Abstract
Hmgb3 is an X-linked member of a family of chromatin-binding proteins that is expressed in primitive hematopoietic cells capable of long-term hematopoietic repopulation. To examine the role of Hmgb3 in adult hematopoiesis, we generated Hmgb3-deficient (Hmgb3–/Y) mice, which are viable but erythrocythemic. Hmgb3–/Y mice contain normal numbers of hematopoietic stem cells (HSCs), which generate fewer than normal numbers of common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs) and greater than normal numbers of more mature progenitors. Although fewer Hmgb3–/Y primitive progenitor cells are in the G2/M cell cycle phase, bromodeoxyuridine (BrdU) incorporation demonstrated enhanced proliferation compared with their wild-type counterparts. Hmgb3–/Y HSCs have increased levels of Gata-2 and c-myb mRNA. We propose that Hmgb3 deficiency leads to a failure of HSCs to expand into normal numbers of CLPs and CMPs. This defect is compensated for by the ability of Hmgb3–/Y progenitors to expand rapidly and differentiate into normal numbers of hematopoietic cells.
Introduction
Hmgb3 is an X-linked member of the high-mobility group (HMG) superfamily of HMG proteins and is classified into the HMG-Box subfamily containing Hmgb1 and Hmgb2.1 Little is known about the function of Hmgb3, but the high homology between Hmgb3 and other HMG-Box family members suggests that Hmgb3 may share some of the same properties with Hmgb1 and 2. Hmgb1 and 2 bind to DNA without sequence specificity and are capable of interacting with and subsequently bending linear DNA,2,3 thereby facilitating nucleoprotein complex formation through alteration of local chromatin architecture.4,5 Hmgb1 and 2 can also directly interact with DNA-binding proteins (eg, Hox and Oct family members, steroid hormone receptors, RAG1 and 2, Rel, and p53) or promote interactions between protein subunits, such as between TATA-binding protein with TFIIB and TFIID with TFIIA.6-8 HMG-Box proteins can assist in either activating or repressing transcription.
In adult vertebrates, Hmgb1 is found in all cell types,9 whereas Hmgb3 mRNA was reported to be absent in most adult tissues.1,10 Previously, we have shown that in the adult mouse, Hmgb3 is expressed in bone marrow, specifically in Ter119+ erythroid cells and lineage-negative (lin–), c-kitHI, stem-cell antigen (Sca)–1HI, interleukin-7 receptor α–negative (IL-7Rα–) cells, which are enriched for hematopoietic stem cells (HSCs).10,11 Other studies have shown similar Hmgb1-related mRNAs to be preferentially expressed in HSCs.12 We demonstrated that Hmgb3 was expressed in both common lymphoid progenitors (CLPs; defined as lin–, c-kitLO, Sca-1LO, IL-7Rα+)11 and common myeloid progenitors (CMPs; defined as lin–, c-kit+, Sca-1–, IL-7Rα–).10,13 Lin–, c-kit+, Hmgb3+ cells were capable of long-term repopulation, whereas minimal repopulation was observed with lin–, c-kit+, Hmgb3– cells. Finally, enforced expression of Hmgb3 in hematopoietic cells prevents myeloid and B-cell differentiation.
To further examine the role of Hmgb3 in hematopoiesis, we have generated an Hmgb3-deficient transgenic mouse (Hmgb3–/Y). Hmgb3–/Y mice contain fewer CLPs and CMPs than wild-type mice. This deficiency is associated with an increased variability in repopulation of bone marrow by Hmgb3–/Y HSCs (defined as lin–, c-kitHI, Sca-1HI, IL-7Rα–)11 despite their normal numbers. We observed a decreased percentage of multipotent and oligopotent progenitors in the G2 cell cycle phase and a 48-4 delay in granulocyte colony-stimulating factor (G-CSF)–mediated mobilization of peripheral white blood cells. Hmgb3–/Y mice exhibited a higher proportion of proliferating CLPs and myeloid progenitors (defined as lin–, c-kitLO, Sca-1–). Together, our data suggest a model in which Hmgb3 deficiency alters the rate of generation and differentiation of primitive hematopoietic progenitor cells.
Materials and methods
Generation of Hmgb3 knock-out mice
The 5′ and 3′ arms for the knock-out vector were excised from a bacterial artificial chromosome containing the entire X-linked Hmgb3 locus. The 5′ arm consists of 8 kb encompassing a region 5′ of the Hmgb3 locus to a site 400 bp 5′ of the second exon. The 3′ arm consists of 2.5 kb located immediately 3′ of exon 5. These arms were subcloned into the pPNT targeting vector on either side of a mouse pgk-1 promoter–neomycin resistance gene cassette flanked by loxP sites. The targeting vector is designed to delete the entire coding sequence of Hmgb3 (Figure 1A).
Generation of Hmgb3 knock-out mice. (A) Generation of Hmgb3 knock-out (Hmgb3–/Y) mice by homologous recombination. (Top) Diagram of Hmgb3 gene locus. (Middle) Targeting construct. A region spanning exons 2-5 (the entire coding region) of the Hmgb3 gene locus is removed. (Bottom) Hmgb3 gene locus after recombination. A gray bar depicts the probe used for Southern blot analysis of recombination events. E and K refer to EcoRI and Kpnl sites, respectively. (B) Representative Southern blot analysis of recombination events. Female F1 mice were mated to male wild-type 129/SvJ mice. Ten μg genomic DNA isolated from the tails of 10-day-old pups was digested with KpnI. A 900-bp EcoRI-HindIII fragment located 3′ of the Hmgb3 locus was used for a probe (see panel A). The wild-type Hmgb3 locus is contained on a 21-kb KpnI fragment. Homologous recombination results in a 16-kb KpnI fragment. Lane 1 shows wild-type female (+/+); lanes 2-3, heterozygous females (–/+); and lanes 4-6, hemizygous males (–/Y). (C) Northern blot analysis of Hmgb3 mRNA in adult bone marrow. (Left) Northern blot analysis of Hmgb3 mRNA in adult bone marrow harvested from male wild-type (+/Y) and Hmgb3–/Y (–/Y) mice. Arrows indicate signals corresponding to Hmgb3 mRNA and 18s and 28s rRNAs. The 3′ untranslated region was used as a probe. (Right) Ethidium-stained agarose gel used to determine RNA loading and integrity. For Northern blotting, 15 μg of bone marrow RNA isolated from mice of each genotype was used.
Generation of Hmgb3 knock-out mice. (A) Generation of Hmgb3 knock-out (Hmgb3–/Y) mice by homologous recombination. (Top) Diagram of Hmgb3 gene locus. (Middle) Targeting construct. A region spanning exons 2-5 (the entire coding region) of the Hmgb3 gene locus is removed. (Bottom) Hmgb3 gene locus after recombination. A gray bar depicts the probe used for Southern blot analysis of recombination events. E and K refer to EcoRI and Kpnl sites, respectively. (B) Representative Southern blot analysis of recombination events. Female F1 mice were mated to male wild-type 129/SvJ mice. Ten μg genomic DNA isolated from the tails of 10-day-old pups was digested with KpnI. A 900-bp EcoRI-HindIII fragment located 3′ of the Hmgb3 locus was used for a probe (see panel A). The wild-type Hmgb3 locus is contained on a 21-kb KpnI fragment. Homologous recombination results in a 16-kb KpnI fragment. Lane 1 shows wild-type female (+/+); lanes 2-3, heterozygous females (–/+); and lanes 4-6, hemizygous males (–/Y). (C) Northern blot analysis of Hmgb3 mRNA in adult bone marrow. (Left) Northern blot analysis of Hmgb3 mRNA in adult bone marrow harvested from male wild-type (+/Y) and Hmgb3–/Y (–/Y) mice. Arrows indicate signals corresponding to Hmgb3 mRNA and 18s and 28s rRNAs. The 3′ untranslated region was used as a probe. (Right) Ethidium-stained agarose gel used to determine RNA loading and integrity. For Northern blotting, 15 μg of bone marrow RNA isolated from mice of each genotype was used.
The targeting vector was linearized with NotI and electroporated into TC1 embryonic stem (ES) cells as described previously.10 Correctly targeted cells were identified by Southern blot analysis using probes outside the targeting region. Five homologous recombination events were identified among 177 G418-resistant and ganciclovir-resistant ES cell clones. Twelve to 18 ES cells from clones 27 and 87 were injected into 3.5-day-old C57BL/6 blastocysts and yielded 5 and 6 chimeras, respectively. One chimera from each clone was bred to 129/SvJ female mice, and F1 generation transgenic mice were identified by Southern blot analysis of tail DNA. Female Hmgb3–/+ mice were bred to wild-type male 129/SvJ mice to produce male hemizygous mice (Hmgb3–/Y). Absence of Hmgb3 mRNA in bone marrow from Hmgb3–/Y mice was confirmed by Northern blot analysis performed as previously described.14 Hematologic analyses of Hmgb3–/Y and littermate wild-type mice were performed in the Clinical Pathology Department, National Institutes of Health (NIH) Clinical Center (Bethesda, MD).
Analysis of clonogenic myeloid and B-lymphoid progenitors
Bone marrow cells harvested from 6- to 8-week-old Hmgb3–/Y and littermate wild-type mice were cultured in Methocult GF M3434 (granulocyte, macrophage–colony-forming units [CFU-GMs]; erythroid–burst-forming units [BFU-Es]; and granulocyte, erythrocyte, macrophage, megakaryocyte–CFUs [CFU-GEMMs]), Methocult M3334 (erythroid-CFUs [CFU-Es]), and MethoCult M3630 (CFU–Pre-Bs; Stem Cell Technologies, Vancouver, BC, Canada) according to the manufacturer's instructions.
Real-time PCR of TRECs
Thymocytes isolated from 6-week-old and 1-year-old Hmgb3–/Y and littermate wild-type mice were stained with fluorescein isothiocyanate (FITC)–conjugated anti–mouse CD3 antibody (145-2C11; Pharmingen, San Diego, CA) followed by staining with anti-FITC microbeads (Miltenyi Biotech, Auburn, CA). CD3+ cells were separated by magnetic bead separation using an AutoMACS Cell Sorter (Miltenyi Biotech). Primers (sense, 5′-CATTGCCTTTGAACCAAGCTG-3′; antisense, 5′-TTATGCACAGGGTGCAGGTG-3′; probe, 5′-CAGGGCAGGTTTTTGTAA AGGTGCTCACTT-3′) used for T-cell receptor excision circle (TREC) amplification are located within the mouse T-cell receptor α/δ (TCRα/δ) locus, amplifying a 92-bp fragment from an excision circle formed by the recombination of ΨJα with δRec1.15 Real-time polymerase chain reactions (PCRs) were performed on an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA) as described in Sempowski et al.15 PCR reactions that amplified the mouse α-globin promoter (sense, 5′-GCTTCTCTGAC CAAGGTAGGAGG-3′; antisense, 5′-GTTGCCCGGACACACTTCTTAC-3′; probe, 5′-TTCTTCCCAAACTGCCATCACTGGAGACGT-3′) were used as DNA-loading controls. Each reaction was performed in triplicate.
Analysis of common lymphoid and myeloid progenitors
Bone marrow cells from 6- to 8-week-old Hmgb3–/Y and littermate wild-type mice were depleted for lineage-positive cells (lin–) as previously described.10 For isolation of CLPs, lin– cells were stained with allophycocyanin (APC)–conjugated anti–c-kit (2B8; Pharmingen), phycoerythrin (PE)–conjugated anti–Sca-1 (E13-161.7; Pharmingen), and PE–cyanin 5 (PE-Cy5)–conjugated anti–IL-7Rα (A734; eBioscience, San Diego, CA) or the recommended isotype antibodies.11 There was no detectable overlap of emission spectra between APC-conjugated c-kit and PE-Cy5–conjugated IL-7Rα antibodies. All cells were analyzed and sorted on a FACS Vantage SE (Becton Dickinson, San Jose, CA) using 488-nm argon and 633-nm helium neon lasers. Lin–, c-kitLO, Sca-1LO, IL-7Rα+ cells were cultured in MethoCult M3630 supplemented with 100 ng/mL stem cell factor (SCF; Amgen, Thousand Oaks, CA) and 20 ng/mL Fms-like tyrosine kinase 3 ligand (Flt3L; Peprotech, Rocky Hill, NJ). CFU–Pre-Bs were scored after 7 days in culture. Lin–, c-kitLO, Sca-1LO, IL-7Rα+ cells cultured in Methocult GF M3434 supplemented with 20 ng/mL Flt3L did not form myeloid colonies.
For isolation of CMPs, lineage depletion was performed with the inclusion of anti–Sca-1 and anti–IL-7Rα antibodies. Lin– bone marrow cells were stained with APC-conjugated anti–c-kit, PE-Cy5–conjugated anti–Sca-1 (D7; eBioscience), PE-Cy5–conjugated anti–IL-7Rα, PE-conjugated anti-FcγRII/III (2.4G2; Pharmingen), and FITC-conjugated anti-CD34 (RAM34; Pharmingen) or the recommended isotype antibodies.13 Staining with FITC-conjugated anti-CD34 was performed prior to staining with the remaining antibodies. Lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, and CD34+ cells were cultured in Methocult GF M3434 supplemented with 20 ng/mL human Flt3L. CFU-GMs, BFU-Es, and CFU-GEMMs were scored after 7, 10, and 14 days, respectively, in culture. Lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, and CD34+ cells cultured in MethoCult M3630 supplemented with 100 ng/mL SCF and 20 ng/mL Flt3L did not form CFU–Pre-B colonies.
Competitive repopulation analysis of HSC function
Bone marrow cells harvested from 6- to 8-week-old Hmgb3–/Y or littermate wild-type mice (129/SvJ strain) were mixed in equal numbers with male C57BL/6 bone marrow cells so that 2 × 106 cells from each strain were injected into lethally irradiated (990 rad, Cs137 source) male 129 × C57BL/6 F1 mice (Jackson Laboratory, Bar Harbor, ME). Sixteen weeks later, Southern blot analyses were performed on recipient bone marrow DNA digested with EcoRI using a BamHI-Pst I fragment of the mouse β-globin gene as a probe.14 Bone marrow cells (2 × 106) from primary recipients were transplanted in lethally irradiated male 129 × C57BL/6 F1 secondary recipients, and Southern blot analyses of bone marrow DNA were performed 16 weeks after transplantation. For both primary and secondary recipients, signals corresponding to 129/SvJ βmaj and βmin and C57BL/6 βS β-globin alleles were quantified by performing densitometry on relative band intensities captured on a PhosphorScreen and resolved on a Typhoon 8600 PhosphorImager (Amersham Pharmacia, Piscataway, NJ). Percent repopulation by either wild-type or Hmgb3–/Y bone marrow was calculated by the formula % 129/SvJ = 100 × [(129βmaj + 129βmin)/(129βmaj + 129βmin + C57BL/6βS)]. Bone marrow cells harvested from 4 secondary recipients (2 originally received transplants of wild-type marrow and 2 of Hmgb3–/Y marrow) were then transplanted into 5 tertiary recipients each using 4 × 106 cells per mouse.
Analysis of cell cycle status in HSCs
Bone marrow cells from groups of three 6- to 8-week-old Hmgb3–/Y and littermate wild-type mice were pooled. Lineage depletion and isolation of the Lin–, c-kitHI, Sca-1HI, IL-7Rα– HSC population (or Lin–, c-kit+ progenitor population) was performed as described under “Analysis of common lymphoid and myeloid progenitors.” Sorted cells were immediately stained with propidium iodide using the NuCycl propidium iodide (PI) staining kit (Exalpha Biologicals, Watertown, MA) according to the manufacturer's instructions before analysis by flow cytometry, which was performed on a FacsCalibur (Becton Dickinson) using a 488-nm argon laser. Cell cycle data analysis was performed using ModFit software (Verity Software, Topsham, ME). P values were generated by Student t test.
Bromodeoxyuridine staining of primitive hematopoietic progenitors
Groups of 3 Hmgb3–/Y or littermate wild-type mice were injected with 1 mg bromodeoxyuridine (BrdU; Pharmingen) intraperitoneally for 3 consecutive days. Twenty-four hours after the final injection, bone marrow was harvested. Lineage depletion and staining with monoclonal antibodies for c-kit, Sca-1, and IL-7Rα was performed as described in “Analysis of common lymphoid and myeloid progenitors.” Cells were fixed and stained with an FITC-conjugated anti-BrdU antibody using the FITC BrdU Flow Kit (Pharmingen) according to the manufacturer's instructions before analysis by flow cytometry on a FacsCalibur using 488-nm argon and 633-nm helium neon lasers.
Cytokine mobilization of peripheral blood cells
Hmgb3–/Y and littermate wild-type mice were splenectomized. Ten days later, mice were injected with 200 μg/kg/d G-CSF (Amgen) subcutaneously for 7 days. Peripheral blood samples were removed by retro-orbital bleeding prior to each round of injections and continuing 24 hours after the last injection. Blood samples were lysed with 2% acetic acid and nucleated cells counted using a hemacytometer. P values were generated by Student t test.
RT-PCR analysis of HSC mRNA
Bone marrow cells were harvested from groups of three 6- to 8-week-old Hmgb3–/Y or littermate wild-type mice and pooled. Lineage depletion and staining with antibodies to c-kit, Sca-1, and IL-7Rα was performed as described in “Analysis of common lymphoid and myeloid progenitors.” RNA was isolated from sorted HSCs (Lin–, c-kitHI, Sca-1HI, IL-7Rα–) using TriZol (Invitrogen) according to the manufacturer's instructions. Reverse transcriptase–PCR (RT-PCR) was performed as described previously using a duplex-PCR reaction with one primer pair amplifying the test gene and the second primer pair amplifying β2-microglobulin as an internal control.10 Limiting dilutions of HSC mRNA were used to ensure amplifications remained within the linear range. Reactions were performed for 32 cycles at 94°C for 15 seconds, 58°C (54°C for CXCR4) for 15 seconds, and 72°C for 30 seconds. Primer pairs used for gene amplification are described as follows: CXCR4 (5′-GGCTGTAGAGCGAGTGTTGC-3′) sense, (5′-GTAGAGGTTGACAGTGTAGAT-3′) antisense16 ; CCR3 (5′-TGGGCAACATGATGGTTGTG-3′) sense, (5′-GCTGTCTTGAGACTCATGGA-3′) antisense16 ; c-kit (5′-GGGCAAGAGTTCCG CCTTCTT-3′) sense, (5′-GCTGCGACCACAAAGCC-3′) antisense17 ; glutamate acetyltransferase 2 (Gata-2; 5′-GGC GTCAAGTACCAAGTGTCAC-3′) sense,17 (5′-CTCCCGGCCTTCTGAGCAGGAG-3′) antisense; Gata-1 (5′-GGAGCCCTCTCAGCTCAGC-3′) sense, (5′-GCCACCAGC TGGTCCTTCAG-3′) antisense17 ; c-myb (5′-GAGCTTGTCCAGAAATATGGT CCGAAG-3′) sense, (5′-GGCTGCCGCAGCCGGCTGAGGGAC-3′) antisense.17 Test gene mRNA levels were measured and normalized to β2-microglobulin by performing densitometry on relative band intensities. Reverse transcriptase reactions performed without enzyme served as negative controls for genomic DNA amplification.
Results
Hematology of Hmgb3–/Y mice
Homologous recombination events that removed the entire coding region of the Hmgb3 locus in male 129/SvJ ES cells (Figure 1A) were identified by Southern blot analysis (data not shown). As Hmgb3 is an X-linked gene, only male mice maintained on a 129/SvJ background hemizygous for the null allele (Hmgb3–/Y) were used for all experiments (Figure 1B). Northern blotting performed on bone marrow RNA confirmed the absence of Hmgb3 mRNA (Figure 1C). Male Hmgb3–/Y mice are grossly normal and fertile. The absolute and differential white blood cell counts were similar in Hmgb3–/Y mice and littermate wild-type mice (Table 1). However, Hmgb3–/Y mice are erythrocythemic with 10% more red blood cells (P < .01), a smaller mean cell volume (P < .01), an elevated hematocrit (P < .01), and higher levels of hemoglobin (P < .05) compared with wild-type mice (Table 1). These data differ significantly from our previous analysis of Hmgb3 knock-in mice,10 which have normal numbers of all hematopoietic cells. In both Hmgb3–/Y and Hmgb3 knock-in animals, the pgk-neo gene is inserted in nearly the same part of the genome indicating that the insertion of the neo gene has no effect on hematopoiesis.
Hmgb3–/Y mice are erythrocythemic
. | Absolute and differential WBC counts . | . | . | . | Characterization of red blood cells . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | WBC count, cells/μL . | % Lymphocytes . | % Neutrophils . | % Monocytes . | RBC count, × 1012/L . | HCT, % . | MCV, fL . | Hb, g/dL . | ||||||
| Wild-type | 7.3 ± 0.6 | 85.5 ± 1.2 | 11.7 ± 1.2 | 2.9 ± 0.6 | 10.0 ± 0.3 | 51.9 ± 0.2 | 51.1 ± 2.1 | 16.1 ± 0.4 | ||||||
| Hmgb3-/Y | 7.8 ± 1.3 | 83.7 ± 2.9 | 12.6 ± 2.3 | 4.1 ± 1.6 | 11.1 ± 0.2 | 53.7 ± 0.3 | 48.1 ± 2.2 | 16.8 ± 0.3 | ||||||
| P | .35 | .11 | .39 | .10 | <.01 | <.01 | <.01 | <.05 | ||||||
. | Absolute and differential WBC counts . | . | . | . | Characterization of red blood cells . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | WBC count, cells/μL . | % Lymphocytes . | % Neutrophils . | % Monocytes . | RBC count, × 1012/L . | HCT, % . | MCV, fL . | Hb, g/dL . | ||||||
| Wild-type | 7.3 ± 0.6 | 85.5 ± 1.2 | 11.7 ± 1.2 | 2.9 ± 0.6 | 10.0 ± 0.3 | 51.9 ± 0.2 | 51.1 ± 2.1 | 16.1 ± 0.4 | ||||||
| Hmgb3-/Y | 7.8 ± 1.3 | 83.7 ± 2.9 | 12.6 ± 2.3 | 4.1 ± 1.6 | 11.1 ± 0.2 | 53.7 ± 0.3 | 48.1 ± 2.2 | 16.8 ± 0.3 | ||||||
| P | .35 | .11 | .39 | .10 | <.01 | <.01 | <.01 | <.05 | ||||||
P values were determined using Student t test.
WBC count indicates white blood cell count; % Lymphocytes, percentage of WBC count that are lymphocytes; % Neutrophils, percentage of WBC count that are neutrophils; % Monocytes, percentage of WBC count that are monocytes; RBC count, red blood cell count; HCT, hematocrit (percent of whole blood volume); MCV, mean cell volume, and Hb, hemoglobin concentration.
For all tests on wild-type mice, the sample size was 10. For all tests on Hmgb3-/Y mice, the sample size was 9.
Analysis of myeloid and lymphoid progenitor cells in Hmgb3–/Y mice
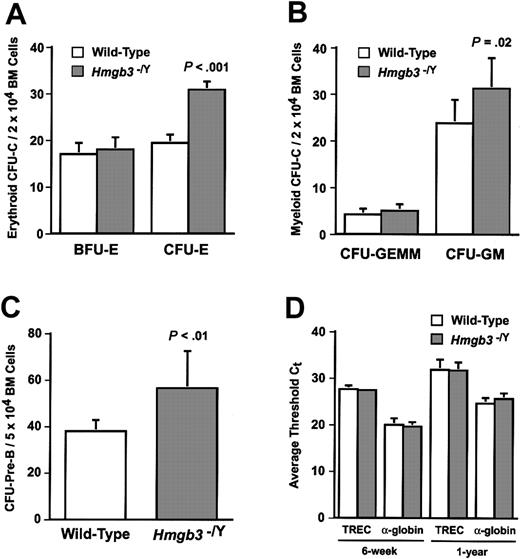
We used clonogenic assays to quantify the numbers of erythroid, myeloid, and B-lymphoid progenitors in wild-type and Hmgb3–/Y bone marrow (Figure 2; Table 2). Hmgb3–/Y mice exhibit a 1.6-fold increase in the frequency of CFU-Es compared with littermate wild-type mice (P < .001; Figure 2A). We also observed a 1.3-fold increase in the frequency of CFU-GMs (P = .02; Figure 2B) and a 1.5-fold increase in the frequency of CFU–Pre-Bs (P < .01) in Hmgb3–/Y mice compared with wild-type mice (Figure 2C). We observed no difference in the frequencies of either BFU-Es (Figure 2A) or CFU-GEMMs (Figure 2B) between wild-type and Hmgb3–/Y mice. Hmgb3 has recently been shown to have a role in the formation of a protein complex that binds to a recognition signal sequence in the TCR α locus.18 To examine T lymphopoiesis, we quantified TRECs in 6-week-old and 1-year-old Hmgb3–/Y and littermate wild-type mice using real-time PCR. There was no difference between Hmgb3–/Y and wild-type mice in the number of total thymocytes, CD3+ thymocytes, or relative numbers of TRECs in either 6-week- or 1-year-old mice (Figure 2D). These results complement and extend our previous studies in which enforced expression of Hmgb3 affected the differentiation of erythroid, myeloid, and B-lymphoid cells, while T lymphopoiesis was unaffected.10 T-cell function was not analyzed.
Analysis of myeloid and lymphoid progenitor cells in Hmgb3–/Y mice. (A) Mean BFU-Es and CFU-Es per 2 × 104 bone marrow cells isolated from wild-type (n = 8; 135 BFU-Es and 1538 CFU-Es counted) and Hmgb3–/Y (n = 8; 144 BFU-Es and 2452 CFU-Es counted) mice. To generate BFU-Es and CFU-Es, 2 × 104 and 2 × 105 bone marrow cells were cultured per sample, respectively. (B) Mean CFU-GEMMs and CFU-GMs per 2 × 104 cultured bone marrow cells isolated from wild-type (n = 8; 33 CFU-GEMMs and 190 CFU-GMs counted) and Hmgb3–/Y (n = 8; 40 CFU-GEMMs and 249 CFU-GMs counted) mice. (C) Mean CFU–Pre-Bs per 5 × 104 cultured bone marrow cells isolated from wild-type (n = 8; 302 CFU–Pre-Bs counted) and Hmgb3–/Y (n = 8; 449 CFU–Pre-Bs counted) mice. For panels A, B, and C, P values were determined by Student t test. The data represent the pooled results of 2 independent experiments. (D) Average Ct values for TRECs and α-globin real-time PCR performed on CD3+ thymocytes harvested from 6-week-old (n = 3) and 1-year-old (n = 3) wild-type and Hmgb3–/Y mice. Ct represents the number of amplification cycles at which the fluorescent signal in a real-time PCR reaction passes a fixed threshold. For all reactions, Ct values were within the linear range of amplification. Reactions were performed in triplicate for each sample. Error bars in all panels represent standard deviations.
Analysis of myeloid and lymphoid progenitor cells in Hmgb3–/Y mice. (A) Mean BFU-Es and CFU-Es per 2 × 104 bone marrow cells isolated from wild-type (n = 8; 135 BFU-Es and 1538 CFU-Es counted) and Hmgb3–/Y (n = 8; 144 BFU-Es and 2452 CFU-Es counted) mice. To generate BFU-Es and CFU-Es, 2 × 104 and 2 × 105 bone marrow cells were cultured per sample, respectively. (B) Mean CFU-GEMMs and CFU-GMs per 2 × 104 cultured bone marrow cells isolated from wild-type (n = 8; 33 CFU-GEMMs and 190 CFU-GMs counted) and Hmgb3–/Y (n = 8; 40 CFU-GEMMs and 249 CFU-GMs counted) mice. (C) Mean CFU–Pre-Bs per 5 × 104 cultured bone marrow cells isolated from wild-type (n = 8; 302 CFU–Pre-Bs counted) and Hmgb3–/Y (n = 8; 449 CFU–Pre-Bs counted) mice. For panels A, B, and C, P values were determined by Student t test. The data represent the pooled results of 2 independent experiments. (D) Average Ct values for TRECs and α-globin real-time PCR performed on CD3+ thymocytes harvested from 6-week-old (n = 3) and 1-year-old (n = 3) wild-type and Hmgb3–/Y mice. Ct represents the number of amplification cycles at which the fluorescent signal in a real-time PCR reaction passes a fixed threshold. For all reactions, Ct values were within the linear range of amplification. Reactions were performed in triplicate for each sample. Error bars in all panels represent standard deviations.
Frequency and total number of erythroid, myeloid, and lymphoid CFU-Cs in wild-type and Hmgb3–/Y mice
. | Average marrow/hind limb . | Average CFU-C frequency: CFU/no. of cells cultured . | Average CFU-Cs from hind limb . | P . |
|---|---|---|---|---|
| BFU-Es | ||||
| Wild-type | 2.1 × 107 | 16.9 ± 2.5/2.0 × 104 | 1.8 × 104 | .4 |
| Hmgb3-/Y | 2.1 × 107 | 18.0 ± 2.5/2.0 × 104 | 1.9 × 104 | |
| CFU-Es | ||||
| Wild-type | 2.1 × 107 | 192.3 ± 19.0/2.0 × 105 | 2.0 × 104 | <.01 |
| Hmgb3-/Y | 2.1 × 107 | 306.5 ± 19.2/2.0 × 105 | 3.2 × 104 | |
| CFU-GEMMs | ||||
| Wild-type | 2.1 × 107 | 4.1 ± 1.2/2.0 × 104 | 4.3 × 103 | .2 |
| Hmgb3-/Y | 2.1 × 107 | 5.0 ± 1.3/2.0 × 104 | 5.3 × 103 | |
| CFU-GMs | ||||
| Wild-type | 2.7 × 107 | 23.8 ± 4.9/2.0 × 104 | 3.2 × 104 | .05 |
| Hmgb3-/Y | 2.4 × 107 | 31.1 ± 6.6/2.0 × 104 | 3.7 × 104 | |
| CFU-Pre-Bs | ||||
| Wild-type | 2.7 × 107 | 37.8 ± 4.9/5.0 × 104 | 2.0 × 104 | .01 |
| Hmgb3-/Y | 2.4 × 107 | 56.1 ± 16.2/5.0 × 104 | 2.7 × 104 |
. | Average marrow/hind limb . | Average CFU-C frequency: CFU/no. of cells cultured . | Average CFU-Cs from hind limb . | P . |
|---|---|---|---|---|
| BFU-Es | ||||
| Wild-type | 2.1 × 107 | 16.9 ± 2.5/2.0 × 104 | 1.8 × 104 | .4 |
| Hmgb3-/Y | 2.1 × 107 | 18.0 ± 2.5/2.0 × 104 | 1.9 × 104 | |
| CFU-Es | ||||
| Wild-type | 2.1 × 107 | 192.3 ± 19.0/2.0 × 105 | 2.0 × 104 | <.01 |
| Hmgb3-/Y | 2.1 × 107 | 306.5 ± 19.2/2.0 × 105 | 3.2 × 104 | |
| CFU-GEMMs | ||||
| Wild-type | 2.1 × 107 | 4.1 ± 1.2/2.0 × 104 | 4.3 × 103 | .2 |
| Hmgb3-/Y | 2.1 × 107 | 5.0 ± 1.3/2.0 × 104 | 5.3 × 103 | |
| CFU-GMs | ||||
| Wild-type | 2.7 × 107 | 23.8 ± 4.9/2.0 × 104 | 3.2 × 104 | .05 |
| Hmgb3-/Y | 2.4 × 107 | 31.1 ± 6.6/2.0 × 104 | 3.7 × 104 | |
| CFU-Pre-Bs | ||||
| Wild-type | 2.7 × 107 | 37.8 ± 4.9/5.0 × 104 | 2.0 × 104 | .01 |
| Hmgb3-/Y | 2.4 × 107 | 56.1 ± 16.2/5.0 × 104 | 2.7 × 104 |
Average marrow per hind limb was determined by dividing the total number of harvested bone marrow cells (from all experiments) by the number of hind limbs. Frequency was determined as the average number of CFU-Cs (see Figure 2) per cells cultured. The total number of CFU-Cs per hind limb was calculated by multiplying the average number of bone marrow cells per hind limb by the average CFU-C frequency.
Analysis of primitive progenitor cells in Hmgb3–/Y mice
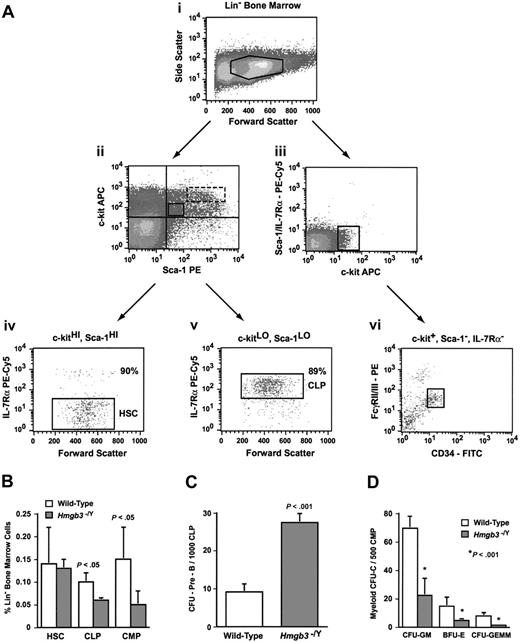
Previously, we demonstrated through functional assays that the majority of HSCs, CLPs, and primitive myeloid progenitors expressed Hmgb3.10 We examined the numbers of CLPs and CMPs in Hmgb3–/Y mice compared with littermate wild-type mice by fluorescence-activated cell sorter (FACS) analysis. Representative data demonstrating isolation of the CLP and CMP populations in wild-type mice are shown in Figure 3A. Mean fluorescence of c-kit, Sca-1, or IL-7Rα on wild-type and Hmgb3–/Y cells was identical.
Analysis of primitive progenitors in Hmgb3–/Y mice. (A) Representative isolation of wild-type common lymphoid and myeloid progenitors by flow cytometry. (Ai) Lin– bone marrow isolated from wild-type mice. (Aii) CLP and HSC populations were isolated from c-kit and Sca-1 double-positive lin– bone marrow cells. Double-positive cells were further segregated into c-kitLO, Sca-1LO (solid box) and c-kitHI, Sca-1HI (dotted box) populations. Regions were drawn based on isotype staining of littermate control cells. (Aiii) The CMP population was isolated from c-kit+, Sca-1–, and IL-7Rα– lin– bone marrow cells. (Aiv) Further isolation of the HSC population based on IL-7Rα expression in c-kitHI, Sca-1HI cells. Approximately 90% of c-kitLO, Sca-1LO cells were IL-7Rα– (HSC phenotype). (Av) Isolation of the CLP population based on IL-7Rα expression in c-kitLO, Sca-1LO cells. Approximately 89% of c-kitLO, Sca-1LO cells were IL-7Rα+ (CLP phenotype). (Avi) Isolation of the CMP population based on FcγRII/III and CD34 expression in c-kit+, Sca-1–, and IL-7Rα– cells. Cells that are FcγRII/IIILO and CD34+ represent the CMP phenotype. (B) Average number of HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–; n = 3), CLPs (lin–, c-kitLO, Sca-1LO, IL-7Rα+; n = 3), and CMPs (lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, CD34+; n = 4) as determined by the percentage of lineage-negative (lin–) bone marrow cells that stained positive for the HSC, CLP, or CMP phenotype. Staining was performed on lineage-depleted cells isolated from groups of 3 Hmgb3–/Y or wild-type mice prior to analysis by flow cytometry. The data represent the pooled results of 3 (HSCs and CLPs) or 4 (CMPs) independent experiments. (C) Mean CFU–Pre-B frequency generated from wild-type and Hmgb3–/Y CLP populations. CFU–Pre-B frequency within the CLP population was determined by scoring pre–B-cell colonies per 1000 cells cultured: wild-type (n = 8; 355 CFU–Pre-Bs counted); Hmgb3–/Y (n = 8; 890 CFU–Pre-Bs counted). The data represent the pooled results of 2 independent experiments. (D) Mean myeloid colony (CFU-C) frequency generated from wild-type and Hmgb3–/Y CMP populations. Myeloid CFU-C frequency within the CMP population was determined by scoring CFU-GM, BFU-E, and CFU-GEMM colonies per 500 cells cultured: wild-type (n = 10; 813 CFU-GMs, 177 BFU-Es, and 91 CFU-GEMMs counted); Hmgb3–/Y (n = 10; 239 CFU-GMs, 47 BFU-Es, and 17 CFU-GEMMs counted). The data represent the pooled results of 2 independent experiments. P values in panels B-D were determined by Student t test. In panels B-D, error bars represent standard deviation.
Analysis of primitive progenitors in Hmgb3–/Y mice. (A) Representative isolation of wild-type common lymphoid and myeloid progenitors by flow cytometry. (Ai) Lin– bone marrow isolated from wild-type mice. (Aii) CLP and HSC populations were isolated from c-kit and Sca-1 double-positive lin– bone marrow cells. Double-positive cells were further segregated into c-kitLO, Sca-1LO (solid box) and c-kitHI, Sca-1HI (dotted box) populations. Regions were drawn based on isotype staining of littermate control cells. (Aiii) The CMP population was isolated from c-kit+, Sca-1–, and IL-7Rα– lin– bone marrow cells. (Aiv) Further isolation of the HSC population based on IL-7Rα expression in c-kitHI, Sca-1HI cells. Approximately 90% of c-kitLO, Sca-1LO cells were IL-7Rα– (HSC phenotype). (Av) Isolation of the CLP population based on IL-7Rα expression in c-kitLO, Sca-1LO cells. Approximately 89% of c-kitLO, Sca-1LO cells were IL-7Rα+ (CLP phenotype). (Avi) Isolation of the CMP population based on FcγRII/III and CD34 expression in c-kit+, Sca-1–, and IL-7Rα– cells. Cells that are FcγRII/IIILO and CD34+ represent the CMP phenotype. (B) Average number of HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–; n = 3), CLPs (lin–, c-kitLO, Sca-1LO, IL-7Rα+; n = 3), and CMPs (lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, CD34+; n = 4) as determined by the percentage of lineage-negative (lin–) bone marrow cells that stained positive for the HSC, CLP, or CMP phenotype. Staining was performed on lineage-depleted cells isolated from groups of 3 Hmgb3–/Y or wild-type mice prior to analysis by flow cytometry. The data represent the pooled results of 3 (HSCs and CLPs) or 4 (CMPs) independent experiments. (C) Mean CFU–Pre-B frequency generated from wild-type and Hmgb3–/Y CLP populations. CFU–Pre-B frequency within the CLP population was determined by scoring pre–B-cell colonies per 1000 cells cultured: wild-type (n = 8; 355 CFU–Pre-Bs counted); Hmgb3–/Y (n = 8; 890 CFU–Pre-Bs counted). The data represent the pooled results of 2 independent experiments. (D) Mean myeloid colony (CFU-C) frequency generated from wild-type and Hmgb3–/Y CMP populations. Myeloid CFU-C frequency within the CMP population was determined by scoring CFU-GM, BFU-E, and CFU-GEMM colonies per 500 cells cultured: wild-type (n = 10; 813 CFU-GMs, 177 BFU-Es, and 91 CFU-GEMMs counted); Hmgb3–/Y (n = 10; 239 CFU-GMs, 47 BFU-Es, and 17 CFU-GEMMs counted). The data represent the pooled results of 2 independent experiments. P values in panels B-D were determined by Student t test. In panels B-D, error bars represent standard deviation.
We observed a 1.6-fold decrease in the number of cells with the CLP phenotype (lin–, c-kitLO, Sca-1LO, IL-7Rα+; P < .05; Figure 3B) in Hmgb3–/Y mice compared with wild-type mice. However, we also observed a 3-fold increase in the ability of Hmgb3–/Y CLPs to generate CFU–Pre-Bs compared with wild-type CLPs (P < .001; Figure 3C), suggesting that although the Hmgb3–/Y CLP population was smaller than the wild-type by phenotypic analysis, it was enriched for cells capable of differentiating into pre–B-cell progenitors. We observed 3-fold fewer cells with the CMP phenotype (defined as lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, and CD34+; P < .05)13 in Hmgb3–/Y mice than in wild-type mice (Figure 3B). Hmgb3–/Y CMPs also exhibited a 3- to 5-fold decrease in the ability to generate clonogenic myeloid colonies (P < .001; Figure 3D) compared with wild-type CMPs.
Determination of Hmgb3–/Y HSC repopulating ability
By FACS analysis, Hmgb3–/Y and wild-type mice were found to contain equivalent numbers of cells with the HSC phenotype (lin–, c-kitHI, Sca-1HI, IL-7Rα–; Figure 3B). To evaluate the ability of HSCs from Hmgb3–/Y mice to repopulate hematopoietic tissues, we mixed equivalent numbers of bone marrow cells from Hmgb3–/Y or littermate wild-type mice (129/SvJ strain) and C57BL/6 mice and transplanted these cells into lethally irradiated 129 × C57BL/6F1 recipients. Sixteen weeks after transplantation, Southern blot analyses of bone marrow DNA were performed. Polymorphisms in the β-globin locus were used to differentiate between the contributions of Hmgb3–/Y/wild-type and C57BL/6 HSCs.19 The mean contribution in bone marrow of Hmgb3–/Y cells is higher as predicted by the erythrocythemic phenotype and the increased number of CFU cells (CFU-Cs; Tables 1 and 2), but due to the high degree of variability in repopulating as reflected by significantly higher standard deviations (Table 3; F test, P = .01), the difference between the means was not significant (t test, P > .05). Previous work by Harrison et al,20 showed that regardless of the mean contribution of donor cells to repopulation, a small number of hematopoietic primitive progenitor cells will result in a greater degree of variability than a large number. The increased variability is consistent with the lower numbers of CLPs and CMPs that we observed.
Analysis of competitive repopulation assays
. | No. . | % Donor-derived bone marrow . | T test, mean . | F test, SD . |
|---|---|---|---|---|
| Primary recipients | ||||
| Wild-type | 17 | 56.6 ± 9.6 | .10 | .01 |
| Hmgb3-/Y | 20 | 64.5 ± 18.9 | ||
| Secondary recipients | ||||
| Wild-type | 12 | 56.2 ± 6.3 | .11 | <.01 |
| Hmgb3-/Y | 19 | 63.5 ± 14.4 |
. | No. . | % Donor-derived bone marrow . | T test, mean . | F test, SD . |
|---|---|---|---|---|
| Primary recipients | ||||
| Wild-type | 17 | 56.6 ± 9.6 | .10 | .01 |
| Hmgb3-/Y | 20 | 64.5 ± 18.9 | ||
| Secondary recipients | ||||
| Wild-type | 12 | 56.2 ± 6.3 | .11 | <.01 |
| Hmgb3-/Y | 19 | 63.5 ± 14.4 |
Southern blot analyses were performed on genomic DNA isolated from bone marrow to determine the contribution to hematopoietic repopulation made by HSCs of either 129/SvJ wild-type or Hmgb3-/Y origin (see “Materials and methods”). For each set of primary and secondary recipients, the mean percent and standard deviation of donor (Hmgb3-/Y or 129/SvJ wild-type) derived bone marrow repopulation was determined as outlined in “Materials and methods.” Statistical analyses were performed using the Student t test (mean percent values) and F test (standard deviation).
Bone marrow cells from primary recipients (either Hmgb3–/Y/C57BL/6 or wild-type/C57BL/6) were also used to transplant lethally irradiated 129 × C57BL/6F1 secondary recipients to measure HSC self-renewal. Southern blot analysis performed on bone marrow 16 weeks after transplantation demonstrated increased variability in repopulation among Hmgb3–/Y recipients (Table 3; F test, P < .01), but the mean values were similar to the primary recipients indicating no defect in self-renewal (t test, P > .05). Tertiary recipients of bone marrow derived from secondary recipients further demonstrated no defect in Hmgb3–/Y HSC self-renewal (data not shown).
Cell cycle and proliferative status of Hmgb3–/Y hematopoietic stem and progenitor cells
To examine the mechanism behind the defect in the ability of Hmgb3–/Y HSCs to generate CLPs and CMPs and initiate hematopoiesis, we performed cell cycle analyses on 2 bone marrow populations; the first was enriched for HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα– cells) and the second, which consisted of multipotent and oligopotent progenitors (MPPs; defined as lin–, c-kit+ cells), included CLPs and CMPs in addition to HSCs. We were unable to observe a difference in the cell cycle status of Hmgb3–/Y HSCs compared with wild-type (Figure 4A). However, we did observe a 2-fold decrease in the number of Hmgb3–/Y MPPs in the G2/M phase of the cell cycle compared with wild-type MPPs (P = .02; Figure 4B-C). The decrease in the number of Hmgb3–/Y MPPs in G2/M phase did not correspond with an increase in the number of cells actively undergoing apoptosis (data not shown).
Cell cycle and proliferative status of Hmgb3–/Y primitive hematopoietic cells. (A) Representative cell cycle histograms obtained through PI staining of wild-type (left) and Hmgb3–/Y (right) HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–). Staining was performed on cells pooled from 3 mice of each genotype. Analysis of histograms performed as described in “Materials and methods.” (B) Representative cell cycle histograms obtained through PI staining of wild-type (left) and Hmgb3–/Y (right) multipotent and oligopotent progenitor cells (MPPs: lin–, c-kit+). Staining was performed on cells pooled from 3 mice of each genotype. (C) Percentages of both wild-type and Hmgb3–/Y Lin–, c-kit+ bone marrow cells in G1, S, and G2 phases (n = 3). The data represent the pooled results of 3 independent experiments. (D) Percentages of wild-type (n = 3) and Hmgb3–/Y (n = 3) HSCs, CLPs (lin–, c-kitLO, Sca-1LO, IL-7Rα+), CMPs (lin–, c-kit+, Sca-1–, IL-7Rα–), and myeloid progenitors (MPs: lin–, c-kitLO, Sca-1–, IL-7Rα–) that incorporated BrdU. The data represent the pooled results of 3 independent experiments using pooled bone marrow from 3 mice. (E) Average concentration of nucleated peripheral blood cells following mobilization of splenectomized wild-type (n = 10, □) and Hmgb3–/Y (n = 10, •) mice with 200 μg/kg/d G-CSF. Peripheral blood cells were counted starting on the day of the first treatment and were counted every day 24 hours after the last treatment. The data represent the pooled data from 2 independent experiments involving at least 5 animals in each group. Error bars in panels C-E represent standard deviation. P values were determined by Student t test.
Cell cycle and proliferative status of Hmgb3–/Y primitive hematopoietic cells. (A) Representative cell cycle histograms obtained through PI staining of wild-type (left) and Hmgb3–/Y (right) HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–). Staining was performed on cells pooled from 3 mice of each genotype. Analysis of histograms performed as described in “Materials and methods.” (B) Representative cell cycle histograms obtained through PI staining of wild-type (left) and Hmgb3–/Y (right) multipotent and oligopotent progenitor cells (MPPs: lin–, c-kit+). Staining was performed on cells pooled from 3 mice of each genotype. (C) Percentages of both wild-type and Hmgb3–/Y Lin–, c-kit+ bone marrow cells in G1, S, and G2 phases (n = 3). The data represent the pooled results of 3 independent experiments. (D) Percentages of wild-type (n = 3) and Hmgb3–/Y (n = 3) HSCs, CLPs (lin–, c-kitLO, Sca-1LO, IL-7Rα+), CMPs (lin–, c-kit+, Sca-1–, IL-7Rα–), and myeloid progenitors (MPs: lin–, c-kitLO, Sca-1–, IL-7Rα–) that incorporated BrdU. The data represent the pooled results of 3 independent experiments using pooled bone marrow from 3 mice. (E) Average concentration of nucleated peripheral blood cells following mobilization of splenectomized wild-type (n = 10, □) and Hmgb3–/Y (n = 10, •) mice with 200 μg/kg/d G-CSF. Peripheral blood cells were counted starting on the day of the first treatment and were counted every day 24 hours after the last treatment. The data represent the pooled data from 2 independent experiments involving at least 5 animals in each group. Error bars in panels C-E represent standard deviation. P values were determined by Student t test.
To analyze cell proliferation, we injected Hmgb3–/Y and littermate wild-type mice with BrdU for 3 consecutive days before measuring BrdU incorporation in hematopoietic cell populations. We did not observe significant differences in BrdU incorporation between Hmgb3–/Y or wild-type HSCs or CMPs (Figure 4D). However, there was a 1.4-fold increase (P = .01) in the percentage of BrdU+ CLPs and a 1.7-fold increase in the percentage of BrdU+ myeloid progenitor cells (lin–, c-kitLO, Sca-1–; P = .01) isolated from Hmgb3–/Y mice compared with wild-type mice. These data are consistent with the observations of increased capacity of Hmgb3–/Y CLPs to form CFU–Pre-B colonies and the increased numbers of Hmgb3–/Y CFU-GMs.
Response of Hmgb3–/Y mice to granulocyte-colony stimulating factor
We compared the response of Hmgb3–/Y and wild-type mice to G-CSF. Splenectomized Hmgb3–/Y and littermate control mice were treated with 200 mg/kg/d G-CSF for 7 days, and the concentration of white blood cells (WBCs) in the peripheral blood was determined. Initially and after 24 hours, WBC concentrations were equivalent between Hmgb3–/Y and wild-type mice (Figure 4E). Between 24 and 48 hours, the WBC concentration of wild-type mice increased 1.4-fold (P < .001) compared with the WBC concentration in Hmgb3–/Y mice, which remained unchanged (P = .36). WBC numbers in Hmgb3–/Y and wild-type mice increased at the same rate after 72 hours and remained statistically similar at later time points.
Increased levels of Gata-2 and c-myb in the Hmgb3–/Y HSC population
We have previously shown that Gata-2 and c-myb mRNA is present at high levels in HSCs17 and both of these genes have been shown to be required for definitive hematopoiesis.21-24 Furthermore, overexpression of Gata-2 and c-myb inhibits differentiation of hematopoietic cells.25-27 We used semiquantitative RT-PCR analysis to compare the levels of these and other mRNAs in wild-type and Hmgb3–/Y HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–; Figure 5A). We observed a 1.7-fold increase in Gata-2 mRNA (P = .03) and a 2.8-fold increase in c-myb mRNA (P < .01) in Hmgb3–/Y HSCs compared with wild-type HSCs (Figure 5B). The mRNA levels for the chemokine receptor genes CXCR4 and CCR3, both of which are expressed in HSCs and are important for HSC homing,16,28 did not differ between wild-type and Hmgb3–/Y HSCs (Figure 5B), leading to the conclusion that the stromal cell–derived factor-1 (SDF-1)/CXCR4 axis required for HSC homing was not affected by an Hmgb3 deficiency. Gata-1 and c-kit mRNA levels were also similar in wild-type and Hmgb3–/Y HSCs (Figure 5; data not shown).
Increased levels of Gata-2 and c-myb in Hmgb3–/Y HSCs. (A) Semiquantitative duplex RT-PCR analysis of selected genes in wild-type and Hmgb3–/Y HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–). Test gene mRNA levels were quantified within the linear range of amplification by densitometry and normalized to β2-microglobulin expression. (B) Test gene mRNA levels relative to β2-microglobulin. The amount of test gene mRNA in wild-type (n = 3) and Hmgb3–/Y (n = 3) HSCs was normalized to β2-microglobulin mRNA (β2) by the formula (densitometry value: test/densitometry value: β2-microglobulin). The data represent the pooled results of 3 independent experiments using mRNA isolated from sorted HSCs pooled from 3 mice. Standard deviation values between independent RT-PCR reactions amplifying CCR3 and Gata-1 mRNA from wild-type HSC RNA are not visible on this scale. Error bars represent standard deviation. P values were determined by Student t test.
Increased levels of Gata-2 and c-myb in Hmgb3–/Y HSCs. (A) Semiquantitative duplex RT-PCR analysis of selected genes in wild-type and Hmgb3–/Y HSCs (lin–, c-kitHI, Sca-1HI, IL-7Rα–). Test gene mRNA levels were quantified within the linear range of amplification by densitometry and normalized to β2-microglobulin expression. (B) Test gene mRNA levels relative to β2-microglobulin. The amount of test gene mRNA in wild-type (n = 3) and Hmgb3–/Y (n = 3) HSCs was normalized to β2-microglobulin mRNA (β2) by the formula (densitometry value: test/densitometry value: β2-microglobulin). The data represent the pooled results of 3 independent experiments using mRNA isolated from sorted HSCs pooled from 3 mice. Standard deviation values between independent RT-PCR reactions amplifying CCR3 and Gata-1 mRNA from wild-type HSC RNA are not visible on this scale. Error bars represent standard deviation. P values were determined by Student t test.
Discussion
Hmgb3, along with Hmgb1 and Hmgb2, comprise the HMG-Box subfamily of the high-mobility group proteins. Although the HMG-Box family members are more than 80% identical to each other,29 mouse knock-out models for these genes have varied phenotypes. Hmgb1 mRNA is present in virtually all tissues and its deficiency results in lethal hypoglycemia in newborn mice.9 Hmgb1 has been shown to interact with RAG1 and RAG2 and to play a role in V(D)J recombination,30-33 yet there are no abnormalities in the Hmgb1–/– thymus and there are normal amounts of serum immunoglobulin and CD4+ and CD8+ thymocytes. Also, there was no overt defect in cellular proliferation, as the growth rate of Hmgb1–/– fibroblasts was comparable to that of control fibroblasts. Hmgb2 is highly expressed in embryonic day 10.5 (E10.5) embryos and in the adult thymus and testes.29 Hmgb2–/– mice are viable, but homozygous males exhibit decreased fertility due to defective spermatogenesis. Similar to Hmgb1–/– mice, Hmgb2–/– mice exhibited a normal thymus, normal serum immunoglobulin levels, and a normal number of peripheral B and T lymphocytes. Our results represent the first demonstration of a role of an HMG-Box protein in hematopoiesis, namely that a deficiency of Hmgb3 leads to a decrease in the ability of HSCs to generate CLPs and CMPs and an increase in erythroid cells and precursors. We propose that any compensation by ubiquitous expression of Hmgb1 must be incomplete, at least in these cell types. Furthermore, our data provide the first direct evidence of genetic control of generation and maturation of common lymphoid and myeloid progenitors.
There are several hypotheses that could explain the impairment in the ability of Hmgb3–/Y HSCs to initiate hematopoiesis. First, homing of Hmgb3–/Y HSCs to the bone marrow may be inhibited. However, the ability of Hmgb3–/Y HSCs to transplant secondary and tertiary recipients indicates that these cells were capable of homing and normal levels of CXCR4 and CCR3 mRNA in Hmgb3–/Y HSCs suggest that the homing axis is intact. A second explanation is that Hmgb3–/Y mice exhibit stem cell depletion or exhaustion. However, transplantations performed with bone marrow from 1-year-old Hmgb3–/Y mice resulted in comparable engraftment to transplantations performed with marrow from 6- to 8-week-old Hmgb3–/Y mice (data not shown) and the differences in repopulating ability between the primary and secondary recipients were not significant. Finally, successful engraftment of secondary and tertiary recipients by Hmgb3–Y HSCs indicates that stem cell renewal was not affected, rendering a stem cell depletion model unlikely.
A third possibility is that Hmgb3–/Y HSCs self-renew normally and maintain normal numbers of HSCs but are impaired in their ability to generate normal numbers of CLPs and CMPs. We propose that reduced numbers of CLPs and CMPs in Hmgb3–/Y mice result in greater variability in the repopulation of the myeloid and lymphoid lineages. Based on our previous result that enforced expression of Hmgb3-inhibited hematopoietic differentiation,10 we predict that CLPs and CMPs differentiate into mature phenotypes more rapidly in the absence of Hmgb3. In this model, the phenotypic HSCs would be more primitive and would be expected to have higher levels of Gata-2 and c-myb mRNA. The decrease in CLPs and CMPs in Hmgb3–/Y mice is not due to enhanced apoptosis since there is no increase in apoptosis of Hmgb3–/Y lin–, c-kit+ cells.
Our data cannot discriminate between the possibility that the decrease in Hmgb3–/Y MPP cells in G2/M phase is due to an increase in the length of the G1 or S phases or cell cycle arrest. We hypothesize that the few dividing Hmgb3–/Y MPP cells differentiate rapidly and acquire a lin+ or c-kit– phenotype. The latter explanation is supported by our findings that Hmgb3–/Y MPPs incorporate BrdU at levels greater than wild-type cells. This is also consistent with our observation that Hmgb3–/Y bone marrow contains increased numbers of more mature myeloid, B lymphoid, and erythroid progenitor cells.
The stage at which differences in progenitor cell numbers appear varies between lineages. In the lymphoid lineage, there are fewer CLPs in Hmgb3–/Y mice, however, they exhibit a greater potential to proliferate and differentiate into CFU–Pre-Bs. In the myeloid lineage, there are fewer CMPs in Hmgb3–/Y mice, which have a decreased capacity to proliferate and differentiate into myeloid colonies. Hmgb3–/Y bone marrow contained equal numbers of CFU-GEMMs, but more CFU-GMs than wild-type marrow, suggesting that the increased proliferation of myeloid cells occurs after the CFU-GEMM stage. Within the erythroid lineage, the stage at which increased proliferation occurs is after the BFU-E stage but prior to (or at) the CFU-E stage, resulting in the erythrocythemic phenotype. Overall, our data support a model in which Hmgb3 plays a significant role in regulating the balance between proliferation and differentiation in primitive stages of hematopoiesis.
Prepublished online as Blood First Edition Paper, September 9, 2004; DOI 10.1182/blood-2004-07-2551.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal