | - |
| HaMDR1 28 | ||||||||
| Impa1 | 55980 | 3A1 | 43360 | R | inositolmonophosphatase | 3 | - | - |
| RP23-57M16 | NA | 4 | NA | NA | unknown | 3 | - | - |
| Cdk5r | 12569 | 11B5 | 84319 | R | cyclin-dependent kinase regulatory subunit | 2(B) | - | - |
| P140 | 56013 | 11D | 41772 | F | negative regulator of cell adhesion | 2(A) | - | - |
| Kctd12 | 239217 | 14E2 | 171438 | NA | potassium channel | 2(A) | - | - |
| Uck2-pending | 80914 | 1H2.3 | 40000 | F | uridine monophosphatase kinase | 3 | - | - |
| Akt1 | 11651 | 12F1-F2 | 46480 | R | thymoma viral proto-oncogene | 1(A) | 3 hits | (AKT2) |
| Ly86 | 17084 | 13A3.3 | 104966 | F | B-cell response to LPS | 2(A) | - | - |
| Lims1 | 110829 | 10B4 | 83293 | F | cell matrix adhesion | 2(A) | - | - |
| Rab3il1 | 74760 | 19A1 | 6329 | R | Rab3a interacting protein | 2(B) | - | - |
| Usp39 | 28035 | 6C1 | -1014 | F | ubiquitin specific protease | 3 | - | - |
| 0610039P13 Rik | 74096 | 5F | 12077 | F | (unknown function) | 3 | - | - |
| HaMDR1 30 | ||||||||
| Rad18 | 58186 | 6F | 9701 | F | DNA replication and recombination | 3 | - | - |
| 1500009M05 Rik | 67006 | 3G3 | 4325 | F | (unknown function) | 3 | - | - |
| HaMDR1 expanded 41 | ||||||||
| LOC384948 | 384948 | 9D | 4668 | R | (similar to mitotic checkpoint protein MAD1a) | 2(B) | - | - |
| A630038D02 | 330305 | 6B2.3 | -544 | R | (unknown function) | 3 | - | - |
| BAC clone RP21-59414 | NA | 10 | NA | NA | unknown | 3 | - | - |
| Lsm4 | 50783 | 8B3.3 | 1396 | F | U6 small nuclear RNA associated | 3 | - | - |
| Evil | 14013 | 3A3 | -155209 | R | transcription factor | 1(B) | 20 hits | + |
| BC042513 | 215113 | 11B5 | 9218 | R | (unknown function) | 3 | - | - |
| Rbbp7 | 245688 | XF4 | -4190 | F | Retinoblastoma binding protein | 2(B) | - | - |
| Abcc3 | 76408 | 11D | -20439 | F | ATP binding cassette, bile transport | 3 | - | - |
| Rnpep | 215615 | 1E4 | 1218 | F | aminopeptidase | 3 | - | - |
| Sox13 | 20668 | 1E4 | -3992 | R | SRY-box transcription factor | 2(B) | - | - |
| BAC clone RP23-373O10 | NA | 17 | NA | NA | unknown | 3 | - | - |
| LOC270174 | 270174 | 9D | -4485 | R | (Gm640 gene model) | 3 | - | - |
| HaMDR1 expanded 42 | ||||||||
| Sema4b† | 20352 | 7D1 | -41046 | R | receptor activity | 1(A) | 4 hits | - |
| Rab31-like‡ | 383272 | 17E1.1 | 48640 | F | GTP binding protein | 2(A) | - | (RAB5EP) |
| Sip1† | 66603 | 12C2 | 10227 | F | splicosome | 3 | - | - |
| Slamf1 | 27218 | 1H3 | 19532 | R | receptor activity, T cell proliferation | 2(A) | - | - |
| HaMDR1 expanded 47 | ||||||||
| RP24-333C3 | NA | 17 | NA | NA | unknown | 3 | - | - |
| RP23-129K10 | NA | 17 | NA | NA | unknown | 3 | - | - |
| no gene around | NA | 12 | NA | NA | unknown | 3 | - | - |
| Tpk1† | 29807 | 6B2.1 | -27024 | F | thiamin pyrophosphokinase | 3 | - | - |
| Gpr43† | 233079 | 7A3 | 13904 | R | GPR; myeloid differentiation | 2(A) | - | - |
| Rab31-like‡ | 383272 | 17E1 + 1 | 48640 | F | (GTP-binding protein) | 2(A) | - | - |
| Evil† | 14013 | 3A3 | -119998 | R | transcription factor | 1(B) | 20 hits | - |
| Lsm10-pending§ | 116748 | 4D2+2 | 12041 | F | mRNA splicing | 3 | - | - |
| LOC383539† | 383539 | 1C5 | -571 | F | (similar to ribosomal protein L30) | 3 | - | - |
| Nfic | 18029 | 10C1 | -11913 | R | p53 regulating nuclear factor | 1(B) | 3 hits | - |
| HaMDR1 67 | ||||||||
| Lak-pending | 71481 | 3G2 | -15 | R | (lymphocyte alfa kinase) | 2(A) | - | - |
| RP23-92G13 | NA | 13 | NA | NA | unknown | 3 | - | - |
| Ccnd2 | 12444 | 6F3 | -14615 | R | CyclinD2 | 1(B) | 18 hits | (CCND1) |
| dsRED 3 | ||||||||
| Bcl11a | 14025 | 11A32 | 167714 | F | zinc finger protein | 1(B) | 6 hits | + |
| Cacnb2 | 12296 | 2(A)2 | 28471 | R | calcium channel | 2(A) | - | - |
| Pdcd1lg1 | 60533 | 19C2 | 34046 | R | programmed cell death ligand | 1(A) | 2 hits | - |
| Phemx (Tssc6) | 27027 | 7F5 | 2431 | F | tetraspanin superfamily member | 2(A) | - | - |
| LOC384063 | 384063 | 4D2+3 | 13364 | R | (sim. to ribosomal protein) | 3 | - | - |
| Runx2 | 12393 | 17B3 | 141712 | R | runt related transcription factor | 1(B) | 6 hits | (RUNX1) |
| Ubb | 22187 | 11B2 | 8464 | F | ubiquitinB | 3 | - | - |
| Cdk6 | 12571 | 5A1 | 157035 | F | Cyclin-dependent kinase | 2(B) | - | (CDK4) |
| dsRED4 | ||||||||
| Mybbp1a | 18432 | 11B4 | 46577 | R | MYB binding protein | 2(B) | - | - |
| Btd | 26363 | 14A3 | 27328 | F | biotinidase | 2(A) | - | - |
| Crsp6 | 234959 | 9A3 | -25697 | R | cofactor of Sp1 transcription factor | 2(B) | - | - |
| Slfn2 | 20556 | 11C | 13391 | R | negative regulation of prolif. | 2(A) | - | - |
No. of the leukemic clone: locus hit, nearest gene* . | Gene ID . | Chromosome . | Distance to transcriptional start . | Forward (F) or reverse (R) orientation . | (Predicted) gene function . | Class (localization) . | RTCGD . | CGD . |
|---|---|---|---|---|---|---|---|---|
| HaMDR1 26 | ||||||||
| Siat4c | 20443 | 9A4 | -1848 | R | sialyltransferase, hemostasis | 3 | - | - |
| Evil | 14013 | 3A3 | -16655 | R | transcription factor | 1(B) | 20 hits | + |
| Nif311 | 65102 | 1C1.3 | 944 | F | transcriptional corepressor | 2(B) | - | - |
| Plcg2 | 234779 | 8E1 | -2770 | R | survival signaling | 2(A) | - | - |
| Gng12 | 14701 | 6C1 | 59426 | F | G protein | 2(B) | - | - |
| Arrb1 | 109689 | 7E1 | -3800 | F | arrestin, IGF-1 pathway, antiapoptotic | 2(A) | - | - |
| HaMDR1 27 | ||||||||
| AW558560 | 106821 | 17C | 8584 | F | (unknown function) | 3 | - | - |
| LOC381313 | 381313 | 1H5 | 610 | F | (similar to Rab3 GTPase activating protein) | 2(A) | - | - |
| LOC381410 | 381410 | 2E1 | -1108 | R | (similar to Zn finger protein) | 2(B) | - | - |
| Neil1 | 72774 | 9B | -854 | F | DNA repair | 3 | - | - |
| RP24-179M18 | NA | 19 | NA | NA | unknown | 3 | - | - |
| LOC381079 | 381079 | 17A3.3 | -27788 | R | (similar to Bcl2/E1B interacting protein) | 2(A) | - | - |
| Dsc2 | 13506 | 18A2 | 39460 | F | cadherin-like, cell adhesion | 2(A) | - | - |
| D630010B17 Rik | 328594 | 15E3 | 7228 | R | (unknown function) | 3 | - | - |
| Xylt1 | 233781 | 7F1 | -93391 | F | xylosyltransferase | 3 | - | - |
| LOC215999 (Gm64) | 215999 | 10B4 | 37233 | R | acetylglucosaminyltransferase | 3 | - | - |
| Mgat4a | 269181 | 1(B) | 48026 | F | metabolic enzyme | 3 | - | - |
| HaMDR1 28 | ||||||||
| Impa1 | 55980 | 3A1 | 43360 | R | inositolmonophosphatase | 3 | - | - |
| RP23-57M16 | NA | 4 | NA | NA | unknown | 3 | - | - |
| Cdk5r | 12569 | 11B5 | 84319 | R | cyclin-dependent kinase regulatory subunit | 2(B) | - | - |
| P140 | 56013 | 11D | 41772 | F | negative regulator of cell adhesion | 2(A) | - | - |
| Kctd12 | 239217 | 14E2 | 171438 | NA | potassium channel | 2(A) | - | - |
| Uck2-pending | 80914 | 1H2.3 | 40000 | F | uridine monophosphatase kinase | 3 | - | - |
| Akt1 | 11651 | 12F1-F2 | 46480 | R | thymoma viral proto-oncogene | 1(A) | 3 hits | (AKT2) |
| Ly86 | 17084 | 13A3.3 | 104966 | F | B-cell response to LPS | 2(A) | - | - |
| Lims1 | 110829 | 10B4 | 83293 | F | cell matrix adhesion | 2(A) | - | - |
| Rab3il1 | 74760 | 19A1 | 6329 | R | Rab3a interacting protein | 2(B) | - | - |
| Usp39 | 28035 | 6C1 | -1014 | F | ubiquitin specific protease | 3 | - | - |
| 0610039P13 Rik | 74096 | 5F | 12077 | F | (unknown function) | 3 | - | - |
| HaMDR1 30 | ||||||||
| Rad18 | 58186 | 6F | 9701 | F | DNA replication and recombination | 3 | - | - |
| 1500009M05 Rik | 67006 | 3G3 | 4325 | F | (unknown function) | 3 | - | - |
| HaMDR1 expanded 41 | ||||||||
| LOC384948 | 384948 | 9D | 4668 | R | (similar to mitotic checkpoint protein MAD1a) | 2(B) | - | - |
| A630038D02 | 330305 | 6B2.3 | -544 | R | (unknown function) | 3 | - | - |
| BAC clone RP21-59414 | NA | 10 | NA | NA | unknown | 3 | - | - |
| Lsm4 | 50783 | 8B3.3 | 1396 | F | U6 small nuclear RNA associated | 3 | - | - |
| Evil | 14013 | 3A3 | -155209 | R | transcription factor | 1(B) | 20 hits | + |
| BC042513 | 215113 | 11B5 | 9218 | R | (unknown function) | 3 | - | - |
| Rbbp7 | 245688 | XF4 | -4190 | F | Retinoblastoma binding protein | 2(B) | - | - |
| Abcc3 | 76408 | 11D | -20439 | F | ATP binding cassette, bile transport | 3 | - | - |
| Rnpep | 215615 | 1E4 | 1218 | F | aminopeptidase | 3 | - | - |
| Sox13 | 20668 | 1E4 | -3992 | R | SRY-box transcription factor | 2(B) | - | - |
| BAC clone RP23-373O10 | NA | 17 | NA | NA | unknown | 3 | - | - |
| LOC270174 | 270174 | 9D | -4485 | R | (Gm640 gene model) | 3 | - | - |
| HaMDR1 expanded 42 | ||||||||
| Sema4b† | 20352 | 7D1 | -41046 | R | receptor activity | 1(A) | 4 hits | - |
| Rab31-like‡ | 383272 | 17E1.1 | 48640 | F | GTP binding protein | 2(A) | - | (RAB5EP) |
| Sip1† | 66603 | 12C2 | 10227 | F | splicosome | 3 | - | - |
| Slamf1 | 27218 | 1H3 | 19532 | R | receptor activity, T cell proliferation | 2(A) | - | - |
| HaMDR1 expanded 47 | ||||||||
| RP24-333C3 | NA | 17 | NA | NA | unknown | 3 | - | - |
| RP23-129K10 | NA | 17 | NA | NA | unknown | 3 | - | - |
| no gene around | NA | 12 | NA | NA | unknown | 3 | - | - |
| Tpk1† | 29807 | 6B2.1 | -27024 | F | thiamin pyrophosphokinase | 3 | - | - |
| Gpr43† | 233079 | 7A3 | 13904 | R | GPR; myeloid differentiation | 2(A) | - | - |
| Rab31-like‡ | 383272 | 17E1 + 1 | 48640 | F | (GTP-binding protein) | 2(A) | - | - |
| Evil† | 14013 | 3A3 | -119998 | R | transcription factor | 1(B) | 20 hits | - |
| Lsm10-pending§ | 116748 | 4D2+2 | 12041 | F | mRNA splicing | 3 | - | - |
| LOC383539† | 383539 | 1C5 | -571 | F | (similar to ribosomal protein L30) | 3 | - | - |
| Nfic | 18029 | 10C1 | -11913 | R | p53 regulating nuclear factor | 1(B) | 3 hits | - |
| HaMDR1 67 | ||||||||
| Lak-pending | 71481 | 3G2 | -15 | R | (lymphocyte alfa kinase) | 2(A) | - | - |
| RP23-92G13 | NA | 13 | NA | NA | unknown | 3 | - | - |
| Ccnd2 | 12444 | 6F3 | -14615 | R | CyclinD2 | 1(B) | 18 hits | (CCND1) |
| dsRED 3 | ||||||||
| Bcl11a | 14025 | 11A32 | 167714 | F | zinc finger protein | 1(B) | 6 hits | + |
| Cacnb2 | 12296 | 2(A)2 | 28471 | R | calcium channel | 2(A) | - | - |
| Pdcd1lg1 | 60533 | 19C2 | 34046 | R | programmed cell death ligand | 1(A) | 2 hits | - |
| Phemx (Tssc6) | 27027 | 7F5 | 2431 | F | tetraspanin superfamily member | 2(A) | - | - |
| LOC384063 | 384063 | 4D2+3 | 13364 | R | (sim. to ribosomal protein) | 3 | - | - |
| Runx2 | 12393 | 17B3 | 141712 | R | runt related transcription factor | 1(B) | 6 hits | (RUNX1) |
| Ubb | 22187 | 11B2 | 8464 | F | ubiquitinB | 3 | - | - |
| Cdk6 | 12571 | 5A1 | 157035 | F | Cyclin-dependent kinase | 2(B) | - | (CDK4) |
| dsRED4 | ||||||||
| Mybbp1a | 18432 | 11B4 | 46577 | R | MYB binding protein | 2(B) | - | - |
| Btd | 26363 | 14A3 | 27328 | F | biotinidase | 2(A) | - | - |
| Crsp6 | 234959 | 9A3 | -25697 | R | cofactor of Sp1 transcription factor | 2(B) | - | - |
| Slfn2 | 20556 | 11C | 13391 | R | negative regulation of prolif. | 2(A) | - | - |
Insertion coordinates for HaMDR healthy mice are given based on the NCBI database, July 2004. There were 3 distinct insertions in Evil, as noted in footnotes †, ‡, and §.
NA indicates not applicable; GPR, G-protein-coupled receptor; A, transmembrane receptor or cytoplasmic protein; B, nuclear protein.
Total number of locus hits was 72. There were 10 (13.9%) class 1 genes, 29 (40.3%) class 2 genes, and 33 (45.8%) class 3 genes
Hits shared with leukemia case no. 40
Hits shared with leukemia cases no. 40 and no. 46
Hits shared with leukemia case no. 46
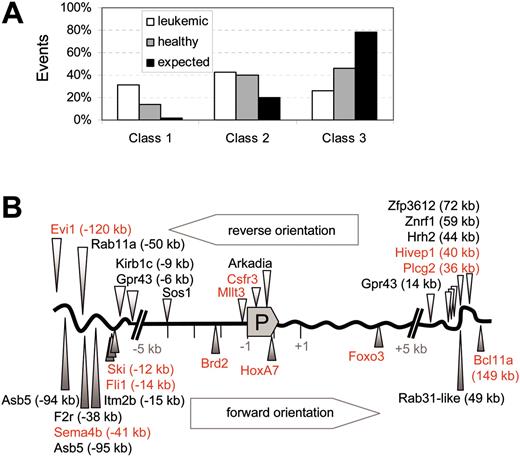
Irrespective of the type of vector involved, we found an overrepresentation of hits in established proto-oncogenes (here summarized as class 1 genes) in leukemias, as compared with healthy controls and predicted gene frequency (Figure 4A). The 2 largest cancer gene databases available (retrovirally tagged [murine] cancer gene database and cancer gene database of human genes)21,35 comprise not more than 2% of the less than 30 000 human or murine genes known to date. Thus, the enrichment in the leukemic samples was more than 15-fold (31.4%), and in healthy animals still about 7-fold (13.9%, Figure 4A). All of the class 1 events found in leukemic clones have previously been implicated in murine or human leukemias or lymphomas (Hivep1, Csfr3, Mllt3, Fli1, Foxo3, HoxA7, Brd2, Bcl11a, Sema4b, Ski, Evi1). If these genes are accepted as the driving force of leukemogenesis, the malignant phenotype was generally consistent with their known mode of action (myeloid leukemias associated with Csfr3, Mllt3, HoxA7, Evi1; erythroid leukemia associated with Fli1). Other “signaling genes” (class 2 genes, including genes involved in cell death, adhesion, proliferation, and developmental decisions), probably accounting for at least 20% of the murine genome,36 were hit with similar frequency in leukemic (43%) and healthy (40%) samples. Those frequencies are in good agreement with recent data that signaling genes may represent 50% of the transcriptome of hematopoietic stem cells.22 Indeed, many of these hits affected genes that belong to pathways with established functions in the regulation of hematopoietic proliferation, differentiation, or apoptosis (Table 3).
Of note, all leukemic clones showed combinations of hits in genes acting as growth factor receptors or in downstream cytoplasmic signal transduction cascades (typically G-protein related) with hits in transcription factors, suggestive of functional cooperations. This was also the case for the single MDR1-associated leukemia (L29, Table 3) in which we could not recover a class 1 hit; in this case, 3 hits escaped database analysis because the flanking sequences were too short.
Leukemia phenotyping. (A) Infiltration of leukemic cells in the spleen (i, ii, iii, viii), thymus (iv), liver (v, vi), and kidney (vii). (i) Erythroid leukemia. Note mitotic figures. (ii) Myeloid leukemia without maturation composed of immature blasts. (iii) Myeloid leukemia with maturation of granulocytes. (iv) T-lymphoblastic lymphoma/leukemia with starry sky cells. (v-vi) Leukemic infiltration as disseminated tumor cells in the sinusoids (v) and/or as cell nests (vi). (vii) T-lymphoblast infiltration. (viii) Myeloid leukemia associated with megakaryocytosis. Original magnifications: × 1000 (i-iv, viii); × 630 (v-vii). Hematoxylin and eosin. A light microscope (Axioplan2, Zeiss, Germany) with 63× and 100× Plan-NEOFLUAR objective lenses was used. The MRGrab (Version 1.0, Zeiss, Germany) with a digital camera (AxioCam MRc, Zeiss, Germany) was used for image processing. (B) FACS profiles of different leukemias. (C) MDR1 activity determined by rhodamine efflux in leukocytes 9 weeks after BMT and in BM cells at the day of sacrifice. Efflux-positive cells are the rhodamine-negative population. Anti-CD3 stain is shown on the y-axis. BM cells of mouse 31 showed no efflux activity after onset of leukemia, except for infiltrating T cells.
Leukemia phenotyping. (A) Infiltration of leukemic cells in the spleen (i, ii, iii, viii), thymus (iv), liver (v, vi), and kidney (vii). (i) Erythroid leukemia. Note mitotic figures. (ii) Myeloid leukemia without maturation composed of immature blasts. (iii) Myeloid leukemia with maturation of granulocytes. (iv) T-lymphoblastic lymphoma/leukemia with starry sky cells. (v-vi) Leukemic infiltration as disseminated tumor cells in the sinusoids (v) and/or as cell nests (vi). (vii) T-lymphoblast infiltration. (viii) Myeloid leukemia associated with megakaryocytosis. Original magnifications: × 1000 (i-iv, viii); × 630 (v-vii). Hematoxylin and eosin. A light microscope (Axioplan2, Zeiss, Germany) with 63× and 100× Plan-NEOFLUAR objective lenses was used. The MRGrab (Version 1.0, Zeiss, Germany) with a digital camera (AxioCam MRc, Zeiss, Germany) was used for image processing. (B) FACS profiles of different leukemias. (C) MDR1 activity determined by rhodamine efflux in leukocytes 9 weeks after BMT and in BM cells at the day of sacrifice. Efflux-positive cells are the rhodamine-negative population. Anti-CD3 stain is shown on the y-axis. BM cells of mouse 31 showed no efflux activity after onset of leukemia, except for infiltrating T cells.
Insertions in (the neighborhood of) other genes (class 3) were more frequently observed in healthy animals than in leukemic animals, indicating that their underrepresentation in leukemias did not reflect the inability to hit such loci in long-term repopulating cells. Among the recipients of cells that were transduced by coculture and expanded prior to transplantation, we found some of the hits recovered from leukemic animals also in healthy controls (mice 42 and 47, Table 4). These may well represent precursor clones that did not accumulate the full series of leukemogenic hits during coculture transduction. Of note, the leukemic samples represented clonal disorders, while samples from healthy mice are likely to represent oligoclonal or polyclonal situations. Thus, healthy controls may have contained “borderline clones,” with hits in proto-oncogenes such as Evi1 (one having the same hit as L40), Sema4b (same hit as L40), Nfic, Ccnd2, Bcl11a, or Pdcdlg1 (Table 4). Even longer observation periods may have revealed the leukemogenic potential of these clones. Vice versa, leukemic samples may contain some “innocent bystander” mutations, especially when associated with a very high vector dose (Table 3).
Vector copy number after engraftment and in leukemias. (A) Transgene copy number in the peripheral blood 9 weeks after transplantation measured by quantitative PCR. Lines indicate the median of each group (Student t test, unpaired, 2-sided). (B) Southern blot analysis of genomic DNA from spleens of leukemic mice. Blots were probed for MDR1 and dsRED2 (marker: Lambda DNA/HindIII marker 2). (C) LM-PCR gel electrophoresis (mice 31.2, 29, 40, 44, and 46). The number of amplified PCR fragments corresponds to the number of insertions detected by Southern blot (marker: GeneRuler DNA ladder mix). The internal band represents the PCR product amplified from the viral 3′ LTR.
Vector copy number after engraftment and in leukemias. (A) Transgene copy number in the peripheral blood 9 weeks after transplantation measured by quantitative PCR. Lines indicate the median of each group (Student t test, unpaired, 2-sided). (B) Southern blot analysis of genomic DNA from spleens of leukemic mice. Blots were probed for MDR1 and dsRED2 (marker: Lambda DNA/HindIII marker 2). (C) LM-PCR gel electrophoresis (mice 31.2, 29, 40, 44, and 46). The number of amplified PCR fragments corresponds to the number of insertions detected by Southern blot (marker: GeneRuler DNA ladder mix). The internal band represents the PCR product amplified from the viral 3′ LTR.
Interestingly, there were also independent insertions in identical loci (Tables 3 and 4). We recovered 3 different hits upstream of Evi1 (L40 = healthy animal 47, and healthy animals 26 and 41), 2 different hits downstream of Bcl11a (L46 and healthy animal dsRED3), 2 different hits in Gpr43 (L31 and L40 = healthy animal 47), and 2 different hits in Plcg2 (L29 and healthy recipient 26). This strongly suggests a selective advantage associated with these insertions.
Higher resolution of target alleles further underlines selective impact of insertional hits
Unselected cell populations have been shown to accumulate around 10% of mouse leukemia virus–derived vector insertions within either of the 5-kb windows upstream or downstream of the transcriptional start site.20 Such insertions may increase or decrease the probability of survival in vivo. Overall, leukemia-associated class 1 hits preferentially occurred 5′ of the promoter, whereas class 3 hits showed a bias for 3′ insertions (Table 3). In contrast, there was no clear preference for forward or reverse orientation of the vector (Figure 4B, showing class 1 and class 2 in leukemias). As shown in Figure 4B, insertions within 5 kb of the transcriptional start site of class 1 genes were overrepresented in leukemias (5 of 11 class 1 hits: 4 upstream, 1 downstream). Such hits were not recovered from healthy animals, suggesting that they may induce clonal extinction unless additional mutations occur. Together, these findings indicated that MDR1 expression itself was not sufficient for clonal selection in leukemogenesis, but rather required combinatorial mutagenesis of crucial growth-regulatory genes.
Evidence of genetic instability with clonal chromosome aberrations
Finally, to address a potential contribution of genetic instability, we employed spectral karyotyping (SKY).37 Metaphase chromosomes were available in 6 leukemias. Within the resolution of SKY, 4 of these showed normal karyotypes (L5.1, L40, L43, L44). In L46 (myeloid leukemia with maturation, hits in HoxA7, Brd2, Bcl11a, and others), 11 of the 14 metaphases analyzed showed an identical balanced translocation between chromosomes 1 and 9 (Figure 5). L31.2 (serially transplanted HaMDR1-associated erythroid leukemia with insertional events in Fli-1, Foxo3, Sos-1, and Gpr43) showed a complex rearrangement involving chromosomes 4 and 11 as well as an additional chromosome 9 derived from an unbalanced translocation T(9;18) (Figure 5). The translocation breakpoints did not coincide with vector insertions. Chromosomal aberrations were thus likely the result of secondary genetic instability in tumor progression rather than due to recombinations between sequence-related transgenes.
Discussion
In the present study, we hypothesized that combinatorial insertional mutagenesis by retroviral vectors may trigger cellular transformation, and concentrated on a model of MDR1 vector–associated leukemias where malignant clones showed multiple vector insertions.12,13 Although our gene transfer rates were even lower than reported in these previous studies, we could demonstrate that MDR1-associated leukemias required clonal selection, and that malignant clones showed the simultaneous presence of at least 3 insertions in potential (class 2) or established (class 1) proto-oncogenes (Table 3). The random nature of such gene hits is a reasonable explanation for the heterogeneous kinetics and phenotypes of the resulting leukemias (Table 2). We observed a similar leukemia using a vector encoding a fluorescent marker protein. Moreover, we demonstrated that an insertional leukemia initiated by retroviral gene transfer could be expanded in vitro and distributed to independent recipients. These findings have important implications for gene therapy.
MDR1 and leukemogenesis
The association of MDR1 vector–induced leukemias with problematic insertional events explains many of the discrepancies between earlier studies. MDR1 did not alter hematopoiesis in carefully controlled studies using transgenic mice or murine FDCP-mix cells, despite clear evidence for functional overexpression.4-6 Also, retroviral MDR1 delivery at low vector dose did not induce hematopoietic alterations in many preclinical studies, including those studying dogs, despite clear evidence of functional overexpression.6,7,16 The only studies revealing an association of MDR1 vector transduction with leukemias have used dose-escalated retroviral transduction conditions.12,13 In retrospect, it remains unclear whether the vector copy number for the control vector was equally high in the nonleukemic control groups of these studies. Our study reveals that MDR1-associated leukemias are closely associated with a transgene copy number of 4 and more, and that MDR1 overexpression is not sufficient for their occurrence. In addition, one leukemia developed after transduction with multiple copies of a vector expressing a fluorescent protein, strongly suggesting that combinatorial vector insertions can be sufficient to induce transformation. Determining the exact contribution of MDR1 to leukemia incidence under conditions of dose escalation, and the potential collaboration of MDR1 overexpression with insertional gene alterations, will require additional studies.
Intriguing combinations and distributions of insertion sites in leukemic clones
Although we cannot provide a formal proof of target gene collaborations on the basis of our present findings, it was intriguing to note a common pattern in all leukemias analyzed for insertion sites: combinatorial events occurred in genes encoding transcription factors along with genes involved in (cytokine) receptor signal cascades. In the context of gene therapy, such combinatorial initiation events are also possible when a vector expressing a signal gene hits a collaborating growth-regulatory gene, as observed in the few previous case reports on vector-induced leukemogenesis.15,17 This is consistent with current concepts of leukemogenesis postulating at least one alteration of a proliferation pathway and another complementing inhibition of a differentiation program.38
Six of the 11 proto-oncogene hits associated with leukemia development in our study (Hivep1, Fli1, HoxA7, Bcl11a, Sema4b, Evi1; Table 3) were independently observed in leukemias elicited by replication-competent retroviruses,35 2 (Mllt3, Foxo3) are represented in the human cancer gene “census,”21 and the remaining 3 (Csfr3, Ski, Brd2) are known from additional literature.39-41 Compared with insertions observed under identical conditions in healthy recipients of gene-modified cells, the accumulation of proto-oncogene hits in leukemias reflects a stringent selection process. This is underlined by the clustering of leukemia-associated class 1 hits in the 5-kb window surrounding the promoter (Figure 4). Moreover, in at least 4 of the 5 cases investigated, the leukemic phenotypes were compatible with the known functions of major proto-oncogenes involved. In keeping with our results in healthy MDR1-modified long-term hematopoiesis, a recent study of MDR1 vector (SF91m3) insertion sites in human hematopoietic cells recovered from transplanted non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice showed an overrepresentation of hits in signaling genes; however, a similar clustering of hits in proto-oncogenes as in the leukemic cases presented here was not observed.42 While these considerations should be sufficient to support the conclusion of combinatorial insertional leukemogenesis as the driving force of MDR1 vector–associated leukemias, additional experiments are required to address the precise consequences of vector insertion on the transcription of the leukemia-associated genes and their mode of action in malignant development.
Class distribution of insertional hits in leukemic animals and healthy controls. (A) The overrepresentation of class 1 hits (established proto-oncogenes) in leukemias. Other signaling genes (class 2) are also more often hit than expected, in both leukemic and healthy mice. Class 3 summarizes hits in other loci. □ indicates leukemic mice; ▦, healthy mice; and ▪, expected value. The expected frequency of hits is calculated on the representation of the gene classes in the mouse genome, as derived from Futreal et al21 and Akagi et al35 for class 1 and Figure 18 (gene ontology) of Waterston et al36 for signaling genes. (B) Schematic representation of the leukemia-associated insertional class 1 hits (red) and class 2 hits (black) with respect to the promoter (P). Hits in reverse orientation are shown above the gene scheme, hits with forward orientation below. There is some preference for the 5-kb window upstream of the transcriptional start site, but not for a particular orientation.
Class distribution of insertional hits in leukemic animals and healthy controls. (A) The overrepresentation of class 1 hits (established proto-oncogenes) in leukemias. Other signaling genes (class 2) are also more often hit than expected, in both leukemic and healthy mice. Class 3 summarizes hits in other loci. □ indicates leukemic mice; ▦, healthy mice; and ▪, expected value. The expected frequency of hits is calculated on the representation of the gene classes in the mouse genome, as derived from Futreal et al21 and Akagi et al35 for class 1 and Figure 18 (gene ontology) of Waterston et al36 for signaling genes. (B) Schematic representation of the leukemia-associated insertional class 1 hits (red) and class 2 hits (black) with respect to the promoter (P). Hits in reverse orientation are shown above the gene scheme, hits with forward orientation below. There is some preference for the 5-kb window upstream of the transcriptional start site, but not for a particular orientation.
Cell expansion and genetic instability
As the leukemia incidence was highest in recipients of ex vivo–expanded cells, our study also demonstrates that prolonged preinfusion culture of insertional mutants may elevate their engraftment potential. Culture conditions may thus select for specific sets of (insertional) mutations. Thus, we found an identical leukemic clone in 2 recipients following ex vivo expansion of cells after dose-escalated gene transfer, and evidence of precursor clones with a subset of the leukemogenic insertions in healthy recipients of expanded cells. Importantly, ex vivo expansion without dose-escalated insertional mutagenesis was not producing a leukemic outcome.
Making use of sensitive SKY analyses,32 we showed clonal chromosome aberrations in 2 insertional leukemias (Figure 5). There was no evidence of recombinations between integrated vector copies. In these cases, we seem to have elicited secondary genetic instability contributing to malignant progression in vivo, as observed in the SCID-X1 patients suffering from insertional lymphoproliferations with clonal chromosome aberrations.17 Molecular characterization of these events may reveal further insights into leukemogenic mechanisms.43 In the remaining leukemias with intact karyotypes, insertional events may have been sufficient for leukemia induction and progression.
Dose-dependent mutagenic toxicity defines a therapeutic window for retroviral transgenesis
The present data indicate that MDR1 vector–associated leukemias require multiple hit insertional mutagenesis as a cooperating factor, opening new perspectives to define a useful therapeutic window for this selectable marker gene. Assuming a risk of at least 10-2 for a single proto-oncogene activation, and an average marking efficiency of at least 5 insertions per cell, the risk for hits in 2 proto-oncogenes of a single cell may become as high as 2.5 × 10-3. In hematopoietic stem cells the risk may even be higher, given the increased accessibility of signal genes.22 However, not every combination of signal alterations may overcome the intrinsic and systemic defense against malignant progression, and not every transplanted cell engrafts. The incidence of leukemogenesis in the worst case scenario tested here (dose escalation of vectors with strong enhancer-promoters plus cell expansion) thus approached 10-6 per treated cell. In addition, our data suggest that the incidence of leukemia development may decline to less than 10-8 per cell with a physiologic vector dose. By introducing a model for reproducible induction of insertional leukemias, our study establishes a platform to define a therapeutic index for integrating vectors, adaptable to the application (and transgene cassette) of interest. Such a model is of great importance for the mechanistic understanding and preclinical development of virus vectors, because it allows to investigate potential specific interactions of insertional signal perturbations with side effects of transgene expression.
SKY analysis of cytogenetically aberrant clones L46 and L31.2. In this classified image, altered chromosome pairs are boxed. Translocated regions are marked with yellow numbers. In L46, a balanced translocation T(1E3-F;9C-D) was present. The karyotype is described as: 37-40, XY, T(1E3-F;9C-D)[11]. L31.2 showed a complex rearrangement involving chromosomes 4 and 11 as well as an additional chromosome 9 derived from an unbalanced translocation T(9B;18A2). The karyotype is described as: 38-40, XX, Der(4)T(4A2;11D), +Der(9)T(9B; 18A2), Der(11)T(11pter->11B3-?4::4A2-A3->4D3::11B3-4->11D::4A2-A3->4A1), -18[13]/40,XX [2.].
SKY analysis of cytogenetically aberrant clones L46 and L31.2. In this classified image, altered chromosome pairs are boxed. Translocated regions are marked with yellow numbers. In L46, a balanced translocation T(1E3-F;9C-D) was present. The karyotype is described as: 37-40, XY, T(1E3-F;9C-D)[11]. L31.2 showed a complex rearrangement involving chromosomes 4 and 11 as well as an additional chromosome 9 derived from an unbalanced translocation T(9B;18A2). The karyotype is described as: 38-40, XX, Der(4)T(4A2;11D), +Der(9)T(9B; 18A2), Der(11)T(11pter->11B3-?4::4A2-A3->4D3::11B3-4->11D::4A2-A3->4A1), -18[13]/40,XX [2.].
Prepublished online as Blood First Edition Paper, February 15, 2005; DOI 10.1182/blood-2004-11-4535.
Supported by grants of the Deutsche Forschungsgemeinschaft (KFO110-A1, FE 568/5-1) and the European Community (QLRT-2001-00 427; LSHB-CT-2004-005 242).
U.M. and O.S.K. contributed equally to this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Christopher Lutz (Cincinnati Children's Hospital Medical Center) for assistance with LAM-PCR and Sabine Knöß (Hannover Medical School) for assistance with RCR assays and cell expansion cultures. Carol Stocking provided us with probes and helpful suggestions in RCR assays and leukemia phenotyping by flow cytometry. We are most grateful to the Heinrich Pette-Institute, in particular Klaus Harbers, Anke Wahlers, and Janine Kraunus for supporting long-term observation of mice.

![Figure 5. SKY analysis of cytogenetically aberrant clones L46 and L31.2. In this classified image, altered chromosome pairs are boxed. Translocated regions are marked with yellow numbers. In L46, a balanced translocation T(1E3-F;9C-D) was present. The karyotype is described as: 37-40, XY, T(1E3-F;9C-D)[11]. L31.2 showed a complex rearrangement involving chromosomes 4 and 11 as well as an additional chromosome 9 derived from an unbalanced translocation T(9B;18A2). The karyotype is described as: 38-40, XX, Der(4)T(4A2;11D), +Der(9)T(9B; 18A2), Der(11)T(11pter->11B3-?4::4A2-A3->4D3::11B3-4->11D::4A2-A3->4A1), -18[13]/40,XX [2.].](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/11/10.1182_blood-2004-11-4535/6/m_zh80110579290005.jpeg?Expires=1767780668&Signature=K4M6nfWw81~eOEQLtvPaS2GrNdyKQDVYlld5dD1fJMVyiCFNycxQq96iMuHf36JLJz9oVbLzIHJvJdo8hbZXBcodbtrwDDhIxPIdOm-xJ~c1MZPRV1n0vAuNNCrEvTb7TcCvgVpxl2phYNfOzeEv3T83pHVrKdFIopSqjLg2QNvOjrAXTejRcdjUw3gWxxRq4k01gcqSWj8yOIpYqNvuw7b2fSLVb9xBthLYU1iLRtvE7I4uRypnOzmlJpOc0kjbM4NxO1SAC5fiGS8TVaYnc0YZzbS4q1YcaY2mk7F~rYTOpkiIiBgJI4Oty9pzu8-6CitucBZz5wDApveSFmbrQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal