Abstract
GATA1 is mutated in patients with 2 different disorders. First, individuals with a GATA1 mutation that blocks the interaction between GATA-1 and its cofactor Friend of GATA-1 (FOG-1) suffer from dyserythropoietic anemia and thrombocytopenia. Second, children with Down syndrome who develop acute megakaryoblastic leukemia harbor mutations in GATA1 that lead to the exclusive expression of a shorter isoform named GATA-1s. To determine the effect of these patient-specific mutations on GATA-1 function, we first compared the gene expression profile between wild-type and GATA-1–deficient megakaryocytes. Next, we introduced either GATA-1s or a FOG-binding mutant (V205G) into GATA-1–deficient megakaryocytes and assessed the effect on differentiation and gene expression. Whereas GATA-1–deficient megakaryocytes failed to undergo terminal differentiation and proliferated excessively in vitro, GATA-1s–expressing cells displayed proplatelet formation and other features of terminal maturation, but continued to proliferate aberrantly. In contrast, megakaryocytes that expressed V205G GATA-1 exhibited reduced proliferation, but failed to undergo maturation. Examination of the expression of megakaryocyte-specific genes in the various rescued cells correlated with the observed phenotypic differences. These studies show that GATA-1 is required for both normal regulation of proliferation and terminal maturation of megakaryocytes, and further, that these functions can be uncoupled by mutations in GATA1.
Introduction
GATA-1 and its cofactor Friend of GATA-1 (FOG-1) are required for proper development of erythroid cells, mast cells, eosinophils, and megakaryocytes (for a review, see Crispino1 ). The role for GATA-1 in megakaryocyte development was initially discovered through the creation and analysis of several lines of mutant mice that express reduced levels of GATA-1. One of these lines, ΔneoΔHS Gata1-targeted mice, expresses normal levels of GATA-1 in erythroid cells but undetectable levels of GATA-1 in megakaryocytes.2,3 As a consequence, these mutant mice display marked, persistent thrombocytopenia, with platelet counts reduced to approximately 15% of normal, and a concomitant expansion of immature megakaryocytes in their bone marrow. GATA-1–deficient megakaryocytes are smaller than wild-type cells and have multiple abnormalities, including large segmented nuclei, scant cytoplasm with a paucity of granules, underdeveloped demarcation membranes, reduced proplatelet processes, and an excess of rough endoplasmic reticulum (rER).3 Further, these cells have been reported to exhibit reduced polyploidization, with an increased proportion of cells having a 2N DNA content and fewer cells reaching a ploidy greater than 8N.4 Similarly, GATA-1–deficient platelets are also abnormal, harboring few α-granules and an excess of rER. In addition to the differentiation block, loss of GATA-1 leads to an abnormal proliferation of immature megakaryocytes, both in vivo and in vitro.4
More recently, definitive evidence of a requirement for GATA-1 in megakaryopoiesis has emerged from studies involving patients with megakaryocytic disorders. Missense mutations in the N-terminal zinc finger of GATA-1, which disrupt the interaction between GATA-1 and FOG-1, lead to several types of congenital dyserythropoietic anemia and thrombocytopenia (for a review, see Crispino1 ). In particular, patients who harbor a mutation that converts valine 205 to methionine (V205M) display severe anemia with thrombocytopenia.5 Within the bone marrow, the affected individuals harbor large, multinucleated erythroid precursors and small, dysplastic megakaryocytes containing an excess of smooth ER, eccentric nuclei, and a paucity of α-granules. Knock-in mice with a FOG noninteracting mutation in GATA-1 (valine 205 to glycine, Gata1V205G mice) displayed defects in hematopoietic cell development similar to those seen in the human disorder.6,7 Of note, Gata1V205G bone marrow progenitors formed similar numbers of megakaryocyte colonies as wild-type bone marrow, but the megakaryocytes within the colonies were significantly larger.7 Similarly, megakaryocytes in humans with X-linked thrombocytopenia, resulting from a glycine 208 to serine (G208S) mutation in GATA-1 that also disrupts the GATA-1/FOG-1 interaction, were found to be large.8
In addition to these rare disorders that involve GATA1 missense mutations, GATA1 is altered in 2 malignancies that are relatively more common: transient myeloproliferative disorder (TMD) and acute megakaryoblastic leukemia (AMKL) in Down syndrome (DS; for a review, see Crispino1 ). The vast majority of patients with these disorders harbor acquired mutations in the sequences encoding the N-terminal activation domain.9,10 Each of the mutations discovered in these patients abolishes translation of full-length GATA-1, but each allows for expression of a shorter, naturally occurring isoform named GATA-1s. This shorter variant binds DNA normally and interacts with FOG-1 to the same extent as the full-length GATA-1, but, because it lacks the N-terminal activation domain, has reduced transcriptional activation potential.9 Previous reports have also shown that GATA-1 molecules lacking the activation domain can promote terminal differentiation of erythroid cells11 and convert the 416B myeloid cell line to megakaryocytes.12 Furthermore, recent studies have shown that overexpression of GATA-1s can rescue the embryonic lethality of GATA-1–deficient mice.13 However, the consequences of GATA-1s expression in megakaryocyte development, and whether the GATA-1s mutation is functionally similar to the GATA-1 ΔneoΔHS mutation, remain to be elucidated.
The present study was undertaken to further delineate the role of GATA-1 in megakaryopoiesis and determine the consequences of GATA1 mutations identified at the molecular level in humans. To do this, we first compared the gene expression profile of wild-type and GATA-1–deficient megakaryocytes to identify genes that were significantly up-regulated or down-regulated in the absence of GATA-1. Next, we reconstituted GATA-1–deficient hematopoietic cells from fetal livers with either wild-type GATA-1, the “leukemic” isoform GATA-1s, or the FOG noninteracting mutant V205G, and analyzed the phenotype as well as the expression of selected genes shown to be misexpressed in GATA-1–deficient megakaryocytes. We show here that reconstitution with GATA-1s resulted in differentiation comparable to that of wild-type GATA-1, although GATA-1s–expressing progenitors continued to proliferate excessively. In contrast, V205G–expressing megakaryocytes displayed reduced proliferation as well as a failure to undergo differentiation. Together, these observations provide insights into how mutations in GATA1 contribute to abnormal megakaryopoiesis.
Materials and methods
Expansion and purification of megakaryocytes
Fetal livers were removed from E12.5 or E13.5 embryos dissected from wild-type and ΔneoΔHS pregnant C57Bl/6 female mice, disaggregated, and cultured as a single-cell suspension for 2 to 3 days in a serum-free expansion media containing stem cell factor (SCF), interleukin 3 (IL-3), and a low dose of erythropoietin (EPO; 2 U/mL) with nutridoma (Roche, Indianapolis, IN) in RPMI/Dulbecco modified Eagle medium (DMEM; 1:1 mixture; Gibco, Grand Island, NY). Cells were differentiated in the absence of SCF and IL-3, but in the presence of higher doses of EPO (10 U/mL) for 4 days.14 In separate experiments thrombopoietin (TPO) was included in the differentiation medium (10 ng/mL; R&D Systems, Minneapolis, MN), and the resulting megakaryocyte outgrowths were identical. Following differentiation, megakaryocytes were enriched on a 1.5%/3.0% discontinuous bovine serum albumin (BSA) gradient.15 Purity was assessed by staining for acetylcholinesterase (AChE) following standard protocols. Experiments involving the use of animals were approved by the University of Chicago Institutional Animal Care and Use Committee.
Ploidy analysis
BSA gradient-enriched megakaryocytes were fixed in 75% ice-cold ethanol and stained with propidium iodide (PI) solution containing 3.8 mM sodium citrate, 0.125 mg/mL RNase A (DNase free), and 0.01 mg/mL PI for 1 hour. DNA content was determined using BD-FACScan (BD Biosciences, Mountain View, CA).
RNA preparation and microarray assays
RNA was extracted from multiple outgrowths of purified wild-type and ΔneoΔHS megakaryocytes using TRIzol reagent (Invitrogen, Carlsbad, CA) and processed in triplicate for hybridization to Affymetrix Mouse Genome 430 2.0 Array GeneChips (Affymetrix, Santa Clara, CA). Based on the high yield, no amplification of the RNA was necessary. Analysis of microarray experiments was performed using dCHIP software.47 Comparative analysis was performed using the average of signals from the 3 replicates of each set of probes. The averaged wild-type and ΔneoΔHS samples were compared to determine genes changed 2-fold or greater with wild-type samples serving as a baseline and default settings. Genes identified in the comparison of baseline to experimental samples had to be above a threshold value of 100, with P less than .05 for the differences to be considered significant.
Retroviral transduction
cDNAs encoding wild-type GATA-1, GATA-1s, or V205G GATA-1 were subcloned into the BglII and HpaI sites of the MIGR1 retroviral vector. Primary murine fetal liver cells were infected by spinoculation as described previously16 on days 2 and 3 of expansion. Differentiation was initiated on the following day, and cells were analyzed by flow cytometry for differentiation after an additional 4 days.
Real-time RT-PCR
RNA from BSA gradient-enriched murine megakaryocytes, expanded ex vivo, was isolated using TRIzol reagent (Invitrogen). cDNA was generated from RNA using reverse transcriptase (RT) according to manufacturer's recommendations. Real-time polymerase chain reaction (PCR) was performed in 25-μL reactions. Relative quantitation of real-time PCR product was performed using the comparative ΔΔCT method (as described in the ABI Prism 7700 Sequence Detection System User Bulletin no. 2) using SYBR green fluorescent labeling and the DNA Engine Opticon 2 continuous fluorescence detection system (MJ Research, Waltham, MA). Samples were amplified in triplicate and normalized by subtracting CT value of the housekeeping gene glyceraldehyde phosphate dehydrogenase (GAPDH) sample value. Primer sequences are provided in Figure S1, available on the Blood website (see the Supplemental Figures link at the top of the online article).
Antibodies and flow cytometry
Surface staining for mouse CD41(glycoprotein IIb [GPIIb], BD PharMingen, San Diego, CA) and CD42b (GPIbα, Emfret Analytics, Wurzburg, Germany) was performed using fluorescein isothiocyanate (FITC)– and phycoerythrin (PE)–conjugated antibodies respectively, in Ca++-free, Mg++-free phosphate-buffered saline (PBS). All flow cytometry was performed on BD-FACScan and data were analyzed using FloJo software (Treestar, Ashland, OR).
Results
Comparison between wild-type and ΔneoΔHS GATA1-deficient megakaryocytes
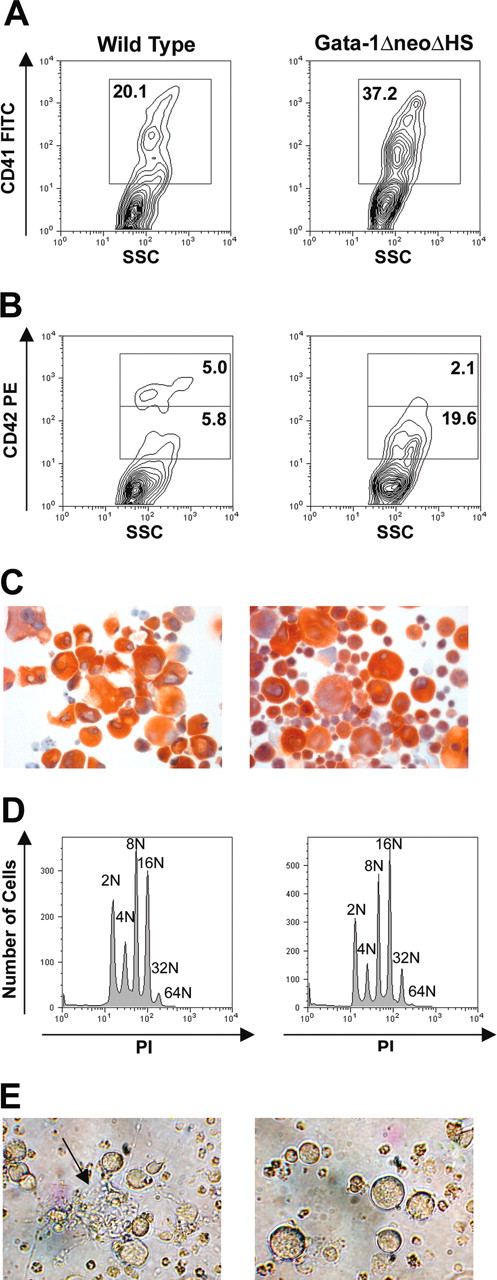
Although several reports have detailed the defects accompanying the loss of GATA-1 from megakaryocytes, to date there has been no comprehensive analysis of the aberrant gene expression program. To determine how the loss of GATA-1 leads to a global change in maturation, we collected fetal livers from E12.5 to E13.5 ΔneoΔHS (knockdown) embryos and cultured them in vitro to generate megakaryocytes. As previously reported, knockdown fetal liver progenitors expanded in vitro to a much greater extent than wild-type cells4 and led to an increased formation of CD41+ cells (Figure 1A). The increased accumulation of megakaryocyte progenitors was further confirmed by another megakaryocyte-specific marker (CD42). Interestingly, the intensity of CD42 expression was much lower in the knockdown cells (Figure 1B). Following enrichment on a BSA gradient, megakaryocyte populations were assessed to be at more than 95% purity by AChE staining (Figure 1C). Gradient-purified megakaryocytes were judged to be comparable in the endomitotic cell cycle; wild-type and ΔneoΔHS megakaryocytes exhibited a similar ploidy (Figure 1D). Finally, as expected, only wild-type megakaryocytes generated proplatelet forms (Figure 1E).
Characterization of ΔneoΔHS Gata1 knockdown megakaryocytes. (A-B) Flow cytometric analysis of megakaryocytes cultured from wild-type or GATA-1 knockdown fetal liver cells stained with the anti-CD41 (A) or CD42 (B) antibody. (C) Following in vitro expansion, wild-type and GATA-1–deficient cells were enriched on a BSA gradient and AChE staining was performed to determine the purity of enriched cells. Images were obtained using a Leica DM4000B light microscope equipped with a 20×/0.04 objective lens (Cambridge, United Kingdom). Original magnification, ×200. Images were captured using a mounted Leica DFC320 camera and Leica Firecam version 1.4 software. (D) Ploidy analysis of BSA gradient-purified megakaryocytes was performed by flow cytometry of cells stained with PI. (E) Light microscopy of differentiated cells in culture. The arrow points to proplatelet forms. Images were obtained using a Zeiss Axiovert S100 inverted microscope equipped with a 20×/0.04 objective lens (Thornwood, NY). Original magnification, ×200. Images were captured with a Nikon Coolpix 990 digital camera (Tokyo, Japan).
Characterization of ΔneoΔHS Gata1 knockdown megakaryocytes. (A-B) Flow cytometric analysis of megakaryocytes cultured from wild-type or GATA-1 knockdown fetal liver cells stained with the anti-CD41 (A) or CD42 (B) antibody. (C) Following in vitro expansion, wild-type and GATA-1–deficient cells were enriched on a BSA gradient and AChE staining was performed to determine the purity of enriched cells. Images were obtained using a Leica DM4000B light microscope equipped with a 20×/0.04 objective lens (Cambridge, United Kingdom). Original magnification, ×200. Images were captured using a mounted Leica DFC320 camera and Leica Firecam version 1.4 software. (D) Ploidy analysis of BSA gradient-purified megakaryocytes was performed by flow cytometry of cells stained with PI. (E) Light microscopy of differentiated cells in culture. The arrow points to proplatelet forms. Images were obtained using a Zeiss Axiovert S100 inverted microscope equipped with a 20×/0.04 objective lens (Thornwood, NY). Original magnification, ×200. Images were captured with a Nikon Coolpix 990 digital camera (Tokyo, Japan).
To examine the gene expression profiles of megakaryocytes with reduced GATA-1 expression, we probed Affymetrix GeneChip Mouse Genome 430 2.0 Arrays, which contain 39 000 mouse genes and expression sequence tags (ESTs). RNA for this study was isolated from BSA gradient-enriched megakaryocytes pooled from multiple litters of wild-type and ΔneoΔHS fetal livers. To minimize random fluctuations in gene expression, samples were hybridized in triplicate, and the average of 3 experiments was used for dCHIP analysis. With P set below .05, we determined that 84 genes were misexpressed 5-fold or greater in knockdown cells; 74 genes were down-regulated in the absence of GATA-1, whereas 10 were up-regulated when GATA-1 was not expressed (Tables 1 and 2; Figure S2). Gene array data have been deposited in the Gene Expression Omnibus database (accession no. GSE2527).
Genes down-regulated 5-fold or greater in ΔneoΔHS megakaryocytes
Probe set . | Gene . | Accession no. . | Description . | Fold change . |
|---|---|---|---|---|
| 1426936_at | cDNA clone 3493956 | BC002257 | EST | -49.47 |
| 1437263_at | A730089K16Rik | BB138441 | EST | -25.83 |
| 1449232_at | Gata1 | NM_008089 | GATA-binding protein 1 | -24.01 |
| 1433600_at | AW122659 | BB262415 | EST | -15.54 |
| 1434725_at | 4921521N14Rik | AV255657 | EST | -14.91 |
| 1425569_a_at | Slam | BB132695 | Signaling lymphocytic activation molecule | -14.67 |
| 1418186_at | Gstt1 | BC012254 | Glutathione S-transferase, θ1 | -14.30 |
| 1450783_at | Ifit1 | NM_008331 | Interferon-induced protein | -13.75 |
| 1449465_at | Reln | NM_011261 | Reelin | -13.74 |
| 1421882_a_at | Elavl2 | BB105998 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 | -13.73 |
| 1439728_at | AW455481 | BB477913 | EST | -13.40 |
| 1421040_a_at | Gsta2 | NM_008182 | Glutathione S-transferase, α2 | -13.03 |
| 1450621_a_at | Hbb-y | NM_008221 | Hemoglobin Y, β-like embryonic chain | -13.00 |
| 1421041_s_at | Gsta2 | NM_008182 | Glutathione S-transferase, α2 | -12.93 |
| 1435906_x_at | Gbp2 | BE197524 | Guanylate nucleotide-binding protein 2 | -12.87 |
| 1434645_at | C530008M17Rik | BB493717 | EST | -12.80 |
| 1446391_at | 12-d embryo cDNA | BB450769 | EST | -12.18 |
| 1422700_at | Alox12 | BB554189 | Arachidonate 12-lipoxygenase | -11.49 |
| 1429184_at | 9130002C22Rik | BM243571 | EST | -11.30 |
| 1422699_at | Alox12 | BB554189 | Arachidonate 12-lipoxygenase | -11.29 |
| 1425570_at | Slam | BB132695 | Signaling lymphocytic activation molecule | -11.17 |
| 1424567_at | 6330415F13Rik | BC007185 | EST | -11.12 |
| 1421144_at | Rpgrip1 | NM_023879 | Retinitis pigmentosa guanosine triphosphatase regulator | -10.89 |
| 1418240_at | Gbp2 | NM_010260 | Guanylate nucleotide-binding protein 2 | -10.85 |
| 1419127_at | 0710005A05Rik | NM_023456 | Neuropeptide Y | -10.78 |
| 1452670_at | Trrp2 | AK007972 | Transient receptor protein 2 | -10.64 |
| 1454137_s_at | 2310035L15Rik | AK009636 | EST | -10.61 |
| 1423294_at | Mest | AW555393 | γ-2 COP | -10.34 |
| 1449591_at | Casp11 | NM_007609 | Caspase 4, apoptosis-related cysteine protease | -10.05 |
| 1449305_at | F10 | NM_007972 | Coagulation factor X | -9.84 |
| 1421977_at | MMP19 | AF153199 | Matrix metalloproteinase 19 | -9.80 |
| 1436717_x_at | Hbb-y | AV156860 | Hemoglobin Y, β-like embryonic chain | -9.76 |
| 1426541_a_at | 2310067E08Rik | BF168366 | EST | -9.72 |
| 1436823_x_at | Hbb-y | AV148191 | Hemoglobin Y, β-like embryonic chain | -9.67 |
| 1452666_a_at | 1110063G11Rik | AK004359 | EST | -9.64 |
| 1451718_at | Plp | BB768495 | Proteolipid protein (myelin) | -9.34 |
| 1418392_a_at | Gbp4 | NM_018734 | Guanylate nucleotide-binding protein 3 | -8.88 |
| 1450947_at | 2610528J11Rik | AK012175 | EST | -8.87 |
| 1418493_a_at | Snca | NM_009221 | Synuclein, α | -8.86 |
| 1421278_s_at | Spna1 | BB740660 | Spectrin α1 | -8.78 |
| 1421883_at | Elavl2 | BB105998 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 | -8.75 |
| 1425571_at | Slam | BB132695 | Signaling lymphocytic activation molecule | -8.72 |
| 1432273_a_at | Dfy | AK010883 | Duffy blood group | -8.69 |
| 1431724_a_at | P2ry12 | AK013804 | Purinergic receptor P2Y, G-protein coupled 12 | -8.52 |
| 1448453_at | Hsd3b1 | NM_008293 | Hydroxysteroid dehydrogenase-1 | -8.37 |
| 1417804_at | Rasgrp2 | NM_011242 | RAS, guanyl-releasing protein 2 | -8.31 |
| 1449989_at | Mcpt2 | NM_008571 | Mast cell protease 2 | -8.14 |
| 1429223_a_at | 2310035L15Rik | AV290575 | EST | -8.14 |
| 1443703_at | Transcribed sequences | BB208080 | EST | -8.14 |
| 1417152_at | 0610020I02Rik | NM_026495 | EST | -7.82 |
| 1421277_at | Spna1 | BB740660 | Erythroid α-spectrin | -7.73 |
| 1415978_at | Tubb3 | NM_023279 | Tubulin, β3 | -7.63 |
| 1416662_at | MGC6279 | BI217574 | Sarcosine dehydrogenase | -7.48 |
| 1426543_x_at | 2310067E08Rik | BF168366 | EST | -7.47 |
| 1450298_at | Tnfsf14 | NM_019418 | Tumor necrosis factor (ligand) | -7.41 |
| 1444214_at | 2810484G07Rik | AW493179 | EST | -7.41 |
| 1437025_at | Cd28 | AV313615 | CD28 antigen | -7.34 |
| 1417597_at | Cd28 | NM_007642 | CD28 antigen | -7.19 |
| 1434900_at | AI852829 | BM196656 | EST | -7.19 |
| 1416702_at | Serpini1 | NM_009250 | Serine (or cysteine) proteinase inhibitor, clade 1 | -7.15 |
| 1442350_at | 4632424N07Rik | AV233462 | EST | -7.02 |
| 1448743_at | AU014939 | NM_138744 | Synovial sarcoma, X breakpoint 2 | -6.90 |
| 1460682_s_at | Ceacam | BC024320 | Carcinoembryonic antigen (CEA) - related cell adhesion molecule 2 | -6.80 |
| 1433796_at | C85344 | BI734389 | EST | -6.77 |
| 1424354_at | 1110007F12Rik | BC020080 | EST | -6.49 |
| 1434109_at | A930014C21 | AV291265 | EST | -6.46 |
| 1454984_at | AW061234 | AV246615 | EST | -6.44 |
| 1421588_at | Tnfsf14 | NM_019418 | Tumor necrosis factor (ligand) | -6.43 |
| 1433765_at | 6530403D07Rik | BB304622 | EST | -6.38 |
| 1428108_x_at | 1110063G11Rik | AK004359 | EST | -6.17 |
| 1448021_at | Transcribed sequences | AA266723 | EST | -6.13 |
| 1417936_at | ScyA9 | AF128196 | Chemokine (C-C motif) ligand 9 | -6.10 |
| 1450981_at | Cnn2 | BI663014 | Calponin 2 | -5.86 |
| 1427428_at | 4930572L20Rik | BC025069 | EST | -5.83 |
Probe set . | Gene . | Accession no. . | Description . | Fold change . |
|---|---|---|---|---|
| 1426936_at | cDNA clone 3493956 | BC002257 | EST | -49.47 |
| 1437263_at | A730089K16Rik | BB138441 | EST | -25.83 |
| 1449232_at | Gata1 | NM_008089 | GATA-binding protein 1 | -24.01 |
| 1433600_at | AW122659 | BB262415 | EST | -15.54 |
| 1434725_at | 4921521N14Rik | AV255657 | EST | -14.91 |
| 1425569_a_at | Slam | BB132695 | Signaling lymphocytic activation molecule | -14.67 |
| 1418186_at | Gstt1 | BC012254 | Glutathione S-transferase, θ1 | -14.30 |
| 1450783_at | Ifit1 | NM_008331 | Interferon-induced protein | -13.75 |
| 1449465_at | Reln | NM_011261 | Reelin | -13.74 |
| 1421882_a_at | Elavl2 | BB105998 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 | -13.73 |
| 1439728_at | AW455481 | BB477913 | EST | -13.40 |
| 1421040_a_at | Gsta2 | NM_008182 | Glutathione S-transferase, α2 | -13.03 |
| 1450621_a_at | Hbb-y | NM_008221 | Hemoglobin Y, β-like embryonic chain | -13.00 |
| 1421041_s_at | Gsta2 | NM_008182 | Glutathione S-transferase, α2 | -12.93 |
| 1435906_x_at | Gbp2 | BE197524 | Guanylate nucleotide-binding protein 2 | -12.87 |
| 1434645_at | C530008M17Rik | BB493717 | EST | -12.80 |
| 1446391_at | 12-d embryo cDNA | BB450769 | EST | -12.18 |
| 1422700_at | Alox12 | BB554189 | Arachidonate 12-lipoxygenase | -11.49 |
| 1429184_at | 9130002C22Rik | BM243571 | EST | -11.30 |
| 1422699_at | Alox12 | BB554189 | Arachidonate 12-lipoxygenase | -11.29 |
| 1425570_at | Slam | BB132695 | Signaling lymphocytic activation molecule | -11.17 |
| 1424567_at | 6330415F13Rik | BC007185 | EST | -11.12 |
| 1421144_at | Rpgrip1 | NM_023879 | Retinitis pigmentosa guanosine triphosphatase regulator | -10.89 |
| 1418240_at | Gbp2 | NM_010260 | Guanylate nucleotide-binding protein 2 | -10.85 |
| 1419127_at | 0710005A05Rik | NM_023456 | Neuropeptide Y | -10.78 |
| 1452670_at | Trrp2 | AK007972 | Transient receptor protein 2 | -10.64 |
| 1454137_s_at | 2310035L15Rik | AK009636 | EST | -10.61 |
| 1423294_at | Mest | AW555393 | γ-2 COP | -10.34 |
| 1449591_at | Casp11 | NM_007609 | Caspase 4, apoptosis-related cysteine protease | -10.05 |
| 1449305_at | F10 | NM_007972 | Coagulation factor X | -9.84 |
| 1421977_at | MMP19 | AF153199 | Matrix metalloproteinase 19 | -9.80 |
| 1436717_x_at | Hbb-y | AV156860 | Hemoglobin Y, β-like embryonic chain | -9.76 |
| 1426541_a_at | 2310067E08Rik | BF168366 | EST | -9.72 |
| 1436823_x_at | Hbb-y | AV148191 | Hemoglobin Y, β-like embryonic chain | -9.67 |
| 1452666_a_at | 1110063G11Rik | AK004359 | EST | -9.64 |
| 1451718_at | Plp | BB768495 | Proteolipid protein (myelin) | -9.34 |
| 1418392_a_at | Gbp4 | NM_018734 | Guanylate nucleotide-binding protein 3 | -8.88 |
| 1450947_at | 2610528J11Rik | AK012175 | EST | -8.87 |
| 1418493_a_at | Snca | NM_009221 | Synuclein, α | -8.86 |
| 1421278_s_at | Spna1 | BB740660 | Spectrin α1 | -8.78 |
| 1421883_at | Elavl2 | BB105998 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 | -8.75 |
| 1425571_at | Slam | BB132695 | Signaling lymphocytic activation molecule | -8.72 |
| 1432273_a_at | Dfy | AK010883 | Duffy blood group | -8.69 |
| 1431724_a_at | P2ry12 | AK013804 | Purinergic receptor P2Y, G-protein coupled 12 | -8.52 |
| 1448453_at | Hsd3b1 | NM_008293 | Hydroxysteroid dehydrogenase-1 | -8.37 |
| 1417804_at | Rasgrp2 | NM_011242 | RAS, guanyl-releasing protein 2 | -8.31 |
| 1449989_at | Mcpt2 | NM_008571 | Mast cell protease 2 | -8.14 |
| 1429223_a_at | 2310035L15Rik | AV290575 | EST | -8.14 |
| 1443703_at | Transcribed sequences | BB208080 | EST | -8.14 |
| 1417152_at | 0610020I02Rik | NM_026495 | EST | -7.82 |
| 1421277_at | Spna1 | BB740660 | Erythroid α-spectrin | -7.73 |
| 1415978_at | Tubb3 | NM_023279 | Tubulin, β3 | -7.63 |
| 1416662_at | MGC6279 | BI217574 | Sarcosine dehydrogenase | -7.48 |
| 1426543_x_at | 2310067E08Rik | BF168366 | EST | -7.47 |
| 1450298_at | Tnfsf14 | NM_019418 | Tumor necrosis factor (ligand) | -7.41 |
| 1444214_at | 2810484G07Rik | AW493179 | EST | -7.41 |
| 1437025_at | Cd28 | AV313615 | CD28 antigen | -7.34 |
| 1417597_at | Cd28 | NM_007642 | CD28 antigen | -7.19 |
| 1434900_at | AI852829 | BM196656 | EST | -7.19 |
| 1416702_at | Serpini1 | NM_009250 | Serine (or cysteine) proteinase inhibitor, clade 1 | -7.15 |
| 1442350_at | 4632424N07Rik | AV233462 | EST | -7.02 |
| 1448743_at | AU014939 | NM_138744 | Synovial sarcoma, X breakpoint 2 | -6.90 |
| 1460682_s_at | Ceacam | BC024320 | Carcinoembryonic antigen (CEA) - related cell adhesion molecule 2 | -6.80 |
| 1433796_at | C85344 | BI734389 | EST | -6.77 |
| 1424354_at | 1110007F12Rik | BC020080 | EST | -6.49 |
| 1434109_at | A930014C21 | AV291265 | EST | -6.46 |
| 1454984_at | AW061234 | AV246615 | EST | -6.44 |
| 1421588_at | Tnfsf14 | NM_019418 | Tumor necrosis factor (ligand) | -6.43 |
| 1433765_at | 6530403D07Rik | BB304622 | EST | -6.38 |
| 1428108_x_at | 1110063G11Rik | AK004359 | EST | -6.17 |
| 1448021_at | Transcribed sequences | AA266723 | EST | -6.13 |
| 1417936_at | ScyA9 | AF128196 | Chemokine (C-C motif) ligand 9 | -6.10 |
| 1450981_at | Cnn2 | BI663014 | Calponin 2 | -5.86 |
| 1427428_at | 4930572L20Rik | BC025069 | EST | -5.83 |
Genes up-regulated 5-fold or greater in ΔneoΔHS megakaryocytes
Probe set . | Gene . | Accession no. . | Description . | Fold change . |
|---|---|---|---|---|
| 1429977_at | 9030425L15Rik | BG797916 | EST | 5.35 |
| 1455177_at | D10Bwg0629e | BQ175532 | Abelson helper integration site | 5.70 |
| 1449876_at | Prkg1 | NM_011160 | Protein kinase, cyclic guanosine monophosphate (cGMP)-dependent, type I | 6.27 |
| 1428942_at | Mt2 | AA796766 | Metallothionein 2 | 6.55 |
| 1452163_at | Ets1 | BB151715 | E26 avian leukemia oncogene 1, 5 domain | 6.62 |
| 1433933_s_at | AI060904 | BB553107 | Solute carrier organic anion transporter | 7.96 |
| 1452318_a_at | Hsp70-1 | M12573 | Heat shock protein 1A | 9.85 |
| 1447927_at | AI595338 | BG092512 | EST | 10.16 |
| 1427127_x_at | hsp68 | M12573 | Heat shock protein 1A | 12.32 |
| 1427126_at | hsp68 | M12573 | Heat shock protein 1A | 15.85 |
Probe set . | Gene . | Accession no. . | Description . | Fold change . |
|---|---|---|---|---|
| 1429977_at | 9030425L15Rik | BG797916 | EST | 5.35 |
| 1455177_at | D10Bwg0629e | BQ175532 | Abelson helper integration site | 5.70 |
| 1449876_at | Prkg1 | NM_011160 | Protein kinase, cyclic guanosine monophosphate (cGMP)-dependent, type I | 6.27 |
| 1428942_at | Mt2 | AA796766 | Metallothionein 2 | 6.55 |
| 1452163_at | Ets1 | BB151715 | E26 avian leukemia oncogene 1, 5 domain | 6.62 |
| 1433933_s_at | AI060904 | BB553107 | Solute carrier organic anion transporter | 7.96 |
| 1452318_a_at | Hsp70-1 | M12573 | Heat shock protein 1A | 9.85 |
| 1447927_at | AI595338 | BG092512 | EST | 10.16 |
| 1427127_x_at | hsp68 | M12573 | Heat shock protein 1A | 12.32 |
| 1427126_at | hsp68 | M12573 | Heat shock protein 1A | 15.85 |
Validation of the microarray data by quantitative RT-PCR
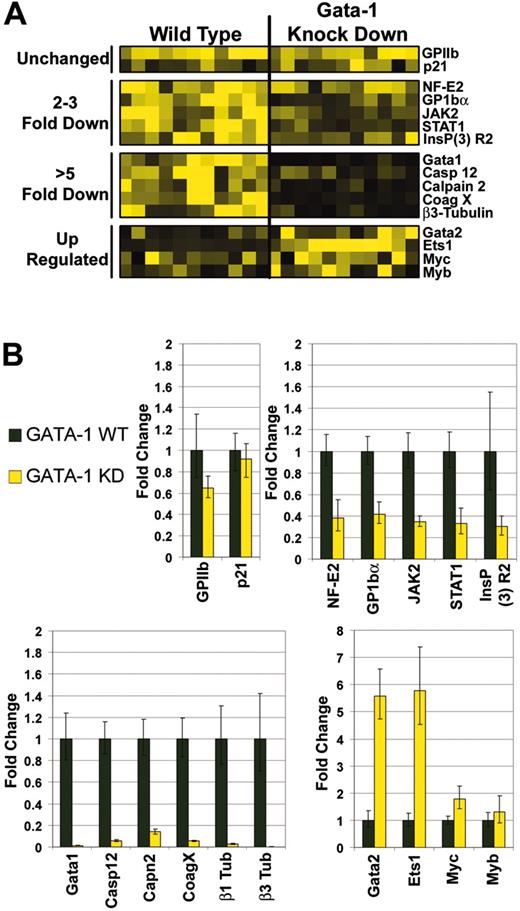
To validate the changes observed in the array study, we selected 17 genes for follow-up real-time quantitative RT-PCR (qRT-PCR) analysis. We selected this subset based on the known relevance of a gene to megakaryopoiesis or because the change detected by the array suggested a novel role in megakaryocyte development. These genes fell into 4 distinct categories: genes that were not significantly affected by the loss of GATA-1, those that were partially reduced (2- to 5-fold), those greatly reduced (> 5-fold), and those whose expression increased in the absence of GATA-1 (Figure 2A). As expected, GATA-1 expression was reduced nearly 100-fold in the knockdown megakaryocytes (Figure 2B bottom left).
Genes that were suggested by microarray analysis to be only modestly affected by the loss of GATA-1 included GPIIb and p21Waf1/Cip1. Follow-up qRT-PCR studies revealed that these genes were expressed at comparable levels in the 2 populations (Figure 2B). It is surprising that GPIIb resides within this group because it has previously been implicated as a GATA-1 target gene.17-19 Note that this is consistent with the normal cell-surface expression of CD41 detected by flow cytometry (Figure 1A). These findings suggest that other transcription factors, possibly GATA-2 or ETS1, may be substituting for GATA-1 in promoting the transcription of a subset of GATA-1 target genes (see “Discussion”).
qRT-PCR also confirmed the down-regulation of each of the other 10 genes assayed. For example, consistent with previous reports, expression of NF-E2 (p45 subunit) and GP1bα was diminished in the absence of GATA-1 (Figure 2B top right). The reduction in GP1bα mRNA expression paralleled the reduced staining by anti-CD42 antibodies detected by flow cytometry (Figure 1B). Of the other genes whose expression was reduced 2- to 3-fold in the absence of GATA-1, Stat1 and Jak2 are notable. Jak2 plays an important role in TPO signaling20 and its reduction suggests that GATA-1–deficient megakaryocytes may have aberrant cytokine signaling. Furthermore, recent work has demonstrated that Stat1 is essential for normal erythropoiesis21 and may also participate in megakaryopoiesis.22
Consistent with a defect in platelet function, levels of expression of several genes involved in calcium signaling and proteolysis were misexpressed in GATA-1–deficient megakaryocytes. For example, expression of the inositol 1,4,5-triphosphate receptor 2 (InsP3-R2), which encodes ER channels that control calcium efflux,23 was markedly reduced (Figure 2B upper right). Furthermore, P2Y receptors, G-coupled 1 and 12, were down-regulated significantly in mutant megakaryocytes (Table 1). These latter genes encode plasma membrane receptors that, when activated, lead to calcium oscillations within the cell.24 Finally, calpain 2, a calcium-dependent cysteine protease that cleaves procaspases 9 and 3 in response to calcium efflux,25 and caspase 12, which has been implicated in inside-out signaling by the αIIb/β3 integrin,26 were down-regulated nearly 10- and 20-fold, respectively (Figure 2B bottom left).
An important participant in proplatelet formation is β1-tubulin. Previous reports have shown that this gene is significantly down-regulated in GATA-1– and p45 NF-E2–deficient megakaryocytes,27,28 consistent with their maturation defect. In line with these reports, we observed that GATA-1 knockdown megakaryocytes expressed nearly 50-fold less β1-tubulin (Figure 2B bottom left). Interestingly, the gene array comparison revealed that β3-tubulin, a family member not previously implicated in megakaryopoiesis, was reduced almost 100-fold (Figure 2B bottom left). One other novel finding to emerge from this study is that expression of coagulation factor X, whose tissue-specific expression has been assigned to the liver,29 was significantly diminished in GATA-1–deficient megakaryocytes (approximately 20-fold; Figure 2B bottom left).
Finally, of the genes that are up-regulated in GATA-1–deficient megakaryocytes, 2 stand out prominently: Gata-2 and Ets-1 (Figure 2B bottom right). GATA-2 expression was elevated 5.5-fold, reminiscent of what has been observed in erythroid cells, in which GATA-2 has been shown to be significantly up-regulated in the absence of GATA-1.30,31 Similarly, Ets-1 expression increased nearly 6-fold when GATA-1 was absent. Because the promoters of many megakaryocyte-specific genes, such as GPIIb, harbor tandem GATA and Ets sites,17,32 it is reasonable to speculate that overexpression of GATA-2 and Ets-1 may substitute for GATA-1 in their expression within the ΔneoΔHS megakaryocytes. Notably, qRT-PCR analysis demonstrated that c-myc and c-myb were changed less than 2-fold in the GATA-1 knockdown cells (Figure 2B bottom right).
Rescue with various alleles of GATA-1 reveals that GATA-1s restores differentiation but does not inhibit the excessive proliferation of mutant megakaryocytes
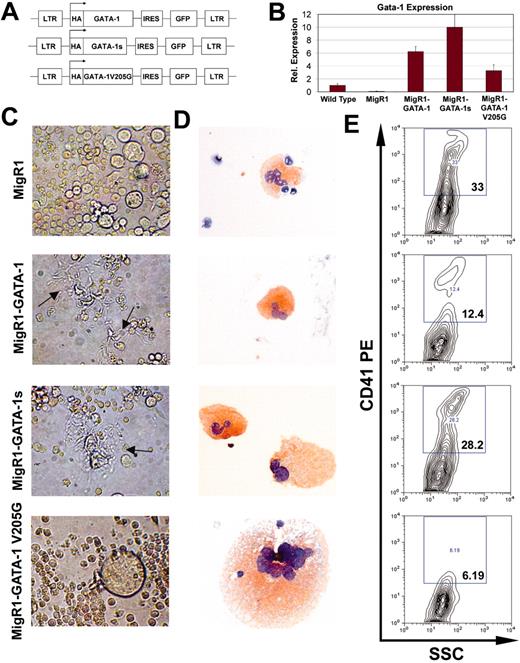
To investigate the requirements for the different functional domains of GATA-1 on megakaryopoiesis, we first introduced cDNAs encoding wild-type GATA-1, GATA-1s, or the V205G mutant, into the MIGR1 retroviral backbone, which includes an internal ribosome entry site-green fluorescent protein (IRES-GFP) cassette (Figure 3A). Following spinoculation of the GATA-1–deficient fetal liver cells with retroviral supernatant, GFP+ cells were isolated by flow cytometry and cultured in vitro in differentiation media for 3 days to generate megakaryocytes. The expression of each of the GATA-1 cDNAs was confirmed by expression in the ϕNX retroviral producer cells (Figure S3) and by qRT-PCR of RNA isolated from BSA-enriched megakaryocytes (Figure 3B). GATA-1 cDNAs were expressed at levels 4- to 10-fold higher than that of endogenous full-length GATA-1 in wild-type megakaryocytes. Notably, RT-PCR studies performed with a primer set that specifically amplifies full-length GATA-1 transcripts revealed that the endogenous ΔneoΔHS GATA-1 locus was not activated by expression of GATA-1s cDNA (data not shown). Next, we examined the effect of wild-type GATA-1 and the GATA-1 mutants on megakaryocyte proliferation and maturation. Differentiated cells were assessed for proplatelet formation in culture as well as morphology and size on AChE-stained cytospin preparations. Flow cytometry was used to determine the number of CD41+ megakaryocytes obtained in culture.
As expected, ΔneoΔHS cells expressing wild-type GATA-1 produced proplatelet forms, whereas those infected with the empty vector did not (Figure 3C). These former cells stained positive for CD41 and AChE and displayed wild-type nuclear and cytoplasmic morphology (Figure 3D-E). Notably, the total number of megakaryocytes formed, as determined by the percentage of CD41+ cells in the culture, was markedly reduced in comparison to the MIGR1-infected knockdown cells. This confirms that expression of GATA-1 can restore normal proliferation and maturation to GATA-1–deficient cells.
Surprisingly, megakaryocytes engineered to express exclusively GATA-1s generated proplatelets (Figure 3C). However, in contrast to cells expressing wild-type GATA-1, GATA-1s–expressing fetal liver progenitors continued to exhibit the hyperproliferative phenotype characteristic of GATA-1–deficient megakaryocytes, as evidenced by the high percentage of CD41+ cells (Figure 3E). These findings were unexpected, because one hallmark of TMD and DS-AMKL (which express exclusively GATA-1s) is the presence of abnormal, immature megakaryocytes within the bone marrow, which resemble, in many ways, ΔneoΔHS megakaryocytes.
Requirements for GATA-1 in megakaryocyte gene expression. (A) Affymetrix probe set hybridization results for 16 representative genes that were differentially affected by the absence of GATA-1. Highest levels of expression are depicted as yellow squares; the lowest levels or no expression are represented by black squares. (B) Quantitative real-time RT-PCR validation of 16 genes selected from the array data. Data were divided into the following groups: genes that were not significantly affected (P > .05, upper left), ones that were down-regulated 2- to 3-fold (P ranged from < .002 to .02, upper right), those that were reduced more than 5-fold (P < .001, lower left), and genes that were induced in the absence of GATA-1 (GATA-2 and Ets-1, P < .001; myc P < .05; myb P < .5; lower right). Fold changes of gene expression in the GATA-1 knockdown cells (yellow bars) are shown relative to the levels detected in wild-type cells (black bars: wild-type expression was set to 1). Means ± SD for 3 experiments are shown.
Requirements for GATA-1 in megakaryocyte gene expression. (A) Affymetrix probe set hybridization results for 16 representative genes that were differentially affected by the absence of GATA-1. Highest levels of expression are depicted as yellow squares; the lowest levels or no expression are represented by black squares. (B) Quantitative real-time RT-PCR validation of 16 genes selected from the array data. Data were divided into the following groups: genes that were not significantly affected (P > .05, upper left), ones that were down-regulated 2- to 3-fold (P ranged from < .002 to .02, upper right), those that were reduced more than 5-fold (P < .001, lower left), and genes that were induced in the absence of GATA-1 (GATA-2 and Ets-1, P < .001; myc P < .05; myb P < .5; lower right). Fold changes of gene expression in the GATA-1 knockdown cells (yellow bars) are shown relative to the levels detected in wild-type cells (black bars: wild-type expression was set to 1). Means ± SD for 3 experiments are shown.
Examination of fetal liver cultures infected with the V205G retrovirus revealed that the FOG interaction is required for the ability of GATA-1 to promote proplatelet formation (Figure 3C). Furthermore, in the absence of the GATA-1/FOG-1 interaction, only 6% of the cells expressed CD41 on day 3 (Figure 3E). Concomitant with the reduced proliferation, V205G megakaryocytes were 3- to 4-fold larger than either ΔneoΔHS, wild-type, or GATA-1s megakaryocytes (Figure 3C-D). These results suggest that the V205G mutation interferes with the proliferation and maturation of megakaryocyte progenitors. Furthermore, the data confirm that the GATA-1/FOG-1 interaction is required for proplatelet development.
Megakaryocyte genes fall into several classes with differential requirements for GATA-1 functionality
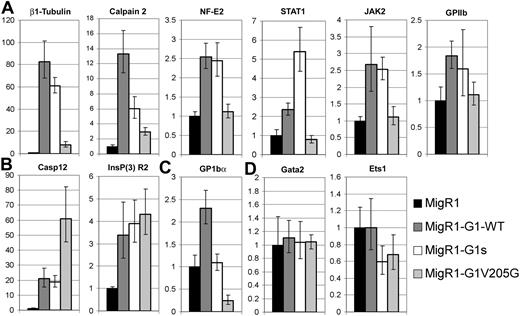
We next isolated mRNA from BSA gradient-enriched megakaryocytes generated from the rescue studies in Figure 3 and analyzed the expression of a number of genes by qRT-PCR. Interestingly, these genes fell into several classes. First, expression of a subset of the genes was restored by GATA-1s in a similar manner or better than wild-type GATA-1 (Figure 4A). This group includes β1-tubulin, NF-E2, Stat1, Jak2, and calpain 2. These genes are all known or predicted to be involved in (or associated with) terminal maturation and are likely concomitant with proplatelet formation. None of these genes was activated to a significant extent in the cells expressing V205G, consistent with the absence of proplatelet formation. Notably, both GATA-1 and GATA-1s induced expression of GPIIb mRNA to a small extent (between 1.5- and 2-fold), whereas V205G did not alter its expression in comparison to the GATA-1 knockdown cells. The second class of genes did not require either the activation domain or the FOG-interaction domain of GATA-1 for their expression (Figure 4B). For example, caspase 12 was induced in GATA-1s–expressing cells to the same extent as the wild-type reconstitution. Interestingly, caspase 12 expression was significantly higher in the cells expressing V205G, suggesting that FOG-1 may act as a repressor of its expression. InsP3-R2 expression was also restored in all rescued megakaryocytes. The third class of genes, represented by GPIbα, requires the N-terminus of GATA-1 for expression. This is a significant finding because it strongly suggests that GATA-1s megakaryocytes are not completely mature. Indeed, there are likely to be many genes in this class, which represents a set of genes most likely to be direct target genes of GATA-1. Finally, the fourth class of genes includes those whose expression was increased in the absence of GATA-1, such as Gata-2, Ets-1, and c-myc (Figure 2B). Surprisingly, we failed to detect repression of Gata-2, Ets-1 (Figure 4B) and c-myc (data not shown) in any of the reconstituted megakaryocytes. The fact that these genes were not down-regulated during the (ostensibly) normal maturation of wild-type reconstituted cells shows that repression of these genes is not an essential step in megakaryopoiesis.
GATA-1s rescues proplatelet formation, but not the hyperproliferative phenotype. (A) Constructs used in this study. Wild-type or mutant GATA-1 cDNAs were introduced into the MIGR1 vector backbone, which includes an IRES-GFP cassette. (B) GATA-1 expression in the rescued population was determined by qRT-PCR. There was no statistically significant difference between full-length GATA-1 and GATA-1s expression in the reconstitutions, but there was a significant difference between GATA-1 and V205G expression (P < .02). Means ± SD for 3 experiments are shown. (C) Light microscopy for the generation of proplatelet forms by the rescued cells. Notable features include the presence of proplatelets in the GATA-1s rescued cells and the presence of very large megakaryocytes in the population expressing V205G. Original magnification × 320. Images were obtained as described in the legend of Figure 1E. (D) AChE-stained cytospins of an aliquot of the 3-day cultures. Note the presence of a very large AChE-stained megakaryocyte in the population expressing V205G. Original magnification × 200. Images were obtained as described in the legend of Figure 1C. (E) Cell surface expression of CD41 on megakaryocytes after 3 days of in vitro differentiation, assayed by flow cytometry.
GATA-1s rescues proplatelet formation, but not the hyperproliferative phenotype. (A) Constructs used in this study. Wild-type or mutant GATA-1 cDNAs were introduced into the MIGR1 vector backbone, which includes an IRES-GFP cassette. (B) GATA-1 expression in the rescued population was determined by qRT-PCR. There was no statistically significant difference between full-length GATA-1 and GATA-1s expression in the reconstitutions, but there was a significant difference between GATA-1 and V205G expression (P < .02). Means ± SD for 3 experiments are shown. (C) Light microscopy for the generation of proplatelet forms by the rescued cells. Notable features include the presence of proplatelets in the GATA-1s rescued cells and the presence of very large megakaryocytes in the population expressing V205G. Original magnification × 320. Images were obtained as described in the legend of Figure 1E. (D) AChE-stained cytospins of an aliquot of the 3-day cultures. Note the presence of a very large AChE-stained megakaryocyte in the population expressing V205G. Original magnification × 200. Images were obtained as described in the legend of Figure 1C. (E) Cell surface expression of CD41 on megakaryocytes after 3 days of in vitro differentiation, assayed by flow cytometry.
Megakaryocyte genes fall into several classes based on their requirement for different GATA-1 functional domains. Quantitative RT-PCR analysis of expression of 10 genes in the different rescued populations. These genes fell into 4 classes: (A) genes rescued by wild-type GATA-1 and GATA-1s but not by V205G, (B) genes rescued by GATA-1 and both GATA-1s and V205G, (C) genes rescued by GATA-1 but not GATA-1s or V205G, and (D) genes not significantly affected by wild-type or either of the GATA-1 mutants. Expression in knockdown cells infected with the MIGR1 retrovirus (black bars, far left) was set to 1, whereas the fold changes observed in the wild-type (dark gray bars), GATA-1s (white bars) and V205G (light gray bars, far right) reconstitutions are shown. Means ± SD for 3 experiments are shown.
Megakaryocyte genes fall into several classes based on their requirement for different GATA-1 functional domains. Quantitative RT-PCR analysis of expression of 10 genes in the different rescued populations. These genes fell into 4 classes: (A) genes rescued by wild-type GATA-1 and GATA-1s but not by V205G, (B) genes rescued by GATA-1 and both GATA-1s and V205G, (C) genes rescued by GATA-1 but not GATA-1s or V205G, and (D) genes not significantly affected by wild-type or either of the GATA-1 mutants. Expression in knockdown cells infected with the MIGR1 retrovirus (black bars, far left) was set to 1, whereas the fold changes observed in the wild-type (dark gray bars), GATA-1s (white bars) and V205G (light gray bars, far right) reconstitutions are shown. Means ± SD for 3 experiments are shown.
Discussion
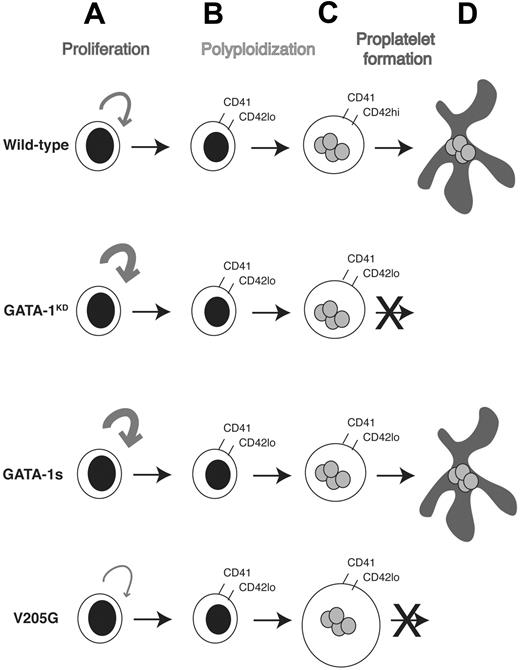
GATA-1 is a key regulator of normal hematopoietic cell development that, when mutated, leads to severe human disorders involving both erythroid cells and megakaryocytes. In this report, we provide several interesting and novel insights into the role of GATA-1 in megakaryopoiesis (summarized in Figure 5). First, we have shown that the loss of GATA-1 does not affect the polyploidization of megakaryocytes grown under our conditions. Furthermore, genes implicated in this process, including cyclin E and cdc6,33,34 were not altered during the ex vivo expansion of megakaryocytes from fetal liver (data not shown). Second, the defect in proplatelet formation observed in GATA-1–deficient cells can be rescued by expression of the leukemic isoform GATA-1s. Thus, the activation domain of GATA-1 is not essential for terminal differentiation of megakaryocytes. Third, the hyperproliferative phenotype seen in GATA-1–deficient cells is not rescued by GATA-1s, suggesting that the activation domain is essential for proper expression of genes that regulate megakaryocyte proliferation. In contrast, expression of the V205G mutant reduced the proliferation of megakaryocyte progenitors to levels substantially below that observed in wild-type cultures. This finding implicates the interaction between GATA-1 and FOG-1 as key for the proper control of megakaryocyte proliferation. Taken together, our work shows that proper regulation of gene expression by GATA-1 in megakaryocytes requires both the N-terminal activation domain and the residues within the N-terminal zinc finger that mediate the interaction with FOG-1. In our studies, the GATA-1 cDNAs were expressed at levels 4- to 10-fold higher than endogenous GATA-1 in wild-type megakaryocytes. Thus, it is possible that the rescue of proplatelet formation by GATA-1s is a consequence of its high expression level. Interestingly, previous studies have shown that a high level of expression of GATA-1s was necessary to rescue the embryonic lethality of GATA-1–deficient mice (GATA-1.05 strain).13 From these studies it can be concluded that GATA-1s may be a less active form of GATA-1 with respect to terminal differentiation of both megakaryocytes and erythroid cells.
Model of the differential requirements for GATA-1 in megakaryocyte gene expression and development. Committed megakaryocyte progenitors (A) normally display limited proliferation. However, in the absence of GATA-1, or in the presence of GATA-1s, these progenitors hyperproliferate and produce a greater number of CD41+ megakaryocytes, which also express low levels of CD42 (B). In contrast, when the ability of GATA-1 to interact with FOG-1 is disrupted, progenitors expand to a lesser degree. Megakaryocytes then undergo repeated rounds of DNA synthesis without cell division (polyploidization) to generate a polyploid cell (C). Mutagenesis of GATA-1 did not affect the endomitosis of megakaryocytes. However, alterations in GATA-1 led to differential effects on terminal maturation. In the absence of GATA-1, or in the presence of the V205G mutant, terminal maturation was blocked. When GATA-1s was expressed, however, megakaryocytes produced proplatelet forms (D) and were similar to wild-type cells in morphology and gene expression.
Model of the differential requirements for GATA-1 in megakaryocyte gene expression and development. Committed megakaryocyte progenitors (A) normally display limited proliferation. However, in the absence of GATA-1, or in the presence of GATA-1s, these progenitors hyperproliferate and produce a greater number of CD41+ megakaryocytes, which also express low levels of CD42 (B). In contrast, when the ability of GATA-1 to interact with FOG-1 is disrupted, progenitors expand to a lesser degree. Megakaryocytes then undergo repeated rounds of DNA synthesis without cell division (polyploidization) to generate a polyploid cell (C). Mutagenesis of GATA-1 did not affect the endomitosis of megakaryocytes. However, alterations in GATA-1 led to differential effects on terminal maturation. In the absence of GATA-1, or in the presence of the V205G mutant, terminal maturation was blocked. When GATA-1s was expressed, however, megakaryocytes produced proplatelet forms (D) and were similar to wild-type cells in morphology and gene expression.
Some implied GATA-1 target genes are not affected by loss of GATA-1
In previous studies, the absence of GATA-1 was reported to lead to reduced expression of several key megakaryocyte-specific genes, including NF-E2, GPIbα, platelet factor 4, and c-mpl.4 In this report, we extend these findings to encompass the entire megakaryocytic genome. Interestingly, we show that not all presumptive GATA-1 target genes are affected by the loss of GATA-1. For example, GPIIb, which has been suggested to be a GATA-1 target gene,17-19 was not significantly reduced. Thus, it is likely that other factors, possibly GATA-2 or Ets-1 or both, which are up-regulated in GATA-1–deficient megakaryocytes, can substitute for GATA-1 on a subset of GATA-1 target genes. Megakaryocytes may harbor at least 2 classes of GATA-1 target genes: one group that harbors a site absolutely dependent on GATA-1 and a second group with a promiscuous site that binds GATA-2. Alternatively, the second class may be controlled by a different set of transcription factors in the absence of GATA-1. Another gene that has been implicated as a GATA-1 target gene in megakaryocytes is cdc6.34 Because expression of this gene was not affected by the loss of GATA-1, other transcription factors likely are responsible for its regulation.
Differences between megakaryocyte and erythroid gene expression and proliferation
It has become clear that GATA-1, in addition to activating target genes, also directly represses gene expression during the course of erythroid differentiation. For example, using a GATA-1 inducible erythroid cell line (G1E cells), Rylski and colleagues35 demonstrated that GATA-1 promotes both terminal erythroid maturation and G1 cell-cycle arrest, in part by directly binding to and repressing expression of the c-myc promoter. Other genes that have been shown to be repressed by GATA-1 during erythroid differentiation include Gata-2 and c-myb.31,36,37 The domain of GATA-1 that most likely mediates transcriptional repression is the FOG-interaction surface because the V205G mutant fails to promote repression of these genes during erythroid differentiation.38 Recent studies have further confirmed that the interaction between GATA-1 and FOG-1 is essential for repression of GATA-2.39,40
Consistent with a role for GATA-1 in repressing GATA-2 expression in hematopoietic cells, we observed that GATA-2 mRNA was significantly up-regulated in megakaryocytes derived from ΔneoΔHS fetal liver progenitors. However, we were surprised to note that ectopic expression of GATA-1 in these fetal liver cells, during their ex vivo expansion and subsequent megakaryocyte differentiation, did not drive repression of GATA-2. This failure occurred despite the ability of GATA-1 to reconstitute normal proliferation and differentiation. Thus, unlike erythroid maturation, repression of GATA-2 expression does not appear to be an essential step in megakaryopoiesis. Consistent with a role for persistent GATA-2 expression in megakaryocytes, overexpression of GATA-2 in K562 cells promoted megakaryocyte differentiation, whereas it inhibited that of erythroid cells.41 Furthermore, GATA-2 has been shown to promote megakaryocytic maturation of the 416B myeloid line.42
Whereas erythroid cells undergo G1 arrest concomitant with terminal differentiation, maturing megakaryocytes enter an endomitotic phase in which cells proceed through prophase and metaphase, but exit anaphase prematurely.33 Hence, the factors that mediate these differences in the cell cycle are likely to have differential effects on megakaryocytes versus erythroid cells. In line with these findings, GATA-1 did not influence p21 expression in megakaryocytes (Figure 2B), despite the observation that GATA-1 directly induces p21 expression in an erythroid cell line.43 Thus, even though erythroid cells and megakaryocytes differentiate from a common precursor, both under the control of GATA-1, insights gained from studies in erythropoiesis cannot be immediately extrapolated to megakaryocytes.
Implications for mechanisms of leukemogenesis
Essentially all cases of TMD and DS-AMKL harbor mutations in GATA1 that block translation of full-length GATA-1, but lead to expression of the shorter GATA-1s isoform.44 It has generally been assumed that the GATA1 mutation contributes to the block in differentiation, whereas other genetic changes, most likely arising from trisomy 21, provide a proliferative signal for the development of leukemia. If this is the case, why do the megakaryocytes in this study, which have been engineered to express GATA-1s without GATA-1, mature to proplatelet forms? There are likely 3 possible explanations for this discrepancy. First, there may be a difference between the activities of GATA-1s in murine versus human progenitors. This would be reminiscent of the finding that a dominant-negative form of CCAAT/enhancer-binding protein α (C/EBPα) inhibits differentiation of human, but not murine, progenitors.45 Second, perhaps GATA-1s is expressed at very low levels in TMD and DS-AMKL blasts and these levels are insufficient to provoke normal maturation. Third, it may be the case that GATA1 mutations do not provide the block in differentiation, but rather the proliferative signal. Overexpression of other genes from chromosome 21, or mutations in other as yet undefined genes, would then be implicated as causing the maturation defect. Interestingly, a recent study has shown that transgenic mice that overexpress Bach1, which resides on human chromosome 21, develop abnormal megakaryocytes with reduced proplatelet formation and impaired endomitosis.46 Although the precise mechanisms by which GATA1 mutations contribute to DS-AMKL are unclear, the finding that cells expressing GATA-1s proliferate excessively suggests that the mutations confer a growth advantage to these malignant cells. Future studies to identify genes that are not properly expressed by GATA-1s will aid in our understanding of this intriguing leukemia.
Prepublished online as Blood First Edition Paper, April 28, 2005; DOI 10.1182/blood-2005-02-0551.
Supported in part by a grant from the National Cancer Institute, R01 CA-101774, and by a junior faculty award from the American Society of Hematology. A.M. designed and performed the research, analyzed the data, and assisted with preparation of the manuscript. J.C. designed the research, analyzed the data, and wrote the manuscript.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Paresh Vyas for supplying the ΔneoΔHS mice and for helpful discussions, and Michael McDevitt, Sandeep Gurbuxani, Yanfei Xu, Gina Mundschau, Cindy Leung, and Monika Stankiewicz for comments on the manuscript. Additional thanks to Hui Liu, Bretton Mularski, and Jed Kim for excellent technical assistance. Finally, special thanks to Xinmin Li and the Functional Genomics Facility, as well as to James Marvin and Ryan Duggan of the Flow Cytometry Facility at the University of Chicago.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal