Abstract
Mutation of the nucleophosmin (NPM) gene has been reported as the most frequent mutation in acute myeloid leukemia (AML), especially in the presence of a normal karyotype. In this subgroup of intermediate-risk AML, the identification of other gene mutations (eg, FLT3, CCAAT/enhancer-binding protein-α [CEBPA]) has helped to refine the prognosis. This study explored the prevalence and the prognostic impact of NPM mutations in a cohort of 106 patients with normal-karyotype AML. NPM exon 12 mutations were detected by polymerase chain reaction (PCR) and fragment analysis for the insertion/deletion globally resulting in a 4-bp insertion. NPM mutations were detected in 47% of patients and were associated with a high white blood cell count, involvement of the monocytic lineage (M4/M5), and a decreased prevalence of CEBPA mutations. Complete remission rate and long-term outcome did not differ between NPM-mutated and -nonmutated patients. Prospective studies are needed to confirm the definitive place of NPM mutation detection to predict AML response to therapy.
Introduction
Mutation of the nucleophosmin (NPM) gene has recently been described as the most frequent mutation in acute myeloid leukemia (AML).1 NPM is a nucleus-cytoplasm shuttling protein already known to be involved in rearrangements in leukemia and lymphoma.2-4 These newly reported mutations are insertions/deletions in NPM exon 12 that encodes the nucleic acid–binding domain (NABD) at the C terminal of the mutation of the nucleophosmin (NPM) protein. The abnormal mutated NPM protein shows an aberrant cytoplasmic localization, whereas the wild-type protein is mainly located in the nucleolus and on the nuclear membrane.
Abnormal NPM cytoplasmic localization (NPMc+) has been described among diverse French-American-British (FAB) subtypes but is strongly associated with a normal karyotype.1,5 No case of NPMc+ profile has been reported among AML with recurrent cytogenetic abnormality (t(15;17), t(8;21), inv(16)/t(16;16), and 11q23 abnormalities) or with complex karyotype. Falini et al reported a higher rate of complete remission in NPMc+ AML, but the long-term prognosis associated with NPM status is not known.
In this study, we evaluated the prevalence, the clinical profile, and the prognosis of NPM mutations (NPMm) in a retrospective cohort of 106 patients with normal-karyotype AML. Our objective was to confirm the reported prevalence of NPMm and to analyze the association between NPMm and the other frequent mutations observed in this subgroup of AML: FMS-like tyrosine kinase 3 internal tandem duplications (FLT3/ITD), CCAAT/enhancer-binding protein-α mutations (CEBPAm), and mixed-lineage leukemia partial tandem duplications (MLL/PTD).
Study design
Patients and samples
We studied 106 patients aged 15 to 65 years with newly diagnosed de novo previously untreated AML and normal karyotype treated according to the French Acute Leukemia French Association-90 (ALFA90) and ALFA9802 protocols between 1990 and 2004.6,7 A normal karyotype was defined as more than 20 normal metaphases. Samples were obtained from blood and bone marrow aspirates (median leukemic blast percentage, 78%) at diagnosis. The main biologic and clinical features are indicated in Table 1.
Characteristics of AML patients with normal karyotype
. | Total . | NPMwt . | NPMm . | P . |
|---|---|---|---|---|
| No. patients | 106 | 56 | 50 | |
| Median age, y (range) | 44 (17-65) | 46 (17-65) | 43 (20-65) | .61 |
| Sex, M/F | 57/49 | 29/27 | 28/22 | .70 |
| Median WBC count, × 109 cells/L (range) | 39 (1-453) | 18 (1-158) | 69 (1-453) | <.001 |
| FAB, no. (%) | ||||
| M0 | 9 | 8 (89) | 1 (11) | .03 |
| M1 | 34 | 17 (50) | 17 (50) | .83 |
| M2 | 19 | 14 (74) | 5 (26) | .07 |
| M4 | 19 | 7 (37) | 12 (63) | .009 |
| M5 | 20 | 7 (35) | 13 (65) | |
| M6 | 2 | 2 (100) | 0 (0) | .50 |
| NC | 3 | 1 (33) | 2 (66) | .60 |
| Mutations, no. (%) | ||||
| FLT3/ITD | 33 (31) | 14 (25) | 19 (38) | .21 |
| CEBPAm | 20 (19) | 15 (27) | 5 (10) | .05 |
| MLL/PTD | 3 (3) | 2 (4) | 1 (2) | .99 |
| Outcome | ||||
| CR, no. (%) | 92 (87) | 49 (88) | 43 (86) | .99 |
| 6-y OS ± SE, % | 41 ± 11 | 37 ± 16 | 43 ± 15 | .83 |
| 6-y RFS ± SE, % | 40 ± 12 | 34 ± 17 | 47 ± 17 | .64 |
| 6-y EFS ± SE, % | 29 ± 10 | 26 ± 14 | 32 ± 14 | .24 |
. | Total . | NPMwt . | NPMm . | P . |
|---|---|---|---|---|
| No. patients | 106 | 56 | 50 | |
| Median age, y (range) | 44 (17-65) | 46 (17-65) | 43 (20-65) | .61 |
| Sex, M/F | 57/49 | 29/27 | 28/22 | .70 |
| Median WBC count, × 109 cells/L (range) | 39 (1-453) | 18 (1-158) | 69 (1-453) | <.001 |
| FAB, no. (%) | ||||
| M0 | 9 | 8 (89) | 1 (11) | .03 |
| M1 | 34 | 17 (50) | 17 (50) | .83 |
| M2 | 19 | 14 (74) | 5 (26) | .07 |
| M4 | 19 | 7 (37) | 12 (63) | .009 |
| M5 | 20 | 7 (35) | 13 (65) | |
| M6 | 2 | 2 (100) | 0 (0) | .50 |
| NC | 3 | 1 (33) | 2 (66) | .60 |
| Mutations, no. (%) | ||||
| FLT3/ITD | 33 (31) | 14 (25) | 19 (38) | .21 |
| CEBPAm | 20 (19) | 15 (27) | 5 (10) | .05 |
| MLL/PTD | 3 (3) | 2 (4) | 1 (2) | .99 |
| Outcome | ||||
| CR, no. (%) | 92 (87) | 49 (88) | 43 (86) | .99 |
| 6-y OS ± SE, % | 41 ± 11 | 37 ± 16 | 43 ± 15 | .83 |
| 6-y RFS ± SE, % | 40 ± 12 | 34 ± 17 | 47 ± 17 | .64 |
| 6-y EFS ± SE, % | 29 ± 10 | 26 ± 14 | 32 ± 14 | .24 |
NPMwt indicates NPM wild-type; NPMm, NPM mutation; NC, not classified; FLT3/ITD, FLT3 internal tandem duplication; CEBPAm, CEBPA mutation; MLL/PTD, MLL partial tandem duplication; CR, complete remission; OS, overall survival; RFS, relapse-free survival; EFS, event-free survival.
Mutation detection
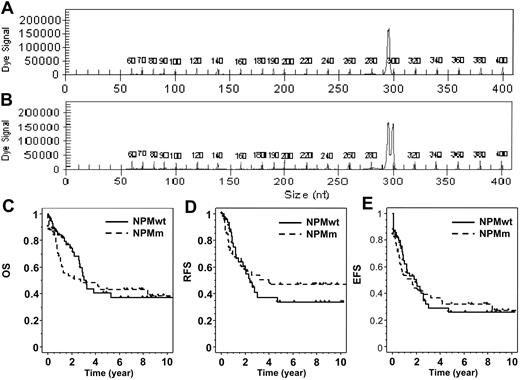
Detection of NPMm was performed on genomic DNA by polymerase chain reaction (PCR) and fragment analysis. To amplify the exon 12 of NPM, we used a fluorescently labeled forward primer 5′-(dyeD4)TTCCATACATACTTAAAACCAAGCA-3′ (Proligo, Boulder, CO) and a reverse primer 5′-TGGTTCCTTAACCACATTTCTTT-3′ (Invitrogen, Cergy-Pontoise, France). Fluorescent PCR products were subjected to capillary electrophoresis on denaturing polyacrylamide gel and analyzed by the CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA). The data were processed using Genetic Analysis System Software (Beckman Coulter). Samples from healthy controls show one peak at 297 bp. As all NPMm result in the insertion of 4 nucleotides, the mutated profile was defined by the presence of an additional peak at 301 bp (Figure 1).
Statistical analysis
Patient characteristic and complete remission (CR) rate comparisons were performed using the Fisher exact test for binary variables and the Mann-Whitney test for continuous variables. Overall survival (OS), relapse-free survival (RFS), and event-free survival (EFS) were estimated by the Kaplan-Meier method and compared using the log-rank test. Survival comparisons were adjusted for covariates using the Cox model and tested by the likelihood ratio test. Because events (relapse, death) occurred earlier in the NPMm group as compared with the NPM wild-type (NPMwt) group, resulting in crossing outcome curves, graphical methods showed that the proportional hazards assumption was violated by the NPM covariate.10 To allow the baseline hazard functions to differ in the early as compared with the later follow-up, Cox proportional hazards models stratified on 2 analysis time subsets were systematically used for all comparisons of OS, RFS, and EFS.11 The optimal time cutoff (0 to 12 months, more than 12 months) was determined from the database.
Detection of NPM exon 12 mutations and patient outcome. PCR fragment analysis of NPM exon 12 amplification showed (A) one peak in patients with NPMwt and (B) an additional peak at 301 bp in mutated patients, corresponding to a 4 bp insertion. (C) Overall survival, (D) relapse-free survival, and (E) event-free survival are shown for NPMwt and NPMm patients.
Detection of NPM exon 12 mutations and patient outcome. PCR fragment analysis of NPM exon 12 amplification showed (A) one peak in patients with NPMwt and (B) an additional peak at 301 bp in mutated patients, corresponding to a 4 bp insertion. (C) Overall survival, (D) relapse-free survival, and (E) event-free survival are shown for NPMwt and NPMm patients.
Results and discussion
Incidence and profile of NPM mutations in adult AML with a normal karyotype
The detection of NPM exon 12 mutations by PCR and fragment analysis revealed 50 mutated cases out of 106 patients (47%). This incidence of NPMm is slightly lower than that reported in a larger cohort by Falini et al (142 of 230; 61.7%). As previously described, all mutated profiles revealed a heterozygous mutation with the expected wild-type fragment and a mutated fragment corresponding to a global insertion of 4 nucleotides. In a previous screening, we had also confirmed the association between NPMm and normal karyotype: among 7 patients with core-binding factor leukemia, none had mutations; among 32 patients with adverse cytogenetic risk according to the Medical Research Council (MRC) classification,12 only 4 presented with a mutation (13%).
As indicated in Table 1, there was no significant difference in age and sex ratio between NPMm and NPMwt patients. White blood cell (WBC) counts were significantly higher in the NPMm group (NPMm: 69 versus 18 × 109 cells/L; P < .001). As previously described, NPMm were significantly more frequent in AML with monocytic component M4 and M5 (P = .009) and less frequent in M0 and M2.
CEBPA mutations were significantly less frequent in the NPMm group (NPMm: 10% versus 27%; P = .05). Contrarily, FLT3/ITD was more frequently associated with NPMm but without reaching statistical significance (NPMm: 38% versus 25%, P = .21). Only 3 patients presented with MLL/PTD, and 1 of them had an NPMm.
Prognosis of NPM mutations in adult AML with a normal karyotype
Of the 106 patients, 92 (87%) achieved CR after induction therapy. In contrast to the previous Italian study, there was no difference in CR rates between NPMm and NPMwt patients (86% versus 88%; P = .99).1 This difference may be related to the higher CR rate we observed, probably related to the intensive induction course approach from our cooperative group. CR rate was not significantly influenced by age, WBC count, CEBPAm, or FLT3/ITD.
The global 6-year OS of this cohort was 41% (±11%). In univariate analysis, age (more than 50 years) but not WBC count (more than 50 × 109 cells/L or as continuous variable) was significantly associated with a shorter OS (P = .03) and EFS (P = .04). Patients with CEBPAm had a longer OS (CEBPAm 6-year OS: 53% versus 38%; P = .17) and EFS (P = .01). As we already reported in the ALFA9000 protocol, the presence of FLT3/ITD was not a prognostic factor for OS (FLT3/ITD 6-year OS: 37% versus 43%; P = .45) but was associated with a shorter EFS in this cohort (FLT3/ITD 5-year EFS: 19% versus 34%; P = .05).8 No difference in outcome was observed between NPMm and NPMwt patients, as shown in Table 1 and Figure 1. However, as illustrated by the Kaplan-Meier representation (Figure 1C), a trend for more early deaths was observed in NPMm patients. This was partly related to a shorter median post-relapse survival in NPMm patients (6.7 versus 9.7 months in the NPMwt group; nonstatistically significant). We performed a multivariate analysis including NPM status and parameters significantly associated with survival in univariate analysis as covariates: age for OS and age, CEBPA, and FLT3 status for EFS. Age (more than 50 years) remained the only parameter independently associated with a shorter OS (P = .03), while no variable influenced EFS in the multivariate setting.
Fifteen patients were screened at relapse for the presence of NPM mutation. Ten of 15 patients had NPM mutations at diagnosis and still displayed NPM mutations at relapse time. None of the 5 NPMwt patients acquired NPM mutation at relapse. This pattern gives evidence for the stability of NPM mutations but should be confirmed in a larger cohort.
Our study did not reveal any association between NPM mutations and patient outcome. On the other hand, improved outcome reported in NPMm patients is quite unexpected because these mutations seem to be associated with numerous bad-prognosis factors, such as high WBC count, FAB M4/5, presence of FLT3/ITD, and absence of CEBPAm. Differences in treatment, and especially induction, strategies might explain these discrepancies. If the stability of NPMm at relapse time is confirmed, these mutations may be useful to monitor residual disease in a large subgroup of AML patients. Further prospective studies are needed to investigate these points and to confirm the prognosis of NPMm in AML.
Appendix
The members of the Acute Leukemia French Association (ALFA) are as follows: S Caillères (Centre Hospitalier [CH], Aix-en-Provence); B Corront, C Martin (CH, Annecy); L Sutton (CH, Argenteuil); G Lepeu (CH, Avignon); M Renoux, F Bauduer (CH, Bayonne); L Ades, P Fenaux, C Gardin (Hôpital Avicenne, Bobigny); O Reman (CH, Caen); M Blanc (CH, Chambéry); T de Revel, T Fagot (Hôpital Percy, Clamart); G Tertian (Hôpital Antoine Beclère, Clamart); JO Bay, B Choufi, M Legros, O Tournilhac (Centre Hospitalier Universitaire [CHU], Clermont-Ferrand); A Devidas, C Petitdidier (CH, Corbeil); D Bories, C Cordonnier, H Jouault, M Kuentz, S Maury, C Pautas (CHU Henri Mondor, Créteil); B Dupriez, P Morel (CH, Lens); F Bauters, S de Botton, JP Jouet, JL Lai, P Lepelley, C Preudhomme, B Quesnel (CHU, Lille); D Bordessoule, P Turlure (CHU, Limoges), E Archimbault, C Charrin, C Dumontet, M Elhamri, D Fière, S Hayette, M Michallet, E Tavernier, A Thiebaut, X Thomas, I Tigaud, D Treille-Ritouet, E Wattel (CHU, Lyon), C Soussain (CH, Meaux); M Schoenwald (CH, Orléans); JM Zini, E Dupuy (Hôpital Lariboisière, Paris); S Choquet, N Dhédin, V Leblond, JP Vernant (Hôpital Pitié Salpêtrière, Paris); V Barbu, NC Gorin, F Isnard, C Perot, J Van den Akker (Hôpital Saint-Antoine, Paris); N Boissel, JM Cayuela, S Chevret, C Chomienne, MT Daniel, L Degos, A de Labarthe, H Dombret, H Espérou, E Gluckman, T Leblanc, V Levy, O Maarek, JM Micléa, E Raffoux, D Réa, G Socié, J Soulier (Hôpital Saint-Louis, Paris); G Auzanneau, G Nedellec (Hôpital du Val de Grâce, Paris); R Leblay, B Grosbois (CHU, Rennes); C Bastard, MP Callat, N Contentin, B Lenormand, S Lepretre, H Tilly, A Stamatoullas (CHU, Rouen); M Janvier, F Turpin (Centre René Huguenin, Saint-Cloud); J Jaubert (Hôpital Saint-Anne, Toulon); B Anglaret (CH, Valence); F Lebaron, M Simon (CH, Valenciennes); S Castaigne, P Rousselot, AL Taksin, C Terré (CHU, Versailles); P Arnaud, JH Bourhis, C Fermé, N Itzhar (Institut Gustave Roussy, Villejuif).
Prepublished online as Blood First Edition Paper, July 26, 2005; DOI 10.1182/blood-2005-05-2174.
A complete list of the members of the Acute Leukemia French Association appears in the “Appendix.”
Supported by the Fondation de France (Comité Leucémie), the Ligue contre le Cancer (Comité Nord), and the Cancéropole Nord-Ouest (Axe Onco-hématologie).
N.B. and A.R. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all participating investigators from the ALFA9000 and ALFA9802 protocols.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal