Abstract
Tissue inhibitor of metalloproteinase 1 (TIMP-1) is a stromal factor with multiple functions. Overexpression of TIMP-1 correlates with aggressive clinical behavior of a spectrum of tumors. Here, for the first time, we address the role of TIMP-1 in the pathogenesis of B-cell lymphomas. An Epstein-Barr virus (EBV)-negative Burkitt lymphoma cell line with ectopic TIMP-1 expression (TIMP-1JD38) was used to identify genes induced/repressed by TIMP-1. Differentially expressed genes were analyzed by cDNA microarray, and they were validated by immunohistochemistry, flow cytometry, and Western blotting. Analysis revealed changes of genes coding for B-cell growth/differentiation, transcription, and cell cycle regulators. TIMP-1 repressed expression of germinal center (GC) markers CD10, Bcl-6, PAX-5 and up-regulated plasma cell-associated antigens CD138, MUM-1/IRF-4, XBP-1, and CD44, suggesting a plasma cell differentiation. This is accompanied by activation of signal transducer and activator of transcription 3 (STAT-3) and switch to cyclin D2 expression. However, TIMP-1JD38 cells expressed an inactive form of XBP-1, lacking antibody production/secretion. This incomplete plasmacytic differentiation occurs without altering cell proliferation, and despite c-Myc deregulation, indicating an arrested plasmacytic/plasmablastic stage of differentiation. Further validation in human lymphoma cell lines and in primary B-cell tumors demonstrated a predominant TIMP-1 expression in tumors with plasmacytic/plasmablastic phenotypes, including multiple myelomas. These findings strongly support TIMP-1 as an important factor in the pathogenesis of plasmacytic/plasmablastic tumors. (Blood. 2005;105:1660-1668)
Introduction
Tissue inhibitor of metalloproteinase 1 (TIMP-1) is a stromal factor that has a wide spectrum of functions in different tissues, and enhanced TIMP-1 expression is associated with poor clinical outcome in many cancer types.1-5 Moreover, we have previously demonstrated that TIMP-1 acts as a modulator of the survival and growth of germinal center B cells.6 Recent cDNA microarray studies have also identified TIMP-1 mRNA in a subset of aggressive lymphomas.7,8 However, these studies have not validated TIMP-1 protein expression or addressed its biologic significance. We have reported that Epstein-Barr virus-positive (EBV+) Burkitt lymphoma cell lines with viral latencies II or III overexpress TIMP-1 and down-regulate germinal center cell surface markers.9 Accordingly, enforced expression of TIMP-1 in EBV- Burkitt lymphoma cells also resulted in a similar phenotype9 while inducing differentiation and expression of interleukin 10 (IL-10). Intriguingly, this TIMP-1 effect on B cells is independent from its matrix metalloproteinase (MMP) inhibitory function.10
Commitment of B cells to differentiate to plasma cells occurs by the sequential expression or silencing of transcription factors (Tfs).11,12 The transition from germinal center cell to plasma cell is not an “all-or-nothing” event; instead, discrete steps in plasma cell development have been identified.13-17 The underlying genetic controls driving plasma cell development, as well as the intermediate plasmablastic precursors, remain largely unknown. One of the first well-characterized Tfs involved in this process was c-Myc, and repression of c-Myc is required for normal plasma cell differentiation. Deregulation of c-Myc prevents B-cell differentiation and is an oncogenic hallmark in Burkitt lymphomas.18,19 The identification of factors controlling this complex process would greatly help in the elucidation of plasma cell disorders. Recently, stromal factors have been proven to be important for the growth, survival, and differentiation of plasma cells.20 Thus, TIMP-1 as a stromal factor involved in B-cell differentiation may also have a potential role in plasma cell development. The aim of this study was to identify the underlying mechanisms mediating the effect of TIMP-1 on B-cell differentiation and the potential role in lymphomagenesis. We used an EBV- Burkitt lymphoma cell line with constitutive expression of TIMP-1 (TIMP-1JD38)6 and cDNA microarray techniques to identify genes induced or repressed by TIMP-1. We also assessed the expression of TIMP-1 in human cell lines and primary tumors. The present study proposes, for the first time, that TIMP-1 is an important factor in plasma cell development and in tumors with plasmacytic/plasmablastic differentiation.
Materials and methods
Cell lines and tumors
TIMP-1JD38, control LXSN-JD38, and JD38 cells were described elsewhere.6 Lymphoma cell lines NCEB, Granta, Ramos, BL-41, SUDHL-4, 5, 6; myeloma cell lines KMM-1, KMS-11, KMS12BM, JIM-3, OPM-2; and primary effusion lymphoma (PEL) BC-2, BC-3, and MCF-7 breast carcinoma cell line were also used. All the cell lines were grown in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics (Gibco, Gaithersburg, MD) at 37°C and 5% CO2.
The LXSN-JD38 and TIMP-1JD38 cells (5-8 × 106) were resuspended in 0.3 mL phosphate-buffered saline (PBS) and injected subcutaneously into the right posterior lateral thoracic wall of athymic nude mice (National Cancer Institute [NCI], Frederick, MD). Tumors were removed 10 days after implantation.
A multiple human lymphoma tissue array containing 9 mantle cell lymphomas (MCLs), 8 follicular lymphomas (FLs), 17 chronic lymphocytic leukemias (CLLs), 2 Burkitt lymphomas (BLs), 7 lymphoplasmacytoid lymphomas (LPLs), 25 diffuse large B-cell lymphomas (DLBCLs), 3 multiple myelomas (MMs), 5 hairy cell leukemias (HCLs), 9 marginal zone lymphomas (MZLs) and 2 tumors with plasmablastic differentiation was obtained from the Tissue Array Facility of the NCI (Advanced Technology Center, Gaithersburg, MD). Additional tumor sections of follicle-center lymphoma, Burkitt lymphoma, and multiple myelomas were also obtained from the NCI Hematopathology Section (Bethesda, MD).
cDNA synthesis and hybridization
Total RNA was isolated from each cell line in TriZol reagent (Invitrogen, Carlsbad, CA) and purified in RNeasy kit (Qiagen, Valencia, CA). RNA quality was assessed by spectrophotometry and on 1% agarose-gel electrophoresis. Total RNA (20 μg) was used to synthesize the labeled cDNA probes following a previously published protocol.21 Microarray chips (NCI, Advanced Technology Center), containing 10 000 human cDNAs (http://nciarray.nci.nih.gov) were used for hybridization. TIMP-1JD38 cDNA was compared with LXSN-JD38 cDNA.
Data analysis
Chips were scanned with the Axon GenePix4000A scanner and analyzed using GenePix Pro 3.0 software (Axon Instruments, Foster City, CA). The intensities on each array were normalized by median centering the log2 ratios to correct for average dye bias, photomultiplier tube (PMT) voltage imbalance, and variations between channels in amounts of samples hybridized. Forward- and reverse-labeled log ratios were averaged to avoid gene-specific dye bias not eliminated by normalization.
Three pairs of forward- and reverse-labeled arrays were used for each cell line. Data were then uploaded to the NCI's Web-based microarray database (mAdb; http://nciarray.nci.nih.gov) and analyzed to identify genes that were differentially expressed by a log2-transformed ratio of more than 2 (equivalent ≥ 4-fold change) in all 3 arrays for each cell line comparison. These genes were then used to generate a heat map (Figure 1). In a separate analysis, the differentially expressed genes were grouped by function and the average of their log ratios calculated. The gene-expression levels were compared by Wilcoxon rank sum test.
Differential expression of genes induced by TIMP-1 in JD38 Burkitt lymphoma cells. The list represents all genes that consistently change with ratio of expression more than 4-fold or less than 0.25-fold in 3 independent cDNA microarrays (1, 2, and 3). The heat map of ratio data (log2 ratio) of hybridization is shown for each gene in each array, with up-regulated genes in red and down-regulated gene expression in green.
Differential expression of genes induced by TIMP-1 in JD38 Burkitt lymphoma cells. The list represents all genes that consistently change with ratio of expression more than 4-fold or less than 0.25-fold in 3 independent cDNA microarrays (1, 2, and 3). The heat map of ratio data (log2 ratio) of hybridization is shown for each gene in each array, with up-regulated genes in red and down-regulated gene expression in green.
Immunohistochemistry
Immunohistochemistry was performed either on frozen or paraffin-embedded sections and stained for TIMP-1 (NeoMarkers, Fremont, CA), Ki-67, MUM-1-/IRF-4, BCL-6, CD10 (Dako Cytomation, Carpinteria, CA), CD30 (Novocastra, New Castle upon Tyne, United Kingdom), CD44, XBP-1 (Santa Cruz Biotechnology, Santa Cruz, CA), CD138 (Serotec, Raleigh, NC), and PAX-5 (BD Transduction, San Diego, CA). In all cases, the EnvisionPlus HRP detection system was used (Dako Cytomation) on an automated immunostainer (Dakoautostainer, Dako Cytomation).
Western blots
Exponentially growing cells were rinsed with cold PBS. After centrifugation, cells were lysed in 50 mM Tris (tris(hydroxymethyl)aminomethane)-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1% Nonidet p-40, 1 mM EDTA (ethylenediaminetetraacetic acid), 100 μg/mL 4-(aminoethyl)-benzene sulfonyl fluoride, 10 μg/mL aprotinin, 1 μg/mL pepstatin A, 0.5 μg/mL leupeptin, 80 mM β-glycerophosphate, 25 mM NaF, and 1 mM sodium orthovanadate for 30 minutes at 4°C. Equal amounts of protein (30 μg) were fractionated by electrophoresis. After transferring, nitrocellulose membranes were incubated overnight in PBS containing 5% fat-free milk and the following antibodies as indicated by manufacturers: STAT-3, phospho-STAT-3 (p-STAT-3), STAT-1, p-STAT-1, cyclin D3 (Santa Cruz Biotechnology), TIMP-1 (NeoMarkers), cyclin D2/D1 (Upstate Biotechnology, Charlottesville, VA), and β-actin for loading control (Santa Cruz Biotechnology). The ECL+Plus chemiluminescence kit (Amersham Pharmacia, Piscataway, NJ) was used for band visualization. After scanning blots, densities were determined using the image analysis software NIH Image 1.63f (National Institutes of Health, Bethesda, MD).
Fluorescence-activated cell sorting analysis
LXSN-JD38 and TIMP-1JD386 cells were incubated for 30 minutes at 4°C in the dark with the following antibodies: CD29, LFA-1, CD49d, CD79a, LFA-2 (Gibco, Rockville, MD). After rinsing in PBS, cells were analyzed by using a FACScan flow cytometer and the CellQuest software (Becton Dickinson, San Jose, CA).
Results
TIMP-1 promotes transcriptional repression
By using global gene expression analysis, we compared the entire transcriptional changes between LXSN-JD38, a Burkitt lymphoma cell line “frozen” in a germinal center stage of differentiation and TIMP-1 plus JD38 cells (TIMP-1JD38). The cDNA microarrays (10 000 genes) clearly identify genes involved in B-cell differentiation (Figure 1). Analysis of genes that consistently change expression by 4-fold or more in each of the 3 independent arrays shown indicate that more than 60% of genes expressed in control LXSN-JD38 cells were consistently repressed by forced expression of TIMP-1, whereas 35% of genes were up-regulated. These TIMP-1-associated effects are not coupled to changes in c-Myc expression because JD38 Burkitt lymphoma cells carry the t(8;14) (q24, q32) translocation involving c-Myc. These results suggest that TIMP-1 functions mainly as a transcriptional repressor in B cells despite c-Myc deregulation. To further determine these TIMP-1 effects, the entire group of genes was regrouped for analysis in 3 main functional categories relevant to the B-cell biology: growth/differentiation, transcription, and cell cycle regulation.
Effects of TIMP-1 on genes associated with B-cell growth/differentiation
Microarray analysis revealed changes in genes that regulate B-cell growth/differentiation (Table 1). TIMP-1 significantly down-regulated genes normally expressed by germinal center B cells. These include CD10, CD38, as well as the B-cell receptor complex (immunoglobulin heavy and light chains, CD79a and b), supporting our previous reports describing reduced B-cell receptor expression and lower immunoglobulin production by TIMP-1+ Burkitt lymphoma cell lines.9 Immunohistochemistry analysis confirmed these changes. Tumor sections from LXSN-JD38 control cell xenografts and overexpressing TIMP-1JD38 cell xenografts demonstrated the repression of CD10 (Figure 2A-B). The cDNA microarray chip used lacks cDNA prints for important plasma cell-associated markers CD138, as well as the Tfs MUM1/IRF-4 and PAX-5 (see “Effects of TIMP-1 on the B-cell transcription regulators”); therefore, as a confirmation of a plasma cell phenotype, tumor sections were also tested for CD138 (syndecan-1) expression by immunohistochemistry. High expression of this marker is also demonstrated in TIMP-1JD38 tumors (Figure 2D). The loss of CD10 and expression of CD138 is the typical phenotype of maturing B cells in postgerminal areas. TIMP-1 also decreased the expression of genes involved in antigen presentation and in B-cell/T-cell interactions (B7, HLA complex, and interferon receptors), as well as genes that regulate adhesion and activation of B cells in the germinal center, such as CD29 and CD49D. In contrast, TIMP-1 increased significantly (≥ 4-fold) CD44, an adhesion protein with homing functions in plasma cells.22 Changes in the extracellular matrix receptors were validated either by immunohistochemistry for high CD44 expression in TIMP-1JD38 xenografts (Figure 2G-H) or fluorescence-activated cell sorting (FACS) analysis for integrins (Figure 2I). This analysis reveals striking changes in the expression of CD49D, β-1/α-3 (CD29) without detecting changes in the expression of others integrins. Moreover, TIMP-1JD38 tumor expressed CD30, a member of the tumor necrosis factor (TNF) receptor family, which indicates an immunologically activated status (Figure 2E-F).23 These results support TIMP-1 as a promoter of a plasmablastic phenotype (CD10-, CD38-, CD138+, CD44+, CD30+).
Changes in B-cell differentiation antigens
Ratio* . | GenBank accession no. . | Gene . | Description . |
|---|---|---|---|
| 14.0713 | X55150 | CD44 | CD44 antigen (homing function and Indian blood group system) |
| 12.0926 | AB006967 | SOCS3 | Suppressor of cytokine signaling 3 |
| 8.877 | K02882 | — | Immunoglobulin heavy-constant δ |
| 8.1159− | L08187 | EB13 | Epstein-Barr virus-induced gene 3 |
| 7.3989 | AB006780 | LGALS3 | Lectin, galactoside-binding, galectin 3 |
| 5.905 | NM002305 | LGALS1 | Lectin, galactoside-binding, soluble, 1 galectin 1 |
| 4.9316 | AL031276 | TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B |
| 4.2297 | NM006847 | LILRB4 | Leukocyte immunoglobulin-like receptor, subfamily B,4 |
| 4.0811 | NM002188 | IL13 | Interleukin 13 |
| 3.4753 | U03397 | TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 |
| 2.8905 | BE386504 | TNFRSF14 | Tumor necrosis factor receptor14 (herpesvirus entry mediator) |
| 2.3185 | NM000565 | IL6R | Interleukin 6 receptor |
| 2.3845 | M57627 | IL10 | Interleukin 10 |
| 2.0553 | NM006709 | BAT8 | HLA-B-associated transcript 8 |
| 0.4942 | U05877 | IFNGR2 | Interferon γ receptor 2 (interferon γ transducer 1) |
| 0.4924 | U72508 | B7 | B7 protein |
| 0.4687 | M29053 | IL7 | Interleukin 7 |
| 0.4644 | AW800443 | IGL@ | Immunoglobulin λ locus |
| 0.4655 | NM000966 | RARG | Retinoic acid receptor γ |
| 0.4562 | NM000874 | IFNAR2 | Interferon (α, β, ω) receptor 2 |
| 0.4053 | NM004233 | CD83 | Activated B lymphocytes, immunoglobulin superfamily |
| 0.4587 | BE270634 | CD79B | CD79B antigen (immunoglobulin-associated β) |
| 0.4157 | M88458 | KDELR2 | KDEL endoplasmic reticulum protein retention receptor 2 |
| 0.4103 | NM006509 | RELB | v-rel homolog B, nuclear factor of κ light polypeptide in B cells |
| 0.4076 | AF006636 | SDCBP | Syndecan-binding protein (syntenin) |
| 0.4129 | M87790 | IGLJ3 | Immunoglobulin λ joining 3 |
| 0.4909 | U83582 | HLA-DQB1 | Major histocompatibility complex, class II, DQ β1 |
| 0.4161 | M14662 | HLA-DRB4 | Major histocompatibility complex, class II, DR β4 |
| 0.3941 | M26628 | MME | Membrane metalloendopeptidase, CALLA, (CD10) |
| 0.385 | AI418605 | HLA-DMA | Major histocompatibility complex, class II, DM α |
| 0.3623 | AW404507 | IGKC | Immunoglobulin κ constant |
| 0.2978 | M13560 | CD74 | CD74 (polypeptide, major histocompatibility complex, class II) |
| 0.3008 | X06026 | CD3G | CD3G antigen, γ polypeptide (TiT3 complex) |
| 0.2724 | NM002211 | ITGB1 | Integrin β1 (fibronectin receptor, antigen CD29) |
| 0.2245 | S75217 | CD79A | CD79A antigen (immunoglobulin-associated α) |
| 0.1995 | NM000885 | ITGA4 | Integrin α4 (CD49D, α4 subunit of VLA-4 receptor) |
| 0.2007 | D84276 | CD38 | CD38 antigen (p45) |
| 0.2489 | NM003242 | TGFBR2 | Transforming growth factor β receptor 2 (70/80 kDa) |
| 0.1623 | AW959214 | CSF2RB | Colony-stimulating factor 2 receptor β |
| 0.0433 | AW172754 | IGJ | J linker protein for immunoglobulin α and μ polypeptides |
Ratio* . | GenBank accession no. . | Gene . | Description . |
|---|---|---|---|
| 14.0713 | X55150 | CD44 | CD44 antigen (homing function and Indian blood group system) |
| 12.0926 | AB006967 | SOCS3 | Suppressor of cytokine signaling 3 |
| 8.877 | K02882 | — | Immunoglobulin heavy-constant δ |
| 8.1159− | L08187 | EB13 | Epstein-Barr virus-induced gene 3 |
| 7.3989 | AB006780 | LGALS3 | Lectin, galactoside-binding, galectin 3 |
| 5.905 | NM002305 | LGALS1 | Lectin, galactoside-binding, soluble, 1 galectin 1 |
| 4.9316 | AL031276 | TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B |
| 4.2297 | NM006847 | LILRB4 | Leukocyte immunoglobulin-like receptor, subfamily B,4 |
| 4.0811 | NM002188 | IL13 | Interleukin 13 |
| 3.4753 | U03397 | TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 |
| 2.8905 | BE386504 | TNFRSF14 | Tumor necrosis factor receptor14 (herpesvirus entry mediator) |
| 2.3185 | NM000565 | IL6R | Interleukin 6 receptor |
| 2.3845 | M57627 | IL10 | Interleukin 10 |
| 2.0553 | NM006709 | BAT8 | HLA-B-associated transcript 8 |
| 0.4942 | U05877 | IFNGR2 | Interferon γ receptor 2 (interferon γ transducer 1) |
| 0.4924 | U72508 | B7 | B7 protein |
| 0.4687 | M29053 | IL7 | Interleukin 7 |
| 0.4644 | AW800443 | IGL@ | Immunoglobulin λ locus |
| 0.4655 | NM000966 | RARG | Retinoic acid receptor γ |
| 0.4562 | NM000874 | IFNAR2 | Interferon (α, β, ω) receptor 2 |
| 0.4053 | NM004233 | CD83 | Activated B lymphocytes, immunoglobulin superfamily |
| 0.4587 | BE270634 | CD79B | CD79B antigen (immunoglobulin-associated β) |
| 0.4157 | M88458 | KDELR2 | KDEL endoplasmic reticulum protein retention receptor 2 |
| 0.4103 | NM006509 | RELB | v-rel homolog B, nuclear factor of κ light polypeptide in B cells |
| 0.4076 | AF006636 | SDCBP | Syndecan-binding protein (syntenin) |
| 0.4129 | M87790 | IGLJ3 | Immunoglobulin λ joining 3 |
| 0.4909 | U83582 | HLA-DQB1 | Major histocompatibility complex, class II, DQ β1 |
| 0.4161 | M14662 | HLA-DRB4 | Major histocompatibility complex, class II, DR β4 |
| 0.3941 | M26628 | MME | Membrane metalloendopeptidase, CALLA, (CD10) |
| 0.385 | AI418605 | HLA-DMA | Major histocompatibility complex, class II, DM α |
| 0.3623 | AW404507 | IGKC | Immunoglobulin κ constant |
| 0.2978 | M13560 | CD74 | CD74 (polypeptide, major histocompatibility complex, class II) |
| 0.3008 | X06026 | CD3G | CD3G antigen, γ polypeptide (TiT3 complex) |
| 0.2724 | NM002211 | ITGB1 | Integrin β1 (fibronectin receptor, antigen CD29) |
| 0.2245 | S75217 | CD79A | CD79A antigen (immunoglobulin-associated α) |
| 0.1995 | NM000885 | ITGA4 | Integrin α4 (CD49D, α4 subunit of VLA-4 receptor) |
| 0.2007 | D84276 | CD38 | CD38 antigen (p45) |
| 0.2489 | NM003242 | TGFBR2 | Transforming growth factor β receptor 2 (70/80 kDa) |
| 0.1623 | AW959214 | CSF2RB | Colony-stimulating factor 2 receptor β |
| 0.0433 | AW172754 | IGJ | J linker protein for immunoglobulin α and μ polypeptides |
Average (log ratio) of 3 arrays (P < .0005)
Effect of TIMP-1 on the plasmablastic differentiation of Burkitt lymphoma cells. Immunostaining panel shows down-regulation of germinal center marker CD10 (B) and up-regulation of the plasma cell marker CD138 (D) in TIMP-1JD38 tumor sections as compared with LXSN-JD38 control sections in panels A and C, respectively. Expression of CD30 (F) and CD44 (H) are also shown in TIMP-1+ tumors, whereas LXSN tumors are negative (E,G); original magnification × 400. (I) FACS analysis of integrin expression profile demonstrates down-regulation of β-1 (CD29), α-3, and CD49 on the cell surface of TIMP-1JD38 tumor cells as indicated in the histograms by a shift to the left of vertical bars by comparison with LXSN-JD38 control cells; no changes in the expression of other integrins are detected. Sections were examined under an Olympus BX41 microscope using UPLAN F1 40 ×/0.75 objective lenses. DAB shows specific staining in brown. Digital images were obtained with an Olympus DP12 (Olympus, Melville, NY) and imported with an Olympus Camedia USB Smartmedia card into AdobePhotoshop 7.0 (Adobe Systems, San Jose, CA) for processing.
Effect of TIMP-1 on the plasmablastic differentiation of Burkitt lymphoma cells. Immunostaining panel shows down-regulation of germinal center marker CD10 (B) and up-regulation of the plasma cell marker CD138 (D) in TIMP-1JD38 tumor sections as compared with LXSN-JD38 control sections in panels A and C, respectively. Expression of CD30 (F) and CD44 (H) are also shown in TIMP-1+ tumors, whereas LXSN tumors are negative (E,G); original magnification × 400. (I) FACS analysis of integrin expression profile demonstrates down-regulation of β-1 (CD29), α-3, and CD49 on the cell surface of TIMP-1JD38 tumor cells as indicated in the histograms by a shift to the left of vertical bars by comparison with LXSN-JD38 control cells; no changes in the expression of other integrins are detected. Sections were examined under an Olympus BX41 microscope using UPLAN F1 40 ×/0.75 objective lenses. DAB shows specific staining in brown. Digital images were obtained with an Olympus DP12 (Olympus, Melville, NY) and imported with an Olympus Camedia USB Smartmedia card into AdobePhotoshop 7.0 (Adobe Systems, San Jose, CA) for processing.
Effects of TIMP-1 on the B-cell transcription regulators
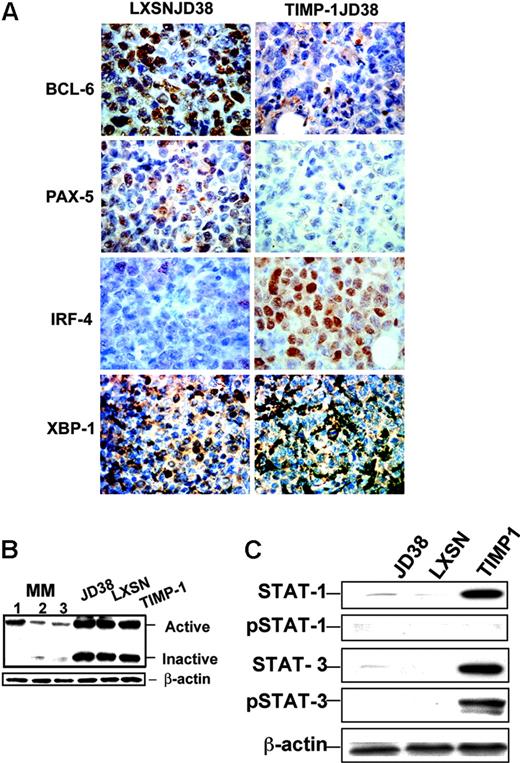
Microarray data show that TIMP-1 drastically down-regulated the main germinal center Tfs such as BCL-6, and up-regulated X-box protein 1 (XBP-1), an important factor required for the development of antibody-producing and -secreting plasma cells24 (Table 2). Immunohistochemistry confirmed down-regulation of BCL-6 in TIMP-1JD38 tumors (Figure 3A). Additional immunohistochemical analysis for cDNAs missing in the microarray chip further demonstrated down-regulation of PAX-5, a broadly expressed B-cell gene that is selectively repressed in plasma cells, as well as a higher expression of MUM-1/IRF-4 in TIMP-1JD38 tumors as compared with the low expression shown by few LXSN-JD38 cells in the control tumor section (Figure 3A). MUM-1/IRF-4 is an important Tf for plasma cell development and its expression depends on BCL-6 repression.25 In the final stages of differentiation, plasma cells are committed under the control of XBP-1 to the synthesis of immunoglobulin light/heavy chains, assembly, and secretion of antibodies.24 cDNA microarray demonstrated XBP-1 mRNA up-regulation in TIMP-1JD38 cells (Table 2). However, immunohistochemistry showed XBP-1 equally expressed in both control LXSN-JD38 and TIMP-1JD38 xenografts in apparent disagreement with the microarray data (Figure 3A). In addition, XBP-1 immunostaining showed exclusion from the nuclei in the majority of both control and TIMP-1+ tumor cells. The cytoplasmic localization indicates that XBP-1 is not transcriptionally active. XBP-1 activation occurs by mRNA splicing in response to unfolded protein response (UPR) in the endoplasmic reticulum, generating 2 different proteins: an active spliced form (371 amino acids) and inactive form (267 amino acids).26 XBP-1 activation depends on the ratio of spliced versus unspliced forms.27 The unspliced form acts as dominant-negative control inhibiting the function of the spliced variant. We used Western blot analysis and an antibody that recognizes both forms to determine the XBP-1 activation status in JD38 cells, LXSN-JD38 cells, and TIMP-1JD38 cells and compared it with 3 antibody-secreting multiple myeloma cell lines (Figure 3B). All JD38 cell lines demonstrated equal expression of both XBP-1 forms; in contrast, the multiple myeloma cell lines (MM 1, 2, and 3) showed higher expression of active XBP-1, which correlates with the amount of antibody production and secretion (data not shown). These findings suggest that TIMP-1 increased transcription, but not activation, of XBP-1 protein. Interestingly, ATF-6 a member of the UPR machinery controlling XBP-1 activation is also down-regulated by TIMP-1 (Table 2).
Changes in B-cell transcription regulators
Ratio* . | GenBank accession no. . | Gene . | Description . |
|---|---|---|---|
| 12.0926 | AB006967 | SOCS3 | Suppressor of cytokine signaling 3 |
| 7.0515 | AF016898 | BATF | Basic leucine zipper transcription factor, ATF-like |
| 2.8815 | L23959 | TFDP1 | Transcription factor Dp-1 |
| 2.6115 | AJ012463 | STAT3 | Signal transducer and activator of transcription 3 |
| 2.3195 | AW021229 | XBP1 | X-box-binding protein 1 |
| 2.2535 | AA478534 | STAT1 | Signal transducer and activator of transcription 1 |
| 2.2981 | D43945 | TFEC | Transcription factor EC |
| 2.1945 | N39944 | ATF3 | Activating transcription factor 3 |
| 0.4845 | U47677 | E2F1 | E2F transcription factor 1 |
| 0.4108 | X12492 | NFIC | Nuclear factor I/C (CCAAT-binding transcription factor) |
| 0.4115 | AW372543 | ATF6 | Activating transcription factor 6 |
| 0.3677 | AA477295 | PC4 | Activated RNA polymerase II transcription cofactor 4 |
| 0.3465 | U49020 | MEF2A | MADS box transcription enhancer factor 2 |
| 0.3507 | AW005941 | BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) |
| 0.3144 | U22376 | MYB | v-myb myeloblastosis viral oncogene homolog (avian) |
| 0.2976 | U18671 | STAT2 | Signal transducer and activator of transcription 2, 113 kDa |
| 0.2455 | AA936434 | TCF4 | Transcription factor 4 |
Ratio* . | GenBank accession no. . | Gene . | Description . |
|---|---|---|---|
| 12.0926 | AB006967 | SOCS3 | Suppressor of cytokine signaling 3 |
| 7.0515 | AF016898 | BATF | Basic leucine zipper transcription factor, ATF-like |
| 2.8815 | L23959 | TFDP1 | Transcription factor Dp-1 |
| 2.6115 | AJ012463 | STAT3 | Signal transducer and activator of transcription 3 |
| 2.3195 | AW021229 | XBP1 | X-box-binding protein 1 |
| 2.2535 | AA478534 | STAT1 | Signal transducer and activator of transcription 1 |
| 2.2981 | D43945 | TFEC | Transcription factor EC |
| 2.1945 | N39944 | ATF3 | Activating transcription factor 3 |
| 0.4845 | U47677 | E2F1 | E2F transcription factor 1 |
| 0.4108 | X12492 | NFIC | Nuclear factor I/C (CCAAT-binding transcription factor) |
| 0.4115 | AW372543 | ATF6 | Activating transcription factor 6 |
| 0.3677 | AA477295 | PC4 | Activated RNA polymerase II transcription cofactor 4 |
| 0.3465 | U49020 | MEF2A | MADS box transcription enhancer factor 2 |
| 0.3507 | AW005941 | BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) |
| 0.3144 | U22376 | MYB | v-myb myeloblastosis viral oncogene homolog (avian) |
| 0.2976 | U18671 | STAT2 | Signal transducer and activator of transcription 2, 113 kDa |
| 0.2455 | AA936434 | TCF4 | Transcription factor 4 |
Average (log ratio) of 3 arrays (P < .0005).
TIMP-1 effect on transcription regulators. (A) Immunohistochemistry showing down-regulation in TIMP-1JD38 tumor of the Tfs BCL-6 and PAX-5 and increasing expression of MUM-1/IRF-4; original magnification × 400. LXSN-JD38 and TIMP-1JD38 xenotransplants express XBP-1; original magnification × 400. (B) Western blot showing equal ratios of XBP-1 active/inactive forms by JD38 control cells and TIMP-1JD38 cells as compared with multiple myeloma cell lines (1, 2, and 3) in which the XBP-1 active form is up-regulated. (C) Western blot analysis confirms STAT-1 and STAT-3 up-regulation by TIMP-1 and demonstrates phosphotyrosine-activated STAT-3 and not STAT-1 in TIMP-1JD38 cells. Equal protein loading was monitored by β-actin Western blot.
TIMP-1 effect on transcription regulators. (A) Immunohistochemistry showing down-regulation in TIMP-1JD38 tumor of the Tfs BCL-6 and PAX-5 and increasing expression of MUM-1/IRF-4; original magnification × 400. LXSN-JD38 and TIMP-1JD38 xenotransplants express XBP-1; original magnification × 400. (B) Western blot showing equal ratios of XBP-1 active/inactive forms by JD38 control cells and TIMP-1JD38 cells as compared with multiple myeloma cell lines (1, 2, and 3) in which the XBP-1 active form is up-regulated. (C) Western blot analysis confirms STAT-1 and STAT-3 up-regulation by TIMP-1 and demonstrates phosphotyrosine-activated STAT-3 and not STAT-1 in TIMP-1JD38 cells. Equal protein loading was monitored by β-actin Western blot.
STATs are important regulators of B-cell transcription.28 STAT-1 and STAT-3 are up-regulated in TIMP-1JD38 cells; in contrast, STAT-2 is down-regulated (Table 2). STAT-3 is an important transducer of survival and growth signals in IL-6 and IL-10 pathways.29-31 Interestingly, cDNA microarray also demonstrated up-regulation in TIMP-1JD38 cells of both IL-6 receptor and IL-10 (Table 1). These findings suggest a TIMP-1 effect on the signal transduction pathways pivotal for the survival and development of plasma cells. Western blot analysis not only confirmed changes in the levels of STAT-1 and STAT-3, but also demonstrated significant differences in their posttranslational protein activation (phosphorylation) as well (Figure 3C). TIMP-1JD38 cells expressed significant levels of p-STAT-3, but not STAT-1, when compared with parental JD38 or vector control cells (LXSN-JD38), indicating that STAT-3 is active in TIMP-1JD38 cells. Taken together, these findings suggest that aberrant TIMP-1 expression concomitant with deregulated c-Myc results in an incomplete or arrested plasmablastic stage of differentiation: down-regulation of B-cell Tfs (PAX-5, BCL-6), up-regulation of plasma cell Tfs (MUM1/IRF-4), but inactive XBP-1 and activated p-STAT-3.
TIMP-1 induces changes in cell cycle regulators
TIMP-1 represses several negative regulators of the cell cycle (Table 3), but does not change c-Myc expression. This is as expected because c-Myc is deregulated in the Burkitt lymphoma JD38 parental cells prior to aberrant expression of TIMP-1 in these cells. However, the most striking change induced by TIMP-1 is a switch in cyclin D usage. Control LXSN-JD38 cells express mRNA for cyclin D3 as normally expressed by germinal center cells, whereas TIMP-1JD38 cells overexpress cyclin D2, the gene with the highest differential expression (Figure 1). Cyclin D2 is mainly expressed by B cells in postgerminal center areas.32
Changes in cell cycle regulators
Ratio* . | GenBank accession no. . | Gene . | Description . |
|---|---|---|---|
| 12.4389 | D13639 | CCND2 | Cyclin D2 |
| 0.46215 | AI803460 | CCND3 | Cyclin D3 |
| 0.46225 | X59798 | CCND1 | Cyclin D1 (PRAD1) |
| 0.45025 | AA455410 | CDKN1B | Cyclin-dependent kinase inhibitor (p27, Kip1) |
| 0.43861 | NM004702 | CCNE2 | Cyclin E2 |
| 0.41005 | AI271688 | CCNG2 | Cyclin G2 |
| 0.35105 | AF041248 | CDKN2C | Cyclin-dependent kinase inhibitor (p18) |
| 0.26465 | AA481712 | CDKN1A | Cyclin-dependent kinase inhibitor (p21, Cip1) |
Ratio* . | GenBank accession no. . | Gene . | Description . |
|---|---|---|---|
| 12.4389 | D13639 | CCND2 | Cyclin D2 |
| 0.46215 | AI803460 | CCND3 | Cyclin D3 |
| 0.46225 | X59798 | CCND1 | Cyclin D1 (PRAD1) |
| 0.45025 | AA455410 | CDKN1B | Cyclin-dependent kinase inhibitor (p27, Kip1) |
| 0.43861 | NM004702 | CCNE2 | Cyclin E2 |
| 0.41005 | AI271688 | CCNG2 | Cyclin G2 |
| 0.35105 | AF041248 | CDKN2C | Cyclin-dependent kinase inhibitor (p18) |
| 0.26465 | AA481712 | CDKN1A | Cyclin-dependent kinase inhibitor (p21, Cip1) |
Average (log ratio) of 3 arrays (P < .0005).
Western blot analysis confirmed the switch in cyclin-D usage (Figure 4A). TIMP-1JD38 cells expressed cyclin D2, whereas control JD38 and LXSN-JD38 cells were positive for cyclin D3. Although control cells (JD38 and LXSN-JD38) expressed cyclin D1 mRNA as detected by microarray analysis (Table 3), these do not express the protein (data not shown). This is expected because cyclin D1 is a gene that is not expressed in normal B cells. Our findings are consistent with those in the molecular signature identifying subsets of large B-cell lymphomas,8 in which mRNAs for cyclin D2, TIMP-1, and CD44 are up-regulated.
TIMP-1 effect in cell proliferation and cell cycle regulators. (A) Western blots demonstrating a switch in cyclin D usage; TIMP-1 up-regulates cyclin D2 and down-regulates cyclin D3 in TIMP-1JD38. β-actin was included as a control for equal protein loading. (B) Immunochemistry of TIMP-1JD38 tumor sections demonstrates differential expression of TIMP-1 (top row) in the cytoplasm and a higher intensity in the immediate cytoplasm membrane perimeter; original magnification × 1000. Aberrant TIMP-1 expression does not affect tumor cell proliferation. Both control LXSN-JD38 and TIMP-1JD38 tumors show equal expression of the cell proliferation marker Ki-67 (bottom row); original magnification × 1000. Sections were examined under an Olympus BX41 microscope using UPLAN F1 40 ×/0.75 objective lenses. DAB shows specific staining in brown. Digital images were obtained with an Olympus DP12 (Olympus, Melville, NY) and imported with an Olympus Camedia USB Smartmedia card into AdobePhotoshop 7.0 (Adobe Systems, San Jose, CA) for processing.
TIMP-1 effect in cell proliferation and cell cycle regulators. (A) Western blots demonstrating a switch in cyclin D usage; TIMP-1 up-regulates cyclin D2 and down-regulates cyclin D3 in TIMP-1JD38. β-actin was included as a control for equal protein loading. (B) Immunochemistry of TIMP-1JD38 tumor sections demonstrates differential expression of TIMP-1 (top row) in the cytoplasm and a higher intensity in the immediate cytoplasm membrane perimeter; original magnification × 1000. Aberrant TIMP-1 expression does not affect tumor cell proliferation. Both control LXSN-JD38 and TIMP-1JD38 tumors show equal expression of the cell proliferation marker Ki-67 (bottom row); original magnification × 1000. Sections were examined under an Olympus BX41 microscope using UPLAN F1 40 ×/0.75 objective lenses. DAB shows specific staining in brown. Digital images were obtained with an Olympus DP12 (Olympus, Melville, NY) and imported with an Olympus Camedia USB Smartmedia card into AdobePhotoshop 7.0 (Adobe Systems, San Jose, CA) for processing.
Importantly, these changes are not accompanied by changes in cell proliferation. Tumor sections of control LXSN-JD38 and TIMP-1JD38 cell xenografts demonstrate differential expression of TIMP-1 in vivo, but no changes in the index of tumor proliferation as assessed by the Ki-67 marker (Figure 4B). This is in agreement with our previous observation in vitro that TIMP-1 up-regulation does not alter cell cycle progression of the TIMP-1-JD38 cells.6
TIMP-1 expression in human lymphoma cell lines, reactive lymphoid tissue, and primary tumors
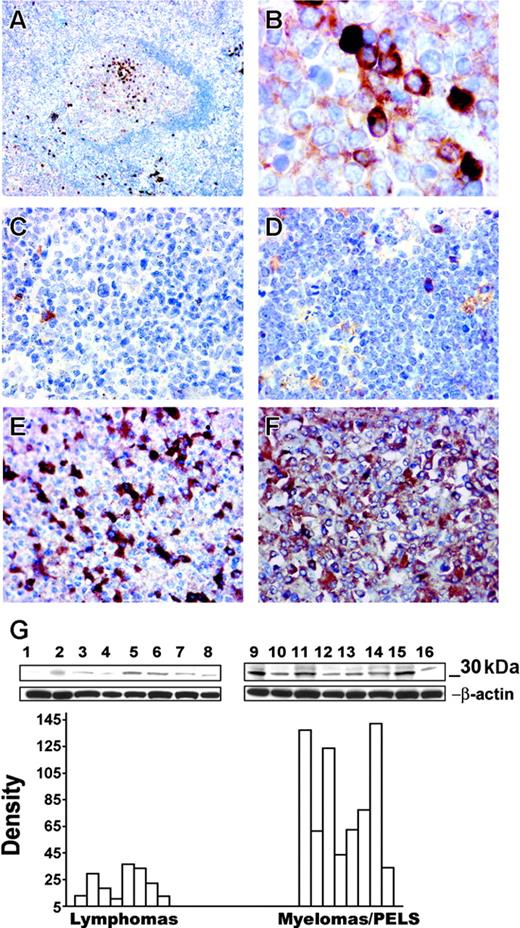
Assessment of the JD38TIMP-1 cell phenotype suggests that TIMP-1 promotes a plasmacytic/plasmablastic differentiation. To confirm the association of TIMP-1 expression with postgerminal center differentiation, we analyzed by Western blotting the expression of TIMP-1 in a series of 15 lymphoma cell lines (Figure 5G). Cell lines were either derived from tumors of a putative germinal center origin or of plasmacytic/plasmablastic differentiation. Multiple myelomas and primary effusion lymphoma cell lines express TIMP-1 at levels comparable with the positive control breast carcinoma cell line MCF-7 (lane 9). Moreover a statistically significant difference in the mean level of TIMP-1 expression (P < .005) is observed when compared with the TIMP-1 expression in the control lymphoma group that includes cell lines of germinal center origin: follicular lymphomas and Burkitt lymphomas.
Correlation of TIMP-1 expression with plasmacytic phenotype. Immunohistochemistry of a normal reactive follicle demonstrates expression of TIMP-1+ cells in the light zone (A); original magnification × 100. Detailed TIMP-1 expression in the cytoplasm of differentiated B cells in the light zone (B); original magnification × 1000. Expression of TIMP-1 in primary tumors. Follicular-center lymphoma (C) and a Burkitt lymphoma (D) do not express TIMP-1; bona fide normal plasma cells are positive in these tumors. Myeloma cases (E-F) demonstrate TIMP-1 overexpression by the majority of tumor cells, with higher expression in panel F and lower TIMP-1 expression in panel E; original magnification × 400. (Image acquisition procedures described in Figure 4 apply.) (G) Western blots demonstrate TIMP-1 (30 kD) expression in different lymphoma cell lines. The y-axis of the graph shows relative density of TIMP-1 versus β-actin bands. A significant difference is noted between the lymphoma cell line group and the cell line group of myelomas and primary effusion lymphomas (PELs; P < .005). Mantle cell lymphomas: NCEB, Granta (lanes 1 and 2); follicular lymphomas: SUDHL-4, SUDHL-5, SUDHL-6 (lanes 3-5); Burkitt lymphomas: Ramos, JD38, BL-41 (lanes 6-8); multiple myelomas: KMM-1, KMS-11, KMS-12BM, JIM-3, OPM-2 and PEL: BC-2, BC-3 (lanes 10-16); MCF-7 breast carcinoma positive control (lane 9).
Correlation of TIMP-1 expression with plasmacytic phenotype. Immunohistochemistry of a normal reactive follicle demonstrates expression of TIMP-1+ cells in the light zone (A); original magnification × 100. Detailed TIMP-1 expression in the cytoplasm of differentiated B cells in the light zone (B); original magnification × 1000. Expression of TIMP-1 in primary tumors. Follicular-center lymphoma (C) and a Burkitt lymphoma (D) do not express TIMP-1; bona fide normal plasma cells are positive in these tumors. Myeloma cases (E-F) demonstrate TIMP-1 overexpression by the majority of tumor cells, with higher expression in panel F and lower TIMP-1 expression in panel E; original magnification × 400. (Image acquisition procedures described in Figure 4 apply.) (G) Western blots demonstrate TIMP-1 (30 kD) expression in different lymphoma cell lines. The y-axis of the graph shows relative density of TIMP-1 versus β-actin bands. A significant difference is noted between the lymphoma cell line group and the cell line group of myelomas and primary effusion lymphomas (PELs; P < .005). Mantle cell lymphomas: NCEB, Granta (lanes 1 and 2); follicular lymphomas: SUDHL-4, SUDHL-5, SUDHL-6 (lanes 3-5); Burkitt lymphomas: Ramos, JD38, BL-41 (lanes 6-8); multiple myelomas: KMM-1, KMS-11, KMS-12BM, JIM-3, OPM-2 and PEL: BC-2, BC-3 (lanes 10-16); MCF-7 breast carcinoma positive control (lane 9).
The in vitro data clearly established a direct correlation between TIMP-1 expression and tumors with plasmacytic/plasmablastic phenotype. We further characterized expression of TIMP-1 in vivo by immunohistochemistry in human reactive lymphoid tissue and in primary tumors (Figure 5A-B). In well-developed reactive lymphoid follicles, TIMP-1+ centrocytes are clustered in the light zone of the germinal center, the site where the B cells committed to a postgerminal differentiation normally accumulate. Scattered plasma cells in the extrafollicular areas are also positive for TIMP-1. We also examined primary tumors that included follicular lymphomas (n = 10), Burkitt lymphomas (n = 8), multiple myelomas (n = 13), and plasmablastic lymphomas (n = 10). All the Burkitt and follicular lymphomas studied were negative for TIMP-1 expression with normal bone fide plasma cell staining as an internal positive control (Figure 5C-D). In contrast, all plasmacytic/plasmablastic tumors examined are strongly positive for TIMP-1. Representative sections are shown in Figure 5E-F. These findings suggest that in malignant B-cell lymphomas TIMP-1 expression is indicative of a postgerminal stage of differentiation.
Discussion
The current view in plasma cell development is of a complex, multistep process that requires transcription repression of the germinal center program to turn on pathways of differentiation.25,33 It is largely unknown if malignant B cells that give rise to plasma cell tumors undergo or are subject to the same transcriptional controls as their normal counterparts. TIMP-1 is a stromal factor with multiple functions and its up-regulation has been associated with the progression of a wide spectrum of tumors including B-cell lymphomas.3,4,34
Here for the first time, we characterize the role of TIMP-1 expression in the plasmacytic/plasmablastic differentiation of a Burkitt lymphoma cell line. TIMP-1 induced important phenotypic and transcriptional changes in Burkitt lymphoma cells, suggesting a new role for TIMP-1 as a transcriptional repressor. In response to TIMP-1 expression, the main germinal center Tfs (PAX-5, BCL-6) were down-regulated in Burkitt lymphoma JD38 cells, whereas those expressed in postgerminal center cells were up-regulated (MUM-1/IRF-4), but XBP-1 was inactive. Repression of PAX-5, BCL-6, and c-Myc is under the control of the B-lymphocyte-induced maturation protein 1 (BLIMP-1).33 However, according to cDNA microarray analysis, BLIMP-1 is not differentially expressed in TIMP-1JD38 tumor cells. A plausible explanation, as previously reported, is that BLIMP-1 represses germinal center transcription in response to antigen stimulation and T-cell interaction.35 Thus, differentiation of B cells by a T-independent mechanism could bypass BLIMP-1 control. However, it remains to be determined if malignant B cells carrying a deregulated c-Myc can undergo similar mechanisms of differentiation independent of BLIMP-1. The result of the present study indicates that TIMP-1 might be inducing differentiation, at least in part, by repressing the germinal center transcription program in B cells and suggests an alternative mechanism of differentiation that can occur even in the absence of antigen stimulation and despite c-Myc deregulation. Whether c-Myc deregulation impaired some TIMP-1 effects on the terminal differentiation of B cells including interference in the transcription of c-Myc targets in the germinal center poses an exciting question that has to be addressed in the future. Collectively, our findings support the notion that despite the c-Myc deregulation, TIMP-1 is capable of inducing a phenotype consistent with a “plasmablastic stage of differentiation,” CD10-CD38-CD138+CD44+CD30+, with an inactive antibody machinery26 (inactive XBP-1 and downregulated ATF-6).
Studies in mice have demonstrated different plasmablastic subsets (plasma cell precursors) with regard to CD138 and CD44 expression.36 The CD138-/CD44+ or CD138+/CD44- plasmablasts will give rise to apoptosis-resistant/long-lived plasma cells. However, CD44+/CD138+ plasmablasts normally give rise to short-lived plasma cells. Thus, the TIMP-1JD38 plasmablast could represent the malignant counterpart of a precursor for short-lived plasma cells. Therefore, our data suggest that the extended lifespan of the TIMP-1JD38 plasmablast could be related to additional alterations in cell cycle regulators and apoptosis induced by TIMP-1. In agreement with this hypothesis is our observation that TIMP-1 expression induces cyclin D2 overexpression and STAT-3 activation. In addition, the plasma cell CD44 and CD138 markers are commonly overexpressed in the bone marrow plasma cells of patients with multiple myeloma. On the other hand, CD44, TIMP-1, and cyclin D2 form part of the molecular signature identifying aggressive subsets of large B-cell lymphomas.8 This is a highly significant finding because cyclin D2 overexpression has been associated in multiple myelomas with secondary translocations involving the genes c-Maf and FRGFR337 and in EBV-infected B-cell lymphomas as well.38 Intriguingly, TIMP-1-induced differentiation is accompanied by down-regulation of cyclin-dependent kinase inhibitors (p18, p27) and despite c-Myc deregulation in, apparently, contradiction with the normal process in which down-regulation of c-Myc and up-regulation of p18 are required for cell cycle arrest and final B-cell differentiation.39 However, inactivation of cyclin-dependent kinase inhibitors through homozygous p18 deletion and p16 methylation, with additional cytogenetic abnormalities in the c-myc locus, are common findings in progressing multiple myelomas.40,41 This aberrant expression of cell cycle regulators is a paradox in the development of plasma cell tumors. Therefore, those TIMP-1 effects down-regulating the cyclin-dependent kinase inhibitors, while inducing differentiation of B cells with deregulated c-Myc, are also conceivable. This is strongly supported by the plasmacytic phenotype induced in vitro by TIMP-1 in the Burkitt lymphoma cell model, as well as the high expression of TIMP-1 in multiple myeloma cell lines and tumors.
We propose a TIMP-1-dependent model of plasmacytic differentiation. In this model, TIMP-1 as a stromal factor can provide further differentiation stimulus and survival signals to highly proliferative plasma cell precursors via activation of STAT-3. We previously reported that TIMP-1 induced BCL-XL expression6 and BCL-XL is a known target of STAT-3. Now, the present findings support the notion that TIMP-1-induced up-regulation of BCL-XL can be STAT-3 dependent.
In summary, our findings suggest an important role for TIMP-1 in plasmacytic/plasmablastic tumors, provide new insights for the diagnosis and treatment of these malignancies,41-43 and offer grounds for future studies to further understand the role of TIMP-1 in the complex process of terminal B-cell differentiation.
Prepublished online as Blood First Edition Paper, October 12, 2004; DOI 10.1182/blood-2004-04-1385.
A.M. and S.Z. authors contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr Carolyn Best (Laboratory of Biosystems, NCI) for her expert advise on cDNA microarray statistical analysis, and Ms Rita Salloum for her assistance in preparing this manuscript.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal