Abstract
Bone remodeling is accompanied by the differentiation of osteoclasts from the monocyte/macrophage lineage of hematopoietic cells. The osteoclast differentiation process requires receptor activator of nuclear factor κB (NF-κB) ligand (RANKL), which causes complex changes in the expression of various genes. In a cDNA microarray study to identify genes targeted by RANKL, we found that monokine induced by the interferon-γ (IFN-γ) (MIG) gene was up-regulated in osteoclast precursor cells. The increase in MIG expression by RANKL was confirmed by reverse transcription–polymerase chain reaction and Western blot analysis. RANKL induction of MIG required the activity of NF-κB, whose binding site is present in the MIG promoter. MIG induction by RANKL was also dependent on p38 mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription 1 (STAT1). RANKL stimulated the phosphorylation of Ser727 of STAT1, which required p38 activity. MIG secreted on RANKL treatment could stimulate the migration and adhesion of osteoclast precursors and osteoclasts that were primed to express CXCR3, the MIG receptor, by macrophage–colony-stimulating factor (M-CSF). Therefore, we provide the first evidence demonstrating that RANKL stimulates the serine phosphorylation of STAT1 through the p38 MAPK pathway, causing MIG gene transcription and secretion, which may have a role in recruiting CXCR3-positive osteoclast precursors and osteoclasts to bone remodeling or inflammatory sites.
Introduction
Overall bone homeostasis is regulated through bone remodeling by the coordinated activity of osteoclasts and osteoblasts. Receptor activator of nuclear factor κB ligand (RANKL) has been identified as a key molecule for osteoclast differentiation and bone remodeling.1 Besides RANKL, the bone remodeling process is accompanied by complex changes in the expression levels of various genes in osteoblasts and osteoclasts. Identifying the genes that are associated with osteoclast differentiation can help elucidate the molecular mechanisms that underlie bone remodeling. We have tried to detect differentially expressed genes during osteoclast differentiation using a cDNA microarray technique. In this effort, we found several up-regulated genes in RANKL-treated osteoclast precursor cells.2,3 Among those genes, monokine induced by interferon-γ (IFN-γ) (MIG) was selected for further investigation in this study. MIG, which is also called CXCL9, is a member of the CXC chemokine family. MIG is mainly produced by activated macrophages. The receptor for MIG, CXCR3, is functionally expressed on activated T and B cells and on endothelial cells.4,5 MIG is chemotactic for activated T cells and induces the adhesion of interleukin-2 (IL-2)–stimulated T lymphocytes through its receptor, CXCR3.6,7
Few studies have investigated chemokines in relation to the regulation of bone. Several chemokines are highly expressed in bone erosive lesions, suggesting a potential role for chemokines in the bone remodeling process. MIP-1α and MIP-1β, produced by multiple myeloma cells, were shown to play critical roles in the development of lytic bone lesions by enhancing osteoclast formation and bone resorption.8 Bendre et al9 reported that IL-8, a member of the α chemokines, stimulated osteoclastogenesis and bone resorption in metastatic bone disease. Osteoclast itself has been shown to highly express CXC chemokines CXCL10 (or IP-10) and CXCL12 (or SDF-1) during the differentiation and maturation process.10
Chemokines are often released from activated macrophages under inflammatory process and viral infection. IFN-γ, an important regulator of macrophage function, induces the CXC chemokines, including MIG and IP-10, in mouse macrophages.11 IFN-γ induction of MIG is mediated by the transcriptional factor, signal transducer and activator of transcription α (STAT1α), which binds to the γRE-1 site of the MIG promoter.12 Interferons bind to a common receptor and activate the receptor-associated Janus kinases (Jaks). The activation of the Jaks and the phosphorylation of tyrosine residues by Jaks on the cytoplasmic domain of the receptors result in the phosphorylation of STAT proteins, which dimerize and translocate into the nucleus to elicit transcriptional activation.13,14
In this study, we found that the chemokine MIG is up-regulated by RANKL in osteoclast precursor cells. STAT1 played a critical role in the induction of MIG by RANKL, and the RANKL-stimulated serine phosphorylation of STAT1 through the p38 MAPK was the signaling event crucial for MIG expression. Furthermore, we present evidence that the expression of MIG induced by RANKL can stimulate the adhesion and migration of macrophage–colony-stimulating factor (M-CSF)–dependent osteoclast precursors and differentiating osteoclasts.
Materials and methods
Reagents and mice
Recombinant human soluble RANKL (sRANKL) and human M-CSF were purchased from PeproTech EC (London, United Kingdom). Antibodies for MIG were from R&D Systems (Minneapolis, MN). Phosphoserine-STAT1, phosphotyrosine-STAT1, and STAT1 antibodies were obtained from Upstate Biotechnology (Waltham, MA). Antiphospho-p38 and anti-p38 were purchased from Cell Signaling Technology (Beverly, MA). SB203580, PD98059, and SP600125 were purchased from Calbiochem (La Jolla, CA). Nuclear factor κB (NF-κB)–specific inhibitor SN50 was from Biomol (Plymouth Meeting, PA). Homozygous STAT1-deficient (STAT1-/-) mice in which the STAT1 gene has been deleted by homologous recombination and wild-type 129/S6 mice were obtained from Taconic (Germantown, NY). ICR mouse strain was used as another mouse source to prepare the osteoclast precursors.
Preparation of osteoclast precursors
Osteoclast precursors were generated from mouse bone marrow cells as previously described.15 Bone marrow cells were obtained by flushing tibiae and femora from 6- to 7-week-old mice, suspended in α-minimum essential medium (α-MEM) containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin in the presence of 1:10 dilution of CMG14-12 culture supernatant to provide M-CSF16 and were cultured for 3 days at 37°C in a humidified atmosphere of 5% CO2. Nonadherent cells were removed by pipetting, and adherent cells were obtained.
RT-PCR analysis
Reverse transcription–polymerase chain reaction (RT-PCR) analysis was performed as previously described.17 Total RNA was prepared using TRI Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Two micrograms total RNA was reverse transcribed with SuperScriptII reverse transcriptase (Invitrogen), and 0.1 of the reverse-transcribed cDNA was amplified by PCR. For PCR amplification, the following primers were used: Mig sense, 5′-ATGAAGTCCGCTGTTCTTTTCCT-3′; Mig antisense, 5′-AGTCTTCCTTGAACGACGACGA-3′; Cxcr3 sense, 5′-GCCACCCATTGCCAGTACAAC-3′; Cxcr3 antisense, 5′-TCCCACAAAGGCATAGAGCAGC-3′; Fra II sense, 5′-CCCTATCCACGCTCACATCCC-3′; Fra II antisense, 5′-CGTTCCTCGGGGCTGATTTT-3′; Mmp-9 sense, 5′-CTGTCCAGACCAAGGGTACAGCCT-3′; Mmp-9 antisense, 5′-GAGGTATAGTGGGACACATAGTGG-3′; glyceraldehyde-3-phosphate dehydrogenase (Gapdh) sense, 5′-CAAGGCTGTGGGCAAGGTCA-3′; Gapdh antisense, 5′-AGGTGGAAGAGTGGGAGTTGCTG-3′.
Western blotting
Cells were lysed in a buffer containing 20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, and protease and phosphatase inhibitors. Protein concentrations of cell lysates were determined using the DC Protein Assay Kit (Bio-Rad, Hercules, CA). Twenty to thirty micrograms cellular protein was resolved by 10% to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and was transferred to a polyvinylidene difluoride membrane (Amersham Biosciences, Piscataway, NJ). After blocking with 5% skim milk, the membrane was probed with anti-MIG, antiphosphoserine-STAT1, antiphosphotyrosine-STAT1, and phospho-p38. The same membrane was stripped and reprobed with anti-STAT1, p38, and actin.
Enzyme-linked immunosorbent assay
MIG was detected in culture supernatants by enzyme-linked immunosorbent assay (ELISA), according to the instructions of the manufacturer (R&D Systems). In brief, an anti-MIG capture antibody (R&D Systems) was diluted to 1 μg/mL in phosphate-buffered saline (PBS) and then added to wells of a microtiter plate. After overnight incubation at room temperature, the capture antibody was removed, and nonspecific binding was blocked by adding 200 μL blocking buffer (3% bovine serum albumin [BSA] in PBS). The plate was incubated at room temperature for 2 hours, after which wells were washed 3 times with PBS containing 0.05% Tween 20 (PBS-T). Sample or standard was added, and the plate was incubated overnight at 4°C. After incubation, the plate was washed 3 times with PBS-T, a biotinylated detection antibody was diluted to 1 μg/mL in PBS and added to the wells, and the plate was incubated for 2 hours at room temperature. After the plate was washed 3 times with PBS-T, streptavidin-conjugated horseradish peroxidase (HRP) was added to the wells. The plate was incubated at room temperature for 20 minutes and then washed 5 times with PBS-T. One hundred microliters substrate solution was added to each well, and the plate was incubated for 20 to 30 minutes. The color reaction was stopped by the addition of 50 μL stopping solution (1 M H2SO4), and the optical density was read with a microtiter ELISA plate reader at 450 nm.
Flow cytometry
Flow cytometry analyses were performed as previously described.18 Briefly, osteoclast precursors were stimulated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated times. Cells were washed with fluorescence-activated cell sorter (FACS) buffer (PBS containing 1% bovine serum albumin and 0.1% sodium azide) and then incubated with anti-CXCR3 antibody (Zymed, South San Francisco, CA) for 1 hour at 4°C. Cells were washed with FACS buffer 3 times and then incubated with a secondary antibody labeled with fluorescein isothiocyanate (FITC) for 1 hour at 4°C. Then cells were fixed with 3.7% formaldehyde and subjected to flow cytometry.
Migration assay
Cell migration assays were performed as previously described with some modification.19 Osteoclast precursors were cultured in the presence of 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to 72 hours. Cells were harvested by treatment with cell dissociation solution (Sigma, St Louis, MO) and then resuspended in serum-free α-MEM. Cells were seeded in the upper well of the Boyden chamber with polycarbonate filters containing 8-μm pore membranes (Corning Costar, Cambridge, MA). The lower well was loaded with α-MEM in the presence or absence of recombinant MIG. In addition, to determine the effect of conditioned media on cell migration, wild-type and STAT1-deficient osteoclast precursors were incubated in serum-free α-MEM in the presence of M-CSF or M-CSF plus RANKL for 24 to 30 hours. Conditioned media were collected and loaded into the lower well as chemoattractant. After 6 to 8 hours of incubation, the migrated cells were fixed in 3.7% formaldehyde for 10 minutes, stained with tartrate-resistant acid phosphatase (TRAP; Sigma), and counted.
Adhesion assay
Adhesion assays were performed as described previously.18 Briefly, 96-well microtiter plates were coated with 20 μg/mL fibronectin or 10 μg/mL vitronectin in PBS for 14 to 16 hours at 37°C. The plates were washed with PBS and incubated with PBS containing 0.2% BSA for 1 hour at 37°C. Osteoclast precursors were cultured in the presence of 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to 72 hours. Cells were detached by treatment with cell dissociation solution and suspended in serum-free α-MEM. Cells were seeded to each well of fibronectin-coated 96-well plates and incubated for 10 minutes at 37°C in the presence of MIG or PBS. Nonadherent cells were removed by washing with PBS. Adherent cells were fixed in 3.7% formaldehyde for 10 minutes and were stained with hematoxylin or TRAP solution. Stained cells were counted.
Fluorescence microscopy
In adhesion experiments with GFP-expressing cells, the bound cells were detected by confocal microscopy. All images are shown at original magnification, × 200. Confocal equipment comprised an LSM 5 PASCAL microscope equipped with a Plan-Neofluor 20 ×/0.5 objective lens, an LSM 5 PASCAL photomultiplier tube, and Laser Scanning Microscope LSM 5 PASCAL software version 3.0 (all from Carl Zeiss, Jena, Germany).
Statistical analysis
Each experiment was performed 3 to 5 times, and all quantitative data are presented as mean plus or minus SD. Statistical differences were analyzed by Student t test.
Results
RANKL induces MIG expression in osteoclast precursors
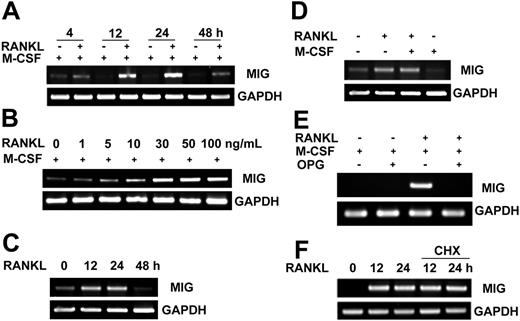
To identify genes regulated by RANKL in osteoclast precursors, we used cDNA microarray technology. RNA from mouse bone marrow–derived osteoclast precursor cells treated for 24 hours with M-CSF or M-CSF plus RANKL were used for hybridization with the cDNA array. One of several genes up-regulated in response to RANKL was MIG. With this array result, we further examined the expression pattern of the MIG gene in osteoclast precursors by RT-PCR and Western blotting analyses. The increase in MIG mRNA level was evident at 12 hours and was maximal at 24 hours after RANKL stimulation (Figure 1A). The induction of MIG mRNA was dependent on RANKL concentrations (Figure 1B). The up-regulation of MIG mRNA by RANKL was observed in the absence of concurrent M-CSF treatment (Figure 1C). M-CSF itself did not increase MIG mRNA and did not have any synergistic effect on the RANKL induction of MIG gene expression (Figure 1D). The induction of MIG by RANKL was blocked by the addition of osteoprotegerin (OPG) (Figure 1E). To exclude the possibility that RANKL could indirectly affect the expression of MIG, osteoclast precursors were stimulated with RANKL in the absence or presence of cycloheximide to block new protein synthesis. The expression pattern of MIG mRNA was not affected by the presence of cycloheximide (Figure 1F), indicating that RANKL directly induces MIG expression in osteoclast precursors. In line with the expression pattern of MIG mRNA, the protein level of MIG was increased at 12 and 24 hours after RANKL treatment (Figure 2A) in a dose-dependent manner (Figure 2B). When the MIG concentration in the culture supernatant was determined by ELISA, 40 to 50 ng/mL was detected (Figure 2C).
Induction of MIG mRNA expression by RANKL. (A) Time course of MIG mRNA induction by RANKL. Osteoclast precursors were stimulated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. Total RNA was extracted from the treated cells. RNA was reverse transcribed and PCR amplified with MIG or GAPDH primers. (B) RANKL dose-dependent increase in MIG mRNA levels. Osteoclast precursors were treated with the indicated concentration of RANKL in the presence of 50 ng/mL M-CSF for 12 hours. (C) RANKL induction of MIG in the absence of M-CSF. Osteoclast precursors were stimulated with 100 ng/mL RANKL for the indicated time in the absence of M-CSF. (D) Lack of synergy between RANKL and M-CSF in MIG induction. Osteoclast precursors were incubated with 50 ng/mL M-CSF, 100 ng/mL RANKL, or both for 24 hours. (E) Inhibition of RANKL induction of MIG by OPG. Osteoclast precursors were pretreated with or without 500 ng/mL OPG and incubated with 50 ng/mL M-CSF and 100 ng/mL RANKL for 12 hours. (F) Effect of cycloheximide (CHX) on RANKL induction of MIG mRNA. Osteoclast precursor cells were pretreated with or without 1 μg/mL cycloheximide and were further stimulated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. Total RNA was extracted from the treated cells. RNA was reverse transcribed and PCR amplified with MIG or GAPDH primers.
Induction of MIG mRNA expression by RANKL. (A) Time course of MIG mRNA induction by RANKL. Osteoclast precursors were stimulated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. Total RNA was extracted from the treated cells. RNA was reverse transcribed and PCR amplified with MIG or GAPDH primers. (B) RANKL dose-dependent increase in MIG mRNA levels. Osteoclast precursors were treated with the indicated concentration of RANKL in the presence of 50 ng/mL M-CSF for 12 hours. (C) RANKL induction of MIG in the absence of M-CSF. Osteoclast precursors were stimulated with 100 ng/mL RANKL for the indicated time in the absence of M-CSF. (D) Lack of synergy between RANKL and M-CSF in MIG induction. Osteoclast precursors were incubated with 50 ng/mL M-CSF, 100 ng/mL RANKL, or both for 24 hours. (E) Inhibition of RANKL induction of MIG by OPG. Osteoclast precursors were pretreated with or without 500 ng/mL OPG and incubated with 50 ng/mL M-CSF and 100 ng/mL RANKL for 12 hours. (F) Effect of cycloheximide (CHX) on RANKL induction of MIG mRNA. Osteoclast precursor cells were pretreated with or without 1 μg/mL cycloheximide and were further stimulated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. Total RNA was extracted from the treated cells. RNA was reverse transcribed and PCR amplified with MIG or GAPDH primers.
MIG protein expression stimulated by RANKL. Osteoclast precursors were treated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time (A) or were incubated with 50 ng/mL M-CSF plus the indicated concentration of RANKL for 24 hours (B). Cell lysates were prepared and subjected to Western blot analysis with anti-MIG antibody. The same membrane was stripped and reprobed with antiactin antibody. (C) Osteoclast precursors were treated with 100 ng/mL RANKL or 1 ng/mL IFN-γ for 24 hours. MIG concentrations in culture supernatants were determined using ELISA as described in “Materials and methods.”
MIG protein expression stimulated by RANKL. Osteoclast precursors were treated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time (A) or were incubated with 50 ng/mL M-CSF plus the indicated concentration of RANKL for 24 hours (B). Cell lysates were prepared and subjected to Western blot analysis with anti-MIG antibody. The same membrane was stripped and reprobed with antiactin antibody. (C) Osteoclast precursors were treated with 100 ng/mL RANKL or 1 ng/mL IFN-γ for 24 hours. MIG concentrations in culture supernatants were determined using ELISA as described in “Materials and methods.”
RANKL induces MIG expression through NF-κB and p38 MAPK-dependent pathways in osteoclast precursors
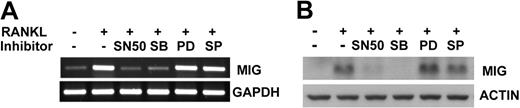
Stimulation of RANK has been shown to activate the NF-κB transcription factor and the extracellular regulated kinases (ERKs), Jun kinases (JNKs), and p38 mitogen-activated protein kinases (MAPKs) in osteoclastogenesis.20 To investigate whether these signaling molecules are involved in the up-regulation of MIG by RANKL, we examined the effect of signaling pathway inhibitors on MIG expression. Osteoclast precursor cells were treated with NF-κB peptide inhibitor SN50, p38 inhibitor SB203580, MEK (ERK upstream kinase) inhibitor PD98059, or JNK inhibitor SP600125 before RANKL stimulation and MIG expression levels were determined by RT-PCR and Western blotting. As shown in Figure 3, the RANKL induction of MIG expression was abolished in the presence of SN50 and SB203580 at mRNA and protein levels. Inhibition of JNK had no effect on MIG expression, whereas inhibition of ERK resulted in a slight increase in the MIG protein level (Figure 3B). These results indicate that NF-κB and p38 MAPK play a crucial role in the expression of MIG in RANKL-treated osteoclast precursors.
Effects of signaling inhibitors on expression of MIG induced by RANKL. Osteoclast precursors were pretreated with 30 μg/mL SN50, 20 μM SB203580, 1 μM PD98059, or 20 μM SP600125 for 30 minutes and were stimulated in the presence of 50 ng/mL M-CSF plus 100 ng/mL RANKL for 12 hours. Expression levels of MIG were analyzed by RT-PCR (A) or Western blot analysis (B).
Effects of signaling inhibitors on expression of MIG induced by RANKL. Osteoclast precursors were pretreated with 30 μg/mL SN50, 20 μM SB203580, 1 μM PD98059, or 20 μM SP600125 for 30 minutes and were stimulated in the presence of 50 ng/mL M-CSF plus 100 ng/mL RANKL for 12 hours. Expression levels of MIG were analyzed by RT-PCR (A) or Western blot analysis (B).
RANKL stimulates serine phosphorylation of STAT1 through a p38-dependent pathway
In the promoter of the MIG gene, NF-κB and STAT1 binding sites are present, and a synergistic regulation of MIG transcription through NF-κB and STAT1 has been reported.21,22 Given that the NF-κB activation by RANK has been well demonstrated in many studies and that SN50 blocked RANKL induction of MIG (Figure 3), it is reasonable to conclude that RANK activation of NF-κB is required for MIG induction in osteoclast precursor cells. In contrast, there have been no reports of the regulation of STAT1 by RANKL. Therefore, we explored the possibility that RANKL modulates STAT1 for MIG expression in osteoclast precursors.
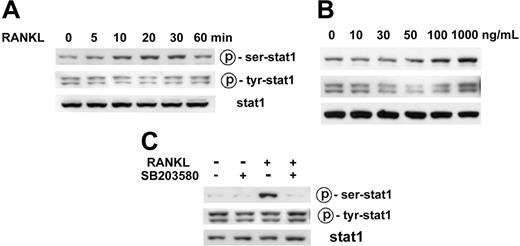
The tyrosine phosphorylation and subsequent SH2 domain–mediated dimerization of STAT is considered an essential prerequisite for its biologic activity because dimerization enables STAT molecules to enter the nucleus and to bind DNA.13 However, serine phosphorylation of STATs also has profound effects on their target gene transcription in certain cases.14 We therefore examined the phosphorylation of STAT1 in RANKL-stimulated osteoclast precursors. As shown in Figure 4A, the serine phosphorylation of STAT1 increased from 10 minutes to 30 minutes and returned to the basal level at 60 minutes in response to RANKL stimulation. This increase in STAT1 serine phosphorylation was RANKL dose dependent (Figure 4B). However, RANKL did not increase the tyrosine phosphorylation level of STAT1 (Figure 4A-B). The basal level of STAT1 tyrosine phosphorylation was more consistently observed than that of serine phosphorylation. Immunostaining of the osteoclast precursor cells with STAT1 antibody showed cytosolic and nuclear distribution of STAT1 in unstimulated and RANKL-stimulated cells (data not shown). Nuclear staining was evident even after serum deprivation for 12 hours (data not shown).
Stimulation of STAT1 serine phosphorylation by RANKL through the p38 MAPK pathway. (A) Time course of STAT1 phosphorylation by RANKL. Osteoclast precursors were serum starved for 12 hours and were stimulated with 100 ng/mL RANKL for the indicated time. Cell lysates were subjected to Western blotting for the phosphorylation of STAT1 at Ser727 and Tyr701 using antiphosphospecific STAT1 antibodies (top and middle rows). The same membrane was stripped and reprobed with anti-STAT1 antibody (bottom row). (B) Serine phosphorylation of STAT1 dependent on RANKL dose. Osteoclast precursors were serum starved for 12 hours and then stimulated with the indicated concentration of RANKL for 10 minutes. Western blot analysis was carried out as described. (C) Effects of p38 inhibitor on RANKL-induced phosphorylation of STAT1. Osteoclast precursors were deprived of serum for 12 hours and were pretreated with 20 μM SB203580, followed by stimulation with 100 ng/mL RANKL for 10 minutes. Cell lysates were prepared and subjected to Western blot analysis, as described.
Stimulation of STAT1 serine phosphorylation by RANKL through the p38 MAPK pathway. (A) Time course of STAT1 phosphorylation by RANKL. Osteoclast precursors were serum starved for 12 hours and were stimulated with 100 ng/mL RANKL for the indicated time. Cell lysates were subjected to Western blotting for the phosphorylation of STAT1 at Ser727 and Tyr701 using antiphosphospecific STAT1 antibodies (top and middle rows). The same membrane was stripped and reprobed with anti-STAT1 antibody (bottom row). (B) Serine phosphorylation of STAT1 dependent on RANKL dose. Osteoclast precursors were serum starved for 12 hours and then stimulated with the indicated concentration of RANKL for 10 minutes. Western blot analysis was carried out as described. (C) Effects of p38 inhibitor on RANKL-induced phosphorylation of STAT1. Osteoclast precursors were deprived of serum for 12 hours and were pretreated with 20 μM SB203580, followed by stimulation with 100 ng/mL RANKL for 10 minutes. Cell lysates were prepared and subjected to Western blot analysis, as described.
The p38 MAPK signaling pathway has been implicated in the serine phosphorylation of STAT1 in some cell types treated with certain stimuli,14 and RANKL-induced expression of MIG was suppressed by the p38 inhibitor SB203580; hence, we assessed the effect of SB203580 on the serine phosphorylation of STAT1 in RANKL-stimulated osteoclast precursor cells. As shown in Figure 4C, SB203580 greatly attenuated the serine phosphorylation of STAT1 by RANKL. These results suggest that the p38 pathway may mediate the RANKL induction of MIG gene expression through the serine phosphorylation of STAT1.
RANKL induction of MIG is absent in osteoclast precursors from STAT1-deficient mice
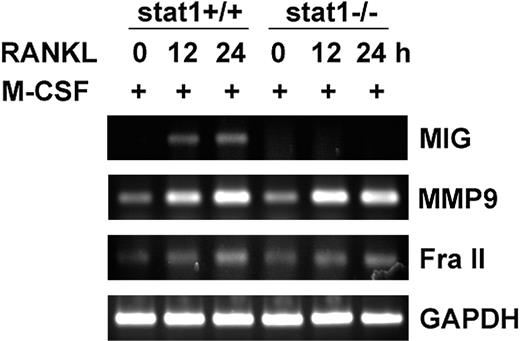
RANKL stimulated the serine phosphorylation of STAT1 in a p38-dependent manner (Figure 4C), and the p38 inhibitor SB203580 blocked RANKL induction of MIG (Figure 3). Therefore, we next examined the requirement of STAT1 in MIG induction by using STAT1-deficient mice. Osteoclast precursors were derived from bone marrow cells of the wild-type and STAT1 knockout mice and were treated with RANKL in the presence of M-CSF. The expression of MIG mRNA was observed in M-CSF plus RANKL-treated wild-type osteoclast precursors, but not in cells derived from STAT1-deficient mice (Figure 5, first panel). The lack of MIG induction in STAT1-deficient osteoclast precursors was not caused by a general defect in cellular responses to RANKL because the expression levels of MMP9 and Fra II were increased in the STAT1-deficient cells as much as in the wild-type cells on treatment with RANKL (Figure 5, second and third panels). These results demonstrate that STAT1 plays an essential and specific role in RANKL-induced expression of MIG in osteoclast precursors.
Lack of MIG induction by RANKL in STAT1-deficient osteoclast precursors. Osteoclast precursors from wild-type and STAT1-deficient mice were stimulated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. RT-PCR was performed with isolated RNA using primers specific for each of MIG, MMP-9, Fra II, and GAPDH. RT-PCR products were separated on a 1.2% agarose gel and were stained with ethidium bromide.
Lack of MIG induction by RANKL in STAT1-deficient osteoclast precursors. Osteoclast precursors from wild-type and STAT1-deficient mice were stimulated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. RT-PCR was performed with isolated RNA using primers specific for each of MIG, MMP-9, Fra II, and GAPDH. RT-PCR products were separated on a 1.2% agarose gel and were stained with ethidium bromide.
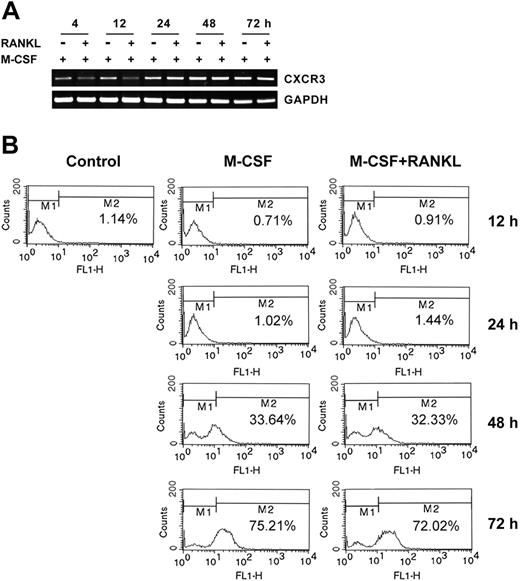
M-CSF induces the expression of CXCR3, the receptor for MIG
We hypothesized that MIG induced by RANKL in osteoclast precursors may play a role in recruiting more osteoclast precursors to form multinucleated cells through fusion. The basic requirement for this hypothesis would be expression of the receptor for MIG in osteoclast precursors or osteoclasts. Therefore, we examined whether the expression of CXCR3, the receptor for MIG, is regulated in osteoclast precursors by M-CSF or M-CSF plus RANKL. The CXCR3 mRNA level was significantly increased from 12 hours and reached maximum at 48 hours in the presence of M-CSF (Figure 6A). Cotreatment with RANKL delayed the M-CSF induction of CXCR3 mRNA expression without reducing the maximum response level (Figure 6A). To determine whether the cell surface expression of CXCR3 was also increased by M-CSF or M-CSF plus RANKL, we checked the CXCR3 receptor expression on the surface by flow cytometry. As shown in Figure 6B, unstimulated bone marrow–derived osteoclast precursor cells contained low numbers (approximately 1.14%) of CXCR3-positive cells. The population of CXCR3-positive cells increased to 33.64% and 75.21% at 48 hours and 72 hours, respectively, after M-CSF treatment. The concomitant presence of RANKL did not influence the surface expression of CXCR3 (Figure 6B). These results demonstrate that the chemokine receptor CXCR3 is expressed on the surfaces of osteoclast precursors (cells incubated with M-CSF) and TRAP+ osteoclasts (cells incubated with M-CSF plus RANKL for 48 hours). Our findings suggest the possibility that MIG produced by RANKL might play a role in an autocrine or a paracrine mode during osteoclastogenesis.
Expression of CXCR3, the MIG receptor, in osteoclast precursors and differentiating osteoclasts incubated with M-CSF. (A) Osteoclast precursors were stimulated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. Total RNA was extracted from the treated cells and subjected to RT-PCR analysis with CXCR3 or GAPDH primers. (B) Cells were prepared as described in panel A. The presence of CXCR3 on the cell surface was analyzed by flow cytometry, as described in “Materials and methods.”
Expression of CXCR3, the MIG receptor, in osteoclast precursors and differentiating osteoclasts incubated with M-CSF. (A) Osteoclast precursors were stimulated with 50 ng/mL M-CSF or 50 ng/mL M-CSF plus 100 ng/mL RANKL for the indicated time. Total RNA was extracted from the treated cells and subjected to RT-PCR analysis with CXCR3 or GAPDH primers. (B) Cells were prepared as described in panel A. The presence of CXCR3 on the cell surface was analyzed by flow cytometry, as described in “Materials and methods.”
MIG stimulates the adhesion of osteoclast precursors and osteoclasts
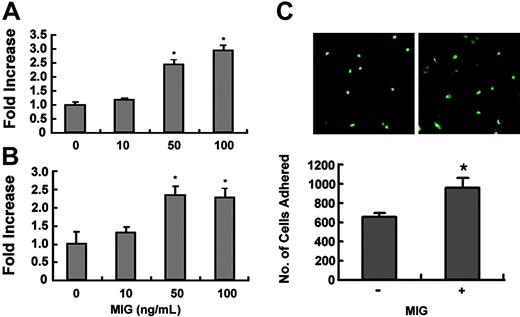
It has been reported that MIG increases the adhesion of CXCR3-expressing hematopoietic progenitor cells in a receptor-dependent manner.23 Because M-CSF treatment induced the surface expression of CXCR3 in osteoclast precursors (Figure 6B), we investigated whether MIG can increase the adhesion of these cells. Osteoclast precursor cells treated with M-CSF for 60 to 72 hours were allowed to attach to the fibronectin-coated substratum in the presence or absence of MIG. The presence of MIG increased the adhesion of osteoclast precursors in a dose-dependent manner (Figure 7A).
Effect of MIG on adhesion of osteoclast precursors and TRAP+ osteoclasts. (A) Osteoclast precursors were treated with 50 ng/mL M-CSF for 60 to 72 hours. Cells were incubated for 10 minutes on fibronectin-coated culture plates supplemented with the indicated concentration of MIG. Nonadherent cells were washed with PBS, and adherent cells were stained with hematoxylin and counted under a light microscope. *Significant difference from the medium control (P < .01). (B) Osteoclast precursors were treated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to 72 hours. Cells were added to the vitronectin-coated culture plates, supplemented with the indicated concentration of MIG, and incubated for 10 minutes. Adhesion assays were performed as described. *Significant difference from the medium control (P < .01). (C) Osteoclast precursors from wild-type mice and GFP-transgenic mice were treated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to72 hours. GFP-expressing cells were then transferred to dishes containing non-GFP cells and were incubated for 1 hour in the presence or absence of 100 ng/mL MIG. Dishes were washed to remove unbound cells, and the number of bound cells was scored under a fluorescence microscope. *Significant difference from the medium control (P < .01).
Effect of MIG on adhesion of osteoclast precursors and TRAP+ osteoclasts. (A) Osteoclast precursors were treated with 50 ng/mL M-CSF for 60 to 72 hours. Cells were incubated for 10 minutes on fibronectin-coated culture plates supplemented with the indicated concentration of MIG. Nonadherent cells were washed with PBS, and adherent cells were stained with hematoxylin and counted under a light microscope. *Significant difference from the medium control (P < .01). (B) Osteoclast precursors were treated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to 72 hours. Cells were added to the vitronectin-coated culture plates, supplemented with the indicated concentration of MIG, and incubated for 10 minutes. Adhesion assays were performed as described. *Significant difference from the medium control (P < .01). (C) Osteoclast precursors from wild-type mice and GFP-transgenic mice were treated with 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to72 hours. GFP-expressing cells were then transferred to dishes containing non-GFP cells and were incubated for 1 hour in the presence or absence of 100 ng/mL MIG. Dishes were washed to remove unbound cells, and the number of bound cells was scored under a fluorescence microscope. *Significant difference from the medium control (P < .01).
We next examined the effect of MIG on the adhesion of TRAP+ osteoclasts. Osteoclast precursor cells were treated with RANKL and M-CSF for 60 to 72 hours and were plated on vitronectin-coated wells. The adhesion of TRAP+ osteoclasts to vitronectin was significantly increased by MIG (Figure 7B). In addition, whether MIG can regulate the adhesion between cells (homophilic adhesion) during osteoclast differentiation was tested. Osteoclast precursor cells from wild-type and green fluorescence protein (GFP)–transgenic mice were treated with RANKL and M-CSF for 60 to 72 hours. Cells from GFP-transgenic mice were then transferred to cultures of non-GFP cells and incubated for 1 hour in the presence or absence of MIG. After removing unbound cells, the bound GFP cells were scored. The homophilic adhesion was significantly increased by MIG (Figure 7C). These results clearly show that MIG can stimulate the adhesion of osteoclast precursors and differentiating osteoclasts induced to express CXCR3 by treatment with M-CSF.
MIG induces the migration of TRAP+ osteoclasts
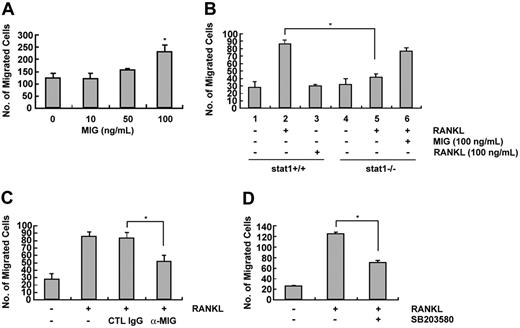
To resorb bone, osteoclasts should be recruited to a local site from bone marrow or peripheral circulation. MIG is known to function as a chemotactic factor for human T cells in inflammatory reactions.4 To determine whether MIG could induce the migration of osteoclasts, we performed migration assays. As shown in Figure 8A, osteoclast migration was significantly increased on treatment with 100 ng MIG. We next examined the effect of conditioned media derived from the osteoclast precursors stimulated by RANKL for 24 hours on cell migration. In addition, because MIG was not expressed in STAT1-deficient mice, we used the conditioned media from STAT1-deficient cells treated with RANKL. As shown in Figure 8B, cell migration was more greatly increased by conditioned medium obtained from wild-type cells stimulated by RANKL (lane 2) than it was by conditioned medium obtained from unstimulated cells (lane 1). The possibility that RANKL itself, which remained in the conditioned medium, might function as a chemotactic factor was excluded because directly adding RANKL to conditioned medium did not stimulate migration (lane 3). In contrast to the situation in wild-type cells, cell migration in response to conditioned media from STAT1-deficient cells was not increased by RANKL treatment (lane 5). Adding MIG directly to the conditioned medium from STAT1-deficient cells restored the cell migration response (lane 6). Furthermore, adding a neutralizing antibody against MIG to the conditioned medium obtained from wild-type cells incubated with RANKL reduced the migration response (Figure 8C). Including the p38 inhibitor SB203580 during RANKL treatment to produce conditioned medium attenuated the extent of migration (Figure 8D). These results demonstrate that MIG induced by RANKL in osteoclast precursors can stimulate the migration of osteoclasts.
Effect of MIG on the migration of TRAP-positive osteoclasts. (A) Osteoclast precursors were cultured in the presence 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to 72 hours. Cells were washed with PBS, suspended in serum-free α-MEM, and loaded to the upper well of transwell chambers. The lower well contained serum-free medium with the indicated concentration of MIG. After 6 to 8 hours, cells migrated onto the lower well were fixed and stained with hematoxylin. *Significant difference from the medium control (P < .01). (B) Conditioned media were obtained from wild-type and STAT1-deficient osteoclast precursors incubated with RANKL for 24 to 30 hours. Conditioned medium was added to the lower well, and osteoclasts were loaded to the upper well. After 6 to 8 hours, cells that had migrated to the lower well were counted. *Significant difference between the indicated groups (P < .01). (C) The lower well of transwell chambers contained conditioned medium from RANKL-treated cells and either a MIG neutralizing antibody or the control immunoglobulin G (IgG). Migrated cells were counted as described. *Significant difference between the indicated groups (P < .01). (D) Conditioned media were obtained from osteoclast precursors treated for 24 hours with 100 ng/mL RANKL in the presence or absence of SB203580. Osteoclasts were added to the upper well and were allowed to migrate toward the conditioned medium added to the lower well. Migrated cells were scored after 8-hour incubation. *Significant difference between the indicated groups (P < .01).
Effect of MIG on the migration of TRAP-positive osteoclasts. (A) Osteoclast precursors were cultured in the presence 50 ng/mL M-CSF plus 100 ng/mL RANKL for 60 to 72 hours. Cells were washed with PBS, suspended in serum-free α-MEM, and loaded to the upper well of transwell chambers. The lower well contained serum-free medium with the indicated concentration of MIG. After 6 to 8 hours, cells migrated onto the lower well were fixed and stained with hematoxylin. *Significant difference from the medium control (P < .01). (B) Conditioned media were obtained from wild-type and STAT1-deficient osteoclast precursors incubated with RANKL for 24 to 30 hours. Conditioned medium was added to the lower well, and osteoclasts were loaded to the upper well. After 6 to 8 hours, cells that had migrated to the lower well were counted. *Significant difference between the indicated groups (P < .01). (C) The lower well of transwell chambers contained conditioned medium from RANKL-treated cells and either a MIG neutralizing antibody or the control immunoglobulin G (IgG). Migrated cells were counted as described. *Significant difference between the indicated groups (P < .01). (D) Conditioned media were obtained from osteoclast precursors treated for 24 hours with 100 ng/mL RANKL in the presence or absence of SB203580. Osteoclasts were added to the upper well and were allowed to migrate toward the conditioned medium added to the lower well. Migrated cells were scored after 8-hour incubation. *Significant difference between the indicated groups (P < .01).
Discussion
To investigate the effect of RANKL on gene expression during osteoclast differentiation, we analyzed gene expression changes in bone marrow–derived osteoclast precursors treated with RANKL using cDNA microarray technology. One of several genes up-regulated by RANKL was MIG, originally isolated as a macrophage product with inflammatory and chemotactic properties.24 The differential expression of MIG was confirmed by RT-PCR and Western blot analysis (Figures 1 and 2). MIG has been known to be induced in macrophages almost exclusively by IFN-γ.4,24 Therefore, RANKL could have increased MIG expression indirectly by inducing another gene, such as IFN-γ, that could up-regulate MIG. However, we could exclude this possibility because the effect of RANKL on MIG expression was not affected by cycloheximide (Figure 1E). These results indicate that RANKL can directly induce MIG gene expression in osteoclast precursors. Our study is the first to show that RANKL can induce MIG gene expression.
We found that NF-κB and p38 inhibitors could suppress the expression of MIG stimulated by RANKL. This result indicated that the expression of MIG was controlled by NF-κB and p38 MAPK signaling pathways. Horton et al12 reported that the MIG proximal promoter has 2 potential NF-κB binding sites and that NF-κB plays an important role in the induction of MIG by IFN-γ. In addition, γRE-1, which is a unique STAT1-binding site, is present in the MIG promoter.11 We hypothesized that p38 may mediate MIG induction by RANKL through the modulation of STAT1. In support of our hypothesis, RANKL stimulated the Ser727 phosphorylation of STAT1 in a p38-dependent manner (Figure 4). This is analogous to IFN-γ– and IL-13–stimulated gene induction, for which p38 was required for STAT1 serine phosphorylation and transcriptional activation.25,26 The lack of MIG induction by RANKL in osteoclast precursor cells from STAT1-deficient mice further supports the requirement of STAT1 in this gene transcription (Figure 5).
To function as transcription factors, STAT proteins should translocate to the nucleus. Tyrosine phosphorylation and subsequent dimerization through interaction with the SH2 domain allows STATs to enter the nucleus and to bind DNA.13 Although tyrosine phosphorylation provides the prerequisite for these transcription factors, the second phosphorylation on serine renders STATs transcriptionally active.14 In our study, RANKL stimulated serine phosphorylation of STAT1, but not tyrosine phosphorylation (Figure 4). What is puzzling is how STAT1 mediates RANKL induction of MIG transcription without the tyrosine phosphorylation required for nuclear localization. One possible answer may derive from the weak but consistently observed basal level of tyrosine phosphorylation (Figure 4), which might have been sufficient to translocate some STAT1 proteins. In confocal microscopic analyses of immunostained osteoclast precursors, a significant amount of STAT1 was observed in the nucleus, but the nuclear localization was not increased by RANKL (data not shown). The tyrosine phosphorylation of STAT1 observed in osteoclast precursors might be a consequence of M-CSF treatment applied to bone marrow cells for 3 days to obtain the osteoclast precursor cells. In fact, M-CSF caused rapid STAT1 tyrosine phosphorylation and increased nuclear localization of STAT1 in osteoclast precursors (data not shown).
MIG is a member of a CXC chemokine superfamily. This secreted low molecular–weight protein has been implicated in the directed migration, adhesion, and activation of macrophages, T cells, and B cells that express CXCR3, the receptor for MIG.27-29 We hypothesized that the RANKL-induced expression of MIG in osteoclast precursors might trigger the adhesion and migration of CXCR3-expressing cells. To gain evidence supporting our hypothesis, we first examined whether osteoclast precursors express CXCR3 along the process of osteoclastogenesis and found that M-CSF treatment induced CXCR3 gene transcription and surface expression. Granulocyte macrophage–colony-stimulating factor (GM-CSF) reportedly induces CXCR3 on CD34+ hematopoietic progenitors from human cord blood, which leads to chemotactic and adhesive responses to MIG and to IP-10, another CXCR3 ligand.23 We report for the first time that M-CSF can induce the expression of CXCR3 in bone marrow–derived hematopoietic progenitors during osteoclastogenesis. We next investigated whether the CXCR3-expressing osteoclast precursors and differentiating osteoclasts show adhesion and migration stimulated by MIG. MIG or the conditioned medium from RANKL-treated osteoclast precursors stimulated the migration and adhesion of osteoclast precursors and differentiating osteoclasts that were induced to express CXCR3 by M-CSF treatment (Figures 7 and 8).
The induction of MIG by RANKL and that of CXCR3 by M-CSF during the differentiation of osteoclasts from bone marrow cells suggest that this chemokine ligand–receptor pair may function in an autocrine or a paracrine mode. Because the time course of the ligand and the receptor expression show more than a 24-hour gap, it is likely that MIG-CXCR3 works in a paracrine rather than an autocrine manner between different cell stages during osteoclastogenesis. In in vitro culture of bone marrow–derived cells for osteoclastogenesis, interactions between fused cells and mononuclear cells (ie, between mature cells and cells at earlier stages) are frequently observed. Another physiologic importance of CXCR3 expression in cells of osteoclast lineage may lie in the possibility that MIG secreted by IFN-γ in inflammatory sites may recruit osteoclast precursors and differentiating osteoclasts, which would result in bone resorption. Osteoclastic bone resorption is commonly associated with inflammatory diseases, such as rheumatoid arthritis and periodontitis.
In summary, we have shown that RANKL directly induces MIG expression during osteoclastogenesis. The RANKL induction of MIG requires STAT1 serine phosphorylation, which is stimulated through p38 MAPK. We have also shown that differentiating osteoclasts express the MIG receptor CXCR3 on the surface and migrate toward MIG. To our knowledge, this paper is the first to reveal that STAT1 is a transcription factor modulated by RANKL and that the MIG-CXCR3 chemokine ligand–receptor pair is regulated during osteoclastogenesis.
Prepublished online as Blood First Edition Paper, December 7, 2004; DOI 10.1182/blood-2004-07-2534.
Supported by the National Research Laboratory Program (Z.H.L., S.H.L.) and the Molecular and Cellular BioDiscovery Research Program (grant M10311000024-03B4500-00410) from the Ministry of Science and Technology, Korea (H.H.K.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal