Abstract
We elucidate the cellular and molecular kinetics of the stepwise differentiation of human embryonic stem cells (hESCs) to primitive and definitive erythromyelopoiesis from human embryoid bodies (hEBs) in serum-free clonogenic assays. Hematopoiesis initiates from CD45 hEB cells with emergence of semiadherent mesodermal-hematoendothelial (MHE) colonies that can generate endothelium and form organized, yolk sac–like structures that secondarily generate multipotent primitive hematopoietic stem progenitor cells (HSPCs), erythroblasts, and CD13+CD45+ macrophages. A first wave of hematopoiesis follows MHE colony emergence and is predominated by primitive erythropoiesis characterized by a brilliant red hemoglobinization, CD71/CD325a (glycophorin A) expression, and exclusively embryonic/fetal hemoglobin expression. A second wave of definitive-type erythroid burst-forming units (BFU-e's), erythroid colony-forming units (CFU-e's), granulocyte-macrophage colony-forming cells (GM-CFCs), and multilineage CFCs follows next from hEB progenitors. These stages of hematopoiesis proceed spontaneously from hEB-derived cells without requirement for supplemental growth factors during hEB differentiation. Gene expression analysis of differentiating hEBs revealed that initiation of hematopoiesis correlated with increased levels of SCL/TAL1, GATA1, GATA2, CD34, CD31, and the homeobox gene-regulating factor CDX4 These data indicate that hematopoietic differentiation of hESCs models the earliest events of embryonic and definitive hematopoiesis in a manner resembling human yolk sac development, thus providing a valuable tool for dissecting the earliest events in human HSPC genesis.
Introduction
Classic and contemporary anatomic studies of human embryos have revealed that human hematopoiesis begins in the second to third embryonic weeks with formation of mesoderm-derived blood islands in the extraembryonic mesoderm of the developing secondary yolk sac.1,2 Blood islands develop foci of nucleated erythroblasts (“megaloblasts”),3 intimately associated with and surrounded by endothelium. Yolk sac (primitive) blood cells consist of nucleated primitive erythrocytes expressing exclusively embryonic hemoglobins (eg, ϵ2ζ2 globin chains) and primitive macrophages that arise without detectable monocytic precursors. Following the onset of circulation at about 21 days of development, yolk sac cells are found in embryonic blood. The fetal liver subsequently replaces the yolk sac as the main hematopoietic organ4,5 with appearance of definitive enucleate, macrocytic erythrocytes expressing fetal hemoglobins (eg, α2γ2 globin chains). Definitive blood cells and hematopoietic stem progenitor cells (HSPCs) can be detected in the fetal liver and embryo beginning at 5 to 6 weeks but have also been assayed as early as 4 to 5 weeks from human yolk sac, suggesting a gradual yolk sac/fetal liver HSPC transition.1,6 The human adult long-term repopulating HSPC that ultimately seeds the fetal bone marrow and thymus is the legacy of fetal liver hematopoiesis.
Both yolk sac–derived and aorta-gonad-mesonephros (AGM)–derived HSPC genesis models have been proposed in murine systems.7-17 Interestingly, in both mouse and humans, there is compelling evidence that both yolk sac– and AGM-derived HSPCs are the direct progeny of a bipotential hemogenic endothelial cell or hemangioblast.18-24
In contrast to zebrafish and mouse, the early developmental biology of human HSPCs remains difficult to approach due to ethical and technical limitations of studying human embryos. In light of fundamental differences between rodent and human developmental hematopoiesis, including anatomically more complex primary and secondary yolk sacs, a relatively shortened yolk sac phase,25 and a much earlier onset of medullary hematopoiesis26 in humans, direct extrapolation of murine development to humans may be inappropriate. Furthermore, although studies with scarce human embryo tissues have provided limited insight into the emergence of HSPCs within the 3- to 5-week human embryo,23,24 it remains impossible to study the initial commitment of mesoderm to angiohematopoiesis because human yolk sac blood islands develop as early as 16 to 17 days of development.
Pluripotent mouse embryonic stem cells (mESCs) have demonstrated capacity to differentiate into hematoendothelial progenitors in a manner recapitulating in vivo murine embryonic hematopoiesis.27-31 In vitro differentiation of human embryonic stem cells (hESCs)32,33 similarly provides opportunities to elucidate the origins of human hematopoiesis. Recent studies have demonstrated that hESCs can differentiate to hematopoietic colony-forming cells (CFCs) using either stromal coculture or human embryoid body (hEB)–based systems.34-37 However, whereas primitive and definitive hematopoiesis have been documented in mESC models,27-31 sequential and spontaneous waves of primitive and definitive hematopoiesis38-44 have not been delineated from hESCs. This obstacle limits interpretation and usefulness of hESCs as tools for dissecting the earliest cellular/molecular events in human embryonic hematopoiesis, including the search for a clonogenic human hemangioblast. Additionally, unlike mouse ES/EB systems, protocols for efficient commitment to hematopoietic cells thus far appear to require pretreatment of differentiating hEBs with complex cocktails of growth factors that induce, augment, but also likely skew spontaneous hematopoietic commitment of hEB progenitors.34,35
The derivation of genetically modified45,46 hematoendothelial progenitors from hESCs may provide a versatile source of both HSPCs and endothelial cells for human tissue engineering. More importantly, the basic cellular and genetic mechanisms of human hematopoietic incipience can now be opened to thorough investigation.
Materials and methods
Culture of hESC and hEB differentiation
The hESC line H1 (National Institutes of Health [NIH] code: WA01) was maintained on irradiated primary murine (CF1) primary murine embryonic fibroblasts (PMEFs) as previously described32,33 and maintained a continuously pluripotent phenotype (more than 80% to 90% GCTM2+CD9+, SSEA4+, SSEA1–, OCT4+, and alkaline phosphatase positive 32,33 ). Freshly thawed hESCs (fewer than 45 passages) with confirmed normal male karyotypes were used for all differentiation experiments. Feeder-free culture on Matrigel prior to hEB formation was avoided.
Our methods for hEB differentiation (Figure S1A, available on the Blood website; see the Supplemental Figures link at the top of the online article) were modified from previous approaches for forming mouse EBs (mEBs) in semisolid and liquid medium.27,31,47 hESC/PMEF cocultures in 6-well gelatinized plates were grown to about 80% confluence. Twenty-four hours prior to hEB formation, serum-free hESC medium was switched to “adaptation medium” consisting of Iscove modified Dulbecco medium (IMDM) containing 20% ES-certified, heat-inactivated fetal calf serum (FCS) (HyClone, Logan, UT), 1 mM glutamine, 0.1 mM 2-mercaptoethanol (2-ME), minimum essential medium (MEM) nonessential amino acids, and 4 ng/mL basic fibroblast growth factor (bFGF) (Invitrogen, Carlsbad, CA). IMDM/FCS-adapted hESC cultures were harvested with 2 mg/mL dispase. hESC clumps were washed and recultured at high density (contents of 2 wells of confluent hESCs into 1 well for hEB culture) in 6-well ultra nonadherent plates (Corning, Corning, NY) in 3 to 4 mL IMDM-based 1% methylcellulose medium (SF H4236; StemCell Technologies, Vancouver, BC, Canada) supplemented with 15% FCS (StemCell Technologies), 50 μg/mL ascorbic acid (Sigma, St Louis, MO), 0.5% cholesterol/lipoprotein supplements (EX-CYTE; Serologicals, Norcross, GA), and 3.5% protein-free hybridoma medium-II PFHM-II (Invitrogen). Semisolid hEB cultures were harvested after 4 days, washed, and resuspended in conical tubes in 50 mL phosphate-buffered saline (PBS). Formed hEBs were collected by gravity settling for 3 to 5 minutes, followed by aspirating the upper two thirds of the hEB suspension. This step efficiently eliminated more than 90% of nonviable, irradiated PMEFs. Formed hEBs were replated (about 300 to 500/mL) in ultra nonadherent 6-well plates in 4 mL “liquid differentiation medium” consisting of serum-free expansion medium (SFEM) (StemCell Technologies) supplemented with 15% FCS, 50 μg/mL ascorbic acid, 0.5% EX-CYTE, 0.5% insulin/transferrin/selenium supplements (Invitrogen), 5% PFHM-II, and penicillin/streptomycin. hEB liquid cultures were fed every 3 to 4 days with 2 to 3 mL fresh liquid differentiation medium (or passaged at a 1:2 ratio).
Replating assays for hematopoietic colony-forming cell (CFC) or endothelial potential
hEBs were harvested at various time points, washed in PBS, and assayed for clonogenic hematopoietic progenitors. Single-cell suspensions of hEBs were made using 0.05% trypsin/EDTA (ethylenediaminetetraacetic acid) (day 3 to 9 hEBs) or Liberase (Roche, Indianapolis, IN) plus 0.05% trypsin/EDTA (day 10 to 30 hEBs). Enzymatically loosened hEBs were passed gently through a 21-gauge needle and triturated into a single-cell suspension. Viable hEB cells were enumerated, plated (1.0 × 105 to 1.5 × 105/mL), and assayed in humidified chambers for hematopoietic CFCs in 2 mL serum-free methylcellulose-based medium (H4436; StemCell Technologies) containing stem cell factor (SCF) 50 ng/mL, erythropoietin (EPO) 3 U/mL, granulocyte-macrophage colony-stimulating factor (GM-CSF) 50 ng/mL, G-CSF 50 ng/mL, interleukin-3 (IL-3) 20 ng/mL, and IL-6 20 ng/mL supplemented with 0.5% EX-CYTE and 5% PFHM-II. Fourteen to 21 days later, colonies were scored and picked for cytospin or real-time quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) analysis.
Erythroid burst-forming units (BFU-e's) were defined for both primitive and definitive colonies by standard definitions (3 or more multiclustered erythroid colonies). Positive control definitive BFU-e's for intracellular hemoglobin fluorescence-activated cell sorter (FACS) analyses were generated by plating CD34+ cord blood cells in H4436 medium.
Plastic-adherent cells from individual mesodermal-hematoendothelial (MHE) colonies were secondarily replated for endothelial differentiation by vigorous washing away of nonadherent cells, trypsinization, passage through a 21-gauge needle, and reculture into EGM2 complete endothelial medium (Cambrex Bioscience, Walkersville, MD) on Matrigel-coated plates. Rapidly proliferating cells appeared 4 to 7 days after replating and were analyzed for endothelial function by overnight incubation with 10 μg/mL acetylated Dil-LDL (Molecular Probes, Eugene, OR) or harvested for RNA for RT-PCR analysis.
Flow cytometry analysis of surface markers and hemoglobins and in situ immunofluorescence
Hematoendothelial markers were evaluated from enzymatically dissociated hEB cells or individually picked and pooled hematopoietic colonies at different time points. Fluorochrome-conjugated monoclonal antibodies included CD31-phycoerythrin (PE), CD34-PE, CD45-PE, CD71–phycoerythrin-cyanin 5 (PeCy5), CD13-PE, glycophorin A (CD235A, GlyA)–PE, vascular endothelial (VE)–cadherin (all from Becton Dickinson, San Diego, CA), antivimentin (Lab Vision, Fremont, CA), and Ulex europaeus agglutinin-1–fluorescein isothiocyanate (UEA1-FITC) (Vector Laboratories, Burlingame, CA). Viable cells were gated for analysis and appropriate isotype and secondary antibody controls compared for each sample using a FACSort flow cytometer and CellQuest software (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA). Hemoglobin F (HbF), hemoglobin A (HbA), gamma chain, and epsilon chain expression was evaluated by intracellular FACS staining of fixed, permeabilized colony cells (FIX and PERM; Caltag, Burlingame, CA) using fluorochrome-conjugated monoclonal antibodies HbF-PE (Caltag), HbA (beta 6-glu epitope [Cortex, San Leandro, CA] plus secondary PE-conjugated antibody [Southern Biotech, Birmingham,AL]), gamma chain–PE (Cortex), and epsilon chain–FITC (Cortex).
Adherent cells of MHE colony cells from CFC assay plates were analyzed by FACS following dissociation with nonenzymatic cell dissociation buffer (Invitrogen) and gentle passage through a 21-gauge needle. Plastic-adherent cells were also analyzed in situ for endothelial function by overnight incubation with 10 μg/mL acetylated Dil-LDL, followed by fixation/permeabilization (FIX and PERM; Caltag) and immunostaining with FITC- or AlexaFluor594-conjugated primary/secondary antibodies/reagents. Nucleii were counterstained with DAPI (4,6 diamidino-2-phenylindole). All images were acquired with a Nikon TE200 microscope equipped with a DXM1200F digital camera and ACTI software (Nikon).
Hematologic stains
Colonies were picked individually from methylcellulose cultures, washed in PBS/2% FCS, and spun onto poly-d-lysine–coated slides with a cytospin apparatus (Shandon; Theims, Waltham, MA). Cells were fixed and stained with Wright-Giemsa reagents (Hema 3 stain; Fisher Scientific, Hampton, NH). Benzidine-hemoglobin and Kleihauer-Betke (K-B) fetal hemoglobin stains with erythrosin-B were done on cytospun cells as described.48
qRT-PCR analysis of hEB cells and hematopoietic colonies
To derive baseline hESC mRNA for hEB qRT-PCR studies, viable, undifferentiated hESCs were purified by FACS (FACSVantage; BDIS) to more than 95% homogeneity from PMEFs after trypsinization of hESC cultures and staining with GCTM2 49 supernatant (a kind gift from Martin Pera) plus anti–mouse immunoglobulin M (IgM)–FITC secondary (Becton Dickinson), and CD9-PE (Becton Dickinson). GCTM2+CD9+ cells represented 80% to 90% of hESCs in healthy, log-phase cultures.
Total RNA from GCTM2+CD9+-sorted (more than 95% purity) H1 hESCs, hEB cells, or picked, pooled colonies from methylcellulose CFC medium was prepared using RNEasy reagents (Qiagen, Valencia, CA). RNA samples were treated with RNAse-free DNAse (Qiagen). First-strand cDNA was reversed transcribed with oligo-dT priming using Superscript reagents (Invitrogen). qRT-PCR was performed using iQ SYBR-Green Supermix reagents and an iCycler thermal cycler and software (Bio-Rad, Hercules, CA). Human gene–specific PCR amplicons of 90 to 300 bp (Table 1) were designed with PRIMER 3.0 software for comparison with human/mouse cDNA GenBank data using basic local alignment sequence tool (BLAST) (National Center for Biotechnology Information [NCBI]) software. All primers were tested and optimized for specificity and mouse nonreactivity with SYBR-Green reagents using reverse-transcribed cDNA from PMEF RNA (negative control), human umbilical vein endothelial cell RNA (positive control), or human CD34+ cord blood RNA (positive controls). Transcripts of target genes and actin controls for each cDNA sample were amplified in duplicate/triplicates. All qRT-PCR reactions were confirmed for specificity of a single PCR product by analysis on 4% agarose gels. Comparative quantification of each target gene was performed based on cycle threshold (CT) normalized to actin using the ΔΔCT method.51 The relative expression of each normalized target gene was compared with the actin-normalized expression of the target gene (ΔCT) in highly purified (GCTM2+CD9+-sorted) undifferentiated hESCs. “Fold change expression from undifferentiated hESCs” was calculated as 2–ΔΔCT, where ΔΔCT = (ΔCT hEB cells)–(ΔCT purified, undifferentiated hESCs). For evaluation of relative expression levels of target genes in individually picked hematopoietic colonies, expression was compared with transcript levels of hEB cells obtained prior to plating in methylcellulose and growth factors. “Fold change in expression from undifferentiated hEB cells” was similarly calculated using actin-normalized methods (ΔCT) and based on the following equation: 2–ΔΔCT, where ΔΔCT = (ΔCT colony-forming cells derived from hEB progenitors)–(ΔCT preplated total hEB cells).
Sequences of human-specific primers used for qRT-PCR
Human-specific genes . | Forward primer, 5′ → 3′ . | Reverse primer, 5′ → 3′ . | Amplicon size, base pairs . | Annealing temperature, °C . |
|---|---|---|---|---|
| Actin | gca cag agc ctc gcc ttt | gga atc ctt ctg acc cat gc | 213 | 59-65 |
| OCT4 | gca gct cgg aag gca gat | tgg att tta aaa ggc aga aga ctt g | 135 | 63 |
| CD31 | gag tcc tgc tga ccc ttc tg | att ttg cac cgt cca gtc c | 107 | 61 |
| CD34 | tgg acc gcg ctt tgc t | ccc tgg gta ggt aac tct ggg | 100 | 61 |
| CD45 | cat ttg gct ttg cct ttc tg | ttc tct ttc aaa ggt gct tgc | 167 | 61 |
| KDR/fik-1 | cca gcc aag ctg tct cag t | ctg cat gtc agg ttg caa ag | 272 | 61 |
| Flt-1 | gac tag ata gcg tca cca gca g | gaa acc gtc aga atc ctc ctc | 101 | 65 |
| VWF | ccc gaa agg cca ggt gta | agc aag ctt ccg ggg act | 288 | 61-63 |
| VE-cadherin | gca gca gca ggt gct aac c | ttg ccc aca tat tct cct ttg | 203 | 61 |
| SCL/TAL1 | atg aga tgg aga tta ctg atg | gcc ccg ttc aca ttc tgc t | 100 | 61 |
| LMO2 | aac tgg gcc gga agc tct | ctt gaa aca ttc cag gtg ata ca | 143 | 65 |
| EKLF | cgg aca cac agg atg act tc | ggc tgg tcc tca gac ttc ac | 115 | 59-60 |
| PU.1 | cac agc gag ttc gag agc tt | gat ggg tac tgg agg cac at | 194 | 61 |
| GATA1 | ggg atc aca ctg agc ttg c | acc cct gat tct ggt gtg g | 176 | 64-65 |
| GATA2 | gcg tct cca gcc tca tct t | gga aga gtc cgc tgc tgt ag | 226 | 61 |
| c-myb | gtc aca aat tga ctg tta caa cac cat | ttc tac tag atg aga ggg tgt ctg agg | 212 | 61 |
| CDX4 | ctg tgg gcg gtg gaa cta | aaa cta cac gbvat act ttt ctt ttg tcc | 186 | 61 |
| (HOXB4)* | gca cgg taa acc cca att a | ggc aac ttg tgg tct ttt tt | 220 | 61-65 |
| AML 1a† | aag aca cag cac cct gga ga | gcc ttc ctc ata acg tgc at | 151 | 60 |
| AML 1a/b‡ | cgt gca cat aca tta gta gca ct acct ttg | ctt cca cga atc ttg ctt gca gag gtt aag | 304 | 60 |
| AML 1c§ | gaa gtc tga acc cag cat agt ggt cag cag | gtg gac gtc tct aga agg att cat tcc aag | 231 | 60 |
| Epsilon | tgc tat taa aaa cat gga caa cc | gcc aga ata atc acc atc acg | 126 | 61 |
| Zeta | att gtc tcc atg tgg gcc | tct gcg ggt ggc tga g | 90 | 61-63 |
| Alpha | aag gtc ggc gcg cac gct | ctc agg tcg aac tgc ggg | 101 | 61 |
| Gamma | gga caa ggc tac tat cac aag c | gga agt cag cac ctt ctt gc | 193 | 63 |
| Beta | ggc acc ttt gcc aca ctg | cac tgg tgg ggt gaa ttc tt | 132 | 61-63 |
Human-specific genes . | Forward primer, 5′ → 3′ . | Reverse primer, 5′ → 3′ . | Amplicon size, base pairs . | Annealing temperature, °C . |
|---|---|---|---|---|
| Actin | gca cag agc ctc gcc ttt | gga atc ctt ctg acc cat gc | 213 | 59-65 |
| OCT4 | gca gct cgg aag gca gat | tgg att tta aaa ggc aga aga ctt g | 135 | 63 |
| CD31 | gag tcc tgc tga ccc ttc tg | att ttg cac cgt cca gtc c | 107 | 61 |
| CD34 | tgg acc gcg ctt tgc t | ccc tgg gta ggt aac tct ggg | 100 | 61 |
| CD45 | cat ttg gct ttg cct ttc tg | ttc tct ttc aaa ggt gct tgc | 167 | 61 |
| KDR/fik-1 | cca gcc aag ctg tct cag t | ctg cat gtc agg ttg caa ag | 272 | 61 |
| Flt-1 | gac tag ata gcg tca cca gca g | gaa acc gtc aga atc ctc ctc | 101 | 65 |
| VWF | ccc gaa agg cca ggt gta | agc aag ctt ccg ggg act | 288 | 61-63 |
| VE-cadherin | gca gca gca ggt gct aac c | ttg ccc aca tat tct cct ttg | 203 | 61 |
| SCL/TAL1 | atg aga tgg aga tta ctg atg | gcc ccg ttc aca ttc tgc t | 100 | 61 |
| LMO2 | aac tgg gcc gga agc tct | ctt gaa aca ttc cag gtg ata ca | 143 | 65 |
| EKLF | cgg aca cac agg atg act tc | ggc tgg tcc tca gac ttc ac | 115 | 59-60 |
| PU.1 | cac agc gag ttc gag agc tt | gat ggg tac tgg agg cac at | 194 | 61 |
| GATA1 | ggg atc aca ctg agc ttg c | acc cct gat tct ggt gtg g | 176 | 64-65 |
| GATA2 | gcg tct cca gcc tca tct t | gga aga gtc cgc tgc tgt ag | 226 | 61 |
| c-myb | gtc aca aat tga ctg tta caa cac cat | ttc tac tag atg aga ggg tgt ctg agg | 212 | 61 |
| CDX4 | ctg tgg gcg gtg gaa cta | aaa cta cac gbvat act ttt ctt ttg tcc | 186 | 61 |
| (HOXB4)* | gca cgg taa acc cca att a | ggc aac ttg tgg tct ttt tt | 220 | 61-65 |
| AML 1a† | aag aca cag cac cct gga ga | gcc ttc ctc ata acg tgc at | 151 | 60 |
| AML 1a/b‡ | cgt gca cat aca tta gta gca ct acct ttg | ctt cca cga atc ttg ctt gca gag gtt aag | 304 | 60 |
| AML 1c§ | gaa gtc tga acc cag cat agt ggt cag cag | gtg gac gtc tct aga agg att cat tcc aag | 231 | 60 |
| Epsilon | tgc tat taa aaa cat gga caa cc | gcc aga ata atc acc atc acg | 126 | 61 |
| Zeta | att gtc tcc atg tgg gcc | tct gcg ggt ggc tga g | 90 | 61-63 |
| Alpha | aag gtc ggc gcg cac gct | ctc agg tcg aac tgc ggg | 101 | 61 |
| Gamma | gga caa ggc tac tat cac aag c | gga agt cag cac ctt ctt gc | 193 | 63 |
| Beta | ggc acc ttt gcc aca ctg | cac tgg tgg ggt gaa ttc tt | 132 | 61-63 |
These primers do not distinguish between human and murine HOXB4.
These primers amplify an amplicon in exon 7A of the AML 1 gene that is specific for the a isoform of AML 1.50
These primers amplify an amplicon in exon 3 of the AML 1 gene that is specific for both the a and b isoforms of AML 1.50
These primers amplify an amplicon that spans exon 1 and 2 of the AML 1 gene and is specific for the c isoform of AML 1.50
Results
hESCs efficiently differentiate into cystic hEBs containing precursors expressing a developmental progression of hematoendothelial surface markers and regulatory genes
Using modified mEB protocols27,31 (Figure S1A), we differentiated hESCs into hEBs capable of spontaneously generating hematopoietic progenitors without need for recombinant hematopoietic growth factors during hEB differentiation.34,35 About 50% to 60% of hESC clumps plated became hEBs and acquired a cystic morphology by 8 to 15 days of differentiation (Figure S1B). Differentiating hEB cells rapidly decreased expression of OCT3/4 (POU5F1; Figure S1C) and lost markers characteristic of undifferentiated hESCs32,33,49 (GCTM2, CD9, and TRA-1-60, data not shown) after 3 to 12 days.
To identify when differentiating hEB cells become competent for generating hematopoietic progenitors, we analyzed hEBs over a 4-week time course for expression of well-characterized hematoendothelial markers using FACS and qRT-PCR. Undifferentiated (day 0) hESCs expressed CD117 (FACS data not shown), CD133 (FACS data not shown), and KDR/flk1 (Figure 1A) but had low/undetectable expression of CD34 and CD31 RNA or surface protein (Figure 1A,C). CD34 and CD31 expression both peaked at about 12 to 15 days of hEB development (Figure 1A,C) and were coexpressed on the same hEB progenitors (Figure S2B). CD45 (Figure 1A,C) was expressed on only 1% to 3% of hEB cells and not until about 15 to 30 days (about 1 week after the onset of CD34/CD31 expression).
hEB cells were next evaluated for hematopoietic gene expression by qRT-PCR. A kinetic analysis revealed a progressive expression pattern of genes known (from murine studies) to initiate and regulate hematopoiesis (Figure 1B). Expression of key hematopoietic transcriptional regulators, including SCL/TAL1, CDX4, GATA1, GATA2, EKLF, and PU.1 were all found to increase dramatically after 1 week of hEB differentiation (Figure 1B). Increases in mRNA levels of these transcription factors coincided with similarly increasing expression levels of CD31, CD34, and KDR/flk-1 (Figure 1A). Interestingly, several genes important for regulating HSPC development in MESCs and embryos (eg, LMO2, AML1, C-MYB [data not shown], KDR/flk-1 [VEGFR2], and FLT-1 [VEGFR1]) were expressed abundantly at the mRNA level in undifferentiated hESCs as well as in hEBs at all time points (Figure 1A). Although we have not confirmed that mRNA levels correlate directly with protein levels for these genes, these data suggest potentially major differences between mouse and human embryonic regulation of hematopoiesis. Expression of KDR/flk-1 (VEGFR2, a receptor for vascular endothelial growth factor), associated with development of hemangioblasts, was only moderately increased but was relatively high at all time points. This is distinct from mEB differentiation, where a more dramatic expression profile has been described.52-55 Expression levels for these genes peaked at days 6 to 10 of hEB differentiation, suggesting a putative coordinated developmental watershed event in hEB hematoendothelial commitment.
Quantitative real-time RT-PCR and FACS expression analysis of key hematoendothelial genes in differentiating hEBs. Human-specific PCR primers (Table 1) were used to amplify indicated target genes. Levels of gene expression differences for (A) hematoendothelial surface markers or (B) hematopoietic regulatory transcription factors were calculated using the 2–ΔΔCT method as described in “Materials and methods,” based on the CT (threshold curve) for each target gene and internal normalizations with actin. Values of fold change in expression are relative to baseline expression levels in FACS-sorted populations of undifferentiated hESCs (day 0 hEBs) and are expressed as “fold expression from undifferentiated hESC.” Shown above graphs are the corresponding agarose gels of PCR products obtained at linear phases of qPCR reactions. Standard deviations between duplicate or triplicate samples are shown. PECAM indicates platelet-endothelial cell adhesion molecule 1. (C) Developmental progression of hematoendothelial surface marker expression on disaggregated hEB cells at various time points. “% positive hEB cell” represents fluorescence value obtained by FACS analysis following subtraction of control sample background. Each time point represents the mean of 3 to 5 independent experiments with indicated standard deviations.
Quantitative real-time RT-PCR and FACS expression analysis of key hematoendothelial genes in differentiating hEBs. Human-specific PCR primers (Table 1) were used to amplify indicated target genes. Levels of gene expression differences for (A) hematoendothelial surface markers or (B) hematopoietic regulatory transcription factors were calculated using the 2–ΔΔCT method as described in “Materials and methods,” based on the CT (threshold curve) for each target gene and internal normalizations with actin. Values of fold change in expression are relative to baseline expression levels in FACS-sorted populations of undifferentiated hESCs (day 0 hEBs) and are expressed as “fold expression from undifferentiated hESC.” Shown above graphs are the corresponding agarose gels of PCR products obtained at linear phases of qPCR reactions. Standard deviations between duplicate or triplicate samples are shown. PECAM indicates platelet-endothelial cell adhesion molecule 1. (C) Developmental progression of hematoendothelial surface marker expression on disaggregated hEB cells at various time points. “% positive hEB cell” represents fluorescence value obtained by FACS analysis following subtraction of control sample background. Each time point represents the mean of 3 to 5 independent experiments with indicated standard deviations.
Day 7 to 12 hEB cells generate organized, mesodermal-hematoendothelial (MHE) colonies and a robust wave of primitive hematopoiesis correlating with increased expressions of SCL/TAL1, GATA1, GATA2, and CDX4
We reproducibly generated colonies of CFC-mixed, M-CFCs, GM-CFCs, and erythroid colony-forming units (CFU-e's) (with morphologies similar to definitive colonies produced from cord blood CD34+ cells) from day 14 to 20 hEBs using standard methylcellulose preparations containing FCS, as has already been reported.34,35 hEB single-cell suspensions obtained at earlier time points, however, when peak expression levels for key regulatory genes were observed (days 6 to 12; Figure 1A-B), failed to generate CFCs using standard methylcellulose media. Because many batches of bovine serum have been reported to inhibit the expansion of primitive murine progenitors,27,31,47 we optimized our hEB CFC assays in serum-free semisolid medium. These conditions revealed vigorous primitive hematopoietic CFCs starting at 7 to 9 days of hEB development, beginning with the detection of unique, laterally expanding semiadherent MHE colonies (Figure 2A-B). CFC generation was not detectable from hEB cells differentiated for fewer than 7 days. MHE colonies expanded either as clonogenic adherent clusters (Figure 2A) or, rarely, from differentiating plastic-adherent secondary hEBs. Early MHE colonies are first detected as adherent clusters of cells with an endothelioid morphology (Figure 2D, left 2 panels) about 1 week after single hEB cell plating, followed by a rapid secondary budding/differentiation of hematopoietic blast colonies several days later (Figures 2A and 3A-B,G). The cellular architecture (Figure 2A) of many MHE cluster colonies differentiated for 2 to 5 weeks was highly reminiscent of human yolk sac blood islands.3
Adherent and nonadherent cells of MHE colonies derived from day 7 to 12 hEBs differentiated fully for more than 2 to 4 weeks in methylcellulose with growth factors were picked and analyzed separately by FACS and in situ immunofluorescence. Adherent cells of mature MHE colonies (obtained after vigorous washing of nonadherent cells) took up acetylated Dil-LDL (Figure 3F) and expressed CD31 (more than 50%; Figure 3G) and VE-cadherin (11%; data not shown), but not CD45 (not shown), while budding, loosely associated cells expressed erythromyeloid markers CD71, CD13, and CD45 (Figure 2C). Wright stains of nonadherent cells (Figure 2A, lower left panel) revealed primitive-type foamy macrophages (without evidence of monocytic precursors) and abundant nucleated, primitive-type erythroblasts. Rare definitive granulocytes were also detected (not shown). Interestingly, endothelial characteristics such as Dil-LDL uptake and VE-cadherin expression were present in early (about 1 week after hEB cell plating; Figure 2D, first left panel) adherent MHE colonies, prior to hematopoietic differentiation/budding.
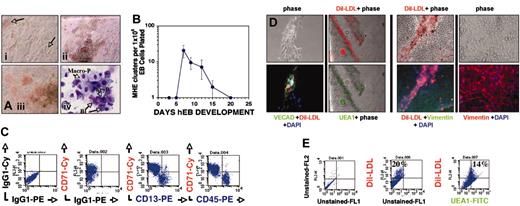
Hematopoietic differentiation from recultured hEB cells initiates with expansion of adherent MHE colonies. Single-cell suspensions of hEB cells differentiated for 3 to 25 days were plated in modified serum-free methylcellulose medium containing hematopoietic growth factors as described in “Materials and methods.” (A) Within 1 to 3 weeks following day 7 to 12 hEB cell suspension CFC assay plating, a population of semiadherent MHE colonies expanded either from plastic-adherent hEB clonogenic progenitors, or rarely from attached secondary hEBs. Plastic-adherent MHE colonies (i; magnification × 200) differentiate by lateral endothelioid expansion accompanied by budding, hemoglobinizing hematopoietic blasts (arrows) that are intimately intermixed with adherent cells (ii; magnification × 40). MHE clusters can become quite prolific after 3 to 5 weeks (iii; magnification × 200) if refreshed with fresh medium with growth factors every 1 to 2 weeks. Wright stains (iv; magnification × 600, oil) of budding, nonadherent cells from prolific MHE clusters revealed abundant hematopoietic blasts (Bl), primitive nucleated erythrocytes (EryP), foamy primitive macrophages (Macro-P), and rare definitive cells including granulocytes and definitive erythroid cells (not shown). (B) Kinetics of MHE colony appearance from differentiating hEBs (mean and standard deviations of 3 independent experiments). Loosely adherent cells from prolific HE clusters were picked and analyzed by FACS (C) and found to express abundant levels of early erythroid (CD71), myeloid (CD13), and panhematopoietic (CD45) markers. Mature (3- to 5-week-old) MHE colonies were extensively washed of budding, nonadherent hematopoietic cells, and remaining plastic-adherent cells were incubated with 10 μg/mL acetylated Dil-LDL overnight and then fixed, permeabilized, and further evaluated for expression of endothelial-specific and mesodermal markers. Approximately 15% to 20% of elongated, plastic-adherent cells that formed the base of MHE colonies were shown by in situ immunofluorescence (D) or FACS analysis (as single cells) (E) to be capable of simultaneously taking up acetylated Dil-LDL and costaining with either Ulex europaeus agglutinin-1–FITC (D; UEA1, magnification × 100) (E) or VE-cadherin plus FITC-conjugated secondary antibody (D; magnification × 200). The remainder of the adherent cells that surrounded intermixed, robust Dil-LDL–positive endothelial clusters stained brightly for intracellular vimentin, thus identifying them as mesodermal-mesenchymal (nonendothelial) in lineage (D, right 4 panels; magnification × 100, × 200, respectively). Shown are merged images of phase and red and/or green immunofluorescent filters.
Hematopoietic differentiation from recultured hEB cells initiates with expansion of adherent MHE colonies. Single-cell suspensions of hEB cells differentiated for 3 to 25 days were plated in modified serum-free methylcellulose medium containing hematopoietic growth factors as described in “Materials and methods.” (A) Within 1 to 3 weeks following day 7 to 12 hEB cell suspension CFC assay plating, a population of semiadherent MHE colonies expanded either from plastic-adherent hEB clonogenic progenitors, or rarely from attached secondary hEBs. Plastic-adherent MHE colonies (i; magnification × 200) differentiate by lateral endothelioid expansion accompanied by budding, hemoglobinizing hematopoietic blasts (arrows) that are intimately intermixed with adherent cells (ii; magnification × 40). MHE clusters can become quite prolific after 3 to 5 weeks (iii; magnification × 200) if refreshed with fresh medium with growth factors every 1 to 2 weeks. Wright stains (iv; magnification × 600, oil) of budding, nonadherent cells from prolific MHE clusters revealed abundant hematopoietic blasts (Bl), primitive nucleated erythrocytes (EryP), foamy primitive macrophages (Macro-P), and rare definitive cells including granulocytes and definitive erythroid cells (not shown). (B) Kinetics of MHE colony appearance from differentiating hEBs (mean and standard deviations of 3 independent experiments). Loosely adherent cells from prolific HE clusters were picked and analyzed by FACS (C) and found to express abundant levels of early erythroid (CD71), myeloid (CD13), and panhematopoietic (CD45) markers. Mature (3- to 5-week-old) MHE colonies were extensively washed of budding, nonadherent hematopoietic cells, and remaining plastic-adherent cells were incubated with 10 μg/mL acetylated Dil-LDL overnight and then fixed, permeabilized, and further evaluated for expression of endothelial-specific and mesodermal markers. Approximately 15% to 20% of elongated, plastic-adherent cells that formed the base of MHE colonies were shown by in situ immunofluorescence (D) or FACS analysis (as single cells) (E) to be capable of simultaneously taking up acetylated Dil-LDL and costaining with either Ulex europaeus agglutinin-1–FITC (D; UEA1, magnification × 100) (E) or VE-cadherin plus FITC-conjugated secondary antibody (D; magnification × 200). The remainder of the adherent cells that surrounded intermixed, robust Dil-LDL–positive endothelial clusters stained brightly for intracellular vimentin, thus identifying them as mesodermal-mesenchymal (nonendothelial) in lineage (D, right 4 panels; magnification × 100, × 200, respectively). Shown are merged images of phase and red and/or green immunofluorescent filters.
The bed of plastic-adherent cells of more mature (about 3 to 5 weeks after plating) MHE colonies was analyzed in situ in more detail following vigorous washing away of nonadherent cells. Overnight incubation of adherent cells with acetylated Dil-LDL and subsequent incubation with endothelial-specific Ulex europaeus agglutinin-1–FITC (UEA1-FITC) revealed that about 20% of cells costained with these markers (Figure 2D; FACS analysis of dissociated adherent cells, Figure 2E). Interestingly, these studies revealed that a significant portion of adherent cells of MHE colonies did not stain for any endothelial-specific markers (compare phase with fluorescent images; Figure 2D). These cells, however, stained abundantly for the mesenchymal marker vimentin (Figure 2D, right 4 panels), thus demonstrating that endothelial cells were intermixed with cells of nonendothelial, mesodermal origin. This important result suggests that MHE colonies likely arise from hEB-derived mesodermal progenitors capable of giving rise to primitive-type erythromyeloid cells, endothelial cells, and mesenchymal stromal cells: the 3 essential cellular components of yolk sac blood islands.
To further characterize these complex MHE colonies, we conducted secondary replating assays of adherent and nonadherent portions of early (about 1 week after replating), developing MHE colonies. Our hypothesis was that the mesodermal progenitor(s), which generates MHE colonies, can secondarily give rise to clonogenic progenitors of either hematopoietic (nonadherent) or endothelial (adherent) lineages. Individual budding hematopoietic blast colonies (Figure 3B) from early MHE colonies (Figure 3A,G) containing few differentiating, hemoglobinizing cells were carefully picked. These colonies were dissociated into single cells and replated for secondary colony analysis (Figure 3D-E). Most (more than 70%) MHE colony-derived hematopoietic blast colonies generated new colonies in methylcellulose with growth factors and readily produced secondary primitive-type mixed erythromyeloid, pure erythroid, or pure macrophage colonies (Figure 3E). This demonstrated that MHE colonies generate multipotent hematopoietic progenitors. In parallel experiments, the adherent cells of individual MHE colonies, which were about 50% to 60% CD31+ (Figure 3G) and CD45– (not shown), were enzymatically dissociated into single cells and recultured in EGM2 medium with endothelial growth factors. Adherent cells from 6 of 6 (100%) individual MHE colonies rapidly grew cells of endothelial morphology, had ability to take up acetylated Dil-LDL, and expressed endothelial genes including von Willebrand factor (VWF) and VE-cadherin but not the hematopoietic marker gene CD45 (Figure 3G).
Colonies with a primitive erythroblast morphology expanded in semisolid medium with growth factors from day 7 to 12 hEB cells in the absence of any obvious MHE source. Primitive erythroblast colonies differentiated into either large primitive multiclustered erythroid colonies (Figure 4A, BFU-e-P) or, less commonly, mixed erythromyeloid colonies (Figure 5C; MIXED-P). These erythroblasts produced distinctive brilliant red hemoglobinized, nucleated erythroid colonies (Figure 4A, EryP or BFU-e-P). Macrophage colonies with a foamy cytoplasmic morphology (Figure 5D, Macro-P), and without associated monocytic precursors, were also abundant at this time and were identical on Wright stains to macrophages arising from HE clusters (Figure 2A). Primitive erythroblast colonies were picked at early stages (7 to 10 days after hEB replating) for benzidine-hemoglobin staining, which demonstrated the presence of nucleated hemoglobin-containing erythroblasts and erythrocytes intermixed with immature blast cells (Figure 4C). These erythroblasts were determined by K-B stains to express abundant amounts of fetal hemoglobins (Figure 4C).
Secondary blast colonies can arise directly from adherent MHE colonies and differentiate directly to primitive erythroblasts, macrophages, or mixed erythromyeloid colonies. During their initial differentiation, adherent MHE clusters (A; magnification × 100) often produced secondary budding, blast colonies (B, magnification × 200) with a Wright-Giemsa stain hematopoeitic blast morphology (C; magnification × 1000, oil) and that differentiated rapidly into hemoglobinizing cells. To further evaluate the clonogenic potential of MHE-derived budding hematopoietic blast colonies derived from day 7 to 9 hEB cells, they were individually picked prior to full differentiation (about 1 week after hEB plating) and recultured in H4436 supplemented with 50 ng/mL bone morphogenetic protein 4 (BMP4) (which greatly improved their replating efficiency). Single, picked, MHE-derived primary blast colonies (B) were demonstrated to be multipotential by their ability to give rise to multiple secondary mixed, macrophage, or erythroid colonies (D,E) in secondary replating experiments ([E] with 5 of 7 primary blast colonies successfully replated in a representative experiment repeated 3 times). After 4 to 6 weeks of continuous culture, prolific, mature MHE clusters (F, left panel; magnification × 40) are composed of nonadherent hematopoietic cells arising from an adherent bed of mesenchymal-endothelial cells. When nonadherent cells are extensively washed away and adherent cells were incubated overnight with acetylated Dil-LDL, approximately 20% to 30% of adherent cells were positive (F, right panel). To further evaluate the endothelial component of MHE colonies, adherent cells from individual colonies (Gi) were picked, disaggregated, analyzed by FACS, or replated in endothelial medium (EGM2) on Matrigel-coated plates. Replated adherent cells from 6 of 6 individual MHE colonies gave rise to cells with endothelial morphology (ii) and ability to take up acetylated Dil-LDL after several days of EGM2 endothelial culture. (iv) These cells were further evaluated for expression of endothelial genes including von Willebrand factor (VWF) and VE-cadherin by qRT-PCR (HU indicates human umbilical vein endothelial cell [HUVEC] RNA control; C, cord blood RNA control; E, endothelial cells from replated MHE adherent layers). More than 50% to 70% of elongated, adherent cells expressed CD34 and/or CD31 (iii, left); EGM2 replated adherent cells continued to express CD31/CD34 and were devoid of CD45 expression (iii, right).
Secondary blast colonies can arise directly from adherent MHE colonies and differentiate directly to primitive erythroblasts, macrophages, or mixed erythromyeloid colonies. During their initial differentiation, adherent MHE clusters (A; magnification × 100) often produced secondary budding, blast colonies (B, magnification × 200) with a Wright-Giemsa stain hematopoeitic blast morphology (C; magnification × 1000, oil) and that differentiated rapidly into hemoglobinizing cells. To further evaluate the clonogenic potential of MHE-derived budding hematopoietic blast colonies derived from day 7 to 9 hEB cells, they were individually picked prior to full differentiation (about 1 week after hEB plating) and recultured in H4436 supplemented with 50 ng/mL bone morphogenetic protein 4 (BMP4) (which greatly improved their replating efficiency). Single, picked, MHE-derived primary blast colonies (B) were demonstrated to be multipotential by their ability to give rise to multiple secondary mixed, macrophage, or erythroid colonies (D,E) in secondary replating experiments ([E] with 5 of 7 primary blast colonies successfully replated in a representative experiment repeated 3 times). After 4 to 6 weeks of continuous culture, prolific, mature MHE clusters (F, left panel; magnification × 40) are composed of nonadherent hematopoietic cells arising from an adherent bed of mesenchymal-endothelial cells. When nonadherent cells are extensively washed away and adherent cells were incubated overnight with acetylated Dil-LDL, approximately 20% to 30% of adherent cells were positive (F, right panel). To further evaluate the endothelial component of MHE colonies, adherent cells from individual colonies (Gi) were picked, disaggregated, analyzed by FACS, or replated in endothelial medium (EGM2) on Matrigel-coated plates. Replated adherent cells from 6 of 6 individual MHE colonies gave rise to cells with endothelial morphology (ii) and ability to take up acetylated Dil-LDL after several days of EGM2 endothelial culture. (iv) These cells were further evaluated for expression of endothelial genes including von Willebrand factor (VWF) and VE-cadherin by qRT-PCR (HU indicates human umbilical vein endothelial cell [HUVEC] RNA control; C, cord blood RNA control; E, endothelial cells from replated MHE adherent layers). More than 50% to 70% of elongated, adherent cells expressed CD34 and/or CD31 (iii, left); EGM2 replated adherent cells continued to express CD31/CD34 and were devoid of CD45 expression (iii, right).
Day 7 to 12 hEB cells thus contained primitive hematopoietic CFCs, which were observed to arise from hEB-derived endothelioid MHE progenitors. These cellular events, indicative of primitive embryonic hematopoiesis, were directly associated with increases of SCL/TAL1, CDX4, GATA1, GATA2, CD31, and CD34 by more than 10- to 10 000-fold in day 7 to 9 cystic hEBs (Figure 1B). Although we have not directly proven it, this critical temporal window may represent a hemangioblastic commitment phase in differentiating hEBs. Importantly, no CD45+ hEB progenitors were detected by FACS or qRT-PCR (Figure 1A,C) during this time period, although large numbers of CD45+ cells were clearly produced from day 9 to 12 hEB-derived MHE colonies following reculture of CD45-hEB cells in semisolid medium with hematopoietic growth factors.
Definitive hematopoietic cells develop from day 12 to 20 hEBs following primitive erythromyelopoiesis from day 7 to 12 hEBs
Kinetic analysis of hematopoietic CFCs generated from day 3 to 20 hEB cells in serum-free CFC assay conditions revealed a rich variety of hematopoietic colonies with both primitive and definitive morphologies. Erythroid colonies observed from day 12 to 20 hEBs cells differed notably in morphology from those observed from day 7 to 12 hEBs. These latter colonies had definitive BFU-e– and CFU-e–derived morphologies, with a salmon red (as opposed to brilliant red) hemoglobinization, similar to erythroid colonies generated from cord blood progenitors.
To further evaluate erythroid colonies scored morphologically as primitive or definitive (Figure 4D), colonies were picked, pooled, and analyzed for hemoglobin expression by qRT-PCR or intracellular FACS analysis. Erythroid colonies from day 7 to 12 hEBs (Figure 4D, BFU-e-P and EryP) expressed embryonic (ϵ2ζ2) and fetal (HbF, α2γ2) but not adult (HbA, α2β2) hemoglobins (Figures 4F and 6B,D). The second wave of salmon red–colored erythroid colonies from day 12 to 20 hEBs scored as definitive (Figure 4D, BFU-e-D, CFU-e-D), however, could be shown to express not only embryonic (ϵ2ζ2) and fetal (HbF) but also adult (HbA) hemoglobins (Figures 4F and 6B). Thus, day 12 to 20 erythroid colonies are more similar to BFU-e– and CFU-e–derived colonies generated from neonatal CD34+ cord blood cells. Intracytoplasmic staining with an anti–gamma-PE hemoglobin chain antibody gave identical results for all erythroid colonies as staining with the anti-HbF antibody (data not shown). CD71 and GlyA coexpression was found to be comparable between primitive and definitive-type erythroid colonies (Figure 4E).
Two types of mixed-lineage colonies were generated from dissociated hEB cells in serum-free CFC assay conditions. Blast colonies from day 7 to 12 hEBs arising either directly from HE colonies (Figure 4A) or from single hEB cells were observed to differentiate into not only BFU-e-P but also into large, mixed primitive colonies (mixed CFC-P) containing nucleated primitive erythroblasts and primitive macrophages (Figure 5A,C). Mixed CFC-P erythromyeloid colonies expressed exclusively embryonic and fetal hemoglobins (Figure 6B) and thus appear to be the progeny of a yolk sac–like hematopoietic progenitor, similar to the blasts budding from MHE colonies. The second type of mixed colony had a more definitive erythromyeloid morphology (Figure 5E, MIXED CFC-D) and predominated in CFC cultures from day 12 to 20 hEBs. These colonies contained a mixture of mature erythroid cells, definitive-type monocyte/macrophages, and rare segmented neutrophils. Colonies derived from mixed CFC-D, unlike mixed CFC-P (Figure 5C), were distinguished by a more compact, colony morphology, salmon red hemoglobinization, and expression of embryonic/fetal and adult (β-globin) hemoglobin chains (Figure 6B). Also generated from day 12 to 20 hEB cultures were definitive-type GM-CFC and M-CFC colonies containing monocytes, macrophages, and rare segmented neutrophils (Figure 5B,F). These definitive myeloid-lineage colonies with abundant monocytic precursors (Figure 5F) contrasted to the primitive macrophage colonies (Figure 5B,D, Macro-P CFC) that had a foamy cytoplasm and no apparent monocytic precursors.
Day 7 to 20 hEB cells contain clonogenic progenitors for both primitive and definitive erythropoiesis. Erythroid colonies with a “brilliant red” hemoglobinization from day 9 hEB-derived BFU-e-P (Ai; magnification × 100) and EryP (ii; magnification × 100) containing nucleated primitive erythrocytes were generated from day 7 to 15 day hEBs. Erythroid colonies containing a brownish, “salmon-red” hemoglobinization were generated from day 12 to 20 day hEBs ([B] BFU-e-D, CFU-e-D). Colonies (3 to 5 pooled) were picked from semisolid medium for staining or FACS analyses. Wright stains from multiclustered BFU-e-P (Aiii; magnification × 600) revealed an increased abundance of erythroblasts, while more mature EryP (iv; magnification × 600) contained primarily differentiated nucleated erythrocytes. Primitive erythroblast (Bl) colonies predominate from day 9 to 12 hEBs differentiate directly into nucleated erythrocytes positive for hemoglobin by benzidine staining (C, top; magnification × 400). Erythroblast colonies stain brightly positive for fetal hemoglobins by erythrosin-B K-B stains (C, bottom; magnification × 200). (Inset) Positive control K-B staining from maternal peripheral blood with 10% fetal erythrocyte cells. (D) Kinetic analysis of primitive and definitive erythroid CFCs. Shown is a representative analysis of 3 independent experiments. Although all types of erythroid CFCs had surface expression of CD71+/GlyA+ cells (F) and expressed various levels fetal hemoglobin (HbF), only day 12 to 20 BFU-e-D and CFU-e-D expressed adult hemoglobin A (HbA). Day 15 to 20 BFU-e-D or CFU-e-D often contained brownish, lysed cells. Coexpression of both HbF and HbA in BFU-e-D from CD34+ cord blood (CB) progenitors was used as a positive control ([F] CB BFU-e).
Day 7 to 20 hEB cells contain clonogenic progenitors for both primitive and definitive erythropoiesis. Erythroid colonies with a “brilliant red” hemoglobinization from day 9 hEB-derived BFU-e-P (Ai; magnification × 100) and EryP (ii; magnification × 100) containing nucleated primitive erythrocytes were generated from day 7 to 15 day hEBs. Erythroid colonies containing a brownish, “salmon-red” hemoglobinization were generated from day 12 to 20 day hEBs ([B] BFU-e-D, CFU-e-D). Colonies (3 to 5 pooled) were picked from semisolid medium for staining or FACS analyses. Wright stains from multiclustered BFU-e-P (Aiii; magnification × 600) revealed an increased abundance of erythroblasts, while more mature EryP (iv; magnification × 600) contained primarily differentiated nucleated erythrocytes. Primitive erythroblast (Bl) colonies predominate from day 9 to 12 hEBs differentiate directly into nucleated erythrocytes positive for hemoglobin by benzidine staining (C, top; magnification × 400). Erythroblast colonies stain brightly positive for fetal hemoglobins by erythrosin-B K-B stains (C, bottom; magnification × 200). (Inset) Positive control K-B staining from maternal peripheral blood with 10% fetal erythrocyte cells. (D) Kinetic analysis of primitive and definitive erythroid CFCs. Shown is a representative analysis of 3 independent experiments. Although all types of erythroid CFCs had surface expression of CD71+/GlyA+ cells (F) and expressed various levels fetal hemoglobin (HbF), only day 12 to 20 BFU-e-D and CFU-e-D expressed adult hemoglobin A (HbA). Day 15 to 20 BFU-e-D or CFU-e-D often contained brownish, lysed cells. Coexpression of both HbF and HbA in BFU-e-D from CD34+ cord blood (CB) progenitors was used as a positive control ([F] CB BFU-e).
Thus, our assay conditions revealed 2 distinct waves of hEB-derived hematopoiesis, with predominance of multipotent primitive hematopoietic CFCs arising from day 7 to 12 hEB cells, followed by a wave of definitive multipotent CFCs arising from day 12 to 20 hEB cells.
hEB-derived primitive versus definitive colonies display distinct molecular phenotypes
To further characterize the various clonogenic cell types generated from hEBs, we picked and pooled individual colonies with identical morphologies for qRT-PCR analysis and compared their gene expression levels to that of pre-CFC assay hEB cells. Colonies characterized and scored as primitive or definitive had different expression profiles of key hematopoietic regulatory genes. Colonies derived from primitive erythroid CFCs (BFU-e-P and EryP) had considerably higher levels of GATA1 and EKLF than did mixed primitive (mixed-P) or mixed definitive (mixed-D) colonies, consistent with the critical roles of these genes in regulating embryonic erythropoiesis (Figure 6A). HOXB4, a homeobox gene associated with self-renewal of HSPCs,56 was expressed in high amounts preferentially in multilineage mixed colonies (either primitive and definitive). Interestingly, although SCL, LMO2, AML1b, and CDX4 were each expressed in abundant amounts in all types of colonies analyzed, the levels of CDX4 were highest in mixed-D colonies. In addition, C-MYB and AML1c (Figure 6A), known for their role in regulating multilineage definitive hematopoiesis, were expressed in high amounts exclusively in mixed erythromyeloid definitive colonies (which also expressed adult β-globin; Figure 6B).
Day 7 to 20 hEB cells contain clonogenic, multilineage erythromyeloid progenitors for both primitive and definitive hematopoiesis. (A) Kinetics of mixed, multipotent erythromyeloid primitive (Mixed CFC-P) or definitive (Mixed CFC-D) colonies. (B) Kinetics of primitive macrophage (Macro-P) colonies and definitive GM-CFC and M-CFC (scored in combination and presented together as “GMCFC-D”). Shown is a representative experiment performed 3 independent times. Large, loosely packed primitive mixed erythromyeloid colonies (C; MIXED-P, magnification × 100) containing foamy macrophages and primitive erythroblasts (C; Wright stain, MIXED CFC-P, magnification × 600, oil) peak from day 9 to 12 hEBs (along with primitive erythroid CFCs) and differentiate from blast colonies similar to those that arise from MHE clusters. (D) Foamy macrophage colonies with no evidence of monocytic precursors on Wright stains (magnification × 1000, oil) also predominated from day 9 to 15 hEBs. (E) Compact colonies containing definitive monocytes/granulocytes (MIXED CFC-D) as well as GMCFC-D (F, left and middle; magnification × 1000, oil) and M-CFC-D (F, right; Wright stain, magnification × 600, oil) arose primarily from day 12 to 20 hEBs. (G) Day 9 hEB-derived MIXED CFCs (C) were picked (2 to 3 colonies) and shown by FACS analysis to express abundant amounts of CD71 and CD45 (as well as CD13; not shown). Shown in the bottom row is the FACS profile of day 9 hEB pooled EryP colonies from the same cultures.
Day 7 to 20 hEB cells contain clonogenic, multilineage erythromyeloid progenitors for both primitive and definitive hematopoiesis. (A) Kinetics of mixed, multipotent erythromyeloid primitive (Mixed CFC-P) or definitive (Mixed CFC-D) colonies. (B) Kinetics of primitive macrophage (Macro-P) colonies and definitive GM-CFC and M-CFC (scored in combination and presented together as “GMCFC-D”). Shown is a representative experiment performed 3 independent times. Large, loosely packed primitive mixed erythromyeloid colonies (C; MIXED-P, magnification × 100) containing foamy macrophages and primitive erythroblasts (C; Wright stain, MIXED CFC-P, magnification × 600, oil) peak from day 9 to 12 hEBs (along with primitive erythroid CFCs) and differentiate from blast colonies similar to those that arise from MHE clusters. (D) Foamy macrophage colonies with no evidence of monocytic precursors on Wright stains (magnification × 1000, oil) also predominated from day 9 to 15 hEBs. (E) Compact colonies containing definitive monocytes/granulocytes (MIXED CFC-D) as well as GMCFC-D (F, left and middle; magnification × 1000, oil) and M-CFC-D (F, right; Wright stain, magnification × 600, oil) arose primarily from day 12 to 20 hEBs. (G) Day 9 hEB-derived MIXED CFCs (C) were picked (2 to 3 colonies) and shown by FACS analysis to express abundant amounts of CD71 and CD45 (as well as CD13; not shown). Shown in the bottom row is the FACS profile of day 9 hEB pooled EryP colonies from the same cultures.
Molecular phenotypes of primitive erythroid and primitive/definitive multipotent colonies by real-time qRT-PCR. Day 9 hEB colonies were scored with primitive/definitive parameters as described in the text. Primitive erythroid (BFU-e-P, EryP) colonies, primitive mixed (MIXED-P) colonies, and definitive mixed (MIXED-D) colonies were pooled (2 to 3 colonies), and RNA was analyzed by qRT-PCR methods as described in text. “Fold change in expression from undifferentiated hEB cells” was calculated using the 2–/ΔΔCT method as described in “Materials and methods,” based on the CT (threshold curve) for each target gene and internal normalizations with actin. Expression of target genes was compared with levels in undifferentiated day 9 hEB cells prior to plating. (A) qRT-PCR analysis of key regulatory genes in primitive and definitive colonies. (B) qRT-PCR expression analysis of primitive (epsilon, zeta), fetal (alpha, gamma), and definitive adult (alpha, beta) hemoglobins. (C) qRT-PCR analysis of AML1 isoforms. (D) Corresponding agarose gels of PCR products obtained at linear phases of qPCR reactions. RNA from erythroleukemia cell line K562 and cord blood were used as positive controls for qRT-PCR reactions.
Molecular phenotypes of primitive erythroid and primitive/definitive multipotent colonies by real-time qRT-PCR. Day 9 hEB colonies were scored with primitive/definitive parameters as described in the text. Primitive erythroid (BFU-e-P, EryP) colonies, primitive mixed (MIXED-P) colonies, and definitive mixed (MIXED-D) colonies were pooled (2 to 3 colonies), and RNA was analyzed by qRT-PCR methods as described in text. “Fold change in expression from undifferentiated hEB cells” was calculated using the 2–/ΔΔCT method as described in “Materials and methods,” based on the CT (threshold curve) for each target gene and internal normalizations with actin. Expression of target genes was compared with levels in undifferentiated day 9 hEB cells prior to plating. (A) qRT-PCR analysis of key regulatory genes in primitive and definitive colonies. (B) qRT-PCR expression analysis of primitive (epsilon, zeta), fetal (alpha, gamma), and definitive adult (alpha, beta) hemoglobins. (C) qRT-PCR analysis of AML1 isoforms. (D) Corresponding agarose gels of PCR products obtained at linear phases of qPCR reactions. RNA from erythroleukemia cell line K562 and cord blood were used as positive controls for qRT-PCR reactions.
Discussion
ESC differentiation has been accepted as a valid in vitro model for murine yolk sac hematopoiesis. Developing mEBs produce yolk sac–like blood islands27 coinciding with cyst formation in the EBs.57 We show herein that a similar developmental process operates in differentiating human EBs, albeit with slower kinetics than for mEBs.27
Although blood elements have recently been reported from differentiated hESCs,34,35,37 neither the delineated phases of primitive and definitive hematopoiesis nor a clonogenic human hemangioblast (which directly precedes primitive erythropoiesis in mEB systems) have yet been described. Evidence for a clonogenic mouse hemangioblast has been provided with characterization of a bipotential yolk sac–like progenitor termed the “blast colony-forming cell” (BL-CFC).38,41,43 This transient, VEGF-responsive, mEB-derived progenitor is believed to arise physiologically from mesoderm in vertebrate yolk sac and has been shown to be a common progenitor for primitive erythroblasts as well as definitive erythromyeloid blood cells.28 In mice, hemangioblasts are likely contained within a subset of KDR/flk-1+ mesoderm cells, and in developing mEBs41 they can be enriched with this marker.
Our studies have not yet demonstrated a clonogenic bipotential human hemangioblast. As a first step toward this goal, however, we have herein described an hEB-based system where both primitive and definitive human hematopoiesis are generated robustly without requirement for supplemental growth factors34,35 or xenogeneic stromal cocultures.37 Using serum-free CFC assay conditions, we have delineated 2 discrete, sequential waves of hematopoiesis resembling the primitive and definitive hematopoietic patterns occurring during human yolk sac development.1-3 This process initiates with primitive erythropoiesis and can be seen to arise secondarily from adherent MHE progenitors. A wave of primitive hematopoiesis arose from day 7 to 12 hEBs, and a distinct second wave of definitive hematopoiesis subsequently arose from day 12 to 20 hEBs. These observations indirectly suggest the development of a hematoendothelial stem cell at days 6 to 9 of hEB differentiation that either precedes or is concomitant with the burst of primitive erythropoiesis. The direct visualization of MHE colony-derived primitive hematopoietic blasts arising in intimate association with adherent CD31+VE-cadherin+ endothelial cells during this time is consistent with this hypothesis. Also in support of this hypothesis, the emergence of MHE colonies and primitive erythroid colonies coincides with a dramatic increase in expression of SCL/TAL1, GATA1, GATA2, EKLF, PU.1, C-MYB, and CDX4 in day 6 to 9 hEB cells (consistent with the known importance of these genes in the commitment of mesoderm to angiohematopoiesis). Furthermore, expression of early hematoendothelial surface markers CD31, CD34, and KDR, which all increased prior to onset of CD45 expression during day 6 to 12 hEB development, also accompanies (or probably closely follows) hematoendothelial transcription factor expression. These events are summarized schematically in Figure 7.
The ontogeny of our novel MHE colonies and their relationship to clonal bipotential hemangioblasts or more upstream mesodermal progenitors currently remains unclear. Because we have observed these semiadherent clusters to occasionally arise from secondary hEBs after attaching to the culture dish, we cannot definitively rule out that some MHE colonies may occasionally arise from oligoclonal cellular aggregates in our CFC assay.
Summary of cellular and molecular events during hEB differentiation. (A) Developmental progression of hematoendothelial gene expression during hEB differentiation. (B) Correlation of key genes and markers to hEB development of MHE, primitive (PRIM), and definitive (DEF) hematopoiesis.
Summary of cellular and molecular events during hEB differentiation. (A) Developmental progression of hematoendothelial gene expression during hEB differentiation. (B) Correlation of key genes and markers to hEB development of MHE, primitive (PRIM), and definitive (DEF) hematopoiesis.
We are intrigued, however, by the morphologic similarity of MHE clusters to Bloom and Bartelmez' classic histologic descriptions of normal intravascular human yolk sac.3 These studies described “hematocytoblasts” in precirculatory day 13 to 24 human yolk sac sections as the primary hematopoietic progenitors for developing yolk sac blood islands, which arose “by direct transformation of mesenchymal cells.” Basophilic “hematocytoblast cells with relatively large nucleoli” (similar to our Wright stains of budding hematopoietic blasts) were observed to give rise to primitive nucleated erythrocytes and also “phagocytes” in early yolk sac (day 13). In the later precirculatory yolk sac (day 24), these same “hematocytoblasts” gave rise to more definitive erythrocytes, megakaryocytes, and granulocytes, thus suggesting a common origin for both primitive and definitive hematopoiesis, as has been noted in the mouse precirculatory yolk sac.58 Interestingly, MHE colonies possess comparable developmental characteristics, including that (1) hematopoietic progenitors arise in intimate association with endothelial-mesenchymal cells, (2) the predominant hematopoietic progeny of both MHE colonies and yolk sac blood islands are nucleated primitive erythroblasts expressing embryonic/fetal hemoglobins, and (3) primitive hematopoiesis is sequentially followed by a wave of definitive erythromyelopoiesis.
Both yolk sac– and AGM-derived hematopoiesis appear to be intimately involved or directly arise from embryonic endothelial cells in all vertebrate species studied, including humans,19,20,23 thus suggesting a common hemangioblast for both locations. A recent study59 has provided evidence that hematopoiesis from hESCs may similarly arise from hEB-derived endothelial progenitors with a CD31+KDR+VE-cadherin+ phenotype.22 Our preliminary data (Figure S2) similarly reveal that day 9 to 10 CD45–CD34+CD31+ hEB cells give rise to (at least) definitive lymphohematopoietic cells.
Our results also provide insight into the role key hematopoietic genes may play in human embryonic development and introduce experimental approaches for studying mechanisms involved in normal and genetically modified hESCs. The highly conserved basic helix-loop-helix (bHLH) transcription factor SCL/TAL1 (stem cell leukemia protein),60 for example, has been shown in mice and zebrafish to play a crucial role in patterning of mesoderm into blood and endothelial lineages by regulating the development of the hemangioblast.60-66 Although its importance in T-cell leukemia67 is established, the role SCL/TAL1 plays in normal human developmental hematopoiesis remains obscure.62 Our data revealed that SCL/TAL1 was the first and most dramatically up-regulated gene coinciding with emergence of primitive hematopoiesis and was expressed abundantly in all hematopoietic colonies. In contrast, other key regulators of mouse hematopoiesis such as LMO2 (LIM domain only protein 2), a LIM-domain binding protein that physically forms a heterodimer with SCL/TAL1,68,69 KDR/flk-1, and acute myeloid leukemia-1 (AML1),70,71 were expressed in large amounts in undifferentiated hESCs, and throughout hEB differentiation, without correlation to CFC emergence. Expression analysis of the 3 isoforms of AML1 further revealed that although AML1b is widely expressed in all CFCs as well as undifferentiated hEB cells, only the AML1c isoform was expressed in mixed-definitive colonies. This result is consistent with the hypothesized role of AML1c in mouse definitive hematopoiesis.50 Another important observation was the burst of CDX4 expression accompanying SCL/TAL1 peak expression at day 9 of hEB development, which in turn coincided with the peak of primitive erythroid CFCs and subsequent emergence of definitive CFCs. Although CDX4 function in humans is unknown, this homeobox gene-regulating factor, whose absence results in complete loss of blood commitment in “kugelig” zebrafish mutants,72 may similarly be important for specifying human hemangioblast differentiation.
In summary, we have described an experimental system for the direct analysis of human embryonic hematopoietic development that proceeds in a manner appearing to model human yolk sac development. The key molecular and cellular events of hematopoietic genesis can be delineated in a manner that was previously impossible due to inaccessibility of human fetal tissue. Furthermore, the potential expansion and transplantation of hematoendothelial stem cells may provide unprecedented translational opportunities for human tissue engineering.
Prepublished online as Blood First Edition Paper, April 14, 2005; DOI 10.1182/blood-2004-11-4522.
Supported by grants from NIH T32 CA60441 (E.T.Z., C.I.C.), National Institutes of Health (NIH) K08-HL077595 (E.T.Z.), the American Society of Clinical Oncology (ASCO) Young Investigators' Award (E.T.Z.), NIH R21-HL077729 (C.I.C.), and the Institute of Cellular Engineering (ICE) at Johns Hopkins Medical Institutions (JHMI) (E.T.Z., F.B., C.I.C.).
The Johns Hopkins University holds patents on CD34 monoclonal antibodies and inventions related to stem cells. Dr Civin is entitled to a share of the sales royalty received by the university under licensing agreements between the university, Becton Dickinson Corp, and Baxter HealthCare Corp. Dr Civin is a paid consultant to Becton Dickinson Corp. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Portions of this work were presented in oral/abstract form at the 45th annual meeting of the American Society of Hematology, San Diego, CA, December 8, 2003.73
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Leslie Meszler and the Cell Imaging Core Facility at Johns Hopkins Medical Institutions (JHMI) for superb graphics support, Ozlem Topaloglu for hESC culture, Jenice D'Costa for enhanced green fluorescent protein (EGFP) lentivirus, Robert Georgantas III for excellent technical consultations in qPCR, Jeanne Wilson and Thomas Kickler for K-B stains, Xiaobing Yu for technical advice, and Linzhao Cheng and Gautam Dravid for consultations on hESC culture methods.


![Figure 3. Secondary blast colonies can arise directly from adherent MHE colonies and differentiate directly to primitive erythroblasts, macrophages, or mixed erythromyeloid colonies. During their initial differentiation, adherent MHE clusters (A; magnification × 100) often produced secondary budding, blast colonies (B, magnification × 200) with a Wright-Giemsa stain hematopoeitic blast morphology (C; magnification × 1000, oil) and that differentiated rapidly into hemoglobinizing cells. To further evaluate the clonogenic potential of MHE-derived budding hematopoietic blast colonies derived from day 7 to 9 hEB cells, they were individually picked prior to full differentiation (about 1 week after hEB plating) and recultured in H4436 supplemented with 50 ng/mL bone morphogenetic protein 4 (BMP4) (which greatly improved their replating efficiency). Single, picked, MHE-derived primary blast colonies (B) were demonstrated to be multipotential by their ability to give rise to multiple secondary mixed, macrophage, or erythroid colonies (D,E) in secondary replating experiments ([E] with 5 of 7 primary blast colonies successfully replated in a representative experiment repeated 3 times). After 4 to 6 weeks of continuous culture, prolific, mature MHE clusters (F, left panel; magnification × 40) are composed of nonadherent hematopoietic cells arising from an adherent bed of mesenchymal-endothelial cells. When nonadherent cells are extensively washed away and adherent cells were incubated overnight with acetylated Dil-LDL, approximately 20% to 30% of adherent cells were positive (F, right panel). To further evaluate the endothelial component of MHE colonies, adherent cells from individual colonies (Gi) were picked, disaggregated, analyzed by FACS, or replated in endothelial medium (EGM2) on Matrigel-coated plates. Replated adherent cells from 6 of 6 individual MHE colonies gave rise to cells with endothelial morphology (ii) and ability to take up acetylated Dil-LDL after several days of EGM2 endothelial culture. (iv) These cells were further evaluated for expression of endothelial genes including von Willebrand factor (VWF) and VE-cadherin by qRT-PCR (HU indicates human umbilical vein endothelial cell [HUVEC] RNA control; C, cord blood RNA control; E, endothelial cells from replated MHE adherent layers). More than 50% to 70% of elongated, adherent cells expressed CD34 and/or CD31 (iii, left); EGM2 replated adherent cells continued to express CD31/CD34 and were devoid of CD45 expression (iii, right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/3/10.1182_blood-2004-11-4522/6/m_zh80150582290003.jpeg?Expires=1767701035&Signature=fCsmdPxHBjcGNLddfnWHzYAHMEBx~cPQ8q3DvWKzenH41YtRlqqUpjRBPUUlHLx3Okt4APjVI0YnLBerFWsXtbQPuVcl8G03qQqyJwqazIDS9fE0-p0RaqQ0NM~hhCwezVuVmEX42Oc6CvnNLx2UZnTk7o~MKeRCqXiJL63ZK-lckHuE2rgUlU6HBZq4H7Yf2G~Fyo4-gkAhE5cy9LAZNtp6fuXEmoLs88HGhhEVyhfafAIrrCIF-dxlMrKx5BLsXPRawgAJaPFmVLSyVeqv9LIuXBknSXYEcqxb3iigg4fUM9wxFxCJmvcYwolDMGY4iDVXxckvpRjtwdv4rLMLPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Day 7 to 20 hEB cells contain clonogenic progenitors for both primitive and definitive erythropoiesis. Erythroid colonies with a “brilliant red” hemoglobinization from day 9 hEB-derived BFU-e-P (Ai; magnification × 100) and EryP (ii; magnification × 100) containing nucleated primitive erythrocytes were generated from day 7 to 15 day hEBs. Erythroid colonies containing a brownish, “salmon-red” hemoglobinization were generated from day 12 to 20 day hEBs ([B] BFU-e-D, CFU-e-D). Colonies (3 to 5 pooled) were picked from semisolid medium for staining or FACS analyses. Wright stains from multiclustered BFU-e-P (Aiii; magnification × 600) revealed an increased abundance of erythroblasts, while more mature EryP (iv; magnification × 600) contained primarily differentiated nucleated erythrocytes. Primitive erythroblast (Bl) colonies predominate from day 9 to 12 hEBs differentiate directly into nucleated erythrocytes positive for hemoglobin by benzidine staining (C, top; magnification × 400). Erythroblast colonies stain brightly positive for fetal hemoglobins by erythrosin-B K-B stains (C, bottom; magnification × 200). (Inset) Positive control K-B staining from maternal peripheral blood with 10% fetal erythrocyte cells. (D) Kinetic analysis of primitive and definitive erythroid CFCs. Shown is a representative analysis of 3 independent experiments. Although all types of erythroid CFCs had surface expression of CD71+/GlyA+ cells (F) and expressed various levels fetal hemoglobin (HbF), only day 12 to 20 BFU-e-D and CFU-e-D expressed adult hemoglobin A (HbA). Day 15 to 20 BFU-e-D or CFU-e-D often contained brownish, lysed cells. Coexpression of both HbF and HbA in BFU-e-D from CD34+ cord blood (CB) progenitors was used as a positive control ([F] CB BFU-e).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/3/10.1182_blood-2004-11-4522/6/m_zh80150582290004.jpeg?Expires=1767701035&Signature=isAqMEoHbTzR7Et4QSK5VYg7EfhQxYKsNTmdbqwc9Z5~s9w96JJkyTATadxu24PDpk-hiN6tEyApqHTs2XXSuIW6y~ZLs8huiLKJbB~53W5Uxbhg0OUGTzm-Hn9kDdtDwdum9g1SLgL7mn~3AliC2DXxdzKvS0VvMcfO2tpSLehd7g~dme~ql9MHARbivBxtU3Ft0z0oEupcurRe135ktEleD7RGDXn9BjbsckJdc6JOPywAfozDuTFTSNg2v1m7QZMWGy6ttUH7D~bfrWkVhJ2GRYxOOJYWK6dWTLyG0BmwvRSKijfVReqd1KYTZZj9ViYdcoupQ8RzKiXEe6VggQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal