Abstract
DNA damage activates the monoubiquitination of the Fanconi anemia (FA) protein, FANCD2, resulting in the assembly of FANCD2 nuclear foci. In the current study, we characterize structural features of FANCD2 required for this intranuclear translocation. We have previously identified 2 normal mRNA splice variants of FANCD2, one containing exon 44 sequence at the 3′ end (FANCD2-44) and one containing exon 43 sequence (FANCD2-43). The 2 predicted FANCD2 proteins differ in their carboxy terminal 24 amino acids. In stably transfected FANCD2—/— fibroblasts, FANCD2-44 and FANCD2-43 proteins were monoubiquitinated on K561. Only FANCD2-44 corrected the mitomycin C (MMC) sensitivity of the transfected cells. We find that monoubiquitinated FANCD2-44 was translocated from the soluble nuclear compartment into chromatin. A mutant form of FANCD2-44 (FANCD2-K561R) was not monoubiquitinated and failed to bind chromatin. A truncated FANCD2 protein (Exon44-T), lacking the carboxy terminal 24 amino acids encoded by exon 44 but retaining K561, and another mutant FANCD2 protein, with a single amino acid substitution at a conserved residue within the C-terminal 24 amino acids (D1428A), were monoubiquitinated. Both mutants were targeted to chromatin but failed to correct MMC sensitivity. Taken together, our results indicate that monoubiquitination of FANCD2 regulates chromatin binding and that D1428 within the carboxy terminal acidic sequence encoded by exon 44 is independently required for functional complementation of FA-D2 cells. We hypothesize that the carboxy terminus of FANCD2-44 plays a critical role in sensing or repairing DNA damage.
Introduction
Fanconi anemia (FA) is a rare autosomal recessive cancer susceptibility syndrome characterized by developmental abnormalities, progressive bone marrow failure, and cellular hypersensitivity to DNA cross-linking agents.1 Eleven FA complementation groups have been identified (A, B, C, D1, D2, E, F, G, I, J, and L)2,3 and 8 FA genes have been cloned.2,4,5 The FANCD1 gene is identical to the breast cancer susceptibility gene, BRCA2.6 The 8 encoded FA proteins (A, C, D1, D2, E, F, G, L) cooperate in a common cellular pathway, the FA/BRCA pathway.7
In this pathway, 6 of the FA proteins (A, C, E, F, G, L)8,9 bind in a constitutive nuclear protein complex (the FA complex). In response to DNA damage10 or during the S phase of the cell cycle,11 the FA complex promotes the monoubiquitination of the downstream FANCD2 protein. This event requires a molecular interaction between the FANCE and FANCD2 proteins.12,13 Monoubiquitination of FANCD2 is required for targeting of FANCD2 into nuclear foci containing BRCA1, FANCD1/BRCA2, and RAD51.11 These subnuclear foci may be sites of homologous recombination-mediated DNA repair, given the known roles of BRCA1, BRCA2, and RAD51 in this process.14,15 Disruption of the FA/BRCA pathway results in the characteristic cellular and clinical features of FA, including hypersensitivity to DNA cross-linking agents.16
A critical regulatory event in the FA/BRCA pathway is the monoubiquitination of FANCD2 on Lysine 561.10 Analysis of FANCD2 monoubiquitination provides a rapid diagnostic screen for the integrity of the FA/BRCA pathway.17 In addition, FANCD2 undergoes an ionizing radiation (IR)–inducible, ataxia telangiectasia (ATM)–dependent phosphorylation on Serine 222.18 Phosphorylation of this serine is required for the establishment of an intra–S-phase checkpoint response but is not required for FANCD2 monoubiquitination, FANCD2 targeting to foci, or FANCD2-mediated DNA repair.
Little is known about the regulation or functional outcome of FANCD2 monoubiquitination. First, the newly cloned FANCL protein has a plant homeodomain (PHD) domain with E3 ubiquitin ligase activity, although its ubiquitination of FANCD2 has not been demonstrated in vitro.2 Interestingly, BRCA1, an 1863–amino acid tumor suppressor protein with an amino terminal E3 RING (Really interesting new gene) finger ubiquitin ligase domain,19 is required for efficient FANCD2 activation following DNA damage, although BRCA1-deficient cells still exhibit some FANCD2 monoubiquitination.20 BRCA1 and/or its heterodimeric E3 ligase partner, BRCA1-associated ring domain 1 (BARD1),21 may therefore contribute directly or indirectly to FANCD2 monoubiquitination. Second, the monoubiquitinated isoform of FANCD2 (FANCD2-L) accumulates in discrete nuclear foci in damaged cells,10 suggesting that it is actively transported to these structures. Accordingly, these foci may contain a specific receptor for FANCD2-L or its ubiquitin moiety. Third, following DNA repair or during the mitotic phase of the cell cycle, FANCD2-L is deubiquitinated, suggesting a reversible and more complex mechanism of regulation.
The nucleus is organized into an integrated structure in which chromatin is associated with a nonhistone scaffold termed the nuclear matrix.22 Various aspects of nucleic acid metabolism, including DNA replication, transcription, and the repair of UV-induced thymidine dimers, require an interaction between chromatin and the nuclear matrix. A previous study indicates that FA proteins required for FANCD2 monoubiquitination are found in the chromatin,23 and we recently reported that monoubiquitinated FANCD2 interacts with the breast cancer susceptibility gene product BRCA2/FANCD1 in chromatin.24
In the current study, we examined structural features of FANCD2 required for monoubiquitination, nuclear foci assembly, and functional activity in the FA/BRCA pathway. We identified a conserved 24–amino acid carboxy terminal region of FANCD2 encoded by exon 44. This region is predicted to form a coiled-coil structure and it is required for FANCD2 functional activity downstream in the FA/BRCA pathway but not for monoubiquitination of FANCD2. Further, monoubiquitination, but not the C-terminal region of the protein, is required for FANCD2 localization to chromatin.
Materials and methods
Cell culture
Epstein-Barr virus (EBV)–transformed lymphoblasts and adherent cells (HeLa and PD20F lines) were maintained in RPMI and Dulbecco modified Eagle medium (DMEM) media, respectively, supplemented with 15% heat-inactivated fetal calf serum (FCS). Cells were grown in a humidified 5% CO2-containing atmosphere at 37°C. The FA-G lymphoblast line, EUFA316, was provided by Dr Hans Joenje (Department of Clinical Genetics and Human Genetics, VU University Medical Center, Amsterdam, The Netherlands).4 The simian virus 40 (SV40)–transformed FA-D2 fibroblast, PD20F, expressing wild-type FANCD2 or FANCD2-K561R or transfected with pMMP vector, has previously been described.5
Retroviral infection of FA cell lines
The indicated pMMP constructs were transfected with Fugene 6 by lipofection into 293-GPG producer cells (human embryonic kidney cells) expressing the vesicular stomatitis virus-G (VSV-G) envelope protein.25 Retroviral supernatants were collected on day 5 following lipofection, as previously described.8 Retroviral supernatants contained 4.6 × 106 infectious units/mL, as estimated by Southern blot analysis of infected NIH-3T3 cells (data not shown).
FA fibroblasts were infected with the various pMMP supernatants by a 6- to 8-hour incubation in the presence of 8 μg/mL polybrene.8 Infected cells were washed free of viral supernatant and resuspended in growth media. After 48 hours, cells were transferred to media containing puromycin (1 μg/mL). Surviving cells were grown under continuous selection in puromycin.
RNA extraction and reverse transcription–polymerase chain reaction (RT-PCR)
Total RNAs were extracted using Trizol reagent (Invitrogen, Carlsbad, CA), measured by optic density, and stored at –80°C. An aliquot (2 μg) of total RNA was placed in 20 μL of room temperature reaction and converted to first-strand cDNA using Superscript II RNase H(-) Reverse Transcriptase (Invitrogen) and oligo(dT)15 (Invitrogen) as instructed by the manufacturer. Negative control reactions without reverse transcriptase were also performed. The shared upstream primer for both exon 43 and exon 44 variants of FANCD2 was DF3862 (5′-CATCCTGTTCTGCATGTATG-3′). The specific downstream primer for exon 43 variant was DR4462 (5′-TCGTTGTTTCTTCTGGGTCC-3′) and the specific downstream primer for exon 44 variant was DSR4360 (5′-GGGTCTAATCAGAGTCATCA-3′). An aliquot (0.4 μL) of cDNA (40 ng RNA equivalent) was placed in 40 μL of 1 × PCR buffer supplied with Taq polymerase, 200 μM of each deoxyribonucleotide triphosphate (dNTP), 0.2 μM of each primer (DF3862, DR4462, and DSR4360), and 2 U of AmpliTaq polymerase (Roche, Indianapolis, IN). PCR was run for 30 cycles and each cycle constituted denaturation (45 seconds at 94°C, first cycle 4 minutes 45 seconds), annealing (1 minute at 55°C), and extension (1 minute at 72°C, last cycle 8 minutes). The PCR reaction (10 μL) was subjected to electrophoresis on a 1.2% agarose gel containing ethidium bromide. The expected sizes of the PCR products are as follows: exon 43 form, 601 base pair (bp); exon 44 form, 499 bp.
MMC sensitivity assay
The mitomycin C (MMC) sensitivity assay was performed essentially as previously described.26 Cells were passaged into 12-well plates at 6 × 103 cells per well. The following morning, a series of concentrations of MMC was added and cells were cultured for 5 days. Cells were then washed with phosphate-buffered saline (PBS) and fixed 10 minutes with 10% (vol/vol) methanol/10% (vol/vol) acetic acid at room temperature. Cells were then stained 10 minutes with 1% (wt/vol) crystal violet (Sigma-Aldrich, St. Louis, MO) in methanol. The plates were washed in an excess volume of dH2O, and absorbed dye was released by incubation with agitation for 1 hour at room temperature in methanol containing 0.1% sodium dodecyl sulfate (SDS). Dye containing solution was then transferred to 96-well microtiter plates, and dilutions (1:2) were prepared. The optical density (OD) at 595 nm was then assayed in a microplate reader (BioRad model 3550; Hercules, CA). Results were normalized based upon the untreated samples (0 ng/mL MMC).
Cell cycle synchronization
HeLa cells were synchronized by the double thymidine block method as previously described, with minor modifications.27 Briefly, cells were treated with 2 mM thymidine for 18 hours, thymidine-free media for 10 hours, and additionally with 2 mM thymidine for 18 hours to arrest the cell cycle at the G1/S boundary. Cells were washed twice with PBS, released in DMEM +15% FCS, and analyzed at various time intervals.
Preparation of cellular fractions for immunoblotting
Cells were grown on 15-cm culture dishes. Cells were either left untreated or were exposed to MMC (170 ng/mL) or IR (15 Gy), trypsinized 12 hours later, and collected by centrifugation. Cells were washed once with cold PBS and aliquoted equally into 4 eppendorf tubes and collected by centrifugation in a Sorvall RT 6000D centrifuge (Kendro, Asheville, NC) at 4°C for 3 minutes at 300 g (1200 rpm). One pellet, representing the whole-cell pellet (P1), was frozen in liquid nitrogen. Remaining pellets were resuspended in cold buffer A (10 mM PIPES [piperazine diethanesulfonic acid], pH 7; 100 mM NaCl; 3 mM MgCl2; 1 mM EGTA [ethyleneglycotetraacetic acid]; 300 mM sucrose; 0.5 mM Na3VO4; 50 mM NaF; 10 μg/mL aprotinin; 10 μg/mL leupeptin; 10 μg/mL pepstatin A; and 1 mM PMSF [phenylmethylsulfonyl fluoride]) containing 0.5% Triton X-100 and incubated at room temperature for 2 minutes to permeabilize cells. The suspension was then recentrifuged at 4°C for 3 minutes at 300 g, and the supernatant (S2), representing cytosol and nucleosol, was collected and frozen. The pellet was washed with cold buffer A. One pellet, representing detergent-insoluble nuclei (P2), was frozen. Nuclei were then digested with RNase-free DNase I (200 U/mL) in buffer A for 30 minutes at room temperature. The residual pellet was collected by centrifugation as described above. The supernatant (S3) was frozen, the pellet was washed with cold buffer A, and one residual pellet (P3) was frozen. The remaining residual pellet was extracted with cold buffer A containing 0.25 M ammonium sulfate for 5 minutes at room temperature to extract remaining chromatin. The residual pellet (P4), representing nuclear matrix, was collected by centrifugation at 1877 g for 3 minutes and the supernatant (S4), containing chromatin as identified by the presence of histones, was frozen. The pellet was frozen following a single wash with cold buffer A and centrifugation at 800 g for 3 minutes. Cell-equivalent volumes from each cell fraction were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted for FANCD2, histone H4 (chromatin marker), and lamin B (nuclear matrix marker).
Nuclear fractionation for immunofluorescence microscopy
HeLa cells were grown for 36 hours on glass coverslips (12-mm diameter) coated with 0.1 mg/mL poly-l-lysine to promote adhesion. Cells were then treated with 15 Gy IR for 12 hours. Fixation was at room temperature with 2% paraformaldehyde in PBS. Cells were fixed either prior to any fractionation (equivalent of P1), following permeabilization for 2 minutes at room temperature with buffer A containing 0.5% Triton X-100 (equivalent of P2), or following permeabilization and sequential treatment with 200 U/mL DNase I for 30 minutes in buffer A and then 250 mM ammonium sulfate for 5 minutes in buffer A (equivalent of P4). Following fixation, cells were washed with PBS. Cells that had not been previously permeabilized were treated 3 minutes with 0.2% Triton X-100 in PBS. Incubation with primary and secondary antibodies and washes were as previously described.10 The nuclear matrix was detected with a goat antibody to lamin B (M-20; Santa Cruz Biotechnology, Santa Cruz, CA).
Immunofluorescence microscopy
Immunofluorescence microscopy of human fibroblasts was performed as previously described,10 with modifications for fractionation as described in the preceding section. In brief, cells were pre-permeabilized with 0.25% Triton X-100 in PBS for 1 minute on ice and then fixed with 4% paraformaldehyde in PBS for 15 minutes, followed by permeabilization with 0.5% Triton X-100 in PBS for 1 minute. For immunofluorescence microscopy, fixed cells were incubated with specific primary antibodies at the appropriate dilution in 3% bovine serum albumin-PBS for 1 hour at room temperature (RT). Phosphorylated histone H2AX (Ser 139) was detected with rabbit polyclonal antibodies (Trevigen, Gaithersburg, MD) diluted 200-fold. For colocalization with H2AX, FANCD2 was detected with mouse monoclonal antibody (FI-17; Santa Cruz Biotechnology) diluted 200-fold. Cells were subsequently washed 3 times in PBS and incubated for 1 hour at RT with species-specific fluorescein (Jackson Immunoresearch) or Texas red–conjugated (Amersham) secondary antibodies diluted in 3% bovine serum albumin in PBS. Cells were counterstained with 4′6-diamidine-2-phenylindole dihydrochloride (Roche) to visualize nuclei. Slides were mounted in Vectashield (Vector Laboratories). Images were acquired using an Axioplan 2 imaging microscope (Carl Zeiss, Thornhill, NY), at 100 ×/1.4 NA (Figure 3) or 60 ×/1.4 NA (Figure 5), with a Hamamatsu Orca CCD camera (Bridgewater, NJ). Images were processed with Openlab software (Improvision, Lexington, MA).
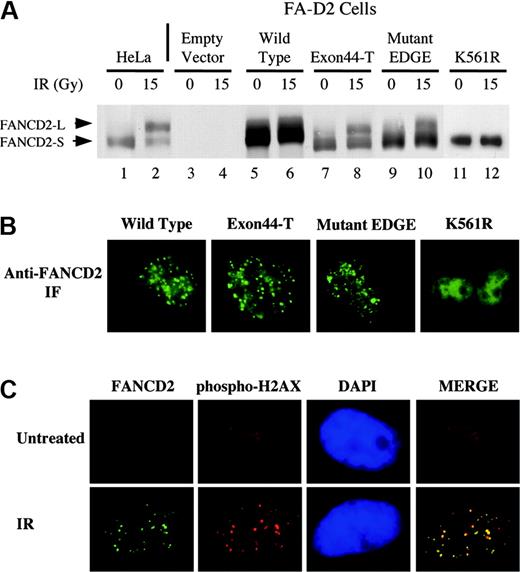
FANCD2 carboxy terminal mutants are monoubiquitinated following ionizing radiation. (A) FA-D2 fibroblasts were stably transduced with either hemagglutinin (HA)–pMMP (empty vector), HA-FANCD2 (wild type), HA–Exon44-T, or HA–mutant-EDGE, as indicated. Cells were irradiated with 15 Gy, as indicated, and whole-cell lysates were immunoblotted with anti-FANCD2 antisera. (B) Immunofluorescence of the transduced cells 8 hours after cellular exposure to IR, using an anti-FANCD2 antiserum. (C) Colocalization of FANCD2 and phosphorylated histone H2AX in DNA damaged-inducible foci. HeLa cells were either untreated (top) or exposed to IR (15 Gy, 15 hours) (bottom), as indicated. HeLa cells were double-stained with monoclonal anti-FANCD2 (FI-17; green) and polyclonal anti–histone γ-H2AX (Ser139; red). Merges of corresponding anti-FANCD2 and anti–histone γ-H2AX images are shown. Original magnifications × 630 (B-C).
FANCD2 carboxy terminal mutants are monoubiquitinated following ionizing radiation. (A) FA-D2 fibroblasts were stably transduced with either hemagglutinin (HA)–pMMP (empty vector), HA-FANCD2 (wild type), HA–Exon44-T, or HA–mutant-EDGE, as indicated. Cells were irradiated with 15 Gy, as indicated, and whole-cell lysates were immunoblotted with anti-FANCD2 antisera. (B) Immunofluorescence of the transduced cells 8 hours after cellular exposure to IR, using an anti-FANCD2 antiserum. (C) Colocalization of FANCD2 and phosphorylated histone H2AX in DNA damaged-inducible foci. HeLa cells were either untreated (top) or exposed to IR (15 Gy, 15 hours) (bottom), as indicated. HeLa cells were double-stained with monoclonal anti-FANCD2 (FI-17; green) and polyclonal anti–histone γ-H2AX (Ser139; red). Merges of corresponding anti-FANCD2 and anti–histone γ-H2AX images are shown. Original magnifications × 630 (B-C).
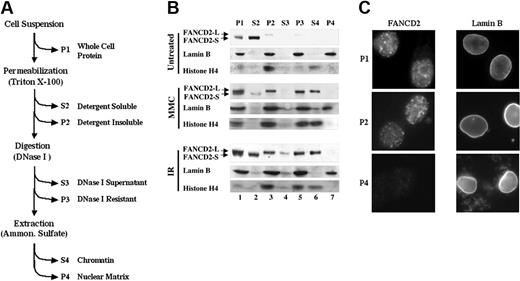
Monoubiquitinated FANCD2 is preferentially retained in the chromatin fraction. (A) Protocol for nuclear fractionation of transduced FA-D2 fibroblasts. Cytoplasm and nucleoplasm were extracted by permeabilization with detergent, and resulting nuclei were nuclease-digested and extracted with NH2SO4. Supernatants (S) and pellets (P) were analyzed for FANCD2, histone H4 (chromatin marker), and lamin B (nuclear matrix marker) by Western blot. (B) Monoubiquitinated FANCD2 is preferentially retained in detergent-insoluble nuclei (P2) and is subsequently extracted with chromatin (S4). (C) FANCD2 nuclear foci are extracted with the chromatin fraction. HeLa cells were grown on coverslips and exposed to IR (15 Gy). After 12 hours, cells were prepared for immunofluorescence microscopy with an antibody to either FANCD2 or lamin B, as indicated. Immunofluorescence microscopy was performed on either unpermeabilized (whole) cells (equivalent to P1 fraction), cells permeabilized with Triton X-100 (equivalent to P2), or cells permeabilized and extracted with DNase I and ammonium sulfate (equivalent to P4 fraction). FANCD2 and lamin B images are from different representative cells. Original magnification × 630.
Monoubiquitinated FANCD2 is preferentially retained in the chromatin fraction. (A) Protocol for nuclear fractionation of transduced FA-D2 fibroblasts. Cytoplasm and nucleoplasm were extracted by permeabilization with detergent, and resulting nuclei were nuclease-digested and extracted with NH2SO4. Supernatants (S) and pellets (P) were analyzed for FANCD2, histone H4 (chromatin marker), and lamin B (nuclear matrix marker) by Western blot. (B) Monoubiquitinated FANCD2 is preferentially retained in detergent-insoluble nuclei (P2) and is subsequently extracted with chromatin (S4). (C) FANCD2 nuclear foci are extracted with the chromatin fraction. HeLa cells were grown on coverslips and exposed to IR (15 Gy). After 12 hours, cells were prepared for immunofluorescence microscopy with an antibody to either FANCD2 or lamin B, as indicated. Immunofluorescence microscopy was performed on either unpermeabilized (whole) cells (equivalent to P1 fraction), cells permeabilized with Triton X-100 (equivalent to P2), or cells permeabilized and extracted with DNase I and ammonium sulfate (equivalent to P4 fraction). FANCD2 and lamin B images are from different representative cells. Original magnification × 630.
Results
Two splice variants of FANCD2
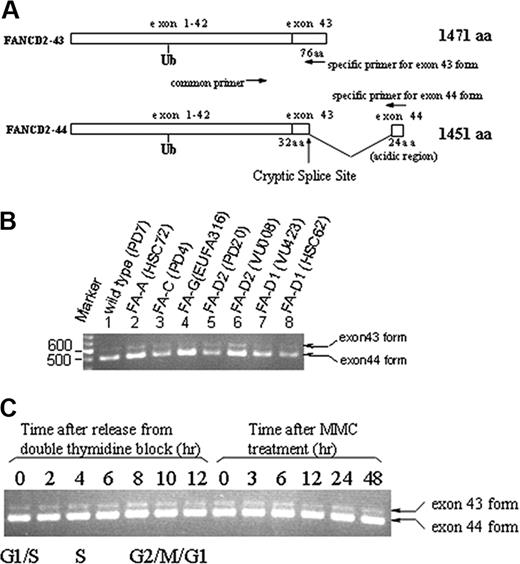
We previously identified 2 normal mRNA splice variants of FANCD2 that differ in their 3′ coding sequence (GenBank accession no. AF273251), as shown schematically in Figure 1A. One splice variant (FANCD2-43), containing exon 43 sequence, is predicted to encode a FANCD2 protein of 1471 amino acids (157 kDa; Figure 1A). The second splice variant (FANCD2-44), containing exon 44 sequence, is predicted to encode a FANCD2 protein of 1451 amino acids (155 kDa). Both splice variants contain the codon in exon 19 encoding lysine 561, the site of FANCD2 monoubiquitination.10 A cDNA corresponding to FANCD2-44 has previously been described and is known to functionally complement the MMC sensitivity of transfected FA-D2 cells.5 FANCD2-44 contains a unique 24–amino acid stretch at its extreme C-terminus.
Expression of 2 splice variants of the FANCD2 gene. (A) Schematic representation of 2 splice variants of FANCD2. A 24–amino acid (aa) sequence, containing 50% acidic residues, is encoded by exon 44 sequence. (B) Detection of FANCD2-44 and FANCD2-43 mRNAs by RT-PCR in various FA cell lines. (C) Detection of FANCD2-44 and FANCD2-43 mRNAs by RT-PCR throughout the cell cycle or following exposure to MMC. The indicated primer pairs (A) were used to amplify the alternatively spliced mRNAs. The mRNA was prepared from either uncorrected lymphoblasts from the indicated FA subtypes, from cells in different stages of the cell cycle, or following exposure to MMC. Ub indicates ubiquitin.
Expression of 2 splice variants of the FANCD2 gene. (A) Schematic representation of 2 splice variants of FANCD2. A 24–amino acid (aa) sequence, containing 50% acidic residues, is encoded by exon 44 sequence. (B) Detection of FANCD2-44 and FANCD2-43 mRNAs by RT-PCR in various FA cell lines. (C) Detection of FANCD2-44 and FANCD2-43 mRNAs by RT-PCR throughout the cell cycle or following exposure to MMC. The indicated primer pairs (A) were used to amplify the alternatively spliced mRNAs. The mRNA was prepared from either uncorrected lymphoblasts from the indicated FA subtypes, from cells in different stages of the cell cycle, or following exposure to MMC. Ub indicates ubiquitin.
We confirmed the expression of these splice variants in human cell lines by RT-PCR (Figure 1B-C). Although this analysis is semiquantitative, the ratio of the 2 mRNAs was approximately 1:10 (exon 43–exon 44 variant). This ratio was constant, regardless of the cell line examined (Figure 1B), the phase of the cell cycle, or whether cells were exposed to MMC (Figure 1C). Northern blot analysis confirmed the expression of these 2 major mRNA splice variants in multiple tissues (data not shown), suggesting that these variants have ubiquitous expression.
Expression of wild-type and mutant forms of FANCD2 in FA-D2 fibroblasts
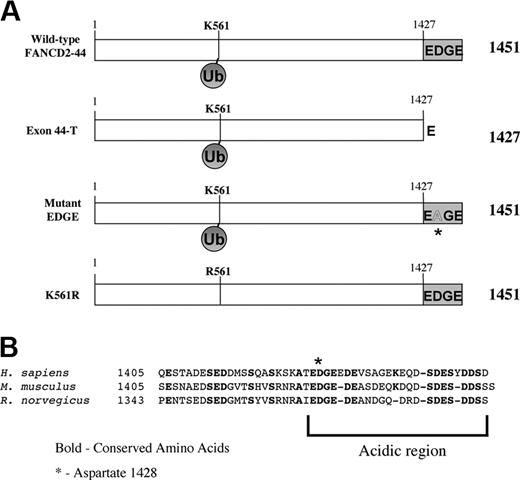
To determine the functional significance of the carboxy terminal sequence of FANCD2, we generated a series of cDNAs encoding the FANCD2 splice variants and mutant FANCD2 proteins (Figure 2A). The carboxy terminus of the FANCD2-44 protein is highly conserved between human and rodent (Mus musculus and Rattus norvegicus;Figure 2B) and may therefore have an important cellular function. This sequence is highly acidic, containing 12 acidic residues (E or D) of 24 residues (50%) in the human sequence. This includes an amino acid stretch (1427-1430) that is conserved in humans and rodents, which we will refer to as the “EDGE” region for the single-letter amino acid code of the corresponding address. A highly conserved aspartic acid residue in the EDGE sequence (D1428) is conserved in the species listed (Figure 2B).
Generation of cDNAs encoding normal splice variants and mutant forms of the FANCD2 protein. (A) Schematic series of FANCD2 proteins. (B) Alignment of amino acids encoded by exon 44 sequence derived from Homo sapiens, M. musculus, and R. norvegicus. The sequence is highly acidic and the D1428 residue is conserved.
Generation of cDNAs encoding normal splice variants and mutant forms of the FANCD2 protein. (A) Schematic series of FANCD2 proteins. (B) Alignment of amino acids encoded by exon 44 sequence derived from Homo sapiens, M. musculus, and R. norvegicus. The sequence is highly acidic and the D1428 residue is conserved.
We generated the Exon44-T mutant of FANCD2, truncated at the carboxy terminus and lacking the EDGE region (Figure 2A). Another construct, the mutant EDGE variant, contains the exon 44 sequence–encoded carboxy terminus but the conserved aspartic acid residue at position 1428 was converted to alanine (D1428A). Another mutant, FANCD2-K561R, has previously been described.10 This mutation disrupts the monoubiquitination of the FANCD2 protein, its targeting to nuclear foci, and its ability to functionally complement FA-D2 cells.
We next expressed the wild-type or mutant FANCD2 proteins in the FANCD2(–/–) fibroblast line, PD20F, which lacks endogenous FANCD2 protein (Figure 3). Previous studies have shown that FANCD2-44 is normally expressed as 2 isoforms, the unubiquitinated isoform (FANCD2-S) and the monoubiquitinated isoform (FANCD2-L).10 Following DNA damage10 or during S phase of the cell cycle,11 FANCD2 was monoubiquitinated. Like the wild-type FANCD2 protein, the Exon44-T and mutant EDGE proteins were monoubiquitinated in response to IR (Figure 3A, compare lanes 6, 8, and 10). The Exon44-T protein exhibited a slightly faster electrophoretic mobility (see bottom bands in Figure 3A lanes 7 and 8), consistent with the removal of 24 amino acids from its carboxy terminus. The FANCD2-43 variant was also monoubiquitinated on K561 (Table 1; data not shown). Taken together, these results indicate that the carboxy terminus of FANCD2 is not required for monoubiquitination.
Characterization of splice variants and mutant forms of the FANCD2 protein compared with wild-type FANCD2
. | Monoubiquitin on K561 . | Detected in chromatin . | Forms nuclear foci . | Corrects MMC sensitivity . |
|---|---|---|---|---|
| FANCD2-43 | + | ND | + | - |
| FANCD2-44 | +* | + | +* | +* |
| Exon44-T | + | + | + | - |
| Mutant EDGE, D1428A | + | + | + | - |
| K561R | -* | - | -* | -* |
. | Monoubiquitin on K561 . | Detected in chromatin . | Forms nuclear foci . | Corrects MMC sensitivity . |
|---|---|---|---|---|
| FANCD2-43 | + | ND | + | - |
| FANCD2-44 | +* | + | +* | +* |
| Exon44-T | + | + | + | - |
| Mutant EDGE, D1428A | + | + | + | - |
| K561R | -* | - | -* | -* |
ND indicates not determined.
Data reported in Garcia-Higuera et al.10
Monoubiquitination of wild-type FANCD2-44 on K561 results in targeting to subnuclear foci.10 Similarly, monoubiquitinated Exon44-T and mutant EDGE were targeted to foci (Figure 3B; Table 1), indicating that the carboxy terminal region of FANCD2-44 is not required for this intranuclear translocation. In contrast, the K561R mutant failed to assemble in foci. In addition, we demonstrate for the first time that wild-type FANCD2 colocalizes with the phosphorylated histone H2AX (Ser139), which is associated with DNA damage foci sites,28,29 following IR (Figure 3C). H2AX does not form prominent foci in randomly cycling HeLa cells.
The carboxy terminus of FANCD2-44 is required for functional complementation of FA-D2 cells
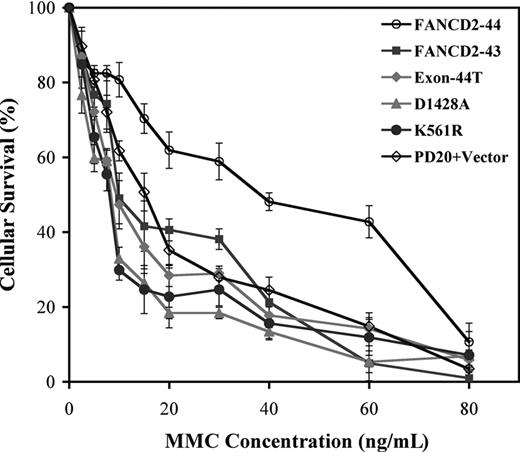
We next examined the FANCD2 variants for their ability to functionally complement the MMC hypersensitivity of FA-D2 cells (Figure 4). Wild-type FANCD2-44 corrected the MMC sensitivity of transduced FA-D2 cells, as measured by a colorimetric assay of cellular viability,5,10,26 consistent with previously reported results.5 Interestingly, the FANCD2-43 variant, the Exon44-T mutant, and the mutant EDGE protein, D1428A, failed to correct MMC hypersensitivity (Figure 4; Table 1). Taken together, these results indicate that the carboxy terminal 24 amino acids of FANCD2-44 are required for MMC resistance.
The carboxy terminal region of FANCD2 is required for MMC resistance. Stably transduced PD20 (FA-D2) fibroblasts, expressing the indicated FANCD2 variants and mutant proteins, were grown in the presence of the indicated concentrations of MMC. Cell survival was assessed by a colorimetric assay using crystal violet. Data points represent the average of 3 experiments, with standard deviation.
The carboxy terminal region of FANCD2 is required for MMC resistance. Stably transduced PD20 (FA-D2) fibroblasts, expressing the indicated FANCD2 variants and mutant proteins, were grown in the presence of the indicated concentrations of MMC. Cell survival was assessed by a colorimetric assay using crystal violet. Data points represent the average of 3 experiments, with standard deviation.
Monoubiquitination of FANCD2 is required for chromatin binding
We next determined whether monoubiquitination of FANCD2 targets the protein to discrete nuclear compartments (Figure 5). Nuclear proteins from HeLa cells were fractionated into soluble nuclear, chromatin, or nuclear matrix components (Figure 5A), and fractions were immunoblotted for FANCD2 (Figure 5B). The unubiquitinated FANCD2 protein fractionated with soluble nuclear proteins (Figure 5B S2 fraction). The FANCD2-L (monoubiquitinated) isoform fractionated with chromatin (Figure 5B S4 fraction). Neither FANCD2-S nor FANCD2-L was detected in the nuclear matrix fraction (P4), a fraction that contains lamin B (Figure 5B lane 7). These results suggest that monoubiquitination of FANCD2 is required for chromatin targeting.
The localization of FANCD2-L to chromatin was consistent with results from immunofluorescence microscopy (Figure 5C). Irradiated HeLa cells were examined by anti-FANCD2 immunofluorescence, either with or without permeablization with Triton X-100. Triton X-100 removed background nuclear fluorescence but failed to remove FANCD2 foci (P2), consistent with the presence of FANCD2-L in the foci. Further, exposure of cells to DNase I and ammonium sulfate resulted in loss of the FANCD2-L immunofluorescence signal but failed to extract lamin B (P4). Taken together, these results confirm that the monoubiquitinated isoform of FANCD2 resides in the chromatin fraction (S4) and that this fraction correlates with FANCD2 observed in the foci. Thus, FANCD2 foci that form following exposure to IR represent monoubiquitinated FANCD2 (FANCD2-L) in chromatin.
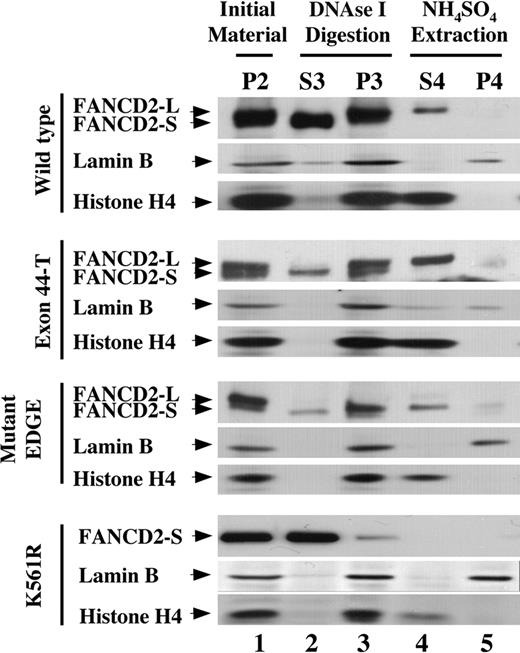
FA-D2 fibroblasts, expressing either wild-type or mutant FANCD2 polypeptides, were next examined by cell fractionation (Figure 6). Cells were exposed to MMC, resulting in expression of approximately equal levels of unubiquitinated (FANCD2-S) and monoubiquitinated (FANCD2-L) isoforms in the residual pellet following detergent permeabilization (lane 1). FANCD2-K561R was not monoubiquitinated and was expressed only as FANCD2-S, which was present in residual amounts following detergent permeabilization (Figure 6 lane 1). The unubiquitinated FANCD2-K561R mutant was not detected in the chromatin fraction (Figure 6 lane 4). The monoubiquitinated isoforms of FANCD2-44, Exon44-T, and mutant EDGE were localized to the chromatin fraction (Figure 6 lane 4), whereas unubiquitinated FANCD2 (FANCD2-S) was present in residual amounts prior to extraction of chromatin with ammonium sulfate (Figure 6 lanes 1-3). The chromatin fraction (S4) also contained histone H4. Again, neither FANCD2-L nor FANCD2-S protein was detected in the nuclear matrix fraction containing lamin B (Figure 6 lane 5).
Monoubiquitination is required for targeting of FANCD2 to the chromatin fraction. PD-20Fs (FA-D2 fibroblasts) were stably transduced with retroviral constructs encoding HA-FANCD2-44 (wild type), HA–Exon44-T, HA–mutant EDGE, or HA–FANCD2-K561R. Cells were pretreated with MMC to increase the level of FANCD2-L to approximately 50% of total cellular FANCD2. Cells were then fractionated as described in Figure 5A, and fractions were immunoblotted with antisera to FANCD2, histone H4, or lamin B. Fractions are shown beginning with the detergent-insoluble pellet following permeabilization (P2).
Monoubiquitination is required for targeting of FANCD2 to the chromatin fraction. PD-20Fs (FA-D2 fibroblasts) were stably transduced with retroviral constructs encoding HA-FANCD2-44 (wild type), HA–Exon44-T, HA–mutant EDGE, or HA–FANCD2-K561R. Cells were pretreated with MMC to increase the level of FANCD2-L to approximately 50% of total cellular FANCD2. Cells were then fractionated as described in Figure 5A, and fractions were immunoblotted with antisera to FANCD2, histone H4, or lamin B. Fractions are shown beginning with the detergent-insoluble pellet following permeabilization (P2).
Discussion
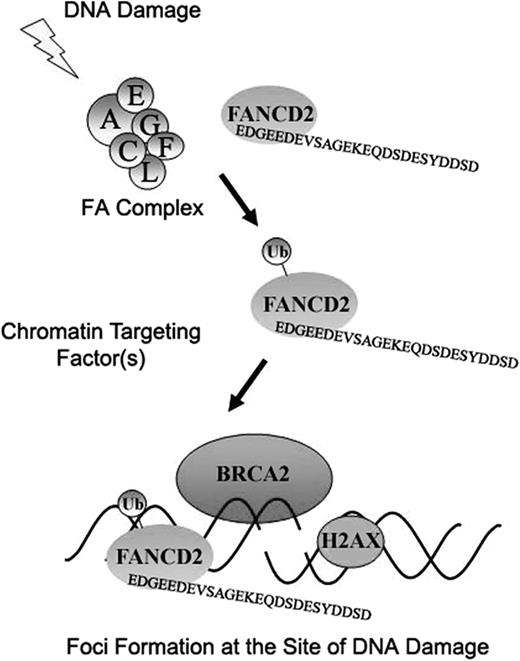
In the current study, we compared the function of the C-terminal acidic region of FANCD2 that differs significantly in 2 endogenous splice variants of FANCD2. Only one of these splice variants, FANCD2-44, encodes a functional protein capable of correcting the FA/BRCA pathway in FA-D2 cells (Figure 4; Table 1). The other variant, FANCD2-43, may have a different cellular function, not identified in the current study. Since FANCD2-44 contains a unique, conserved 24–amino acid sequence at the carboxy terminus, we focused our attention on this sequence. Several conclusions are possible. First, the carboxy terminal region of FANCD2 is not required for FANCD2 monoubiquitination on K561. Second, monoubiquitination of FANCD2 is required for accumulation of FANCD2 into chromatin. Third, the carboxy terminal 24 amino acids of FANCD2-44 is required for the function of FANCD2 in the FA/BRCA pathway (Figure 7).
Schematic model indicating the downstream function of the FANCD2 protein in the FA/BRCA pathway. Following cellular exposure to DNA damaging agents (IR or MMC, for example), FANCD2 is monoubiquitinated in an FA complex–dependent manner. Monoubiquitinated FANCD2 is then targeted to the chromatin, perhaps by a chromatin targeting factor. The carboxy terminal EDGE region of FANCD2, required for its functional activity (resistance to MMC), may then interact with other downstream proteins involved in DNA repair. Also, the monoubiquitin moiety of FANCD2 may also interact with other chromatin factors(s) containing a ubiquitin binding domain.
Schematic model indicating the downstream function of the FANCD2 protein in the FA/BRCA pathway. Following cellular exposure to DNA damaging agents (IR or MMC, for example), FANCD2 is monoubiquitinated in an FA complex–dependent manner. Monoubiquitinated FANCD2 is then targeted to the chromatin, perhaps by a chromatin targeting factor. The carboxy terminal EDGE region of FANCD2, required for its functional activity (resistance to MMC), may then interact with other downstream proteins involved in DNA repair. Also, the monoubiquitin moiety of FANCD2 may also interact with other chromatin factors(s) containing a ubiquitin binding domain.
The carboxy terminal 24 amino acids of FANCD2-44, containing the EDGE region, act independently from FANCD2 monoubiquitination at K561. Truncation or point mutation of this C-terminal region did not alter the efficiency of FANCD2 monoubiquitination, its targeting to foci, or its distribution to chromatin. By contrast, monoubiquitination at K561 is required both for FANCD2 foci formation10 and FANCD2 fractionation with chromatin24 (Figure 6).
Perhaps local sequence determinants, such as amino acids adjacent to K561, control the sequence-specific monoubiquitination of FANCD2. For instance, FANCD2 contains several conserved serine residues in its primary sequence adjacent to K561. One or more of these serine residues may become phosphorylated in vivo, leading to FANCD2 monoubiquitination. Analogously, protein polyubiquitination at an internal lysine residue is often preceded by nearby serine phosphoryation.30
Monoubiquitination may alter the conformation of FANCD2, thereby revealing a chromatin binding motif. Alternatively, a monoubiquitin receptor in the chromatin, perhaps assembled at sites of damaged chromatin, may mediate FANCD2-L accumulation. Ubiquitin binding proteins may contain a ubiquitin-associated (UBA) domain31 or some other ubiquitin binding activity.32 Other examples of protein monoubiquitination in protein sorting are known.32 The Ste2p protein in yeast is internalized from the cell surface after monoubiquitination of its cytoplasmic tail.33,34 Also, PCNA is monoubiquitinated following cellular exposure to DNA damage and perhaps targeted to sites of DNA repair.35 Monoubiquitination may therefore play a more general role in the targeting of DNA repair proteins to sites of damaged chromatin.
Increasing evidence suggests a role of chromatin modification in DNA repair. H2AX, which assembles in nucleosomes,36 is phosphorylated in response to double-strand breaks and is believed to recruit other DNA damage response machinery to these sites.28,29 Also, chromatin assembly, mediated by modification of histones by either acetylation or methylation, is required for DNA repair following intrastrand cross-links or double-strand breaks.37,38 Besides FANCD2, other chromatin-associated proteins, including the MRE11/NBS1/RAD50 complex, play a role in the DNA damage response.39,40
The finding that nucleotide excision repair (NER) requires chromatin relaxation involving acetyltransferase activity41 is of particular interest in the context of our finding that activated FANCD2 is chromatin associated. Along with homologous recombination, nucleotide excision repair pathways are required for the repair of DNA interstrand cross-links.42,43 FA cells are specifically sensitive to DNA interstrand cross-linkers, such as MMC.
According to the model (Figure 7), DNA damage activates the monoubiquitination of FANCD2 by an E3 ligase, which is probably the FANCL subunit of the FA complex.44 Monoubiquitinated FANCD2 subsequently binds to chromatin.24 There it may interact with other protein components of the DNA repair response or with damaged DNA itself in a manner that requires the C-terminal 24–amino acid EDGE region. In the current study, we show that DNA damage activates the colocalization of FANCD2-Ub with histone γ-H2AX foci. Recent studies indicate that monoubiquitinated FANCD2 may also interact with either ATM,18 the FANCD1/BRCA2 protein,6 or the MRE11/NBS1/RAD50 complex.45 Other proteins involved in the DNA repair response, such as MDC146,47 and 53BP1,48,49 may also have interactions with FANCD2. Whether the EDGE region of FANCD2 binds directly to any of these chromatin components remains to be determined.
Since FAcells are hypersensitive to DNAcross-linking agents, it will be important to determine whether the EDGE region plays a specific role in the DNA cross-link response. While both monoubiquitination of FANCD2 and the C-terminal EDGE region are required for a normal cellular response to MMC, the observation that monoubiquitination occurs in FANCD2 polypeptides lacking the EDGE region suggests that the EDGE region may be involved in sensing or repairing damaged DNA after FANCD2 is targeted into chromatin by monoubiquitination. These 2 elements of FANCD2 function could therefore act by interaction with different sets of proteins.
Prepublished online as Blood First Edition Paper, September 28, 2004; DOI 10.1182/blood-2003-11-3997.
Supported by National Institutes of Health grants RO1HL52725, RO1DK43889, P01DK50654, and PO1HL54785 (A.D.D.). P.R.A. is a Special Fellow of the Leukemia and Lymphoma Society. S.P.M. is supported in part by the Amy Clare Potter Fund. T.T. is a Scholar Fellow of the American Society of Hematology. R.C.G. is supported by the American Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank H. Joenje for EUFA316 cells.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal