Abstract
There is now considerable in vitro evidence that tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) is involved in HIV-1 pathogenesis by inducing CD4+ T-cell death characteristic of AIDS. Therefore, we have tested levels of TRAIL in plasma samples from 107 HIV-1–infected and 53 uninfected controls as well as in longitudinal plasma samples from patients who started antiret-roviral therapy (ART). TRAIL was elevated in plasma of HIV-1–infected patients compared with uninfected individuals, and patients receiving ART showed decreased plasma TRAIL levels that correlated with reduction in viral load. In vitro exposure to infectious and noninfectious HIV-1 induced TRAIL in monocytes and marginally in dendritic cells (DCs) but not in macrophages or T cells. Interestingly, the HIV-1 entry inhibitor, soluble CD4, blocked HIV-1–induced production of TRAIL. Furthermore, production and gene expression of TRAIL by monocytes were regulated by type I interferon via signal transducer and activator of transcription-1 (STAT1)/STAT2 signaling molecule. Ex vivo HIV-1 infection of human tonsil lymphoid tissue also resulted in increased TRAIL production. We demonstrate here that plasma TRAIL is elevated in HIV-1–infected patients and is decreased by ART therapy. The high production of TRAIL by antigen-presenting cells may contribute to the death of CD4+ T cells during progression to AIDS.
Introduction
The depletion of CD4+ T cells that results from HIV-1 infection remains unsolved despite more than 20 years of investigation of AIDS pathogenesis. The direct cytopathic effect resulting from HIV-1 infection has been considered to be responsible for CD4+ T-cell depletion.1 However, the low percentage of CD4+ T cells that are HIV-1 infected prior to and during the early decrease in CD4+ T-cell numbers cannot account for the severe loss of this cell population.2,3 This observation led to the suggestion of infection-induced indirect mechanisms that destroy uninfected T cells by apoptosis, such as activation-induced cell death.4
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL), a member of the TNF superfamily,5 may be involved in CD4+ T-cell depletion during progression to AIDS.6,7 TRAIL induced selective apoptosis of uninfected CD4+ T cells in a human peripheral blood lymphocyte–transplanted nonobese diabetic severe combined immunodeficient (hu-PBL-NOD-SCID) mouse model,8 and TRAIL produced by monocytes exposed to the HIV-1 Tat protein killed uninfected CD4+ T cells.9 Moreover, soluble TRAIL was found in HIV-1–infected patients10,11 and may be responsible for the death of neurons in AIDS patients leading to dementia.12 TRAIL induces apoptosis in human tumor cell lines13 and in infected cells14 but not in normal cells.15 The 2 biologically active forms of TRAIL, membrane-bound TRAIL (mTRAIL) and soluble TRAIL (sTRAIL), are regulated by type I interferon (interferon-alpha and -beta [IFN-α and IFN-β]).16-18 TRAIL is secreted by leukocytes, including T lymphocytes,19 natural killer cells,20 dendritic cells,21,22 and monocytes and macrophages.23 The lack of involvement of Fas-FasL in HIV-1–induced CD4+ T-cell direct killing24 implies that other death molecules are involved. Indeed, a recent report indicates that noninfectious HIV-1 induced T-cell death that is not mediated by Fas-FasL and suggested a potential role of TRAIL.25 However, the role of TRAIL in CD4+ T-cell depletion in vivo during progression to AIDS remains to be established. Several reports have shown that macrophages and activated monocytes can play a role in CD4+ T-cell depletion that may be due to TRAIL-mediated death.26-28 HIV-1 infection activates circulating monocytes by inducing IFN-α29 and secretion of apoptotic cytokines such as tumor necrosis factor alpha (TNF-α), CD30 ligand (CD30L), and FasL.30 Furthermore, antigen-presenting cells such as monocytes and dendritic cells have been implicated in AIDS pathogenesis,4,28 and macrophages found in brain tissue of AIDS patients were recently shown to express TRAIL.31
Two studies reported sTRAIL in the serum of HIV-1–infected patients.10,11 However, the TRAIL-producing cells were not identified, and no mechanism of TRAIL activation or association with HIV-1 disease was provided. Therefore, we compared levels of plasma sTRAIL from HIV-1–infected patients with those of uninfected control donors and tested whether sTRAIL levels in AIDS patients were associated with viral load. We recently found that exposure of peripheral blood T cells from HIV-1–uninfected donors to infectious HIV-1 or to HIV-1 that had been rendered noninfectious by chemical treatment (aldrithiol-2) (AT-2 HIV-1 particles) resulted in the expression of membrane TRAIL on CD4+ but not CD8+ T cells (J.P.H. et al, manuscript in preparation). Therefore, we also investigated whether mTRAIL and/or sTRAIL would be produced by monocytes, macrophages, and/or dendritic cells from HIV-1–infected and uninfected blood donors upon exposure to AT-2 HIV-1 as well as to infectious HIV-1. We used both HIV-1MN (CXCR4-tropic) and HIV-1Ada (CCR5-tropic) to determine if TRAIL production was HIV-1 coreceptor dependent. Because type I interferons (IFN-α and IFN-β) are required for TRAIL expression and production, we also tested (1) whether inhibition of this pathway would interfere with TRAIL expression and (2) whether agents that inhibit HIV-1 infection would block HIV-1–induced production of sTRAIL. Finally, we used the ex vivo tonsil lymphoid tissue histoculture model to determine whether HIV-1 would induce TRAIL production in primary human lymphoid tissue.
Patients, materials, and methods
Patients
Peripheral blood was collected from 107 HIV-1–infected patients who were involved in the United States Air Force (USAF) Natural History Study, of the USAF Wilford Hall Medical Center, Lackland Air Force Base (AFB), TX. Peripheral blood was collected from 8 HIV-1–infected patients who were involved in a longitudinal study of antiretroviral therapy (ART) at the Children's Hospital Medical Center, Cincinnati, OH. Approval was obtained from the institutional boards of the National Cancer Institute, the USAF Wilford Medical Center, and the Children's Hospital Medical Center. The voluntary, fully informed consent of the subjects used in this research was obtained as required by Air Force Regulation (AFR) 169-9 and by Children's Hospital Medical Center. Informed consent was provided according to the Declaration of Helsinki. Patients involved in the longitudinal study were followed for 40 weeks. Patient I was not on antiretroviral therapy prior starting the study and was then started on tenofovir, D4T, and Kaletra (lopinavir and ritonavir). The patient went off ART and did not start another regimen prior to the end of the study. Patient II was naive prior to starting the study and was enrolled into a study that included Trizivir (abacavir, zidovudine, and lamivudine), Combivir (zidovudine and lamivudine), and Sustiva (efavirenz). Therapy continued through week 40. Patient III was naive prior to starting the study and was enrolled in a study that included Trizivir, Combivir, and Sustiva. The patient was switched to didanosine (DDI), Combivir, and Sustiva, which continued through week 40. Patient IV was on ART prior starting the study and was switched to Kaletra and Sustiva at the start of TRAIL study.
Preparation of AT-2–inactivated virions
HIV-1MN (X4-tropic) and HIV-1Ada (R5-tropic) were propagated as described previously.32 Viral supernatants were inactivated with 1 mM AT-2 for 18 hours at 4°C before purification. Microvesicles, used as a control reagent, were isolated from supernatants of uninfected cell cultures in a method identical to that used for virus preparation from infected cells.33 The AT-2 HIV-1 glycoproteins remain functional after AT-2 treatment as previously described.34
Isolation and culture of PBMCs and T cells
Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation (Ficoll-Hypaque; Pharmacia, Uppsala, Sweden) from peripheral blood from healthy, HIV-1–seronegative donors by the Department of Blood Transfusion, National Institutes of Health (NIH), Bethesda, MD. Cells were cultured in RPMI medium (Invitrogen, Gaithersburg, MD) with 10% fetal bovine serum (Sigma, St Louis, MO.). CD4+ and CD8+ cells were purified from PBMCs by positive selection with anti-CD4+ or anti-CD8+ beads (Miltenyi Biotec, Auburn, CA). Macrophages and dendritic cells were generated using monocytes as previously described.35 Cells were incubated with AT-2 HIV-1 at a final concentration of 50 ng/mL p24 equivalent. Infectious HIV-1MN and HIV-1LAV were used at the same concentration.
Ex vivo human lymphoid tissue preparation and infection
Human tonsils were obtained from patients undergoing routine tonsillectomy within 6 hours from surgery. Tissues were kept in phosphate-buffered saline (PBS) and washed in complete culture medium composed of RPMI 1640 (Invitrogen) containing 15% heat-inactivated fetal calf serum (FCS) (Sigma), nonessential amino acids (1 mM), sodium pyruvate (1 mM), l-glutamine (292 μg/mL), amphotericin B (2.5 μg/mL; Gibco, Grand Island, NJ), ticarcillin disodium/clavulanate potassium (Timentin; 310 μg/mL; SmithKline Beckman, Philadelphia, PA), and gentamicin (50 μg/mL; Quality Biologics, Gaithersburg, MD). Tissues were then sectioned into 2 mm3 blocks and placed on top of medium-hydrated collagen sponge gel (Gelfoam; Pharmacia & Upjohn, Kalamazoo, MI) in complete medium at the air-liquid interface in 6-well plates (Costar, Cambridge, MA) with 9 blocks per sponge in each well in 4 mL medium. Twenty-four hours later, the culture supernatant was replaced with fresh medium containing 1% penicillin/streptomycin (Gibco), and each tissue block was infected with 1 ng p24gag of an HIV-1 LAV.04 viral stock (3 μL, 120 median tissue culture infective dose [TCID50]). Culture medium was sampled and changed every 3 days.
Plasma quantification of HIV-1 RNA
Plasma HIV-1 RNA was measured by quantitative reverse transcriptase–polymerase chain reaction (RT-PCR), and all data are expressed in copies per milliliter (Amplicor Monitor; Roche Diagnostic Systems, Branchburg, NJ; detection limit, 50 copies per milliliter).
Membrane TRAIL detection
Membrane TRAIL expression was determined by incubating cells for 20 minutes with phycoerythrin (PE)–conjugated mouse immunoglobulin G1 (IgG1) anti–human TRAIL monoclonal antibody (clone B-S23; Diaclone, Besançon France) or with control isotype-matched antibodies (at 5 μg/mL each) in PBS containing 2% mouse serum (Sigma). Cells were acquired on FACSCalibur flow cytometer using CellQuest software (Becton Dickinson Bioscience Immunocytometry Systems, San Jose, CA). Samples were gated on viable cells by forward and side light scatter, and at least 50 000 live cell events were acquired for each sample.
Soluble TRAIL and type I interferon measurement
Levels of soluble TRAIL were measured in serum of HIV-positive or HIV-negative individuals or in cell supernatants with the commercial TRAIL enzyme-linked immunosorbent assay (ELISA) kit (Diaclone), according to the manufacturer's instructions. IFN-α/β levels were evaluated using ELISA kit (PBL Biomedical Laboratories, Piscataway, NJ).
Blocking assay
To block TRAIL production, monocytes were cultured with AT-2 HIV-1MN/Ada in presence of CD4 binding inhibitor soluble CD4 (2 μg/mL), the fusion inhibitor T20 (2 μg/mL), the CXCR4 inhibitor AMD-310 (2 μg/mL) (AIDS Research and Reagents Program, National Institute of Allergy and Infectious Diseases [NIAID], Bethesda, MD), or in presence of both sheep polyclonal anti–human IFN-α (2000 IFN-α neutralizing U/mL) and sheep polyclonal anti–human IFN-β antibodies (500 IFN-β neutralizing U/mL) (BioSource International, Camarillo, CA). Recombinant IFN-α and IFN-β (Peprotech, Rocky Hill, NJ) were used at 200 U/mL.
Real-time quantitative RT-PCR
Total RNA was extracted from monocytes with the acid guanidine thiocyanatephenol-chloroform method,36 modified for TRIzol (Invitrogen). One microgram of total RNA was reverse transcribed in a reaction containing 1 μM random hexanucleotide primers, 1 μM oligo(dT), and 200 units of Moloney murine leukemia virus reverse transriptase. (Promega, Madison, WI).
Real-time PCR was conducted with the ABI Prism 7900HT (Applied Biosystems, Foster City, CA). All reactions were performed using a SYBR green PCR mix (QuantiTect SYBR Green PCR; Qiagen, Valencia, CA). The primer sequences were designed using the Primer Express Software v2.0 (TRAIL: forward 5-GCTCTGGGCCGCAAAAT-3, reverse 5-TGCAAGTTGCTCAGGAATGAA-3; glyceraldehyde-3-phosphate dehydrogenase [GAPDH]: forward 5-CCACCCATGGCAAATTCC-3, reverse 5-TGGGATTTCCATTGATGACAAG-3). All reactions were performed in triplicate (denaturation at 95°C for 15 seconds, annealing at 60°C [61°C for TRAIL] for 15 seconds, extension at 72°C for 15 seconds). Data analysis was performed with the SDS2.1 software (Applied Biosystems). Results are presented as ratios between the target gene and the GAPDH mRNA.
Western blot
Monocytes were cultured with AT-2 HIV-1MN, AT-2 HIV-1Ada, recombinant IFN-α (10 ng/mL), and IFN-β (10 ng/mL), anti–IFN-α, and anti–IFN-β (R&D Systems, Minneapolis, MN). After 24 hours, the cells were pelleted, washed in PBS, and lysed (1% Nonidet P-40 [NP-40], 200 mM NaCl, 50 mM Tris [tris(hydroxymethyl)aminomethane] pH 7.5, supplemented with protease inhibitor cocktail [Roche]). Total protein was quantified by bicinchoninic acid (BCA) (Pierce, Rockford, IL), and 10 μg protein was mixed 1:1 with SDS Protein Gel Buffer (Quality Biologics, Gaithersburg, MD). Samples were run on a 10% or 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel (Bio-Rad, Hercules, CA). After transfer the blot was blocked in 3% milk/Tris-buffered saline Triton (TBST) and then incubated with anti–signal transducer and activator of transcription-1 (anti-STAT1) or anti-STAT2 (Upstate Biotechnology, Lake Placid, NY) antibodies followed by anti–rabbit horseradish peroxidase (HRP) secondary antibody (Jackson ImmunoResearch Labs, West Grove, PA). Enhanced chemiluminescence (ECL) was performed, and bands were visualized on Hyperfilm (Amersham, Piscataway, NJ); β-actin was visualized as a loading control using a mouse monoclonal antibody (Sigma).
Statistical analysis
All experiments were repeated a minimum of 3 times. All comparisons of data were made with a 2-sided Student t test (data meet the requirements for a Student t test).
Differences in plasma TRAIL between HIV-1–infected patients and healthy donors were also analyzed using the F test to compare variances. All the statistical data were analyzed using Prism software (GraphPad, San Diego, CA). The correlation between viral load and TRAIL in the longitudinal study was statistically analyzed as follows. The variations in TRAIL concentration (Δ[TRAIL]) and viral load (Δviral load) between each sampling time for each patient were calculated. A linear regression analysis between Δ[TRAIL] and log (Δviral load) was performed using Excel software.
Results
TRAIL level is elevated in plasma of HIV-1–infected patients
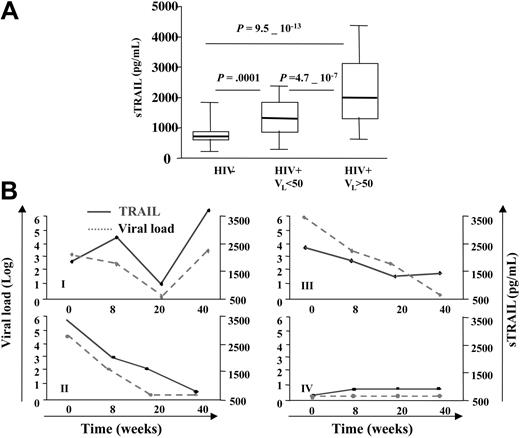
We tested whether the plasma of HIV-1–infected patients contains higher levels of sTRAIL than plasma from healthy donors. As shown in Figure 1A, mean values of plasma TRAIL were 852 ± 52 pg/mL for 53 control donors, 1339 ± 79 pg/mL for 49 HIV-1–infected patients with undetectable viral load (less than 50 HIV-1 RNA copies per milliliter of blood), and 2242 ± 131 pg/mL for 58 HIV-1–infected patients with detectable load (more than 50 HIV-1 RNA copies per milliliter of blood). These results suggest that in vivo TRAIL production is significantly increased in HIV-1–infected patients with high viral loads. However, there appeared to be a saturation of the sTRAIL level in patients (12 of 58) with viral loads exceeding 40 000 RNA copies per milliliter, because higher viral loads were not associated with further increases in sTRAIL. The data of Figure 1B provide a longitudinal study of 4 HIV-1–infected patients who began highly active ART (HAART) therapy and were subsequently followed for 40 weeks with measurement of plasma viral load and sTRAIL. Patient I exhibited an initial parallel drop in viral load and sTRAIL, followed by a concomitant rebound in both of these parameters. This rebound in viral load may reflect development of HIV-1 drug resistance. Patient II showed a continuous parallel drop in viral load and sTRAIL. Patient III showed 3 of 4 points in which changes in viral load and sTRAIL levels were similar. Patient IV, who had received ART prior to enrollment into this study, exhibited a parallel flat and low profile for both viral load and sTRAIL throughout the 40 weeks. This patient's initial low viral may have been due to successful ART prior to enrollment, which continued during this ART protocol. Thus, we observed both cross-sectional and longitudinal association between higher plasma viral loads and elevated sTRAIL levels in 2 different cohorts of HIV-1–infected patients and a parallel between HAART-induced reduction in viral load and sTRAIL levels. The correlation between the fluctuations in viral load and TRAIL in the longitudinal study was statistically analyzed by performing a linear regression between the variations of TRAIL concentration (Δ[TRAIL]) and viral load (Δviral load) between each sampling time for each patient. This analysis showed a positive correlation between Δ[TRAIL] and log (Δviral load) in these 4 patients (r = 0.627 and P = .029).
Increased plasma levels of TRAIL in HIV-1–infected patients. (A) Plasma samples from 107 HIV-1–infected patients and 53 uninfected controls were tested for their soluble TRAIL content by ELISA. Two groups of HIV-1–infected patients were defined depending on their viral load. The mean values of plasma TRAIL were 852 ± 52 pg/mL for 55 control donors, 1339 ± 79 pg/mL for 49 HIV-1–infected patients with undetectable viral load (less than 50 RNA copies per milliliter of blood), and 2242 ± 131 pg/mL for 58 HIV-1–infected patients with higher viral load (more than 50 RNA copies per milliliter of blood). P indicates P values from unpaired t test. Boxes indicate 25th and 75th percentiles, and error bars indicate 10th and 90th percentiles. (B) Longitudinal study of 4 HIV-1–infected patients who, at time 0, began antiretroviral therapy. Patients were followed for 40 weeks. TRAIL level was measure by ELISA. Data are representative of 4 different groups of the 8 patients tested.
Increased plasma levels of TRAIL in HIV-1–infected patients. (A) Plasma samples from 107 HIV-1–infected patients and 53 uninfected controls were tested for their soluble TRAIL content by ELISA. Two groups of HIV-1–infected patients were defined depending on their viral load. The mean values of plasma TRAIL were 852 ± 52 pg/mL for 55 control donors, 1339 ± 79 pg/mL for 49 HIV-1–infected patients with undetectable viral load (less than 50 RNA copies per milliliter of blood), and 2242 ± 131 pg/mL for 58 HIV-1–infected patients with higher viral load (more than 50 RNA copies per milliliter of blood). P indicates P values from unpaired t test. Boxes indicate 25th and 75th percentiles, and error bars indicate 10th and 90th percentiles. (B) Longitudinal study of 4 HIV-1–infected patients who, at time 0, began antiretroviral therapy. Patients were followed for 40 weeks. TRAIL level was measure by ELISA. Data are representative of 4 different groups of the 8 patients tested.
Noninfectious HIV-1 induces TRAIL production by monocytes
To study the effects of noninfectious viruses on TRAIL production, we cultured PBMCs from healthy HIV-1–uninfected donors and AIDS patients in the presence of microvesicles (negative control), AT-2 HIV-1MN (CXCR4-tropic), or AT-2 HIV-1Ada (CCR5-tropic) for 3 to 6 days. Soluble TRAIL levels in the culture supernatants were determined by ELISA. We did not detect TRAIL production in untreated cultures or in cultures of PBMCs from healthy donors treated with microvesicles (n = 10). In contrast, unstimulated PBMCs from AIDS patients produced 190 ± 10 pg/mL sTRAIL (n = 16). Exposure to AT-2 HIV-1MN induced sTRAIL production by PBMCs from control donors (800 ± 160 pg/mL) as well as from HIV-1–infected patients (760 ± 140 pg/mL). Similarly, exposure to AT-2 HIV-1Ada induced sTRAIL production by PBMCs from healthy donors (900 ± 300 pg/mL) and HIV-1–infected patients (640 ± 300 pg/mL) (Figure 2A).
Effects of AT-2 HIV-1 particles on TRAIL secretion. (A) PBMCs from HIV-1–uninfected or HIV-1–infected individuals were cultured for 3 days in presence or not of AT-2 HIV-1MN or HIV-1Ada. Levels of TRAIL were detected using ELISA. (B) CD4+, CD8+ T cells, DCs, macrophages, and monocytes from infected (□) or noninfected individuals (•) were cultured 3 days in presence of AT-2 HIV-1MN. Mean values (± standard errors) are shown for 6 independent experiments for each condition tested. (C) Monocytes from HIV-1–uninfected donors were cultured for 1 day in presence of microvesicles (controls) or AT-2 HIV-1MN. Cells were analyzed for mTRAIL by fluorescence-activated cell sorting (FACS). Iso indicates isotype control. Data are representative of 6 independent experiments. (D) Monocytes from HIV-1–uninfected individuals were cultured for 1 day in presence of microvesicles or AT-2 HIV-1MN. TRAIL mRNA level was quantified by real-time PCR. Mean values with standard errors are shown for 4 independent experiments. (E) Monocytes from HIV-1–uninfected donors were cultured in presence of a different concentration of AT-2 HIV-1MN, and sTRAIL was quantified by ELISA. Mean values with standard errors are shown for 6 independent experiments.
Effects of AT-2 HIV-1 particles on TRAIL secretion. (A) PBMCs from HIV-1–uninfected or HIV-1–infected individuals were cultured for 3 days in presence or not of AT-2 HIV-1MN or HIV-1Ada. Levels of TRAIL were detected using ELISA. (B) CD4+, CD8+ T cells, DCs, macrophages, and monocytes from infected (□) or noninfected individuals (•) were cultured 3 days in presence of AT-2 HIV-1MN. Mean values (± standard errors) are shown for 6 independent experiments for each condition tested. (C) Monocytes from HIV-1–uninfected donors were cultured for 1 day in presence of microvesicles (controls) or AT-2 HIV-1MN. Cells were analyzed for mTRAIL by fluorescence-activated cell sorting (FACS). Iso indicates isotype control. Data are representative of 6 independent experiments. (D) Monocytes from HIV-1–uninfected individuals were cultured for 1 day in presence of microvesicles or AT-2 HIV-1MN. TRAIL mRNA level was quantified by real-time PCR. Mean values with standard errors are shown for 4 independent experiments. (E) Monocytes from HIV-1–uninfected donors were cultured in presence of a different concentration of AT-2 HIV-1MN, and sTRAIL was quantified by ELISA. Mean values with standard errors are shown for 6 independent experiments.
To determine which cell types in PBMCs produce sTRAIL in response to AT-2 HIV-1, isolated monocytes as well as CD4+ and CD8+ T cells and monocyte-derived macrophages and dendritic cells were tested for TRAIL production after culture with AT-2 HIV-1. Monocytes from HIV-1–infected patients but not from healthy donors produced sTRAIL (130 ± 40 pg/mL) when cultured without AT-2 HIV-1. Monocytes produced high levels of sTRAIL after exposure to AT-2 HIV-1 (MN or Ada) (Figure 2B). No significant differences were observed between monocytes from uninfected donors and HIV-1–infected patients. Exposure of monocyte-derived dendritic cells from HIV-1–infected but not from uninfected donors produced significant amounts (210 ± 20 pg/mL), although lower levels, of sTRAIL than monocytes (1100 ± 210 pg/mL) (Figure 2B). Because membrane and soluble TRAIL are 2 forms of active TRAIL, the expression of mTRAIL was also studied. We found increased expression of mTRAIL in AT-2 HIV-1–exposed monocytes (P < .0001) compared with microvesicle-treated or untreated monocytes (controls). After AT-2 HIV-1MN exposure, 80% ± 8% of monocytes expressed mTRAIL with a mean fluorescence intensity (MFI) of 19 ± 3 compared with 7% (MFI, 6 ± 1) in control monocytes (Figure 2C). Similar results were obtained using AT-2 HIV-1Ada (data not shown).
Monocytes from uninfected donors treated with AT-2 HIV-1MN were tested for TRAIL mRNA expression using real-time quantitative PCR. TRAIL mRNA was barely detectable in untreated or microvesicle-treated monocytes, whereas exposure to AT-2 HIV-1MN induced 20-fold increase in TRAIL mRNA expression (P = .0045) (Figure 2D). These results indicated that monocytes expressed TRAIL mRNA and produced sTRAIL and mTRAIL upon exposure to AT-2 HIV-1. In contrast, sTRAIL was not produced by CD4+ or CD8+ T cells or by macrophages. However, CD4+ T cells express mTRAIL but not sTRAIL after exposure to AT-2 HIV-1 (J.P.H. et al, manuscript in preparation).
To investigate whether low amounts of virus induce sTRAIL production, we performed a dose response experiment in which monocytes were cultured for 3 days with dilutions of AT-1 HIV-1 spanning 4 logs of concentration. Very low concentrations of AT-2 HIV-1MN induced measurable production of sTRAIL (Figure 2E). Optimal concentrations of AT-2 HIV-1 mn for sTRAIL production were in the 5 to 50 ng/mL (p24CA equivalents) range, with no further increase in TRAIL at higher virus concentrations. Therefore, AT-2 HIV-1 was used at 50 ng/mL for subsequent experiments.
Noninfectious HIV-1 induces TRAIL production by the type I IFN pathway
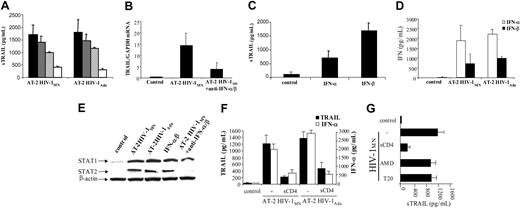
To test whether type I IFN was required for TRAIL production by monocytes isolated from PBMCs of HIV-1–uninfected donors, we attempted to block TRAIL production using anti–IFN-α and/or anti–IFN-β antibodies. Anti–IFN-α antibody (Ab) or anti–IFN-β Ab inhibited the sTRAIL production of AT-2 HIV-1–exposed monocytes by 25% ± 11% and 40% ± 3%, respectively. When used together, these antibodies reduced TRAIL production by 76% ± 4% in monocytes exposed to AT-2 HIV-1MN (P < .000 01) (Figure 3A). Similar results were obtained using HIV-1Ada. We also tested whether Ab against type I interferons would affect AT-2 HIV-1MN–induced TRAIL gene expression. Addition of anti–IFN-α and -β blocking monoclonal antibody (mAb) decreased TRAIL mRNA expression by 70% ± 30% (P = .046) (Figure 3B). To determine whether IFN-α and IFN-β were both involved in TRAIL production, we cultured monocytes in the presence of recombinant IFN-α or IFN-β. IFN-α induced 730 ± 200 pg/mL and IFN-β 1650 ± 300 pg/mL of sTRAIL (Figure 3C). Because AT-2 HIV-1 induction of TRAIL is IFN-α/β dependent, we verified that monocytes from uninfected donors cultured with AT-2 HIV-1MN or AT-2 HIV-1Ada produced IFN-α (Figure 3D). Moreover, we demonstrated that AT-2 HIV-1MN and AT-2 HIV-1Ada as well as recombinant IFN-α/β increased monocyte expression of STAT1 and STAT2 (Figure 3E), the principal signaling molecules for type I IFN.37,38 We also observed a decrease of STAT1 expression and a strong inhibition of STAT2 expression when monocytes were cultured with HIV-1 and antibodies against type I interferons. Taken together, these findings indicate that AT-2 HIV-1–induced expression and production of TRAIL by monocytes is dependent on STAT1/STAT2 type I interferons.
Effect of IFN type I antibodies on TRAIL production by monocytes from HIV-1–uninfected donors. (A) Monocytes were cultured 3 days in presence of AT-2 HIV-1MN alone (•) or with blocking antibodies against IFN-α (2000 neutralizing U/mL; ▦), IFN-β (500 neutralizing U/mL; ▨), or both (□). The level of sTRAIL was quantified by ELISA. (B) Monocytes were cultured for 1 day in presence of AT-2 HIV-1MN and with or without blocking antibodies against IFN-α and IFN-β. The level of TRAIL mRNA was quantified by real-time PCR analysis. (C) Monocytes were cultured for 3 days in presence of recombinant IFN-α or IFN-β (10 ng/mL). The level of sTRAIL was quantified by ELISA. (D) IFN-α (□) and IFN-β production (•) by monocytes cultured for 3 days in presence of microvesicles (control), AT-2 HIV-1MN, or AT-2 HIV-1Ada. Mean values with standard errors are shown for 4 independent experiments in panels A-C for each condition tested. (E) STAT1 and STAT2 expression. Monocytes were cultured 24 hours in presence of AT-2 HIV-1MN, AT-2 HIV-1Ada, recombinant IFN-α/β (10 ng/mL), or HIV-1MN plus anti–IFN-α/β antibodies. Production of STAT1 and STAT2 was analyzed by Western blot; β-actin was used as a loading control. (F) Blocking assay. Monocytes were cultured for 3 days with AT-2 HIV-1MN or HIV-1Ada and in presence of isotype control antibody or the CD4 binding inhibitor soluble CD4 (2 μg/mL). IFN-α (□) and TRAIL levels (•) were quantified by ELISA. (G) Monocytes were cultured for 3 days with AT-2 HIV-1MN and in presence of isotype control antibody; the CD4 binding inhibitor soluble CD4 (2 μg/mL); the CXCR4 inhibitor AMD-3100 (2 μg/mL), or the fusion inhibitor T20 (2 μg/mL). Monocytes cultured without AT-2 HIV-1MN were used as control. TRAIL level was quantified by ELISA. Mean values with standard errors are shown for 3 independent experiments for each condition tested (panels F-G).
Effect of IFN type I antibodies on TRAIL production by monocytes from HIV-1–uninfected donors. (A) Monocytes were cultured 3 days in presence of AT-2 HIV-1MN alone (•) or with blocking antibodies against IFN-α (2000 neutralizing U/mL; ▦), IFN-β (500 neutralizing U/mL; ▨), or both (□). The level of sTRAIL was quantified by ELISA. (B) Monocytes were cultured for 1 day in presence of AT-2 HIV-1MN and with or without blocking antibodies against IFN-α and IFN-β. The level of TRAIL mRNA was quantified by real-time PCR analysis. (C) Monocytes were cultured for 3 days in presence of recombinant IFN-α or IFN-β (10 ng/mL). The level of sTRAIL was quantified by ELISA. (D) IFN-α (□) and IFN-β production (•) by monocytes cultured for 3 days in presence of microvesicles (control), AT-2 HIV-1MN, or AT-2 HIV-1Ada. Mean values with standard errors are shown for 4 independent experiments in panels A-C for each condition tested. (E) STAT1 and STAT2 expression. Monocytes were cultured 24 hours in presence of AT-2 HIV-1MN, AT-2 HIV-1Ada, recombinant IFN-α/β (10 ng/mL), or HIV-1MN plus anti–IFN-α/β antibodies. Production of STAT1 and STAT2 was analyzed by Western blot; β-actin was used as a loading control. (F) Blocking assay. Monocytes were cultured for 3 days with AT-2 HIV-1MN or HIV-1Ada and in presence of isotype control antibody or the CD4 binding inhibitor soluble CD4 (2 μg/mL). IFN-α (□) and TRAIL levels (•) were quantified by ELISA. (G) Monocytes were cultured for 3 days with AT-2 HIV-1MN and in presence of isotype control antibody; the CD4 binding inhibitor soluble CD4 (2 μg/mL); the CXCR4 inhibitor AMD-3100 (2 μg/mL), or the fusion inhibitor T20 (2 μg/mL). Monocytes cultured without AT-2 HIV-1MN were used as control. TRAIL level was quantified by ELISA. Mean values with standard errors are shown for 3 independent experiments for each condition tested (panels F-G).
We tested whether noninfectious HIV-1–induced production of sTRAIL would be inhibited by (1) soluble CD4, which blocks the interaction between viral glycoprotein 120 (gp120) and CD4 expressed on CD4+ cells (monocytes, macrophages, dendritic cells [DCs], or T cells); (2) AMD-3100, which blocks CXCR4 coreceptor binding; and (3) T20, which prevents fusion of the virus with the CD4 cell membrane. The results (Figure 3F) demonstrate that soluble CD4 inhibited HIV-1MN and HIV-1Ada from inducing sTRAIL production by monocytes. Furthermore, soluble CD4 (sCD4) also inhibited IFN-α production. In contrast, AMD-3100 or T20 did not statistically block TRAIL production by monocytes cultured with HIV-1MN (Figure 3G). These results are consistent with a triggering mechanism that is dependent on very early CD4/gp120 binding events but that does not require early post-binding events such as coreceptor engagement or membrane fusion.
All of the results presented above were reproduced using the infectious counterparts of HIV-1MN and HIV-1Ada. Infectious HIV-1LAV, which is commonly used in tonsil model,39 also induced TRAIL production in monocytes.
Infection with HIV-1 induces TRAIL production in tonsil tissue culture
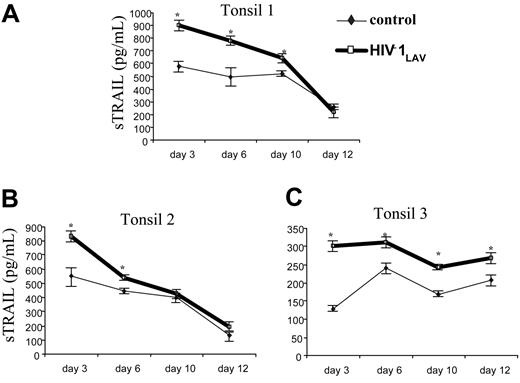
To investigate whether TRAIL could be produced in primary lymphoid organs, we infected human tonsils with infectious HIV-1Lav, the HIV-1 strain used previously in this model,39 and measured the TRAIL production at different times ranging from 1 to 12 days. As shown in Figure 4, for each of 3 independent experiments using tissues from 3 different individuals, we observed increased TRAIL secretion after infection with HIV-1Lav compared with uninfected tonsils. These results suggest that sTRAIL can be found in the primary lymphoid organs of HIV-1–infected patients as well as in their blood (Figure 1).
Effect of infectious HIV-1 on sTRAIL production in tonsil culture. Ex vivo human lymphoid tissues were cultured for 12 days in absence ( and thin line) or presence of HIV-1LAV (bold line and □). Soluble TRAIL was measured at days 3, 6, 9, and 12 by ELISA. Culture medium was changed at 3-day intervals. The statistically different (P < .05) samples from control are indicated by an asterisk. Error bars indicate standard deviation derived from 3 different ELISAs for each condition tested.
and thin line) or presence of HIV-1LAV (bold line and □). Soluble TRAIL was measured at days 3, 6, 9, and 12 by ELISA. Culture medium was changed at 3-day intervals. The statistically different (P < .05) samples from control are indicated by an asterisk. Error bars indicate standard deviation derived from 3 different ELISAs for each condition tested.
Effect of infectious HIV-1 on sTRAIL production in tonsil culture. Ex vivo human lymphoid tissues were cultured for 12 days in absence ( and thin line) or presence of HIV-1LAV (bold line and □). Soluble TRAIL was measured at days 3, 6, 9, and 12 by ELISA. Culture medium was changed at 3-day intervals. The statistically different (P < .05) samples from control are indicated by an asterisk. Error bars indicate standard deviation derived from 3 different ELISAs for each condition tested.
and thin line) or presence of HIV-1LAV (bold line and □). Soluble TRAIL was measured at days 3, 6, 9, and 12 by ELISA. Culture medium was changed at 3-day intervals. The statistically different (P < .05) samples from control are indicated by an asterisk. Error bars indicate standard deviation derived from 3 different ELISAs for each condition tested.
Discussion
In the present study, we observed elevated sTRAIL levels in the plasma of HIV-1–infected patients compared with healthy control donors. After in vitro exposure to either infectious or noninfectious HIV-1, we found that both sTRAIL and mTRAIL were produced by monocytes. Monocyte-derived dendritic cells from HIV-1–infected patients also produced low levels of sTRAIL. In contrast, sTRAIL was not produced by macrophages or by CD4+ or CD8+ T cells upon exposure to HIV-1. TRAIL production was associated with increased transcription of the TRAIL gene in monocytes exposed to HIV-1 particles and was mediated by the type I IFN pathway. Induction of sTRAIL by HIV-1 is likely to be dependent on CD4-gp120 interaction because it was blocked by soluble CD4. Finally, the ex vivo tonsil model demonstrates that TRAIL can be produced in primary lymphoid tissue after HIV-1 infection.
Soluble TRAIL was reported in HIV-1–infected patients,10 and in vitro studies linked TRAIL to the depletion of T cells from HIV-1–infected patients.6,8,9 However, the identification of TRAIL-producing cells in vivo, the association of viral load with the level of sTRAIL produced, and the mechanism responsible for TRAIL-mediated T-cell death remain to be established. The present study demonstrates that sTRAIL can be detected in the plasma of HIV-1–infected patients. Furthermore, the amount of sTRAIL is higher in patients with elevated viral load than in patients with low or undetectable viral loads. However, plasma from all HIV-1 patients tested showed higher TRAIL content than plasma from healthy donors. Our 40-week longitudinal study of patients on HAART showed striking parallel changes between viral load and TRAIL levels. Thus, when HAART decreased viral load, we observed a concomitant decrease in plasma TRAIL, which may be one of the reasons for the efficacy of antiviral therapy.
More than 95% of HIV-1 virions detected in HIV-1–infected patients' plasma do not possess culturable infectivity,40 which raises the possibility that noninfectious virus particles contribute to HIV-1 pathogenesis. We demonstrate here that the major cellular source of TRAIL is monocytes, which produce TRAIL in response to both infectious and chemically inactivated X4 (MN) and R5 (Ada) HIV-1. Interestingly, PBMCs and DCs from HIV-1–infected donors produced sTRAIL without further stimulation, suggesting that previous contact with HIV-1 was sufficient to induce sTRAIL secretion. However, after 3 days of culture, sTRAIL production disappeared if the patients' PBMCs were not stimulated by HIV-1 particles (data not shown), suggesting that continuous cell-virus interaction is required to maintain sTRAIL production.
Our results do not conflict with the effect of Tat on TRAIL production by monocytes or macrophages.9 We confirmed that Tat induced TRAIL production; however, the range of concentration at which Tat was efficient was narrower (data not shown) than the range at which HIV-1 was inducing TRAIL. We think that both Tat and HIV-1 particles likely induce TRAIL production in HIV-1–infected patients and these 2 nonmutually exclusive but rather concomitant mechanisms contribute together to the high level of TRAIL measured in the serum of HIV-1–infected patients.
Type I interferons are produced as a result of viral infection and can exert antiviral effects.41 Type I interferons activate the signaling molecules STAT1 and STAT2, which bind to the interferon-stimulated response element (ISRE), which activates IFN-inducible genes16 such as TRAIL.42,43 Here we demonstrate that in monocytes, type I interferons, STAT1 and STAT2 expressions are induced by exposure to HIV-1 virions. Recombinant IFN-α or IFN-β induced STAT1 and STAT2 and sTRAIL production, while antibodies against these interferons greatly reduced STAT2 expression and both TRAIL gene transcription and production in monocytes exposed to AT-2 HIV-1. We showed that sCD4 blocked the effects of HIV-1MN and HIV-1Ada on TRAIL and IFN-α production by monocytes. This blockade strongly suggests that the interaction between viral gp120 and the CD4 molecule is required for HIV-1 induction of type I interferons and sTRAIL production. Conversely, our findings that AMD-310 and T20 did not significantly inhibit sTRAIL production suggest that coreceptor binding and viral fusion with the target cell membrane are not required for HIV-1 activation of sTRAIL production by monocytes. Thus, only the earliest interaction between HIV-1 and CD4 on monocytes is essential for initiation of TRAIL production.
We also tested whether HIV-1 induced TRAIL production in an ex vivo human tonsil model that resembles human lymphoid organs where critical events of HIV disease occur. Increased TRAIL secretion was seen in tonsil organ cultures from 3 different donors after HIV-1 infection. TRAIL production was maintained over an extended time, because we still observed higher TRAIL production in infected than in uninfected tonsil cultures after 10 days. Detection of TRAIL in these HIV-1–infected cultures is consistent with increased apoptosis of CD4+ T cells after HIV-1 infection of tonsil cultures.2 Because CD4+ T cells express mTRAIL but do not produce sTRAIL upon exposure to HIV-1,18 the sTRAIL that we detected in plasma of HIV-1–infected patients is unlikely to have been secreted by CD4+ T cells and may have been produced by monocytes and/or dendritic cells after exposure to HIV-1. Furthermore, because B cells, present in large number in tonsil, did not produce TRAIL (data not shown), it is possible that dendritic cells in the tonsil were the source of sTRAIL in our HIV-1–infected tonsil cultures, a possibility that we are now investigating.
The findings of the present study, in combination with our observation that the membrane DR5 death receptors required for induction of cell death via TRAIL are expressed on CD4+ but not CD8+ T cells from HIV-1–infected patients (J.P.H. et al, manuscript in preparation), suggest a plausible mechanism for preferential loss of CD4+ T cells in HIV-1 infection. In this model, sTRAIL and/or mTRAIL produced by monocytes contribute to TRAIL-mediated death of CD4+ T cells in HIV-1–infected patients by directly interacting with the DR5 death receptor expressed on CD4+ T cells.
Establishment of the in vivo relevance of our in vitro findings to the loss of CD4+ T cells in HIV-1–infected patients may point toward novel therapeutic approaches for maintaining immune cell numbers and function. In particular, we found that sCD4 was the most efficient inhibitor of TRAIL production, suggesting that this reagent might provide effective therapy that would inhibit the interaction of either infectious or noninfectious HIV-1 with the CD4 molecule that results in TRAIL production by CD4-expressing cells.
Prepublished online as Blood First Edition Paper, December 7, 2004; DOI 10.1182/blood-2004-08-3058.
Supported by the Intramural Program of the Centers for Cancer Research, NCI, NIH, and by the NIH Intramural AIDS Targeted Antiviral Program. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. N01-CO-12400 (Article H.36 of the Prime Contract). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States government.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Leonid Margolis, Laboratory of Cellular and Molecular Biophysics, NICHD, NIH, Bethesda, MD, for reviewing the manuscript; Julian Bess, AIDS Vaccine Program, NCI-Frederick, MD, for preparing AT-2 HIV-1; and Dr David Venzon, Biostatistics and Data Management Section, National Cancer Institute, NIH, Bethesda, MD, for statistical analysis and helpful suggestions concerning the longitudinal study.
The views expressed herein are those of the authors and do not reflect the official policy of the Department of Defense or other departments of the US Government.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal