Abstract
Embryonic stem (ES) cells homozygous for a Shp-2 mutation (Shp-2Δ46-110) demonstrate leukemia inhibitory factor (LIF) hypersensitivity and increased LIF-stimulated phosphorylation of signal transducer and activator of transcription (STAT3). We hypothesized that LIF-responsive genes in Shp-2Δ46-110 cells would represent potential candidates for molecules vital for ES cell self-renewal. Using microarray analysis, we detected 41 genes whose expression was modified by LIF in Shp-2Δ46-110 ES cells. Induction of 2 significantly up-regulated genes, suppressor of cytokine signaling–3 (SOCS-3) and Krüppel-like factor 4 (Klf4), was verified using Northern blotting. ES cells overexpressing SOCS-3 had an increased capacity to differentiate to hematopoietic progenitors, rather than to self-renew. In contrast, ES cells overexpressing Klf4 had a greater capacity to self-renew based on secondary embryoid body (EB) formation. Klf4-transduced d6 EBs expressed higher levels of Oct-4, consistent with the notion that Klf4 promotes ES cell self-renewal. These findings verify the negative role of SOCS-3 on LIF signaling and provide a novel role for Klf4 in ES cell function.
Introduction
The mechanisms that control self-renewal of somatic and embryonic stem (ES) cells are unknown; however, identification of regulating molecules is essential for developing stem cell-based therapeutic strategies. Leukemia inhibitory factor (LIF) promotes murine ES cell self-renewal and maintains pluripotency in culture.1 LIF binds the heterodimeric LIF receptor-glycoprotein 130 (gp130) complex and activates the Jak kinases with recruitment of Shp-2 and signal transducer and activator of transcription (STAT3).2 Mutation of STAT3-interacting tyrosines on gp130 reduces LIF-stimulated phospho-STAT3 and STAT3-DNA–binding activity as well as ES cell self-renewal, providing strong evidence that LIF-stimulated STAT3 activation is critical for ES cell self-renewal.3,4 Mutation of the Shp-2–binding tyrosine on gp130 prolongs STAT3 activation and increases self-renewal, whereas it diminishes Erk activation, indicating that Shp-2 functions to balance LIF-stimulated STAT3 and Erk activation, thus modulating ES cell self-renewal and differentiation, respectively.5
Shp-2Δ46-110 ES cells, which are homozygous for an exon 3 deletion from the murine SHP-2 locus,6 have increased sensitivity to LIF and increased self-renewal capacity as assayed by secondary embryoid body (EB) formation, both of which are normalized upon reintroduction of wild-type (WT) Shp-2.7,8 Consistently, Shp-2Δ46-110 ES cells manifest increased LIF-stimulated phospho-STAT3 and decreased LIF-stimulated phospho-Erk activity.7
Because LIF-stimulated STAT3 is crucial for ES cell self-renewal, we hypothesized that LIF-induced genes would be fundamental in ES cell self-renewal. Additionally, because LIF-induced phospho-STAT3 levels are higher in Shp-2Δ46-110 cells than in WT cells,7 we reasoned that LIF stimulation of Shp-2Δ46-110 cells would provide a sensitive system for the detection of STAT3-regulated genes. Therefore, we identified LIF-regulated genes by comparing expression profiles of Shp-2Δ46-110 ES cells either unstimulated or stimulated with LIF. The cDNAs for 2 LIF-stimulated genes, which encode suppressor of cytokine signaling-3 (SOCS-3) and the transcription factor, Krüppel-like factor 4 (Klf4), were introduced into WT ES cells for functional analysis.
Study design
ES cell culture and microarray analysis
The Shp-2Δ46-110 cell line, IC3, was generated as described.7 IC3 cells were cultured in serum-free, LIF-free media for 6 hours followed by treatment with phosphate-buffered saline (PBS) or LIF (1000 U/mL) for 45 minutes. Total RNA was extracted using QIAamp RNA Mini kit (Qiagen, Valencia, CA) and submitted to the Center for Medical Genomics at the Indiana University School of Medicine for labeling and hybridization to Affymetrix Murine Genome U74A GeneChips (Affymetrix, Santa Clara, CA). LIF stimulation, RNA preparation, and microarray analysis were performed in triplicate and analyzed with MAS5 software (Affymetrix, Santa Clara, CA). The t tests using the signal were performed using CyberT9 for genes present in at least one half of the arrays for either condition.10 Confirmatory Northern blots were conducted as described.11
Retroviral plasmids, ES cell transduction, and in vitro differentiation
The SOCS-3 cDNA and the Klf4 cDNA (from Dr Vincent Yang, Emory University School of Medicine, Atlanta, GA) were epitope-tagged with FLAG at the 5′ end, subcloned into the retroviral vector, pMIEG3, in tandem with enhanced green fluorescent protein (EGFP), and confirmed by sequencing. Retroviral supernatants were prepared using Eco-Phoenix packaging cells by the Indiana University Vector Production Facility.
R1 (WT) ES cells were transduced using polybrene (40 mg/mL) over 48 hours. EGFP+ cells were collected using fluorescence-activated cell sorting and subjected to hematopoietic in vitro differentiation assays, as described.7,12 Transduction, sorting, and in vitro differentiation assays were conducted in 2 independent experiments.
Immunoprecipitation and immunoblotting
Proteins from EGFP+ cells were immunoprecipitated as described13 and probed with anti–SOCS-3, anti-Klf4 (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-FLAG (Sigma, St Louis, MO). For Oct-4 detection, 50 μg protein extract was used and probed with anti–Oct-4 (Santa Cruz Biotechnology) or with anti–β-actin to control for protein loading. Signals were detected by enhanced chemiluminescence and quantitated using ImageJ (National Institutes of Health, Bethesda, MD).
Results and discussion
To define an appropriate time to evaluate LIF-stimulated gene expression, we determined that c-fos mRNA induction, known to be regulated by LIF-induced STAT3 activation,14,15 was maximal following LIF treatment for 45 minutes (not shown). Total cellular RNA from Shp-2Δ46-110 ES cells treated with PBS or LIF 1000 U/mL for 45 minutes was submitted for microarray analysis, which revealed that 41 genes or expressed sequence tags were significantly modified (P < .01, determined by t test on the signal value, Tables S1 and S2; see the Supplemental Tables link at the top of the online article link at the top of the online article on the Blood website). The 2 most significantly up-regulated genes were SOCS3 (increase 34.4-fold; P < .000001) and KLF4 (increase 2.1-fold; P = .000057). Using Northern blot, we examined the effect of LIF on SOCS3 and KLF4 expression in both WT (R1) and Shp-2Δ46-110 cells. In the Shp-2Δ46-110 cells, SOCS3 expression was increased dramatically and KLF4 expression was increased approximately 2-fold, in agreement with the microarray results (Figure 1A). SOCS3 was similarly induced in the WT cells. However, at baseline, KLF4 expression was elevated in the WT ES cells compared to the Shp-2Δ46-110 cells and was not induced by LIF treatment (Figure 1A).
SOCS-3 promotes, whereas Klf4 inhibits, ES cell differentiation. (A) Northern blot analysis of LIF stimulation of SOCS3 and KLF4 in WT and Shp-2Δ46-110 ES cells (representative of 2 independent experiments). (B) Schematic diagram of retroviral vectors used to conduct functional studies. IRES indicates internal ribosome entry site; LTR, long terminal repeat. (C) Transduced and sorted ES cells were subjected to in vitro differentiation for hemangioblasts, primitive erythroid (EryP) progenitors, and secondary EBs; data represent 2 independent experiments with cultures plated in triplicate; *P = .02 for pMIEG3-SOCS-3 versus pMIEG3; **P = .03 for pMIEG3-SOCS-3 versus pMIEG3; ***P = .0002 for pMIEG3-Klf4 versus pMIEG3. Error bars represent SEM. Statistics were analyzed using the 2-tailed, unpaired, Student t test.
SOCS-3 promotes, whereas Klf4 inhibits, ES cell differentiation. (A) Northern blot analysis of LIF stimulation of SOCS3 and KLF4 in WT and Shp-2Δ46-110 ES cells (representative of 2 independent experiments). (B) Schematic diagram of retroviral vectors used to conduct functional studies. IRES indicates internal ribosome entry site; LTR, long terminal repeat. (C) Transduced and sorted ES cells were subjected to in vitro differentiation for hemangioblasts, primitive erythroid (EryP) progenitors, and secondary EBs; data represent 2 independent experiments with cultures plated in triplicate; *P = .02 for pMIEG3-SOCS-3 versus pMIEG3; **P = .03 for pMIEG3-SOCS-3 versus pMIEG3; ***P = .0002 for pMIEG3-Klf4 versus pMIEG3. Error bars represent SEM. Statistics were analyzed using the 2-tailed, unpaired, Student t test.
To examine SOCS-3 and Klf4 function within ES cells, each cDNA was introduced into R1 ES cells using retroviral transduction (Figure 1B). Overexpression of SOCS-3 promoted ES cell differentiation to both hemangioblasts and primitive erythroid progenitors, whereas overexpression of Klf4 demonstrated a trend toward reduced differentiation (Figure 1C). Klf4-transduced ES cells retained a high capacity to generate secondary EBs, suggesting that Klf4 promotes ES cell self-renewal in addition to inhibiting ES cell differentiation (Figure 1C). These data support previous studies indicating that SOCS-3 acts as a negative regulator of LIF signaling15-17 and provide a novel role for Klf4 in ES cell differentiation and self-renewal.
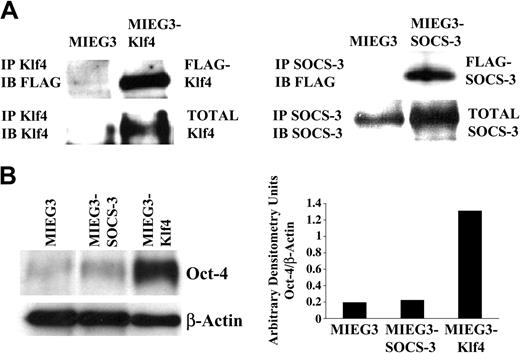
Based on the differences in ES cell function induced by SOCS-3 or Klf4 overexpression, we determined the exogenously introduced and total SOCS-3 and Klf4 protein levels achieved in the transduced ES cells. Immunoblotting with anti-FLAG demonstrated that the exogenously introduced SOCS-3 and Klf4 were expressed in the transduced ES cells (Figure 2A). Densitometric measurements demonstrated a 3-fold increase in total SOCS-3 protein and a 6-fold increase in total Klf4 protein compared to cells transduced with vector alone (Figure 2A).
Overexpression of Klf4 results in sustained Oct-4 expression. (A) Immunoprecipitation studies demonstrating that the exogenously introduced (FLAG epitope-tagged) Klf4 and SOCS-3 proteins are expressed in transduced ES cells and that, consistently, total Klf4 and SOCS-3 protein levels are increased compared to cells transduced with vector alone. (B) Evaluation of Oct-4 protein levels in d6 EBs derived from pMIEG3-, pMIEG3-SOCS-3–, and pMIEG3-Klf4–transduced ES cells. Oct-4 band intensities normalized to β-actin band intensities are shown graphically. IP indicates immunoprecipitation; IB, immunoblotting.
Overexpression of Klf4 results in sustained Oct-4 expression. (A) Immunoprecipitation studies demonstrating that the exogenously introduced (FLAG epitope-tagged) Klf4 and SOCS-3 proteins are expressed in transduced ES cells and that, consistently, total Klf4 and SOCS-3 protein levels are increased compared to cells transduced with vector alone. (B) Evaluation of Oct-4 protein levels in d6 EBs derived from pMIEG3-, pMIEG3-SOCS-3–, and pMIEG3-Klf4–transduced ES cells. Oct-4 band intensities normalized to β-actin band intensities are shown graphically. IP indicates immunoprecipitation; IB, immunoblotting.
Because the secondary EB assay suggested that Klf4 promotes ES cell self-renewal, we examined the level of Oct-4 protein, which is essential for ES cell self-renewal,18,19 in day-6 EBs. We observed that Oct-4 was significantly elevated (7-fold) in the Klf4-transduced d6 EBs compared to that of vector alone or SOCS-3–transduced d6 EBs (Figure 2B). These results support a role for Klf4 in modulating ES cell self-renewal.
This study has defined a battery of LIF-sensitive genes within murine ES cells that provide potential mediators of ES cell self-renewal and differentiation. Functional evaluation demonstrated a novel role for Klf4 in ES cell differentiation and self-renewal. Klf4 is abundant in epithelial cells of the intestine, colon, skin, and thymus20 and has been implicated as a negative prognosticator in breast cancer21,22 and as a tumor suppressor gene in gastrointestinal malignancies.23,24 Based on the differential effect of LIF-stimulated KLF4 expression in the WT and Shp-2Δ46-110 cells, it is possible that altered Klf4 expression partially accounts for the increased self-renewal phenotype in the Shp-2Δ46-110 cells. In contrast, we observed that SOCS-3 promotes ES cell differentiation, likely by diminishing the self-renewing effects of LIF. Our findings support the conclusions of others that SOCS-3 is regulated at the transcriptional level by LIF17,25 and functions as a feedback inhibitor on LIF signaling.16,17 These data provide feasible candidate genes for ES cell self-renewal, support a negative role of SOCS-3 on LIF signaling in ES cells, and present a novel role for Klf4 in ES cell function.
Prepublished online as Blood First Edition Paper, September 9, 2004; DOI 10.1182/blood-2004-07-2681.
Supported by F32CA84677 (R.J.C.), RO1HL63169 (M.C.Y.), the Indiana 21st Century Research and Technology Fund (M.C.Y, H.J.E.), and the Indiana Genomics Initiative (M.C.Y., H.J.E.). RJC is a recipient of a March of Dimes Basil O'Connor Starter Scholar Award (5-FY03-136).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Chris Starkey at the Indiana University Vector Production Facility, Susan Rice and Denessa Luckett at the Indiana University Flow Cytometry Facility, Arliene K. Britt for assistance in manuscript preparation, and Edward Chan for critical review of the manuscript.
Supplemental data
This table lists the genes up-regulated by LIF treatment of Shp-2Δ46-110 cells as determined by microarray analysis (P < .01). Field descriptions are as follows: (1) probe set name: Affymetrix probe set name; (2) Control mean/std. dev.: mean and standard deviation of the signal for the untreated samples; (3) LIF treated mean/std. dev.: mean and standard deviation of the signal for the LIF treated samples; (4) Cyber-T p-value: P value of Cyber-T t test using the signal; (5) Fold-change: fold change calculated by CyberT; (6) GenBank, Unigene ID, Locus Link ID, Gene Symbol and Gene Description: Affymetrix annotations from May 2004 (www.affymetrix.com).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal