Mutations in nucleophosmin NPM1 are the most frequent acquired molecular abnormalities in acute myeloid leukemia (AML). We determined the NPM1 mutation status in a clinically and molecularly well-characterized patient cohort of 275 patients with newly diagnosed AML by denaturing high-performance liquid chromatography (dHPLC). We show that NPM1 mutations are significantly underrepresented in patients younger than 35 years. NPM1 mutations positively correlate with AML with high white blood cell counts, normal karyotypes, and fms-like tyrosine kinase-3 gene (FLT3) internal tandem duplication (ITD) mutations. NPM1 mutations associate inversely with the occurrence of CCAAT/enhancer-binding protein-α (CEBPA) and NRAS mutations. With respect to gene expression profiling, we show that AML cases with an NPM1 mutation cluster in specific subtypes of AML with previously established gene expression signatures, are highly associated with a homeobox gene–specific expression signature, and can be predicted with high accuracy. We demonstrate that patients with intermediate cytogenetic risk AML without FLT3 ITD mutations but with NPM1 mutations have a significantly better overall survival (OS) and event-free survival (EFS) than those without NPM1 mutations. Finally, in multivariable analysis NPM1 mutations express independent favorable prognostic value with regard to OS, EFS, and disease-free survival (DFS).
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease with diverse genetic abnormalities and variable responsiveness to therapy. Cytogenetic analyses and molecular analyses are currently used to risk-stratify AML. For instance, the translocations inv(16), t(8;21), and t(15;17) herald a favorable prognosis, whereas certain other cytogenetic aberrations indicate leukemia with intermediate or high risk of relapse.1-5 Nevertheless, the classification of AML on the basis of karyotyping is still far from satisfactory. In recent years extended molecular analyses have yielded novel molecular markers important for proper diagnostics of AML. The internal tandem duplication (ITD) in the fms-like tyrosine kinase-3 gene (FLT3),6,7 partial tandem duplication (PTD)8,9 of the mixed lineage leukemia gene (MLL), and increased expression of the transcription factor ecotropic virus integration site 1 (EVI1)10 are indicative of poor prognosis. In contrast, mutations in the transcription factor CCAAT/enhancer-binding protein-α (CEBPA) have been associated with a favorable response to therapy.11,12 A recent study showed mutations in exon 12 of the gene encoding nucleophosmin NPM1 in approximately 35% of cases of de novo AML.13 Mutations in NPM1 were found to be mutually exclusive with certain common recurrent chromosomal aberrations and are predominantly seen in AML with normal karyotypes and FLT3 ITD mutations.
NPM1 is predominantly localized in the nucleolus and is thought to function as a molecular chaperone of proteins, facilitating the transport of ribosomal proteins through the nuclear membrane.14-16 Disruption of NPM1, either by chromosomal translocation or by mutation, results in the cytoplasmic dislocation of NPM1. The high frequency of NPM1 mutations in AML with normal karyotypes and the observation that cytoplasmic NPM1 cannot exert its normal functions as binding partner and transporter protein lead to the notion that NPM1 mutation may be an early event in leukemogenesis.
An important role for NPM1 in leukemias and lymphomas has been proposed previously. NPM1 has been found to be part of several fusion proteins that are formed as a result of chromosomal translocation and in which only the NPM1 N-terminal region is conserved. A t(2;5)(p23;q35) chromosomal translocation occurs in approximately 8% of non-Hodgkin lymphomas in children and young adults and results in the chimeric fusion of NPM1 to ALK.17 In rare cases of acute promyelocytic leukemia (APL), characterized by chromosomal translocations that disrupt the gene encoding the retinoic acid receptor (RARA), fusion of NPM1 to RARA was shown.18 A t(3;5)(q25.1;q34) chromosomal translocation, infrequently seen in myelodysplastic syndrome and AML, gives rise to a fusion transcript of NPM1 and MLF1.19
Gene expression profiling is a powerful way to comprehensively classify individuals with AML and to further resolve the heterogeneous nature of AML.20 Using this technique, new prognostically relevant AML subtypes have been identified, while the presence of recurrent chromosomal abnormalities such as inv(16), t(15;17), and t(8;21) as well as other molecular aberrations (eg, C- and N-terminal mutations in CEBPA) could be predicted with high accuracy by unique expression patterns.21-23 In a recent study, novel subtypes of AML have also been defined based on gene expression profiling; however, the common molecular abnormalities in these AML subtypes are largely unknown.21 Because NPM1 is mutated in approximately one third of AML patients, this molecular abnormality may drive the clustering of these AML subtypes. The effect of mutant NPM1 has been studied using gene expression profiling and revealed a distinctive signature for NPM1 mutations.24 Among players in this signature were several homeodomain-containing family members of homeobox (HOX) transcription factors and CD34, both observations being indicative of hematopoietic development.24 However, it is currently not known whether NPM1 mutations are predictable on the basis of a gene expression signature.
Cytoplasmic NPM1 has been positively associated with remission rate13 ; however, the relation of mutant NPM1 with survival outcome parameters remains to be elucidated.
We have studied a well-characterized cohort of 275 cases of de novo AML for the presence of a NPM1 mutations to (1) validate denaturing high-performance liquid chromatography (dHPLC) as a rapid approach to determine NPM1 mutations; (2) investigate the relation of NPM1 mutations with regard to clinical parameters, cytogenetics, and various molecular abnormalities; (3) determine the relation of NPM1 mutations in subtypes of AML, recently identified by gene expression profiling21 ; (4) derive NPM1 mutation–specific and predictive gene expression signatures; and (5) determine the prognostic value of mutated NPM1.
Patients, materials, and methods
Patients and cell samples
Patients had a diagnosis of primary AML confirmed by cytologic examination of blood and bone marrow (median age, 44 years; range, 15-78 years); median bone marrow blast count, 65% (range, 0% [for APL] to 98%); median white blood cell (WBC) count, 32 × 109/L (range, 0.3 × 109/L to 263 × 109/L). All patients had been treated according to the Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON) protocols.25-27 After informed consent, bone marrow aspirates or peripheral blood samples were taken at diagnosis. Blasts and mononuclear cells were purified by Ficoll-Hypaque (Nygaard, Oslo, Norway) centrifugation and cryopreserved. The AML samples contained 80% to 100% blast cells after thawing, regardless of the blast count at diagnosis.
PCR, WAVE, and sequence analyses
RNA isolation and cDNA synthesis were performed as described.21,28 Complementary DNA prepared from 50 ng RNA was used for all polymerase chain reaction (PCR) amplifications. NPM1 mutations in exon 12 were determined by cDNA amplification using the primers NPM1-FOR 5′-CTTCCGGATGACTGACCAAGAG-3′ and primer NPM1-REV 5′-CCTGGACAACATTTATCAAACACG-3′ (25 mM deoxyribonucleoside triphosphate [dNTP], 15 pmol primers, 2 mM MgCl2, Taq polymerase, and 10 × buffer [Invitrogen Life Technologies, Breda, The Netherlands]). Cycling conditions for NPM1 mutation detection were as follows: 1 cycle, 5 minutes at 94°C; 30 cycles, 1 minute at 94°C, 1 minute at 58°C, and 1 minute at 72°C; and 1 cycle, 7 minutes at 72°C. PCR products were subsequently subjected to dHPLC using a Transgenomics (Omaha, NE) WAVE dHPLC system.29 Samples were run at 56°C and 58°C. The exact NPM1 mutant sequence was confirmed for all samples showing an abnormal dHPLC profile. PCR products were purified using the Multiscreen-PCR 96-well system (Millipore, Bedford, MA) followed by direct sequencing with NPM1-REV using an ABI-PRISM3100 genetic analyzer (Applied Biosystems, Foster City, CA).
Gene expression profiling and unsupervised cluster analyses
A total of 285 AML cases were analyzed using Affymetrix HGU133A GeneChips (Affymetrix, Santa Clara, CA).21 Unsupervised cluster analysis on the basis of the gene expression profiles of the 285 cases of AML was performed using the correlation view tool (version 3.6) of OmniViz (Maynard, MA).21 The Pearson correlation values calculated in OmniViz were subsequently imported into the MicroArray Data Explorer (MADEx), which was developed in our laboratory. MADEx was used to visualize the relations between the OmniViz unsupervised clustering results and other parameters, such as clinical and molecular characteristics of the AML patients (Figure 1). MADEx is a database system that stores, mines, and visualizes microarray data in a secure and scalable manner.
Significance analysis of microarrays (SAM)
All supervised analyses were performed using significance analysis of microarrays (SAM; version 1.21).32 A threshold was set for a minimum change in expression of at least 1.5-fold. A false discovery rate (FDR) of 0.01 was used to select the differentially expressed genes.
Prediction analysis of microarrays (PAM)
All supervised class prediction analyses were performed by applying prediction analysis of microarrays (PAM; version 2.0).33 The gene signature was selected based on the smallest prediction error in the training set and was subsequently tested using the test set. The positive predictive value was calculated as follows: true positives/(true positives + false positives).
Statistical analyses of survival
Cytogenetic abnormalities were categorized in 3 cytogenetic groups for statistical analyses. Patients with inv(16)/t(16;16), t(8;21), and t(15;17) abnormalities were considered as being in the favorable-risk category. The unfavorable-risk category was defined by the presence of –5/del(5q), –7del(7q), t(6;9), t(9;22), 3q26 abnormality, or complex karyotype (more than 3 abnormalities). All other patients were classified as intermediate risk. Statistical analyses were performed with Stata Statistical Software, Release 7.0 (Stata, College Station, TX). Actuarial probabilities of overall survival (OS) (with failure defined as death due to any cause) and event-free survival (EFS) (with failure defined as not achieving complete remission [set at day 1], relapse, or death in first complete remission) were estimated by the method of Kaplan and Meier. The Cox proportional hazards model was applied to determine the association of NPM1 mutation with OS, EFS, and disease-free survival (DFS) without and with adjustment for other factors such as cytogenetic risk, age, WBC count, and FLT3 ITD. All tests were 2-sided, and a P of less than .05 was considered statistically significant.
Results
Different NPM1 variant mutations in AML
The presence of NPM1 mutations in 275 cases of primary AML was rapidly and reliably detected by dHPLC WAVE. Nucleotide sequencing was performed on those cases with an abnormal dHPLC profile (Table 1). Each NPM1 mutation variant reveals a specific dHPLC WAVE profile. Thus, each type of NPM1 mutation could be predicted on the basis of a specific dHPLC WAVE profile.
NPM1 mutation frequencies in 275 cases of de novo AML
NPM1 variant . | Nucleotide sequence* . | Protein sequence† . | No. of NPM1 mutants (%) . |
|---|---|---|---|
| WT | GATCTCTG GCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLWQWRKSL | NA |
| Mutant A | GATCTCTGTCTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCLAVEEVSLRK | 72 (26) |
| Mutant B | GATCTCTGCATGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCMAVEEVSLRK | 12 (4) |
| Mutant D | GATCTCTGCCTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCLAVEEVSLRK | 4 (1) |
| Mutant I‡ | GATCTCTGCAGAGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCRAVEEVSLRK | 1 (< 1) |
| Mutant J‡ | GATCTCTGCTTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCLAVEEVSLRK | 1 (< 1) |
| Mutant K‡ | GATCTCTGTATGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCMAVEEVSLRK | 1 (< 1) |
NPM1 variant . | Nucleotide sequence* . | Protein sequence† . | No. of NPM1 mutants (%) . |
|---|---|---|---|
| WT | GATCTCTG GCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLWQWRKSL | NA |
| Mutant A | GATCTCTGTCTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCLAVEEVSLRK | 72 (26) |
| Mutant B | GATCTCTGCATGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCMAVEEVSLRK | 12 (4) |
| Mutant D | GATCTCTGCCTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCLAVEEVSLRK | 4 (1) |
| Mutant I‡ | GATCTCTGCAGAGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCRAVEEVSLRK | 1 (< 1) |
| Mutant J‡ | GATCTCTGCTTGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCLAVEEVSLRK | 1 (< 1) |
| Mutant K‡ | GATCTCTGTATGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG | -DLCMAVEEVSLRK | 1 (< 1) |
Number of NPM1 mutants with undetermined variants, 4 (1%); total number of NPM1 mutants, 95 (35%).
Inserted nucleotides are underlined and italicized.
The C-terminal tryptophane residues (W) are underlined for wild-type (WT) NPM1.
NPM1 mutants I, J, and K are novel variants identified in addition to the 6 known NPM1 variants.13
In addition, 3 novel NPM1 mutant variants were identified (NPM1 mutants I to K [Table 1]). These rare variants have comparable 4 bp insertions, like NPM1 variant mutations A to D,13 resulting in a frame shift and replacement of the 7 C-terminal amino acids of the NPM1 protein by 11 different residues (Table 1).
NPM1 mutations in relation to clinical and molecular features in AML
The NPM1 mutation frequencies of the 275 cases of primary AML with regard to clinical parameters, morphology, cytogenetics, and molecular characteristics are shown in Table 2. NPM1 mutations are significantly less frequently present in patients of younger age (less than 35 years, P < .001). The mean age of patients with AML and NPM1 mutations is 47.3 years (±10.7 years), whereas the mean age of patients with AML and wild-type NPM1 is 39.7 years (±13.3 years). NPM1 mutations are seen in AML French-American-British (FAB) subtypes M1 to M6 but are absent in AML FAB M0. The mutations are relatively frequently found in AML FAB M5 as well as in all 3 cases of AML FAB M6. In AML with recurrent translocations (ie, t(8;21), inv(16), and t(15;17)) no NPM1 mutations were demonstrated. In cases with various other cytogenetic abnormalities, mutations in NPM1 were also rare. As a result, there appears to be a positive correlation between NPM1 mutations and AML with normal karyotypes (P < .001). The analysis of NPM1 mutations reveals interesting relationships with particular common molecular abnormalities. The presence of NPM1 mutations significantly correlates with the presence of FLT3 ITD mutations (P < .001). A correlation between NPM1 mutations and FLT3 TKD mutations is not apparent. AML with NRAS mutations showed a significantly lower percentage of NPM1 mutations (P = .024). Among 5 of 8 cases of AML with NPM1 mutations, KRAS mutations were found. No mutations in NPM1 were found in cases of AML with CEBPA mutations (P = .001).
NPM1 mutation frequencies in relation to clinical parameters, morphology, cytogenetics, and molecular characteristics of the 275 patients with de novo AML
. | No. . | No. of NPM1 mutants (%) . | P . |
|---|---|---|---|
| Sex | .100 | ||
| Male | 135 | 40 (30) | |
| Female | 140 | 55 (39) | |
| Age, y | < .001 | ||
| Younger than 35 | 74 | 11 (15) | |
| 35 to 60 | 169 | 69 (41) | |
| 60 and older | 32 | 15 (47) | |
| WBC count, × 109/L | < .001 | ||
| 20 or below | 113 | 28 (25) | |
| Above 20 | 157 | 66 (42) | |
| ND | 5 | 1 (20) | |
| FAB | |||
| M0 | 6 | 0 (0) | ND |
| M1 | 62 | 21 (34) | > .999 |
| M2 | 63 | 18 (29) | .296 |
| M3 | 17 | 1 (6) | .008 |
| M4 | 49 | 15 (31) | 062 |
| M5 | 65 | 32 (49) | .007 |
| M6 | 3 | 3 (100) | ND |
| ND | 10 | 5 (50) | |
| Cytogenic abnormalities* | |||
| t(15;17) | 16 | 0 (0) | .002 |
| t(8;21) | 21 | 0 (0) | < .001 |
| inv(16)/t(16;16) | 17 | 0 (0) | .001 |
| +8 | 24 | 5 (21) | ND |
| +11 | 5 | 0 (0) | ND |
| +21 | 1 | 1 (100) | ND |
| -5 | 2 | 0 (0) | ND |
| -5(q) | 1 | 0 (0) | ND |
| -7 | 13 | 0 (0) | .005 |
| -7(q) | 7 | 0 (0) | ND |
| 3q | 5 | 1 (20) | ND |
| t(6;9) | 4 | 0 (0) | ND |
| t(9;22) | 2 | 1 (50) | ND |
| t(11q23) | 16 | 1 (6) | 014 |
| Complex; more than 3 abnormalities | 11 | 0 (0) | .018 |
| Other | 55 | 10 (18) | .004 |
| Normal | 116 | 74 (64) | < .001 |
| ND | 10 | 6 (60) | |
| Molecular abnormalities | |||
| FLT3 ITD | 78 | 47 (60) | < .001 |
| FLT3 TKD | 32 | 14 (44) | .243 |
| NRAS | 25 | 3 (12) | .024 |
| KRAS | 8 | 5 (63) | .130 |
| CEBPA | 17 | 0 (0) | .001 |
. | No. . | No. of NPM1 mutants (%) . | P . |
|---|---|---|---|
| Sex | .100 | ||
| Male | 135 | 40 (30) | |
| Female | 140 | 55 (39) | |
| Age, y | < .001 | ||
| Younger than 35 | 74 | 11 (15) | |
| 35 to 60 | 169 | 69 (41) | |
| 60 and older | 32 | 15 (47) | |
| WBC count, × 109/L | < .001 | ||
| 20 or below | 113 | 28 (25) | |
| Above 20 | 157 | 66 (42) | |
| ND | 5 | 1 (20) | |
| FAB | |||
| M0 | 6 | 0 (0) | ND |
| M1 | 62 | 21 (34) | > .999 |
| M2 | 63 | 18 (29) | .296 |
| M3 | 17 | 1 (6) | .008 |
| M4 | 49 | 15 (31) | 062 |
| M5 | 65 | 32 (49) | .007 |
| M6 | 3 | 3 (100) | ND |
| ND | 10 | 5 (50) | |
| Cytogenic abnormalities* | |||
| t(15;17) | 16 | 0 (0) | .002 |
| t(8;21) | 21 | 0 (0) | < .001 |
| inv(16)/t(16;16) | 17 | 0 (0) | .001 |
| +8 | 24 | 5 (21) | ND |
| +11 | 5 | 0 (0) | ND |
| +21 | 1 | 1 (100) | ND |
| -5 | 2 | 0 (0) | ND |
| -5(q) | 1 | 0 (0) | ND |
| -7 | 13 | 0 (0) | .005 |
| -7(q) | 7 | 0 (0) | ND |
| 3q | 5 | 1 (20) | ND |
| t(6;9) | 4 | 0 (0) | ND |
| t(9;22) | 2 | 1 (50) | ND |
| t(11q23) | 16 | 1 (6) | 014 |
| Complex; more than 3 abnormalities | 11 | 0 (0) | .018 |
| Other | 55 | 10 (18) | .004 |
| Normal | 116 | 74 (64) | < .001 |
| ND | 10 | 6 (60) | |
| Molecular abnormalities | |||
| FLT3 ITD | 78 | 47 (60) | < .001 |
| FLT3 TKD | 32 | 14 (44) | .243 |
| NRAS | 25 | 3 (12) | .024 |
| KRAS | 8 | 5 (63) | .130 |
| CEBPA | 17 | 0 (0) | .001 |
P values were calculated using the Fisher exact test (2-tailed; ND indicates not determined).
All patients with a specific abnormality were considered irrespective of the presence of additional abnormalities.
NPM1 mutations occur within specific AML subtypes defined by gene expression profiling
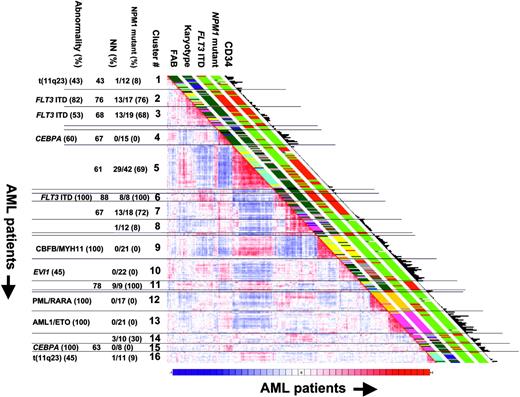
Of the cohort of 285 cases of primary AML that had previously been profiled using the Affymetrix HGU133A GeneChip21 and for which 16 distinct expression signatures had been defined following unsupervised cluster analyses, we have now examined 275 cases for the presence of NPM1 mutations. Among these pre-established signatures, the AML cases with NPM1 mutations aggregate within particular clusters (Figure 1). Most AML cases in cluster nos. 2, 3, 5, and 7 carry NPM1 mutations. In fact, all cases of AML of cluster nos. 6 (100% FLT3 ITD) and 11 (78% normal karyotypes) carried mutations in NPM1. Although cluster nos. 7 and 8 have comparable expression profiles (Figure 1), 13 of the 18 AML cases in cluster no. 7 (72%) and only 1 of 12 cases of cluster no. 8 (8%) reveal NPM1 mutations. The clusters merely consisting of AML with inv(16) (no. 9), t(15;17) (no. 12), or t(8;21) (no. 13) as well as the clusters predominantly containing cases with CEBPA mutations (nos. 4 and 15) all lack NPM1 mutations. The subset of AML patients in cluster no. 10, with adverse prognosis and an expression profile comparable to CD34+ cells,21 did not present with NPM1 mutations.
Falini and colleagues13 had shown a negative correlation between the presence of NPM1 mutations and CD34 expression levels. In fact, by plotting the CD34 mRNA expression levels of the 285 AML cases, as determined by the GeneChip analyses, along with the NPM1 mutation status (Figure 1), a distinguishable association of CD34 mRNA expression and NPM1 mutation is apparent (ie, CD34 mRNA expression is low or absent in cases of AML with NPM1 mutations, while CD34 mRNA levels are high in AML cases without NPM1 mutations).
HOX gene–specific gene expression signature of NPM1 mutant cases
To identify genes with significant differential expression between primary AML samples with NPM1 mutations (n = 95) and samples without NPM1 mutations (n = 180) the SAM algorithm was used.32 A fold change threshold of 1.5 for up-regulation of gene expression and 0.667 for down-regulation of gene expression was applied. With an FDR of 0.01, 569 probe sets representing 440 unique genes were identified as being significantly differentially expressed (Figure S1 and Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The identity of the top 50 genes, in some cases represented by multiple probe sets, is depicted in Table 3.
NPM1 mutation-associated gene expression in 275 patients with de novo AML
Probe set ID . | Gene symbol . | Fold change . |
|---|---|---|
| Up-regulated in AML with mutant NPM1 | ||
| 213844_at | HOXA5 | 4.2 |
| 205366_s_at | HOXB6 | 2.6 |
| 208414_s_at | HOXB3 | 1.8 |
| 204082_at | PBX3 | 2.8 |
| 205600_x_at | HOXB5 | 2.1 |
| 206289_at | HOXA4 | 2.1 |
| 205453_at | HOXB2 | 2.1 |
| 213150_at | HOXA10 | 2.6 |
| 204069_at | MEIS1 | 2.6 |
| 209905_at | HOXA9 | 2.8 |
| 201664_at | SMC4L1 | 2.1 |
| 209439_s_at | PHKA2 | 1.6 |
| 206847_s_at | HOXA7 | 1.5 |
| 63825_at | ABHD2 | 1.6 |
| 219304_s_at | PDGFD | 1.9 |
| 212820_at | RC3 | 2.4 |
| 213110_s_at | COL4A5 | 2.9 |
| 207111_at | EMR1 | 2.1 |
| 208557_at | HOXA6 | 1.6 |
| 203471_s_at | PLEK | 1.6 |
| 203680_at | PRKAR2B | 2.3 |
| 202729_s_at | LTBP1 | 3.6 |
| 210145_at | PLA2G4A | 1.6 |
| 220162_s_at | CARD9 | 1.5 |
| 206298_at | ARHGAP22 | 2.0 |
| 219602_s_at | FAM38B | 1.9 |
| Down-regulated in AML with mutant NPM1 | ||
| 209543_s_at | CD34 | -5.4 |
| 206896_s_at | GNG7 | -1.7 |
| 209583_s_at | MOX2 | -3.1 |
| 200953_s_at | CCND2 | -2.1 |
| 221004_s_at | ITM2C | -3.0 |
| 205330_at | MN1 | -4.3 |
| 200602_at | APP | -2.6 |
| 200665_s_at | SPARC | -3.8 |
| 201015_s_at | JUP | -3.1 |
| 218899_s_at | BAALC | -4.3 |
| 209679_s_at | LOC57228 | -1.9 |
| 219694_at | FLJ11127 | -2.1 |
| 206042_x_at | SNRPN | -2.5 |
| 211535_s_at | FGFR1 | -2.6 |
| 214582_at | PDE3B | -1.8 |
| 221523_s_at | RRAGD | -1.8 |
| 213618_at | CENTD1 | -1.7 |
| 218589_at | P2RY5 | -3.3 |
| 202016_at | MEST | -2.9 |
| 208116_s_at | MAN1A1 | -3.1 |
| 205240_at | GPSM2 | -1.9 |
| 202747_s_at | ITM2A | -4.1 |
| 206622_at | TRH | -9.1 |
| 206726_at | PGDS | -4.7 |
Probe set ID . | Gene symbol . | Fold change . |
|---|---|---|
| Up-regulated in AML with mutant NPM1 | ||
| 213844_at | HOXA5 | 4.2 |
| 205366_s_at | HOXB6 | 2.6 |
| 208414_s_at | HOXB3 | 1.8 |
| 204082_at | PBX3 | 2.8 |
| 205600_x_at | HOXB5 | 2.1 |
| 206289_at | HOXA4 | 2.1 |
| 205453_at | HOXB2 | 2.1 |
| 213150_at | HOXA10 | 2.6 |
| 204069_at | MEIS1 | 2.6 |
| 209905_at | HOXA9 | 2.8 |
| 201664_at | SMC4L1 | 2.1 |
| 209439_s_at | PHKA2 | 1.6 |
| 206847_s_at | HOXA7 | 1.5 |
| 63825_at | ABHD2 | 1.6 |
| 219304_s_at | PDGFD | 1.9 |
| 212820_at | RC3 | 2.4 |
| 213110_s_at | COL4A5 | 2.9 |
| 207111_at | EMR1 | 2.1 |
| 208557_at | HOXA6 | 1.6 |
| 203471_s_at | PLEK | 1.6 |
| 203680_at | PRKAR2B | 2.3 |
| 202729_s_at | LTBP1 | 3.6 |
| 210145_at | PLA2G4A | 1.6 |
| 220162_s_at | CARD9 | 1.5 |
| 206298_at | ARHGAP22 | 2.0 |
| 219602_s_at | FAM38B | 1.9 |
| Down-regulated in AML with mutant NPM1 | ||
| 209543_s_at | CD34 | -5.4 |
| 206896_s_at | GNG7 | -1.7 |
| 209583_s_at | MOX2 | -3.1 |
| 200953_s_at | CCND2 | -2.1 |
| 221004_s_at | ITM2C | -3.0 |
| 205330_at | MN1 | -4.3 |
| 200602_at | APP | -2.6 |
| 200665_s_at | SPARC | -3.8 |
| 201015_s_at | JUP | -3.1 |
| 218899_s_at | BAALC | -4.3 |
| 209679_s_at | LOC57228 | -1.9 |
| 219694_at | FLJ11127 | -2.1 |
| 206042_x_at | SNRPN | -2.5 |
| 211535_s_at | FGFR1 | -2.6 |
| 214582_at | PDE3B | -1.8 |
| 221523_s_at | RRAGD | -1.8 |
| 213618_at | CENTD1 | -1.7 |
| 218589_at | P2RY5 | -3.3 |
| 202016_at | MEST | -2.9 |
| 208116_s_at | MAN1A1 | -3.1 |
| 205240_at | GPSM2 | -1.9 |
| 202747_s_at | ITM2A | -4.1 |
| 206622_at | TRH | -9.1 |
| 206726_at | PGDS | -4.7 |
The top 50 unique most discriminating genes and fold change in expression with regard to NPM1 mutation as determined by SAM (up-regulated: increased expression in NPM1 mutant cases; down-regulated: decreased expression in NPM1 mutant cases).
A dominant homeobox (HOX) gene–specific signature is strongly associated with AML carrying an NPM1 mutation. Moreover, the expression of members of the HOXA and HOXB gene families, but also the HOX gene–related three–amino acid loop extension (TALE) genes, PBX3 and MEIS1, is increased. In contrast, the CD34 gene is the strongest significantly down-regulated gene with regard to NPM1 mutation in the AML patient cohort.
NPM1 mutant cases are predicted with high accuracy based on their gene expression signature
NPM1 mutation prediction analyses were performed using the PAM algorithm.33 All 275 primary AML samples were randomly assigned to a training set, consisting of 122 samples without NPM1 mutations and 62 samples with NPM1 mutations, and a validation series, consisting of 58 samples lacking the NPM1 mutation and 33 samples with mutations in NPM1. Cross-validation to predict the mutation status of NPM1 on the training set resulted in 100% correct predictions of presence of mutation (sensitivity) and 80.3% correct predictions of absence of mutation (specificity) (Table 4). Prediction of an independent validation set also resulted in 100% correct prediction of presence of mutation and 82.7% correct prediction of absence of mutation. The positive predictive value in this cohort is 72% in the training set and 74% overall. As expected, the genes included in the PAM gene signature were among the most significant differentially expressed genes as determined by SAM, thereby validating both algorithms. Of note, NPM1 mRNA expression did not correlate with mutation status (Figure 2). The NPM1 mutant AML cases have a distinct signature with regard to the 18 selected genes (Figure 2) and are therefore predicted with high accuracy (Table 4). However, these 18 genes seem to be expressed at comparable levels in a subset of AML cases with wild-type NPM1 (Figure 2).
NPM1 mutation prediction by using PAM
. | Predicted genotype . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | 10-fold CV error* . | . | Error validation set* . | . | |||
| Genotype . | WT . | Mutant . | WT . | Mutant . | |||
| WT NPM1 | 98 | 24 | 48 | 10 | |||
| Mutant NPM1 | 0 | 62 | 0 | 33 | |||
. | Predicted genotype . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | 10-fold CV error* . | . | Error validation set* . | . | |||
| Genotype . | WT . | Mutant . | WT . | Mutant . | |||
| WT NPM1 | 98 | 24 | 48 | 10 | |||
| Mutant NPM1 | 0 | 62 | 0 | 33 | |||
Most optimal result for NPM1 mutation prediction using a cohort of 275 cases of AML divided in a training and test set.21 The number of probe sets used in this prediction was 22, which represents 18 unique genes. For the 10-fold CV error set, n = 184; for the error validation set, n = 91.
Ten-fold cross-validation (CV) prediction error on training set (n = 184); prediction error on validation set (n = 91). In 10-fold cross-validation, the model is fitted on 90% of the samples and the class of the remaining 10% is predicted. This procedure is repeated 10 times, with each part playing the role of the test samples and the error of all 10 parts added together to compute the overall error. The error within the validation set reflects the number of samples wrongfully predicted in this set.
NPM1 mutation is an independent favorable prognostic marker
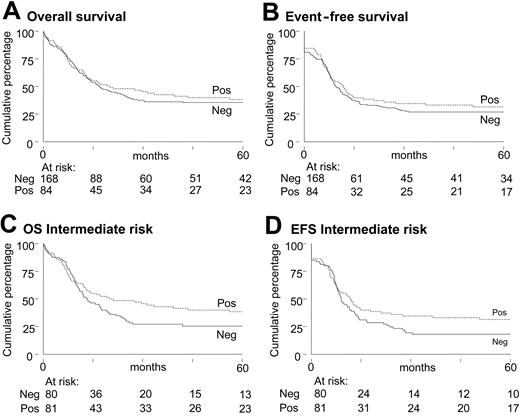
To investigate the prognostic value of NPM1 mutation, 252 AML patients with long-term follow-up following therapy completion were included for survival analysis. The EFS, OS, and probability of relapse at 60 months for the AML patients with or without NPM1 mutations were similar (Figure 3A-B). Likewise, among the subgroup with cytogenetics of intermediate prognostic risk, EFS, OS, and probability of relapse at 60 months were not different (Figures 3C-D), although there appears a trend for more favorable outcome for patients with AML with NPM1 mutations. Because NPM1 mutations are significantly associated with both normal karyotype and presence of a FLT3 ITD (Table 2), we investigated the prognostic value of NPM1 mutations within the intermediate cytogenetic risk group in relation to FLT3 ITD status. Patients in the intermediate cytogenetic risk group without FLT3 ITD mutations but with NPM1 mutations have a significantly better OS and EFS than those without NPM1 mutations (P = .05) (Figure 4A-C). In intermediate cytogenetic risk AML with FLT3 ITD mutations, NPM1 mutations do not significantly distinguish prognosis (Figure 4B,D).
NPM1 mutations are asynchronously associated with particular karyotypes and molecular abnormalities that might express additional positive or negative prognostic value and therefore might obscure the prognostic significance of NPM1 mutations (Table 2; Figure 1). Therefore we investigated the prognostic value of NPM1 mutations in both univariable and multivariable analyses. Univariable and multivariable Cox regression analyses were applied to assess the prognostic significance of NPM1 mutation, cytogenetic risk class, WBC count below or above 20 × 109/L, age, and FLT3 ITD mutation for OS, DFS, and EFS (Table 5). In univariable analyses NPM1 mutation showed no statistical significance with respect to the end points. However, in multivariable analyses mutated NPM1 was significantly associated with a much lower hazards ratio (HR), which was statistically significant for all end points (EFS: HR = 0.59, P = .005; DFS: HR = 0.52, P = .003; and OS: HR = 0.49, P < .001). Mutant NPM1 appeared as an independent prognostic marker in addition to cytogenetic risk, age, and FLT3 ITD mutations.
Univariable and multivariable analyses of NPM1 mutation as prognostic factor for EFS, DFS, and OS in AML
. | EFS . | . | DFS . | . | OS . | . | |||
|---|---|---|---|---|---|---|---|---|---|
. | HR (95% CI) . | P* . | HR (95% CI) . | P* . | HR (95% CI) . | P* . | |||
| Univariable | |||||||||
| Intermediate† | 2.06 (1.33-3.20) | .001* | 2.82 (1.63-4.87) | <.001* | 2.67 (1.62-4.41) | <.001* | |||
| Poor† | 3.88 (2.29-6.56) | <.001* | 5.09 (2.61-9.91) | <.001* | 4.44 (2.49-7.93) | <.001* | |||
| WBC count‡ | 1.14 (0.84-1.53) | .39 | 1.18 (0.84-1.68) | .34 | 1.19 (0.87-1.62) | .29 | |||
| Age, decades | 1.05 (0.94-1.19) | .38 | 1.06 (0.92-1.22) | .41 | 1.17 (1.03-1.32) | .014* | |||
| FLT3ITD§ | 1.76 (1.28-2.41) | .001* | 1.71 (1.17-2.50) | .006* | 1.89 (1.36-2.62) | <.001* | |||
| NPM1 mutation∥ | 0.88 (0.64-1.21) | .43 | 0.93 (0.64-1.34) | .69 | 0.90 (0.65-1.27) | .57 | |||
| Multivariable | |||||||||
| Intermediate† | 2.18 (1.36-3.50) | .001* | 3.11 (1.74-5.55) | <.001* | 2.87 (1.69-4.86) | <.001* | |||
| Poor† | 3.96 (2.34-6.74) | <.001* | 5.73 (2.93-11.22) | <.001* | 4.68 (2.60-8.43) | <.001* | |||
| WBC count‡ | 1.06 (0.78-1.44) | .72 | 1.10 (0.77-1.58) | .60 | 1.18 (0.85-1.63) | .32 | |||
| Age, decades | 1.07 (0.95-1.21) | .27 | 1.10 (0.95-1.27) | .21 | 1.19 (1.05-1.35) | .008* | |||
| FLT3ITD§ | 1.96 (1.39-2.76) | <.001* | 2.00 (1.31-3.06) | .001* | 2.13 (1.49-3.05) | <.001* | |||
| NPM1 mutation∥ | 0.59 (0.41-0.85) | .005* | 0.52 (0.34-0.80) | .003* | 0.49 (0.33-0.72) | <.001* | |||
. | EFS . | . | DFS . | . | OS . | . | |||
|---|---|---|---|---|---|---|---|---|---|
. | HR (95% CI) . | P* . | HR (95% CI) . | P* . | HR (95% CI) . | P* . | |||
| Univariable | |||||||||
| Intermediate† | 2.06 (1.33-3.20) | .001* | 2.82 (1.63-4.87) | <.001* | 2.67 (1.62-4.41) | <.001* | |||
| Poor† | 3.88 (2.29-6.56) | <.001* | 5.09 (2.61-9.91) | <.001* | 4.44 (2.49-7.93) | <.001* | |||
| WBC count‡ | 1.14 (0.84-1.53) | .39 | 1.18 (0.84-1.68) | .34 | 1.19 (0.87-1.62) | .29 | |||
| Age, decades | 1.05 (0.94-1.19) | .38 | 1.06 (0.92-1.22) | .41 | 1.17 (1.03-1.32) | .014* | |||
| FLT3ITD§ | 1.76 (1.28-2.41) | .001* | 1.71 (1.17-2.50) | .006* | 1.89 (1.36-2.62) | <.001* | |||
| NPM1 mutation∥ | 0.88 (0.64-1.21) | .43 | 0.93 (0.64-1.34) | .69 | 0.90 (0.65-1.27) | .57 | |||
| Multivariable | |||||||||
| Intermediate† | 2.18 (1.36-3.50) | .001* | 3.11 (1.74-5.55) | <.001* | 2.87 (1.69-4.86) | <.001* | |||
| Poor† | 3.96 (2.34-6.74) | <.001* | 5.73 (2.93-11.22) | <.001* | 4.68 (2.60-8.43) | <.001* | |||
| WBC count‡ | 1.06 (0.78-1.44) | .72 | 1.10 (0.77-1.58) | .60 | 1.18 (0.85-1.63) | .32 | |||
| Age, decades | 1.07 (0.95-1.21) | .27 | 1.10 (0.95-1.27) | .21 | 1.19 (1.05-1.35) | .008* | |||
| FLT3ITD§ | 1.96 (1.39-2.76) | <.001* | 2.00 (1.31-3.06) | .001* | 2.13 (1.49-3.05) | <.001* | |||
| NPM1 mutation∥ | 0.59 (0.41-0.85) | .005* | 0.52 (0.34-0.80) | .003* | 0.49 (0.33-0.72) | <.001* | |||
CI indicates confidence interval.
P values < .05.
Cytogenetic risk versus cytogenetic good risk.
More than 20 × 109/L versus less than 20 × 109/L.
FLT3ITD versus no FLT3ITD.
NPM1 mutation versus no NPM1 mutation.
OmniViz correlation view of 285 AML patients. The correlation view displays pairwise correlations between AML patients. The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors indicating higher positive (red) or negative (blue) correlations. The scale bar indicates maximum positive correlation (red) toward maximum negative correlation (blue). The 16 clusters identified in the cohort of 285 AML patients using 2856 probe sets on the basis of the correlation view are indicated (1 to 16).21 Clinical and molecular data are depicted in the columns along the original diagonal of the correlation view.21 FAB classification and karyotype based on cytogenetics are depicted in the first 2 columns (FAB M0, red; M1, green; M2, purple; M3, orange; M4, yellow; M5, blue; M6 gray; and karyotype: normal, green; inv(16), yellow; t(8;21), purple; t(15;17), orange; 11q23 abnormalities, blue; 7(q) abnormalities, red; +8, pink; complex, black; other, gray). FLT3 ITD and NPM1 mutations are depicted in the same set of columns (red bar, positive; green bar, negative). The expression levels of CD34 (probe set: 209543_s_at) in the 285 AML patients are plotted in the last column (bars are proportional to the level of expression). The percentages of the most common (more than 40%) abnormalities, NPM1 mutations, as well as normal karyotypes (NN) for each cluster are indicated.
OmniViz correlation view of 285 AML patients. The correlation view displays pairwise correlations between AML patients. The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors indicating higher positive (red) or negative (blue) correlations. The scale bar indicates maximum positive correlation (red) toward maximum negative correlation (blue). The 16 clusters identified in the cohort of 285 AML patients using 2856 probe sets on the basis of the correlation view are indicated (1 to 16).21 Clinical and molecular data are depicted in the columns along the original diagonal of the correlation view.21 FAB classification and karyotype based on cytogenetics are depicted in the first 2 columns (FAB M0, red; M1, green; M2, purple; M3, orange; M4, yellow; M5, blue; M6 gray; and karyotype: normal, green; inv(16), yellow; t(8;21), purple; t(15;17), orange; 11q23 abnormalities, blue; 7(q) abnormalities, red; +8, pink; complex, black; other, gray). FLT3 ITD and NPM1 mutations are depicted in the same set of columns (red bar, positive; green bar, negative). The expression levels of CD34 (probe set: 209543_s_at) in the 285 AML patients are plotted in the last column (bars are proportional to the level of expression). The percentages of the most common (more than 40%) abnormalities, NPM1 mutations, as well as normal karyotypes (NN) for each cluster are indicated.
Discussion
Recently, we established a comprehensive classification of AML based on gene expression profiling.21 In this classification several clusters of AML signatures correlated with distinct (cyto)genetic abnormalities, such as t(8;21), t(15;17), inv(16), and C- and N-terminal mutations in CEBPA. However, the common underlying molecular abnormality for the other subtypes of AML was unknown. In the present study we show that 2 clusters consist entirely of AML cases with NPM1 mutations (ie, cluster nos. 6 and 11), whereas cluster nos. 2, 3, 5, and 7 predominantly include patients with NPM1 mutations. Thus, we identify mutant NPM1 as a common molecular abnormality in these subtypes of AML.
Falini and colleagues13 have shown that mutations in NPM1 are present in 35% of patients with AML. In this study, we confirm that NPM1 mutations are frequently present in AML (ie, in 35% of all cases). NPM1 mutant is less frequently represented in patients with AML of age younger than 35 years. This seems consistent with the tendency of the NPM1 mutation to be more frequently present in older children with AML.34 Furthermore, NPM1 mutations are significantly associated with AML with high WBC count.
The most predictive molecular signature with regard to NPM1 mutation in AML assessed with 22 probe sets representing 18 unique genes. The levels of expression of the probe sets in the 275 cases of AML are depicted (scale bar indicates an increase [red] or a decrease [green] in the level of expression of at least 4 relative to the geometric mean).21 The 3 probe sets representing the NPM1 gene are also depicted.
The most predictive molecular signature with regard to NPM1 mutation in AML assessed with 22 probe sets representing 18 unique genes. The levels of expression of the probe sets in the 275 cases of AML are depicted (scale bar indicates an increase [red] or a decrease [green] in the level of expression of at least 4 relative to the geometric mean).21 The 3 probe sets representing the NPM1 gene are also depicted.
Survival analyses of AML patients with and without NPM1 mutations. Kaplan-Meier estimates of OS (A) and EFS (B) among patients with AML and OS (C) and EFS (D) among patients with AML with intermediate-risk karyotypes.
Survival analyses of AML patients with and without NPM1 mutations. Kaplan-Meier estimates of OS (A) and EFS (B) among patients with AML and OS (C) and EFS (D) among patients with AML with intermediate-risk karyotypes.
We detected the various mutation variants of NPM1 at similar frequencies as was described recently13 and also identified 3 novel mutant variants. These novel variants carry, like the other NPM1 mutant variants in our study, an insertion of 4 bp, resulting in a protein with an altered C-terminus (Table 1). Falini and colleagues13 suggested that the disruption of 1 of the 2 C-terminal tryptophan residues and the last 5 residues (ie, VSLRK) are important for NPM1 mutant function. Our study suggests that the final 9 amino acids (ie, AVEEVSLRK) may in general be required for mutant NPM1 function.
NPM1 mutations often coincide with mutations in FLT3, particularly with the ITD-type mutations. Our data may suggest an association of mutations in KRAS and NPM1, but the study has limited power because of the small number of mutant KRAS AMLs. In contrast, mutations in NRAS were not associated with mutant NPM1, because NRAS mutations are found in AML with inv(16),21,30 a subclass of AML that lacks NPM1 mutations. In addition, we did not find NPM1 mutations in clusters of AML patients, previously identified by gene expression profiling,21 characterized by C- and N-terminal mutations in CEBPA. These observations might perhaps suggest that constitutive active FLT3 or K-RAS provides the proliferative signal, whereas mutant NPM1 might serve to impair differentiation in the multistep pathogenesis model of AML.35
Notably, the discriminative genes identified by SAM and PAM revealed a strong HOX and TALE gene–specific signature associated with AML cases with mutant NPM1, as was published recently.24 Thus, although CD34 has generally been used as marker for immature hematopoietic progenitor cells, the NPM1 mutant CD34– cells display a molecular signature similar to that of hematopoietic stem cells (HSCs). Recent studies have shown that CD34– HSCs exist as well.36,37 These CD34– cells also possess HSC-specific characteristics, including the ability for hematopoietic engraftment.36,37 This brings up the question as to whether the NPM1 mutant AML cells in fact represent a more primitive population of HSCs with a HOX gene signature.
The sharp distinction between CD34 positivity and NPM1 mutations in AML cases based on the HOX gene signature is notable because 22 of the 39 HOX genes are expressed in human CD34+ cells, whereas during normal differentiation HOX gene expression declines.38 Extensive studies have demonstrated for a number of HOX genes that sustained overexpression in murine bone marrow results in perturbations in the stem cell pools, and coexpression of certain HOX gene family members with their protein binding partner, such as MEIS1, results in leukemia.38 Thus, in AML with NPM1 mutations the hematopoietic progenitor cells may have arrested at a differentiation stage with endogenous coexpression of the HOX genes (ie, HOXA5, -A9, -A10, -B2, -B3, -B5, and -B6) and their TALE partner genes (ie, MEIS1 and PBX3) or as a result of the increased expression of these genes.
SAM and PAM analyses were highly concordant for the genes identified with differential expression between AML with mutant and AML with wild-type NPM1, where, in both cases, CD34 was the most discriminating gene, down-regulated in NPM1 mutants. Patients harboring an NPM1 mutation can be predicted with high accuracy; however, a subset of patients with wild-type NPM1 is wrongly predicted. Interestingly, within this subgroup the percentage of patients with 11q23 abnormalities is significantly increased. In fact, this may not be surprising because MLL has been implicated as a HOX gene regulator39 and selective expression of HOX genes in ALL cases with mutant MLL has been shown.40
By univariate analyses we show that there is a tendency that the presence of an NPM1 mutation, and concomitant low CD34 mRNA expression, is a favorable marker for clinical outcome (Figure 3). This is in agreement with CD34 expression as an indicator for poor response to induction therapy.41-43 In addition, we demonstrate by multivariate analyses that NPM1 mutations are a strong independent favorable predictive marker for EFS, DFS, and OS in AML. The finding that the effect of NPM1 mutation is much more pronounced in multivariable than in univariable analyses can be explained by the strong correlation between NPM1 and FLT3 ITD mutations and additionally by the association with high WBC count and age. NPM1 mutations are more frequently present in AML patients with FLT3 ITD, high WBC count, and older age. These factors are unfavorable determinants of prognosis,44 while NPM1 mutation seems to express a favorable prognostic impact. This implies that in univariable analysis the positive effect of NPM1 mutations, as measured by the method of Kaplan and Meier or by the HR, is less pronounced, because the adverse effects of FLT3 ITD, high WBC count, and age might mask this effect. In fact, NPM1 mutations distinguish a favorable subgroup among intermediate cytogenetic risk FLT3 ITD–negative AML that shows comparatively better OS and EFS (Figure 4). In addition, in the multivariable analyses the effect of NPM1 mutations is more pronounced than is apparent in the univariable analyses. This is not only true for NPM1 mutations but also for the independent prognostic effect of FLT3 ITD mutations, age, and cytogenetic risk (Table 5).
Survival analyses of intermediate cytogenetic risk AML patients with and without FLT3 ITD and/or NPM1 mutations. Kaplan-Meier estimates of OS (panels A and B) and EFS (panels C and D) for patients with intermediate-risk AML and FLT3 ITD mutations (FLT3 ITD positive [panels B and D]) versus those with intermediate-risk AML without FLT3 ITD mutations (FLT3 ITD negative [panels A and C]).
Survival analyses of intermediate cytogenetic risk AML patients with and without FLT3 ITD and/or NPM1 mutations. Kaplan-Meier estimates of OS (panels A and B) and EFS (panels C and D) for patients with intermediate-risk AML and FLT3 ITD mutations (FLT3 ITD positive [panels B and D]) versus those with intermediate-risk AML without FLT3 ITD mutations (FLT3 ITD negative [panels A and C]).
The data presented here demonstrate that the frequent C-terminal insertion mutations in NPM1 correlate with favorable outcome for patients with AML. Because NPM1 mutations are predominantly found in patients with standard-risk AML, determination of the NPM1 mutation status, in combination with other prognostically relevant markers, will be useful for further risk stratification of adult patients with de novo AML.
Prepublished online as Blood First Edition Paper, August 18, 2005; DOI 10.1182/blood-2005-05-2168.
Supported by grants from the Dutch Cancer Society (Koningin Wilhelmina Fonds) and the Erasmus University Medical Center (Revolving Fund).
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to our colleagues from the bone marrow transplantation group and molecular diagnostics laboratory of the Department of Hematology at the Erasmus Medical Center for storage of the samples and molecular analyses, respectively.

![Figure 2. The most predictive molecular signature with regard to NPM1 mutation in AML assessed with 22 probe sets representing 18 unique genes. The levels of expression of the probe sets in the 275 cases of AML are depicted (scale bar indicates an increase [red] or a decrease [green] in the level of expression of at least 4 relative to the geometric mean).21 The 3 probe sets representing the NPM1 gene are also depicted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/12/10.1182_blood-2005-05-2168/2/m_zh80240587560002.jpeg?Expires=1765888310&Signature=PS6E~QwQwZhdC3hLDzt5abNrDubNJA7dC3DjTeNL1e-g-S6fOEMf2oohzdQ9wXL3np8qd8pma3TbTE~Ou0OKu-hN5jVPMleUwzuAf1CC-EmyGcVrTDUtQ0XH0Di40cmSgbHVF6FxVQX0Ld1qHtnp8hWY1ke5aNHdYVABYuwYAvoW4jC~GxDLiPt4HWxVRgFL5FtqV1k5i~fOh9ds-nPX0fov56HoTikjjQnkyRx5KKAKEFlwKuWeC~XqpEIOsiw7Ov4LcC8GqeHdui-19B1KK4Yra038Ul2mdhqg1b0buXCbtexSwgs9W9CDezx9TR3Qk0OgoHj9p4TG4U6AxMWaMg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Survival analyses of intermediate cytogenetic risk AML patients with and without FLT3 ITD and/or NPM1 mutations. Kaplan-Meier estimates of OS (panels A and B) and EFS (panels C and D) for patients with intermediate-risk AML and FLT3 ITD mutations (FLT3 ITD positive [panels B and D]) versus those with intermediate-risk AML without FLT3 ITD mutations (FLT3 ITD negative [panels A and C]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/12/10.1182_blood-2005-05-2168/2/m_zh80240587560004.jpeg?Expires=1765888310&Signature=2cHyESHciTzmeBBccihFN4mG1IjXCaU4FfQ6semwXCmJFR4AcbkYLKYQUxI11Hqh-ogNoNudQYQcbT1cP7Dm31wVB1KMMHuhyJmHx4NnDdeZAxDSpqCzxaEQPD7YGLMVd-RWxFwLo4h78H82XQXTnIJLpPTUIqBEJeu4Bh6~1POZG3WvmpvWbarwWWcGcwEVNWQfYFIuIRehOpXYxQddbuFC-doHo0TmGgTB-2x3G3bF8lTlnMLObp1xJuZ2Mihe29UJMWGgCIzchZ1u4Yi4FF8g3coVEKl8WQhSZHEl6cdWvw9Up2uwY5C-VmXJcFOydi6iMHj11D8sVEMKoUudQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal