Abstract
Acquired genetic characteristics of acute lymphoblastic leukemia (ALL) cells are used to individualize therapy, whereas germ line genetic characteristics generally are not. We determined whether ALL outcome was related to 16 genetic polymorphisms affecting the pharmacodynamics of antileukemic agents. Of 246 children, 116 were treated on the lower-risk (LR) and 130 on the higher-risk (HR) arms of a St Jude protocol. Patients in the HR group with the glutathione S-transferase (GSTM1) nonnull genotype had greater risk of hematologic relapse (P = .03), which was further increased by the thymidylate synthetase (TYMS) 3/3 genotype (P = .03). These genotypes remained predictive in multivariate analyses (P < .001 and .003, respectively). No genotypes were predictive in the LR arm. Expression of these 2 genes in ALL blasts was lower in those with low-activity genotypes. For central nervous system relapse, among the HR group, the vitamin D receptor start site (P = .02) and intron 8 genotypes (P = .04) predisposed, whereas for LR patients the TYMS 3/3 genotype predisposed (P = .04). The GSTM1 non-null and TYMS 3/3 genotypes are plausibly linked to drug resistance. Polymorphisms interact to influence antileukemic outcome and represent determinants of response that can be used to optimize therapy. (Blood. 2005;105:4752-4758)
Introduction
Childhood acute lymphoblastic leukemia (ALL) is cured in approximately 80% of patients.1-4 One treatment strategy for improving cure has been to modify the intensity of therapy based on the acquired genetic characteristics of the leukemia, using more intensive therapy for ALL with molecular markers (eg, the t(9;22) or MLL translocations), indicating a resistant leukemia.5 Other than age, host characteristics are usually not used for assigning patients with ALL to risk groups or to determine the intensity of therapy.
ALL regimens consist of combination chemotherapy (eg, vincristine, glucocorticoid, methotrexate, a thiopurine, and asparaginase with or without anthracyclines, topoisomerase II inhibitors, cytarabine, and cyclophosphamide) administered continuously over 2 to 3 years.1-4,6 Outcome of ALL may be influenced by modest changes in drug dose or exposure.7-10 Thus, if the determinants of interpatient variability in drug pharmacodynamics were better defined, tailoring drug therapy based on these factors might further improve outcome.
Germ line polymorphisms in genes that code for the proteins involved in the pharmacodynamics of antileukemic agents are common, with the frequency of the “variant” allele ranging from 5% to 50%.11,12 Although we and others have explored whether germ line polymorphisms may relate to ALL outcomes,13-19 such studies have examined only a few polymorphisms at a time, have failed to adjust for other risk factors, have not distinguished among relapse risk versus other types of adverse outcome, or have potential for selection bias by including only a small fraction of patients enrolled on the treatment protocols. Most prior studies have not assessed whether gene-gene interactions could influence the relationships between germ line polymorphisms and outcomes.
Herein, we have used a candidate gene approach to assess whether ALL outcome is related to 16 common polymorphisms in genes plausibly linked to the pharmacodynamics of the drug therapy, accounting for known prognostic factors, types of ALL outcome, competing risks, and gene-gene interactions.
Patients, materials, and methods
Study population
Of the 247 children with newly diagnosed ALL treated on the St Jude Children's Research Hospital Total XIIIB study for ALL,20 246 were included; one patient died within a few weeks after diagnosis and a germ line DNA sample could not be obtained. The research was approved by our Institutional Review Board, and informed consent was obtained from parents, guardians, or patients (as appropriate). Patients were prospectively assigned to risk groups that determined the intensity of therapy; those with at least one of the following were assigned to the higher-risk group: age younger than 1 year or older than 10 years (except cases with DNA index ≥ 1.16 and ≤ 1.60), initial leukocyte count greater than 50 × 109/L (except cases with DNA index ≥ 1.16 and ≤ 1.60), central nervous system (CNS) involvement (defined as ≥ 5 white cells/mm3 in cerebrospinal fluid and the presence of leukemic blasts on cytocentrifuge preparation from a nontraumatic spinal tap, or cranial palsies), testicular involvement, greater than 5% leukemic blasts in bone marrow on day 19, T-cell ALL, presence of BCR-ABL [t(9;22)], MLL-AF4 [t(4;11)], E2A-PBX1 [t(1;19)] in pre-B ALL, or MLL rearrangement by karyotype and/or molecular probes. All others were enrolled on the lower-risk arm.21 Minimal residual disease on day 43 was assessed by flow cytometric assays as previously described.21,22
Race/ethnicity was classified by a hospital staff member at the time of diagnosis (using 1994-1998 policies). Parents (or patients if older than 18 years of age) were asked which of the race categories (American Indian, black, Chinese, Filipino, Hispanic, Japanese, white, or unknown) that they considered applied to their child. Race and/or ethnicity was assessed in this study, because of the prior association of race with ALL outcome in some studies,23,24 and because of the considerable racial differences in frequencies of genotypes.11,12,25
In summary, treatment consisted of high-dose methotrexate, mercaptopurine, or both, followed by therapy with prednisone, vincristine, daunorubicin, asparaginase, cytarabine, and etoposide.20 At complete remission, patients received 2 weeks of high-dose methotrexate and mercaptopurine, followed by 120 weeks of risk-directed postremission rotating pairs of chemotherapy. For lower-risk cases, postremission therapy consisted of daily mercaptopurine and weekly methotrexate, high-dose methotrexate and mercaptopurine every 8 weeks for the first year, and dexamethasone plus vincristine every 4 weeks. For higher-risk cases, postremission therapy consisted of drug pairs (etoposide, cyclophosphamide, mercaptopurine, methotrexate, cytarabine, dexamethasone, vincristine, and mercaptopurine) administered in weekly rotation, interrupted by reinduction therapy from weeks 16 to 21.21 Events were defined as any relapse, death, or secondary malignancies, using criteria previously published.26
Genotyping
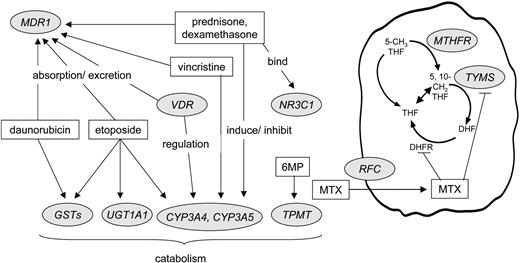
DNA was extracted from normal blood cells. Candidate genes exhibited polymorphisms and encoded proteins that were involved in the pharmacokinetics or pharmacodynamics of antileukemic agents (Figure 1). Priority was given to polymorphisms that were demonstrated to be associated with phenotypes in both clinical and preclinical studies.9,11-14,27-33 Vincristine, prednisone, dexamethasone, etoposide, and daunorubicin are substrates for cytochrome P4503A4 (CYP3A4), cytochrome P4503A5 (CYP3A5), and/or p-glycoprotein (MDR1), which are partially regulated by the vitamin D receptor (VDR).27,34,35 The parent drug or metabolites of etoposide, daunorubicin, and cyclophosphamide are substrates for glutathione S-transferases (GSTM1, GSTP1, and GSTT1).28 Etoposide is glucuronidated by UDP-glucuronosyltransferase 1A1 (UGT1A1).36 Mercaptopurine is a substrate for thiopurine S-methyltransferase (TPMT),9 and methotrexate interacts with methylenetetrahydrofolate reductase (MTHFR), thymidylate synthetase (TYMS), and the reduced folate carrier (RFC, or SLC19A1).11 Prednisone and dexamethasone bind to the glucocorticoid receptor (NR3C1).29
Interaction of primary antileukemic agents (in rectangles) with products of polymorphic genes (in circles) that were subject of study. GMP indicates mercaptopurine; MTX, methotrexate.
Interaction of primary antileukemic agents (in rectangles) with products of polymorphic genes (in circles) that were subject of study. GMP indicates mercaptopurine; MTX, methotrexate.
Genotyping was performed for the 16 polymorphic loci using methods as previously described: CYP3A4*1B (A>G at position -392) and CYP3A5*3 (G>A at position 22893)37 ; GSTP1 313A>G,37 GSTM1 deletion and GSTT1 deletion16 ; MDR1 exon 21 (2677G>T/A) and MDR1 exon 26 (3435C>T)37 ; MTHFR 677C>T38 and MTHFR 1298A>C30 ; NR3C1 1088A>G31 ; RFC 80G>A38 ; TPMT 238G>C, TPMT 460G>A, and TPMT 719A>G32 ; TYMS enhancer repeat33 ; UGT1A1 promoter repeat polymorphism37 ; VDR intron 8 G>A, and VDR FokI (start-site) T>C.37
Gene expression
In all children with available RNA in their diagnostic ALL blasts, gene expression levels were analyzed using the Affymetrix GeneChip array HG_U95Av2, as described.39 Expression signals were scaled to a target average intensity and analyzed using Affymetrix Microarray Suite (MAS) version 5.0 (Affymetrix, Santa Clara, CA). The expression levels of GSTM1 and TYMS were based on probe sets 39054_at and 37899_at, respectively. Patients whose blasts had numeric or structural abnormalities of chromosomes 1 and 18 were excluded from the analysis of blast expression of the GSTM1 and TYMS genes, located on 1p13.3 and 18p11.32, respectively.
Statistical analysis
The distribution of genotypes between racial groups or between risk groups was compared using chi-square test with Yates correction. The cumulative incidences of hematologic and CNS relapse by risk group were compared using the Gray estimator with incorporation of competing events.40 Death in remission and second malignancy were treated as competing risks. Survival time was the time between diagnosis and the date of last follow-up.
Genotypes for each of the 16 polymorphisms were pooled into binary groups for the statistical analysis. For GSTM1 deletion, GSTT1 deletion, and NR3C1 1088A>G, the genotypic groups were originally measured as binary (Table 1). For CYP3A4*1B, CYP3A5*3, GSTP1 313A>G, MDR1 exon 21 (2677G>T/A), MDR1 exon 26 (3435C>T), MTHFR 677C>T, MTHFR 1298A>C, RFC 80G>A, VDR intron 8 G>A, and VDR FokI T>C loci, it was not clear a priori how to pool genotypes, and so the analysis was conducted using both possible major genotypic groups (eg, RFC 80 GG versus GA or AA, and RFC 80 GG or GA versus AA). For each locus, the groupings that yielded the most significant predictors of outcome are indicated in Table 1. For TYMS enhancer and UGT1A1 promoter repeats, genotypes were merged according to prior functional data into 2 distinct groups (TYMS 3/3 versus others, and UGT1A1 7/7 versus others); rare genotypes (ie, TYMS 2/9 [n = 1] and UGT1A1 5/7 [n = 2]) were excluded from the analysis. For TPMT, common haplotypes that combined TPMT 460G and TPMT 719A, which constitute the *3A allele, were assumed, so that none of the patients were homozygous deficient for TPMT (Table 1).32
Genotype frequencies by race and risk group
. | No. patients, higher-risk arm (%) . | . | . | . | No. patients, lower-risk arm (%) . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci and genotypes . | Whites . | Blacks . | Others . | Total . | Whites . | Blacks . | Others . | Total . | ||||||
| CYP3A4*1B | ||||||||||||||
| AA | 88 (96.7) | 4 (13.8) | 8 (80) | 100 (76.9) | 75 (93.7) | 3 (18.7) | 19 (95) | 97 (83.6) | ||||||
| AG and GG | 3 (3.3) | 25 (86.2) | 2 (20) | 30 (23.1) | 5 (6.3) | 13 (81.3) | 1 (5) | 19 (16.4) | ||||||
| CYP3A5*3 | ||||||||||||||
| GG | 75 (82.4) | 4 (13.8) | 7 (70) | 86 (66.1) | 67 (83.7) | 1 (6.2) | 15 (75) | 83 (71.5) | ||||||
| AG and AA | 16 (17.6) | 25 (86.2) | 3 (30) | 44 (33.9) | 13 (16.3) | 15 (93.8) | 5 (25) | 33 (28.5) | ||||||
| GSTM1 deletion | ||||||||||||||
| Null | 42 (46.1) | 12 (41.4) | 5 (50) | 59 (45.4) | 37 (46.2) | 4 (25) | 5 (25) | 46 (39.7) | ||||||
| Nonnull | 49 (53.9) | 17 (58.6) | 5 (50) | 71 (54.6) | 43 (53.8) | 12 (75) | 15 (75) | 70 (60.3) | ||||||
| GSTP1 313A>G | ||||||||||||||
| GG | 9 (9.9) | 9 (31) | 3 (30) | 21 (16.2) | 15 (18.7) | 4 (25) | 1 (5) | 20 (17.2) | ||||||
| AG and AA | 82 (90.1) | 20 (69) | 7 (70) | 109 (83.8) | 65 (81.3) | 12 (75) | 19 (95) | 96 (82.8) | ||||||
| GSTT1 deletion | ||||||||||||||
| Null | 16 (17.6) | 3 (10.3) | 1 (10) | 20 (15.4) | 10 (12.5) | 5 (31.2) | 5 (25) | 20 (17.2) | ||||||
| Nonnull | 75 (82.4) | 26 (89.7) | 9 (90) | 110 (84.6) | 70 (87.5) | 11 (68.8) | 15 (75) | 96 (82.8) | ||||||
| MDR1 exon 21 G>T/A | ||||||||||||||
| GG | 19 (20.9) | 25 (86.2) | 5 (50) | 49 (37.7) | 24 (30) | 12 (75) | 6 (30) | 42 (36.2) | ||||||
| GT and others | 72 (79.1) | 4 (13.8) | 5 (50) | 81 (62.3) | 56 (70) | 4 (25) | 14 (70) | 74 (63.8) | ||||||
| MDR1 exon 26 C>T | ||||||||||||||
| CC | 14 (15.4) | 22 (75.9) | 2 (20) | 38 (29.2) | 20 (25) | 7 (43.7) | 5 (25) | 32 (27.6) | ||||||
| CT and TT | 77 (84.6) | 7 (24.1) | 8 (80) | 92 (70.8) | 60 (75) | 9 (56.3) | 15 (75) | 84 (72.4) | ||||||
| MTHFR 677C>T | ||||||||||||||
| CC | 40 (44) | 22 (75.9) | 6 (60) | 68 (52.3) | 36 (45) | 9 (56.3) | 9 (45) | 54 (46.5) | ||||||
| CT and TT | 51 (56) | 7 (24.1) | 4 (40) | 62 (47.7) | 44 (55) | 7 (43.7) | 11 (55) | 62 (53.5) | ||||||
| MTHFR 1298A>C | ||||||||||||||
| AA | 49 (53.9) | 19 (65.6) | 2 (20) | 70 (53.9) | 39 (48.7) | 11 (68.8) | 10 (50) | 60 (51.7) | ||||||
| AC and CC | 42 (46.1) | 10 (34.4) | 8 (80) | 60 (46.1) | 41 (51.3) | 5 (31.2) | 10 (50) | 56 (48.3) | ||||||
| NR3C1 1088A>G | ||||||||||||||
| AA | 86 (94.5) | 29 (100) | 9 (90) | 124 (95.4) | 78 (97.5) | 16 (100) | 20 (100) | 114 (98.3) | ||||||
| AG | 5 (5.5) | 0 (0) | 1 (10) | 6 (4.6) | 2 (2.5) | 0 (0) | 0 (0) | 2 (1.7) | ||||||
| RFC 80A>G | ||||||||||||||
| GG | 33 (36.3) | 8 (27.6) | 3 (30) | 44 (33.8) | 25 (31.2) | 4 (25) | 4 (20) | 33 (28.4) | ||||||
| AG and AA | 58 (63.7) | 21 (72.4) | 7 (70) | 86 (66.2) | 55 (68.8) | 12 (75) | 16 (80) | 83 (71.6) | ||||||
| TPMT combined genotypes | ||||||||||||||
| 238GG, 460GG and 719AA | 84 (92.3) | 28 (96.5) | 9 (90) | 121 (93.1) | 77 (96.3) | 15 (93.7) | 18 (90) | 110 (94.8) | ||||||
| Others | 7 (7.7) | 1 (3.5) | 1 (10) | 9 (6.9) | 3 (3.7) | 1 (6.3) | 2 (10) | 6 (5.2) | ||||||
| TYMS enhancer repeat | ||||||||||||||
| 3/3 | 23 (25.3) | 15 (51.7) | 5 (50) | 43 (33.1) | 19 (23.7) | 4 (25) | 12 (60) | 35 (30.2) | ||||||
| 2/2, 2/3 and others* | 68 (74.7) | 14 (48.3) | 5 (50) | 87 (66.9) | 61 (76.3) | 12 (75) | 8 (40) | 81 (69.8) | ||||||
| UGT1A1 promoter repeat | ||||||||||||||
| 7/7 | 8 (8.8) | 2 (6.9) | 1 (10) | 11 (8.5) | 7 (8.7) | 3 (18.7) | 4 (20) | 14 (12.1) | ||||||
| 6/6, 6/7 and others† | 83 (91.2) | 27 (93.1) | 9 (90) | 119 (91.5) | 73 (91.3) | 13 (81.3) | 16 (80) | 102 (87.9) | ||||||
| VDR intron 8 G>A | ||||||||||||||
| GG | 39 (42.9) | 18 (62.1) | 4 (40) | 61 (46.9) | 30 (37.5) | 9 (56.3) | 12 (60) | 51 (44) | ||||||
| GA and AA | 52 (57.1) | 11 (37.9) | 6 (60) | 69 (53.1) | 50 (62.5) | 7 (43.7) | 8 (40) | 65 (56) | ||||||
| VDR Fokl | ||||||||||||||
| CC | 37 (40.7) | 17 (58.6) | 5 (50) | 59 (45.4) | 30 (37.5) | 10 (62.5) | 13 (65) | 53 (45.7) | ||||||
| TC and T T | 54 (59.3) | 12 (41.4) | 5 (50) | 71 (54.6) | 50 (62.5) | 6 (37.5) | 7 (35) | 63 (54.3) | ||||||
. | No. patients, higher-risk arm (%) . | . | . | . | No. patients, lower-risk arm (%) . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci and genotypes . | Whites . | Blacks . | Others . | Total . | Whites . | Blacks . | Others . | Total . | ||||||
| CYP3A4*1B | ||||||||||||||
| AA | 88 (96.7) | 4 (13.8) | 8 (80) | 100 (76.9) | 75 (93.7) | 3 (18.7) | 19 (95) | 97 (83.6) | ||||||
| AG and GG | 3 (3.3) | 25 (86.2) | 2 (20) | 30 (23.1) | 5 (6.3) | 13 (81.3) | 1 (5) | 19 (16.4) | ||||||
| CYP3A5*3 | ||||||||||||||
| GG | 75 (82.4) | 4 (13.8) | 7 (70) | 86 (66.1) | 67 (83.7) | 1 (6.2) | 15 (75) | 83 (71.5) | ||||||
| AG and AA | 16 (17.6) | 25 (86.2) | 3 (30) | 44 (33.9) | 13 (16.3) | 15 (93.8) | 5 (25) | 33 (28.5) | ||||||
| GSTM1 deletion | ||||||||||||||
| Null | 42 (46.1) | 12 (41.4) | 5 (50) | 59 (45.4) | 37 (46.2) | 4 (25) | 5 (25) | 46 (39.7) | ||||||
| Nonnull | 49 (53.9) | 17 (58.6) | 5 (50) | 71 (54.6) | 43 (53.8) | 12 (75) | 15 (75) | 70 (60.3) | ||||||
| GSTP1 313A>G | ||||||||||||||
| GG | 9 (9.9) | 9 (31) | 3 (30) | 21 (16.2) | 15 (18.7) | 4 (25) | 1 (5) | 20 (17.2) | ||||||
| AG and AA | 82 (90.1) | 20 (69) | 7 (70) | 109 (83.8) | 65 (81.3) | 12 (75) | 19 (95) | 96 (82.8) | ||||||
| GSTT1 deletion | ||||||||||||||
| Null | 16 (17.6) | 3 (10.3) | 1 (10) | 20 (15.4) | 10 (12.5) | 5 (31.2) | 5 (25) | 20 (17.2) | ||||||
| Nonnull | 75 (82.4) | 26 (89.7) | 9 (90) | 110 (84.6) | 70 (87.5) | 11 (68.8) | 15 (75) | 96 (82.8) | ||||||
| MDR1 exon 21 G>T/A | ||||||||||||||
| GG | 19 (20.9) | 25 (86.2) | 5 (50) | 49 (37.7) | 24 (30) | 12 (75) | 6 (30) | 42 (36.2) | ||||||
| GT and others | 72 (79.1) | 4 (13.8) | 5 (50) | 81 (62.3) | 56 (70) | 4 (25) | 14 (70) | 74 (63.8) | ||||||
| MDR1 exon 26 C>T | ||||||||||||||
| CC | 14 (15.4) | 22 (75.9) | 2 (20) | 38 (29.2) | 20 (25) | 7 (43.7) | 5 (25) | 32 (27.6) | ||||||
| CT and TT | 77 (84.6) | 7 (24.1) | 8 (80) | 92 (70.8) | 60 (75) | 9 (56.3) | 15 (75) | 84 (72.4) | ||||||
| MTHFR 677C>T | ||||||||||||||
| CC | 40 (44) | 22 (75.9) | 6 (60) | 68 (52.3) | 36 (45) | 9 (56.3) | 9 (45) | 54 (46.5) | ||||||
| CT and TT | 51 (56) | 7 (24.1) | 4 (40) | 62 (47.7) | 44 (55) | 7 (43.7) | 11 (55) | 62 (53.5) | ||||||
| MTHFR 1298A>C | ||||||||||||||
| AA | 49 (53.9) | 19 (65.6) | 2 (20) | 70 (53.9) | 39 (48.7) | 11 (68.8) | 10 (50) | 60 (51.7) | ||||||
| AC and CC | 42 (46.1) | 10 (34.4) | 8 (80) | 60 (46.1) | 41 (51.3) | 5 (31.2) | 10 (50) | 56 (48.3) | ||||||
| NR3C1 1088A>G | ||||||||||||||
| AA | 86 (94.5) | 29 (100) | 9 (90) | 124 (95.4) | 78 (97.5) | 16 (100) | 20 (100) | 114 (98.3) | ||||||
| AG | 5 (5.5) | 0 (0) | 1 (10) | 6 (4.6) | 2 (2.5) | 0 (0) | 0 (0) | 2 (1.7) | ||||||
| RFC 80A>G | ||||||||||||||
| GG | 33 (36.3) | 8 (27.6) | 3 (30) | 44 (33.8) | 25 (31.2) | 4 (25) | 4 (20) | 33 (28.4) | ||||||
| AG and AA | 58 (63.7) | 21 (72.4) | 7 (70) | 86 (66.2) | 55 (68.8) | 12 (75) | 16 (80) | 83 (71.6) | ||||||
| TPMT combined genotypes | ||||||||||||||
| 238GG, 460GG and 719AA | 84 (92.3) | 28 (96.5) | 9 (90) | 121 (93.1) | 77 (96.3) | 15 (93.7) | 18 (90) | 110 (94.8) | ||||||
| Others | 7 (7.7) | 1 (3.5) | 1 (10) | 9 (6.9) | 3 (3.7) | 1 (6.3) | 2 (10) | 6 (5.2) | ||||||
| TYMS enhancer repeat | ||||||||||||||
| 3/3 | 23 (25.3) | 15 (51.7) | 5 (50) | 43 (33.1) | 19 (23.7) | 4 (25) | 12 (60) | 35 (30.2) | ||||||
| 2/2, 2/3 and others* | 68 (74.7) | 14 (48.3) | 5 (50) | 87 (66.9) | 61 (76.3) | 12 (75) | 8 (40) | 81 (69.8) | ||||||
| UGT1A1 promoter repeat | ||||||||||||||
| 7/7 | 8 (8.8) | 2 (6.9) | 1 (10) | 11 (8.5) | 7 (8.7) | 3 (18.7) | 4 (20) | 14 (12.1) | ||||||
| 6/6, 6/7 and others† | 83 (91.2) | 27 (93.1) | 9 (90) | 119 (91.5) | 73 (91.3) | 13 (81.3) | 16 (80) | 102 (87.9) | ||||||
| VDR intron 8 G>A | ||||||||||||||
| GG | 39 (42.9) | 18 (62.1) | 4 (40) | 61 (46.9) | 30 (37.5) | 9 (56.3) | 12 (60) | 51 (44) | ||||||
| GA and AA | 52 (57.1) | 11 (37.9) | 6 (60) | 69 (53.1) | 50 (62.5) | 7 (43.7) | 8 (40) | 65 (56) | ||||||
| VDR Fokl | ||||||||||||||
| CC | 37 (40.7) | 17 (58.6) | 5 (50) | 59 (45.4) | 30 (37.5) | 10 (62.5) | 13 (65) | 53 (45.7) | ||||||
| TC and T T | 54 (59.3) | 12 (41.4) | 5 (50) | 71 (54.6) | 50 (62.5) | 6 (37.5) | 7 (35) | 63 (54.3) | ||||||
All 16 genotypes are presented in the binary groups used for analysis, and the number of patients carrying those genotypes is shown in absolute numbers and in percentage (in parentheses). The most common allele for each locus is underlined. For CYP3A4*1B, CYP3A5*3, GSTP1 313A>G, MDR1 exon 21 (2677G>T/A), MDR1 exon 26 (3435C>T), MTHFR 677C>T, MTHFR 1298A>C, RFC 80G>A, VDR intron8 G>A, and VDR Fokl T>C loci, genotypes were pooled according to their significant association with outcome, as described in “Patients, materials, and methods.”
Rare genotypes: TYMS 2/9 (n = 1), 3/4 (n = 3), 3/7 (n = 1).
Rare genotypes: UGT1A1 5/6 (n = 6), 5/7 (n = 2), 6/8 (n = 2), 7/8 (n = 2).
To assess possible pharmacogenetic determinants of relapse, a classification and regression tree (CART) approach was used.41 Statistical analysis was performed on each polymorphism individually to make a decision tree for prediction of relapses. For each outcome, the most significant polymorphism determined the second node of the CART (after treatment arm), and polymorphisms for the remaining 15 loci were then analyzed, and the process was repeated. At each node, all the loci that had not yet been assessed for association between genotypes with outcome were analyzed, and the polymorphism with the most significant association (by the smallest P value using Gray test) was used to define that node and the downward branches. The CARTs were stopped either when no further significant genotypes were observed or when the number of patients in the terminal nodes was equal to 10. In these univariate analyses, Gray test was used to compare the cumulative incidences of relapse by genotypic groups.40
To assess whether genetic variants had independent prognostic significance for outcome, we performed multiple regression analyses adjusting for the established risk factors in this protocol21 : initial leukocyte count (< 100 × 109/L versus ≥ 100 × 109/L), DNA index (1.16-1.6 versus others), presence versus absence of BCR-ABL [t(9;22)] or of MLL-AF4 [t(4;11)], percentage of blasts in the bone marrow on day 19 (< 5% blasts versus ≥ 5% blasts), and minimal residual disease on remission date (< 0.01% versus ≥ 0.01%). For the analysis of CNS relapse, CNS status at diagnosis was also included as a possible prognostic factor (no blasts in cerebrospinal fluid in an atraumatic or traumatic lumbar puncture versus other CNS status). Fine and Gray estimator with incorporation of competing events was used to compare the cumulative incidence of relapse by genotypic groups.42
To assess the possible correlation between genotypes at the 16 polymorphic loci, pair-wise associations were performed using Fisher exact or chi-square tests. We used Wilcoxon rank-sum test to compare the expression levels of GSTM1 and TYMS among genotypic groups.
Results
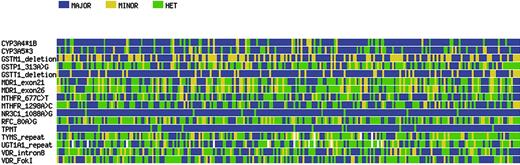
After a median follow-up of 5.3 years, there have been a total of 47 adverse events (relapses, second malignancies, and deaths in remission), including 9 relapses in the lower-risk arm and 23 in the higher-risk arm. Because the type of treatment, as well as relapse incidence and event-free survival, differed by risk group (P = .01 and P = .002, respectively), we performed the analysis separately in the lower- and higher-risk groups. Although many allele frequencies differed significantly between blacks and whites (Table 1), and race has been associated with ALL cure rates in other studies,23,24 race was not a prognostic factor in this single-institution protocol.21,43 Thus, the genotype/phenotype association analysis was not stratified by race. Genotypes of the 16 loci in 246 patients are summarized in Figure 2.
Visual genotype graph for 16 polymorphic loci (rows) among the 246 patients (columns) with ALL treated in the Total XIIIB Protocol. Those homozygous for the major allele are depicted in blue, homozygous for the minor allele in yellow, and heterozygous are in green. For GSTM1 and GSTT1 deletions, the nonnull and the null genotypes are represented in blue and in yellow, respectively. The rare genotypes for TYMS and UGT1A1 were excluded from the plot (in white).
Visual genotype graph for 16 polymorphic loci (rows) among the 246 patients (columns) with ALL treated in the Total XIIIB Protocol. Those homozygous for the major allele are depicted in blue, homozygous for the minor allele in yellow, and heterozygous are in green. For GSTM1 and GSTT1 deletions, the nonnull and the null genotypes are represented in blue and in yellow, respectively. The rare genotypes for TYMS and UGT1A1 were excluded from the plot (in white).
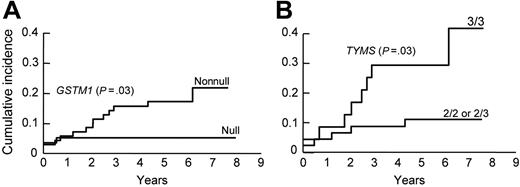
Among those on the higher-risk arm, the nonnull GSTM1 genotype (Figure 3A) was associated with an increased risk of hematologic relapse (isolated or combined) (5-year cumulative incidence, 17.1% ± 4.5% versus 5.1% ± 2.9% for GSTM1 null genotype, P = .03). Among those with the GSTM1 nonnull genotype, the presence of the TYMS 3/3 genotype was associated with a higher-risk of hematologic relapse (5-year cumulative incidence, 29.2% ± 9.5% versus 10.9% ± 4.7% for TYMS 2/3 or 2/2 genotypes, P = .02) (Figure 3B), whereas among those with GSTM1 null genotype, the NR3C1 AG genotype was associated with an increased risk of hematologic relapse (5-year cumulative incidence, 33.3% ± 33.3% versus 3.6% ± 2.5% for NR3C1 AA genotype, P = .02). In univariate analyses, the only other significant predictors of hematologic relapse were the presence of BCR-ABL [t(9;22)] at diagnosis (P < .001), the percentage of blasts in the day 19 bone marrow (P < .001), and minimal residual disease status on remission date (P < .001). In multivariate analyses, including other features predictive of hematologic relapse, either the GSTM1 nonnull genotype alone (P < .001) or the combined GSTM1 nonnull and TYMS 3/3 genotypes (P = .003), were independent prognostic factors for hematologic relapse (Tables 2 and 3). No genotype was associated with hematologic relapse among those patients on the lower-risk arm.
Risk of hematologic relapse among patients on the higher-risk arm, based on GSTM1 genotype, by multivariate analysis
Feature* . | Coefficient . | SE . | Hazard ratio (95% CI) . | P† . |
|---|---|---|---|---|
| GSTM1 (nonnull vs null) | 2.90 | 0.784 | 18.1 (3.9-84.3) | .0002 |
| Leukocyte count (fewer than 100 × 109/L vs at least 100 × 109/L) | -1.74 | 0.844 | 0.18 (0.03-0.92) | .04 |
| t(9;22)/BCR-ABL (present vs absent) | 0.83 | 0.949 | 2.29 (0.36-14.7) | .38 |
| t(4;11)/MLL-AF4 (present vs absent) | 2.94 | 0.972 | 19.0 (2.82-128) | .0025 |
| Day 19 marrow (at least 5% blasts vs less than 5% blasts) | -0.46 | 0.888 | 0.63 (0.11-3.58) | .60 |
| Minimal residual disease on remission date (at least 0.01% vs less than 0.01%) | 2.69 | 0.972 | 14.7 (2.18-98.5) | .0057 |
Feature* . | Coefficient . | SE . | Hazard ratio (95% CI) . | P† . |
|---|---|---|---|---|
| GSTM1 (nonnull vs null) | 2.90 | 0.784 | 18.1 (3.9-84.3) | .0002 |
| Leukocyte count (fewer than 100 × 109/L vs at least 100 × 109/L) | -1.74 | 0.844 | 0.18 (0.03-0.92) | .04 |
| t(9;22)/BCR-ABL (present vs absent) | 0.83 | 0.949 | 2.29 (0.36-14.7) | .38 |
| t(4;11)/MLL-AF4 (present vs absent) | 2.94 | 0.972 | 19.0 (2.82-128) | .0025 |
| Day 19 marrow (at least 5% blasts vs less than 5% blasts) | -0.46 | 0.888 | 0.63 (0.11-3.58) | .60 |
| Minimal residual disease on remission date (at least 0.01% vs less than 0.01%) | 2.69 | 0.972 | 14.7 (2.18-98.5) | .0057 |
This analysis was performed in the 64 patients with all features evaluable; because all patients in the higher-risk group who relapsed had an unfavorable DNA index, it was not included as a potential predictive feature.
SE indicates standard error for the coefficient estimate; CI, confidence interval.
Hazard of relapse for the first feature compared with those with the second feature.
P value was determined by Fine and Gray estimator.
Risk of hematologic relapse among patients on the higher-risk arm, based on combined GSTM1 nonnull and TYMS genotypes, by multivariate analysis
Feature* . | Coefficient . | SE . | Hazard ratio (95% CI) . | P† . |
|---|---|---|---|---|
| Combined GSTM1 and TYMS (GSTM1 nonnull + TYMS 3/3 vs others) | 2.67 | 0.905 | 14.5 (2.46-85.2) | .0031 |
| Leukocyte count (fewer than 100 × 109/L vs at least 100 × 109/L) | -1.95 | 0.964 | 0.14 (0.02-0.94) | .043 |
| t(9;22)/BCR-ABL (present vs absent) | 0.38 | 0.906 | 1.47 (0.25-8.66) | .67 |
| t(4;11)/MLL-AF4 (present vs absent) | 3.13 | 0.985 | 22.8 (1.20-157) | .0015 |
| Day 19 marrow (at least 5% blasts vs less than 5% blasts) | 1.07 | 0.551 | 2.92 (0.99-8.61) | .052 |
| Minimal residual disease on remission date (at least 0.01% vs less than 0.01%) | 2.75 | 1.00 | 15.7 (2.20-112) | .0060 |
Feature* . | Coefficient . | SE . | Hazard ratio (95% CI) . | P† . |
|---|---|---|---|---|
| Combined GSTM1 and TYMS (GSTM1 nonnull + TYMS 3/3 vs others) | 2.67 | 0.905 | 14.5 (2.46-85.2) | .0031 |
| Leukocyte count (fewer than 100 × 109/L vs at least 100 × 109/L) | -1.95 | 0.964 | 0.14 (0.02-0.94) | .043 |
| t(9;22)/BCR-ABL (present vs absent) | 0.38 | 0.906 | 1.47 (0.25-8.66) | .67 |
| t(4;11)/MLL-AF4 (present vs absent) | 3.13 | 0.985 | 22.8 (1.20-157) | .0015 |
| Day 19 marrow (at least 5% blasts vs less than 5% blasts) | 1.07 | 0.551 | 2.92 (0.99-8.61) | .052 |
| Minimal residual disease on remission date (at least 0.01% vs less than 0.01%) | 2.75 | 1.00 | 15.7 (2.20-112) | .0060 |
This analysis was performed in the 64 patients with all features evaluable; because all patients in the higher-risk group who relapsed had an unfavorable DNA index, it was not included as a potential predictive feature.
SE indicates standard error for the coefficient estimate; CI, confidence interval.
Hazard of relapse for the first feature compared to those with the second feature.
P value was determined by Fine and Gray estimator.
Cumulative incidence curves for the risk of hematologic relapse for ALL patients assigned in the higher-risk arm of the Total XIIIB Protocol. Children with the GSTM1 nonnull genotype (n = 71) experienced a higher risk of relapse compared with children with the null genotype (n = 59) (P = .03) (A). Among patients with the GSTM1 nonnull genotype, those with the TYMS 3/3 genotype (n = 24) were at even higher risk of relapse compared with those with the TYMS 2/2 or 2/3 genotypes (n = 47) (P = .03) (B).
Cumulative incidence curves for the risk of hematologic relapse for ALL patients assigned in the higher-risk arm of the Total XIIIB Protocol. Children with the GSTM1 nonnull genotype (n = 71) experienced a higher risk of relapse compared with children with the null genotype (n = 59) (P = .03) (A). Among patients with the GSTM1 nonnull genotype, those with the TYMS 3/3 genotype (n = 24) were at even higher risk of relapse compared with those with the TYMS 2/2 or 2/3 genotypes (n = 47) (P = .03) (B).
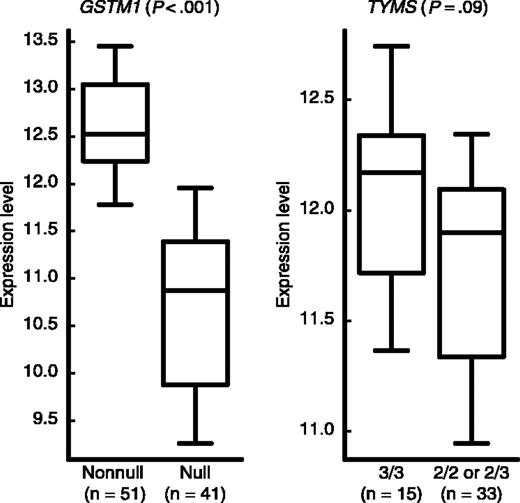
To explore whether genotypes were related to expression of those genes in the target tissue, we compared gene expression levels in the same set of patients in whom GSTM1 and TYMS genotypes were prognostic. In diagnostic ALL blasts from children treated with the higher-risk therapy, those with the GSTM1 nonnull genotype had higher GSTM1 gene expression than those with the null genotype (P < .001), and among those with the GSTM1 nonnull genotype, there was a trend for higher TYMS expression in those with the TYMS 3/3 compared with 2/3 or 2/2 genotypes (P = .09) (Figure 4).
Because different polymorphisms might be important for drug penetration into the CNS, we performed a separate analysis of genetic determinants of CNS relapse (isolated or combined), although only 6 and 2 CNS relapses occurred on the higher- and lower-risk arms, respectively. On the higher-risk arm, the VDR FokI T allele was associated with an increased risk of CNS relapse (5-year cumulative incidence, 8.6% ± 3.4% versus 0% for VDR FokI CC genotype, P = .02). All CNS relapses occurred among those with at least 1 VDR FokI T allele, but the frequency of VDR FokI genotypes did not differ (P > .14 for all) between patients with high- versus low-risk features (by initial leukocyte count, CNS status at diagnosis, DNA index, BCR-ABL [t(9;22)], MLL-AF4 [t(4;11)], day 19 bone marrow blasts, and minimal residual disease). Among those with at least 1 VDR FokI T allele, the VDR intron 8 GG genotype was associated with a greater risk of CNS relapse (5-year cumulative incidence, 15.6% ± 6.6% versus 2.7% ± 2.7% for VDR intron 8 AA genotype, P = .04), and the combined VDR FokI and VDR intron 8 genotypes were significantly associated with CNS relapse (P = .01) in a multivariate analysis including other prognostic features. On the lower-risk arm, the TYMS 3/3 genotype was associated with an increased risk of CNS relapse (5-year cumulative incidence, 5.7% ± 4% versus 0% for TYMS 2/3 or 2/2 genotypes, P = .04).
We compared the frequencies of significantly predictive genotypes in the higher- and lower-risk arms: no predictive polymorphisms differed in frequency between risk groups. Within risk groups, the only association of predictive genotypes with other patient features was among the higher-risk group: GSTM1 null genotype was more common (P = .03) among those with initial leukocyte count at least 100 × 109/L (57.9%) than those with less than 100 × 109/L (36.8%), but GSTM1 remained a significant predictor of hematologic relapse even after adjustment for presenting leukocyte counts and other prognostic features (Table 2). To assess the possibility that the association of genotypes with outcome was confounded by associations among genotypes, we assessed the correlations among genotypes at the 16 polymorphic loci. Although there was the expected linkage disequilibrium within some loci (eg, CYP3A4 with CYP3A5, MDR1 exon 21 with MDR1 exon 26 polymorphisms), there was no significant association between any of the predictive genotypes (eg, GSTM1 was not associated with TYMS or NR3C1 genotypes) (data not shown).
Discussion
The most common reason for failure in childhood ALL is hematologic relapse. We previously found that the only prognostic features in this protocol were risk group (treatment arm), initial leukocyte count, DNA index, BCR-ABL [t(9;22)], MLL-AF4 [t(4;11)], and early treatment response.21 Herein, after accounting for the difference in relapse hazard because of treatment arms, risk group, and other prognostic features (Tables 2 and 3), the incidence of hematologic relapse was significantly associated with common germ line genetic polymorphisms. In the higher-risk arm, the GSTM1 deletion was a determinant for the risk of hematologic relapse: 13 of the 16 children who experienced a hematologic relapse had the nonnull genotype. Our analysis allowed us to identify gene-gene interactions that modified the relapse risk. For patients with the GSTM1 nonnull genotype, the presence of the higher activity TYMS 3/3 genotype was associated with greater risk of hematologic relapse (P = .03). That GSTM1 and TYMS genotypes remained prognostic even after adjustment for factors reflective of early treatment response suggests that these genotypes continue to have an impact on prognosis for treatment administered beyond the remission induction period. This finding is plausible, in that drugs that are substrates for glutathione S-transferases (GSTs; eg, cyclophosphamide, etoposide), or that target thymidylate synthetase (TYMS; eg, methotrexate) were important components of the therapy given after induction. Although confidence intervals are large, the hazard ratios associated with unfavorable GSTM1 or combined GSTM1/TYMS genotypes are on par with those associated with features such as MLL translocations or the presence of minimal residual disease (Tables 2 and 3). Among those with the GSTM1 null genotype, the NR3C1 G allele was associated with an increased relapse risk, although a very small number of patients carried the G allele.
No polymorphisms were associated with hematologic relapse among patients treated on the lower-risk arm. That the GSTM1 germ line polymorphism was important for relapse among patients in the higher-risk arm, but not in the lower-risk arm, is consistent with more intensive therapy given to the former patients.20 GSTs catalyze the inactivation of many antileukemic agents and their metabolites (eg, cyclophosphamide, anthracyclines, etoposide, and steroids)28 and protect against oxidative stress. Null GST genotypes have been linked with improved outcome in some ALL trials14,16 but not in others.17 Our study is distinguished by the fact that virtually all patients on the trial were included in this analysis. Because cyclophosphamide and etoposide constituted a major proportion of therapy in the higher-risk patients, it is plausible that GST genotype was of greater importance in the higher-risk than in the lower-risk arm. Using pretreatment ALL blasts, we confirmed that the germ line nonnull GSTM1 genotype is associated with higher GSTM1 expression, in support of the hypothesis that the inferior outcome associated with the nonnull genotype is related to the increased detoxification conferred by GSTM1 expression.
The TYMS enhancer repeat 3/3 genotype causes higher expression and activity of one of the major targets of methotrexate, thymidylate synthetase.11,33 Higher thymidylate synthetase requires higher concentrations of methotrexate and its metabolites for inhibition33 and cytotoxicity.44,45 Although one single-gene study reported shorter event-free survival in children with ALL with the 3/3 genotype,18 another case-control study of 80 patients failed to find such an association.15 Such disparate findings may be due to differences in intensity of methotrexate therapy between studies. In our study, among patients on the higher-risk arm with an unfavorable GSTM1 status, the TYMS expression tended to be higher (Figure 4), and the TYMS 3/3 genotype was associated with a higher risk of hematologic relapse. Our findings illustrate the potential for gene-gene interactions: in the patients with low glutathione transferase activity, it is possible that methotrexate was a less critical element of therapy than among those with high glutathione transferase; thus, a polymorphism in the target of methotrexate (thymidylate synthetase) was of less importance in those with a favorable (low activity) than an unfavorable (high activity) GSTM1 genotype.
Expression levels (natural log-scaled signal intensities) of GSTM1, based on GSTM1 genotype among patients in the higher-risk arm, and of TYMS, based on TYMS genotype, among those with GSTM1 nonnull genotype. The boxes and central horizontal define the quartiles and median; the whiskers represent the 10th and 90th percentiles. The number of patients (n) included in each group is indicated below the plots.
Expression levels (natural log-scaled signal intensities) of GSTM1, based on GSTM1 genotype among patients in the higher-risk arm, and of TYMS, based on TYMS genotype, among those with GSTM1 nonnull genotype. The boxes and central horizontal define the quartiles and median; the whiskers represent the 10th and 90th percentiles. The number of patients (n) included in each group is indicated below the plots.
Only 2 CNS relapses occurred in the lower-risk group, and both patients were homozygous for the high-activity TYMS 3/3 genotype. For those on the lower-risk arm, methotrexate (given as systemic high doses and as intrathecal injections) was the primary form of CNS prophylaxis; thus, it is plausible that this unfavorable genotype might constitute a therapeutic disadvantage for this subset of patients.
In the higher-risk arm, 2 polymorphisms in the VDR locus were associated with risk of CNS relapse, which remained significant predictors after adjusting for other prognostic features. The VDR regulates the expression of CYP3A4 and possibly p-glycoprotein,34,35 which are important in the disposition of vincristine, etoposide, daunorubicin, prednisone, and dexamethasone. Both VDR polymorphisms have been associated with many clinical phenotypes.46-48 If the VDR FokI T allele and intron 8 GG genotype were associated with higher expression of p-glycoprotein, their association with an increased risk of relapse might be consistent with worse penetration of active drugs into the CNS (due to higher p-glycoprotein in the blood-brain barrier) and with higher risk of relapse because of greater p-glycoprotein-mediated biliary excretion of antileukemic drugs. Whether polymorphisms in VDR directly affect drug sensitivity or expression of downstream targets (CYP3A4 and/or p-glycoprotein) is not clear.
Relationships between genotypes and outcome could be influenced by a causative relationship between the polymorphism and response to therapy, or by an a priori relationship between the polymorphism and the development of specific subtypes of ALL. Our data favor the former rather than the latter. First, no polymorphisms differed in allele frequency between risk groups. Second, among the higher-risk group, although the GSTM1 genotype was associated with a high presenting leukocyte count, GSTM1 remained a significant predictor of relapse after adjusting for this feature.
This single-institution protocol was an ideal platform in which to assess how genotypes predicted outcomes in a racially diverse patient group, because, although genotypes differed by race, this is one of the first studies in which ALL cure rates did not differ by race.21;43 Thus, the genotype/outcome relationships we identified maintained prognostic significance in a therapeutic setting in which race itself was unimportant. Moreover, the same genotypes (GSTM1 nonnull and TYMS 3/3) were predictive for hematologic relapse within the major subgroups of whites and blacks (data not shown). Dosing based on pharmacogenetics holds the promise for delivery of “color-blind” therapy: genotyping allows for individualized dosing according to genetic rather than racial characteristics.49
Improved outcomes in patients with ALL depend on interactions among drugs, ALL blast sensitivity, and host factors. Currently, patients whose ALL blasts have unfavorable acquired genetic characteristics receive more intensive therapy. In our study of 16 target polymorphic loci, several common polymorphisms were found to be prognostic, most of which have pharmacologic plausibility. However, we highlight that prognostic genotypes are highly treatment dependent; hence, caution should be used in attempting to generalize these results to other protocols. Both of the primary polymorphisms associated with relapse, the high activity TYMS 3/3 and the GSTM1 nonnull genotypes, would be expected to cause drug resistance and were linked with higher gene expression in the target tissue of ALL blasts. We acknowledge that it is likely that additional pharmacogenetic prognostic factors will be identified in other protocols and that genotypes that were not prognostic in this study might be important in other protocols: as with all prognostic features, they depend on treatment. In the future, patients may be classified not only on the genetic characteristics of their blasts but also on host genetic characteristics, so that therapy could be intensified according to a pharmacogenetic index of drug resistance. We conclude that pharmacogenetics influences the outcome of antileukemic therapy and may provide a tool to improve individualization of therapy.
Prepublished online as Blood First Edition Paper, February 15, 2005; DOI 10.1182/blood-2004-11-4544.
Supported by the National Cancer Institute (CA 51001, CA 78224, CA 60419, CA21 765) and the National Institute of General Medical Sciences (NIGMS) Pharmacogenetics Research Network and Database (U01 1GM61374) from the National Institutes of Health, by a Center of Excellence grant from the State of Tennessee, and by American Lebanese Syrian Associated Charities (ALSAC).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank our protocol coinvestigators, clinical staff, research nurses (Sheri Ring, Lisa Walters, Margaret Edwards, Terri Kuehner, and Paula Condy), the patients and their parents for their participation. We also thank Jean Cai, Pam McGill, and Nancy Duran for laboratory assistance and Wenjian Yang, Nancy Kornegay, Carl Panetta, and Mark Wilkinson for computing assistance.
All the authors contributed substantially to the manuscript. M.V.R. and W.E.E. designed the research; J.C.C.R., S.K., S.D., E.H.C., J.T.S., J.R., R.R., D.C., C.-H.P., W.E.E., and M.V.R. performed the research; J.C.R., C.C., and W.L. analyzed the data; and J.C.R. and M.V.R. were primary authors of the paper.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal