Abstract
Heterozygous mutations of the human telomerase RNA template gene (TERC) have been described in patients with acquired aplastic anemia and the autosomal dominant form of dyskeratosis congenita (DKC). Patients with mutations in both TERC alleles have not yet been reported. Here, we report a patient with DKC who inherited 2 distinct TERC sequence variants from her parents; a deletion (216_229del) in one and a point mutation (37A>G) in the other allele of the TERC gene. Her marrow was hypocellular and showed an abnormal clone [46, XX t(7; 21)(q34;q22)]. The telomere lengths in leukocytes of the patient and her relatives were shorter than those of the age-matched controls and were progressively shorter in subsequent generations of family members with the 216_229del allele. Telomerase enzymatic levels in lymphocytes from the patient were approximately half of those measured in healthy controls. The 216_229del mutation failed to reconstitute telomerase activity in transfected cells, but, when coexpressed with the 37A>G variant, telomerase activity was only modestly suppressed. These clinical and laboratory findings support the concept that telomerase levels in human hematopoietic stem cells are tightly controlled as even moderately reduced levels result in accelerated telomere shortening and eventual marrow failure.
Introduction
Dyskeratosis congenita (DKC) is an inherited bone marrow failure syndrome characterized by abnormal skin pigmentation, nail dystrophy, and mucosal leucoplakia.1,2 The main causes of death are bone marrow failure, immunodeficiency, pulmonary complications, and malignancies.3 X-linked recessive, autosomal recessive, and autosomal-dominant forms of DKC have been reported. While the genetic basis for the autosomal recessive form of the disease is unknown, the X-linked form has been linked to mutations in the dyskerin protein,4,5 whereas mutations in the gene that encodes the human telomerase RNA template component (TERC) have been shown to cause the autosomal-dominant form of the disease.5-7 Such patients with DKC carry one normal and one mutant TERC gene allele.6
Telomerase is a ribonucleoprotein (RNP) complex with 2 main components: a protein (telomerase reverse transcriptase; TERT) with RNA-dependent–DNA-polymerase activity and an integral TERC RNA, which provides a template to synthesize telomeric DNA repeats.8 Recent data have shown that DKC-associated mutations of the TERC gene can cause changes in the delicate interplay between telomerase RNA conformational states9-11 and produce varying degrees of impairment in telomerase enzymatic activity when tested in a cell-based transfection assay.10,12 These observations may contribute to the differences in the severity of the disease in the families of various patients with DKC.6
Whereas mutation in 1 of the 2 copies of the telomerase RNA gene in humans is often associated with either DKC or marrow failure, complete loss of telomerase RNA is tolerated for at least several generations in laboratory mice.13 To explain these differences, it has been proposed that telomerase levels and telomere length are more tightly controlled in human than in murine cells, effectively providing an additional barrier to tumor growth in humans that does not exist in mice.14,15 To further delineate the role of telomerase in human biology, we studied a patient with inborn errors in both copies of the telomerase RNA gene.
Patient, materials, and methods
Cloning of TERC mutations 37A>G and 216_229del
The study described in this article was approved by the University of British Columbia institutional review board. Informed consent was provided according to the Declaration of Helsinki. DNA was isolated from peripheral blood mononuclear cells using DNAzol (Invitrogen, Burlington ON, Canada). Genomic DNA was amplified with Taq Polymerase (Boehringer Ingelheim, Laval, QC, Canada) for 35 cycles at 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 45 seconds. Oligos used were o550 (3′ primer) TGGCCGACTTTGGAGGTGCC and o536 (5′ primer) TCATGGCCGGAAATGGAACT. Polymerase chain reaction (PCR) products were subcloned into pGEM/T (Promega, Madison, WI), and selected clones were sequenced bidirectionally using SP6 and T7 primers (Nucleic Acid Protein Service Unit at UBC, Vancouver, BC, Canada).
X-chromosome inactivation studies
Quantitative PCR amplification of the CAG repeat in the human androgen receptor with and without prior digestion of the DNA with the enzyme HpaII was carried out as described.16 PCR products from leukocyte DNA were analyzed using an Applied Biosystems 373A DNA sequencer (Applied Biosystems, Foster City, CA). Both the patient and her mother have 2 alleles of the androgen receptor gene which can be distinguished by size (PCR products 291 base pair [bp] and 300 bp). In the undigested specimen of the patient, both alleles were represented, whereas in the HpaII-digested sample 100% of the PCR product was from the 291-bp allele.
Telomere length measurements
The average telomere length in various nucleated cells from peripheral blood was measured by flow fluorescence in situ hybridization (FISH)17 with modifications as described.18 Measurements of 392 individuals with blood counts in the normal range over the entire age range were used to develop telomere length distribution profiles encompassing less than 1%, less than 10%, 50%, greater than 90%, and greater than 99% of the telomere length measurements for each cell type at a given age (Gabriela Baerlocher and P.M.L., unpublished data).
Telomerase reconstitution assay in human VA13+ TERT cells
Recombinant murine retrovirus containing the TERT transcript and puromycin-resistant gene was used to infect VA13 human lung fibroblasts transformed by simian virus 40 large T-antigen (American Type Culture Collection [ATCC], Manassas, VA) expressing neither the TERT nor the TERC component of the human telomerase complex. About 100 puromycinresistant colonies were pooled at the end of the selection and expanded for use in the subsequent experiments.
Wild-type or mutant TERC DNAs (2 μg) in either the pBud-CE 4.3 vector backbone (Stratagene, La Jolla, CA) or in the pcDNA3 plasmid (Invitrogen) were transfected into VA13– TERT cells (at ∼ 70% confluence) in a 6-well polystyrene dish using SuperFect transfection reagent (Qiagen, Valencia, CA). Approximately 48 hours after transfection, cellular extracts were prepared in 1 × cyclic hydroxamic acid–containing peptide (CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) lysis buffer (Intergen, Atlanta, GA). Telomerase activity of the cellular extracts at 2 × 104 cells was monitored using the TRAPeze Telomerase Detection Kit as per manufacturer's directions (Intergen), except that the polymerase chain reaction was performed as follows: 1 cycle of 95°C for 2 minutes, 25 cycles of 94°C for 10 seconds, 50°C for 30 seconds, 72°C for 30 seconds, and 1 cycle of 72°C for 5 minutes. The telomeric DNA repeats were separated on a 12% native polyacrylamide gel and visualized by phospho-imaging (Molecular Dynamics, Sunnyvale, CA).
Telomerase assay in primary cells
Approximately 3 × 105 primary lymphocytes were grown in RPMI 1640 medium (Gibco, Gaithersburg, MD) containing 10% fetal calf serum (Gibco), 1% human serum (Sigma, St Louis, MO), 100 U/mL interleukin 2 (StemCell Technologies, Vancouver, Canada), and 5 × 105 irradiated feeder RJK B cells. After 4 days of incubation, 1 mL medium was replaced with fresh medium. At day 7, cell lysate was prepared in 1 × CHAPS lysis buffer and used in the fluorescent telomeric repeat amplification (fluorescent-TRAP) assay as described by the manufacturer (Chemicon International, Temecula, CA).
Telomerase reconstitutions in rabbit reticulocyte lysates
In vitro telomerase reconstitution assay was carried out as described.19 Briefly, an in vitro transcription-translation system (Promega) was used to synthesize TERT protein from the pCR3-TERT vector in the presence of 100 ng, 10 ng, and 1 ng of the various in vitro–transcribed and gel-purified TERC RNAs. Whenever 2 different TERC RNA variants were used, approximately half the amount of each RNA species was used to equal the final concentrations to either 100 ng, 10 ng, or 1 ng. One microliter of the reconstituted lysates was used for telomerase measurements using the TRAP assay.20
Northern blotting analysis
Wild-type or mutant pBud-TERC DNAs (3 μg) were transfected into VA13– TERT cells using SuperFect transfection reagent as described above. Approximately 48 hours after transfection, Trizol reagent was used to extract total cellular RNA (Invitrogen). Northern blot analysis was performed essentially as described previously.10 The amounts of TERC RNAs and those of the β-actin in all samples were quantified using the ImageQuant analysis software (Molecular Dynamics). The expression levels of the TERC RNA were normalized to those of the housekeeping β-actin gene by dividing the TERC values in each of the lanes with the amounts of the β-actin gene in the respective lanes. The ratios of TERC gene expression for each of the samples (Figure 3E, lanes 1-5) to that of the wild-type TERC sample (lane 6) were calculated.
Results
Clinical presentation and family history
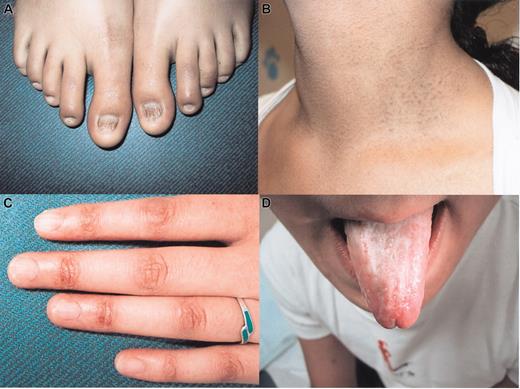
A 17-year-old female patient presented with a history of easy bruising, nail dystrophy (Figure 1A), atypical skin pigmentation (Figure 1B-C), and oral leukoplakia (Figure 1D). She had no hepatosplenomegaly or adenopathy. The peripheral blood film showed abnormal erythrocyte morphology with marked macrocytosis, oval macrocytes, and teardrop forms. No circulating blasts were seen and the granulocytes appeared normally granulated. Blood counts of the patient and several of her relatives were low (Table 1).
Results of blood and marrow examination
Person, age at time of analysis, y . | WBC count, × 109/L (range)* . | Neutrophils, × 109/L (range)* . | Lymphocytes, × 109/L (range)* . | Hemoglobin level, g/L (range)* . | MCV, fL (range)* . | Platelets, × 109/L (range)* . | BM . |
|---|---|---|---|---|---|---|---|
| Patient, 17† | 1.37 (3.9-10.2) | 0.62 (1.9-7.3) | 0.52 (0.9-3.5) | 68 (117-149) | 105.2 (80-95) | 13 (165-397) | Abnormal |
| Mother, 45 | 2.5 (4-11) | 1.3 (2-8) | 0.9 (1-4) | 117 (115-160) | 102 (80-100) | 203 (150-400) | Normal |
| Father, 46 | 4.7 (4-11) | 2.4 (2-8) | 1.8 (1-4) | 145 (135-180) | 91 (80-100) | 186 (150-400) | Unavailable |
| Brother, 16 | 5.7 (3.9-10.2) | 2.8 (1.9-7.3) | 2.1 (0.9-3.5) | 144 (131-169) | 90 (80-95) | 187 (165-397) | Unavailable |
| Sister, 12 | 4.4 (3.9-10.2) | 2.0 (1.6-7.9) | 1.5 (0.9-3.5) | 126 (117-149) | 92 (77-92) | 260 (180-440) | Unavailable |
| Maternal cousin, 12 | 3.1 (3.9-10.2) | 0.93 (1.5-5.6) | 1.88 (0.9-3.5) | 108 (118-146) | 94 (77-92) | 65 (180-440) | Unavailable |
| Maternal aunt, 44 | 5.42 (4.2-10.8) | 3.4 (1.9-7.4) | 1.52 (1-3.3) | 121 (114-143) | 98 (83-98) | 141 (160-390) | Unavailable |
| Maternal grandmother, 67 | 4.9 (4.2-10.8) | 2.3 (1.9-7.4) | 1.6 (1-3.3) | 99 (114-143) | 96 (83-98) | 209 (160-390) | Unavailable |
Person, age at time of analysis, y . | WBC count, × 109/L (range)* . | Neutrophils, × 109/L (range)* . | Lymphocytes, × 109/L (range)* . | Hemoglobin level, g/L (range)* . | MCV, fL (range)* . | Platelets, × 109/L (range)* . | BM . |
|---|---|---|---|---|---|---|---|
| Patient, 17† | 1.37 (3.9-10.2) | 0.62 (1.9-7.3) | 0.52 (0.9-3.5) | 68 (117-149) | 105.2 (80-95) | 13 (165-397) | Abnormal |
| Mother, 45 | 2.5 (4-11) | 1.3 (2-8) | 0.9 (1-4) | 117 (115-160) | 102 (80-100) | 203 (150-400) | Normal |
| Father, 46 | 4.7 (4-11) | 2.4 (2-8) | 1.8 (1-4) | 145 (135-180) | 91 (80-100) | 186 (150-400) | Unavailable |
| Brother, 16 | 5.7 (3.9-10.2) | 2.8 (1.9-7.3) | 2.1 (0.9-3.5) | 144 (131-169) | 90 (80-95) | 187 (165-397) | Unavailable |
| Sister, 12 | 4.4 (3.9-10.2) | 2.0 (1.6-7.9) | 1.5 (0.9-3.5) | 126 (117-149) | 92 (77-92) | 260 (180-440) | Unavailable |
| Maternal cousin, 12 | 3.1 (3.9-10.2) | 0.93 (1.5-5.6) | 1.88 (0.9-3.5) | 108 (118-146) | 94 (77-92) | 65 (180-440) | Unavailable |
| Maternal aunt, 44 | 5.42 (4.2-10.8) | 3.4 (1.9-7.4) | 1.52 (1-3.3) | 121 (114-143) | 98 (83-98) | 141 (160-390) | Unavailable |
| Maternal grandmother, 67 | 4.9 (4.2-10.8) | 2.3 (1.9-7.4) | 1.6 (1-3.3) | 99 (114-143) | 96 (83-98) | 209 (160-390) | Unavailable |
WBC indicates white blood cells; MCV, mean corpuscular volume, HBF, fetal hemoglobin.
Normal range for given age.
HBF of patient was 16%, less than 1.2% of the normal range.
Bone marrow biopsy and aspirate of the patient showed a hypocellular marrow with cellularity estimated between 5% and 10%. There was relative erythroid hyperplasia with dyserythropoietic changes that included multinuclear erythroblasts, defective hemoglobinization, Howell Jolly bodies, and vacuolization. Granulopoietic activity was sparse with large band forms and occasional pelgeroid nuclei. Blasts were not increased, and no blast-cell clusters were found on the biopsy. Megakaryocytes were markedly decreased. The iron stain showed modest amounts of stainable iron in the scant bone marrow particles. No ring sideroblasts were seen, but rare sideroblasts were observed. Cytogenetic studies of the bone marrow revealed a chromosomally abnormal clone with an apparently balanced translocation between the long arms of chromosomes 7 and 21 [46, XX, t(7;21) (q34;q22)] in 23 of 25 cells analyzed. X-chromosome inactivation studies confirmed complete (100:0) clonality of the bone marrow. The number of colony-forming cells (CFCs) in the bone marrow measured was reduced (< 10 CFC/105 nucleated cells). However, no blast-cell colonies or clusters were identified. Taken together, these findings are consistent with a hypoplastic marrow failure syndrome.
The patient has been followed closely for the past 2 years. She required several blood transfusions during the first year but eventually responded to erythropoietin injections 3 times a week with her hemoglobin rising to 110 g/L. Her platelet count and neutrophil count have remained stable. Repeat bone marrow examination has continued to show no increase in blasts, suggesting no evolution of the abnormal clone. The morphology of the bone marrow has remained stable. Clinically, she has noticed an increase in her skin pigmentation, particularly on her neck and the extensor surfaces of her extremities. Bone marrow transplantation is being considered as a treatment option. Her siblings are not HLA-matched and would not have been suitable as donors even if they were HLA-matched.7 A search through the unrelated bone marrow registries has not identified any potential matched donors or closely matched cord blood units.
Clinical features of a patient (F17) with dyskeratosis congenita. (A) Nail dystrophy, (B-C) abnormal skin pigmentation, and (D) mucosal leucoplakia.
Clinical features of a patient (F17) with dyskeratosis congenita. (A) Nail dystrophy, (B-C) abnormal skin pigmentation, and (D) mucosal leucoplakia.
The patient has a younger sister and a brother who are clinically well. However, her mother had previously been found to have mild neutropenia (Table 1) and lyonization studies showed a 65:35 X-chromosome inactivation pattern (within the normal range). A bone marrow examination was done on the mother and the morphology was normal. The parents are nonconsanguineous and of multiethnic heritage. A maternal male first cousin in Holland also had previously presented with neutropenia and thrombocytopenia that has been persistent over 2 years when rechecked on a visit to Canada.
Sequence analysis shows that the patient carries 2 different alleles of the TERC gene
To study possible involvement of the TERC gene in the disease of this female patient, leukocyte DNA encompassing the 451-base TERC coding region was amplified by PCR, cloned, and sequenced. This analysis revealed the presence of 2 abnormalities in the TERC coding sequence in the patient (Figure 2A-C): a substitution at position 37 (37A>G) in one set of the DNA clones and a deletion (216_229del) in the other. Further analysis of family members showed that the deletion allele was inherited from the mother and the point mutation from the father. A pedigree showing the distribution of the TERC haplotypes in available family members is shown in Figure 2B. The location of the mutations identified in this study relative to the predicted secondary structure of the human TERC RNA21 is shown in Figure 2C.
Molecular analysis and telomere length in a family with hTERC mutations. (A) Sequence and PCR analysis of the hTERC gene in patient F17 and family members F43, M44, M14, and F10. The 37A>G substitution is shown in yellow and the nucleotide (nt) 216 to 229 deletions are shown in blue. The presence of both abnormalities is represented by the black symbol. (B) Family pedigree showing inheritance of variant TERC genes (color codes as in panel A). Numbers indicate the ages of the individuals at the time of enrollment into the study. Question marks indicate the absence of clinical information for these individuals. (C) Cartoon illustration of the 2 sequence variants detected in this study (37A>G and 216_229del) relative to the predicted secondary structure of the human telomerase RNA (TERC), based on the phylogenetic analysis of more than 30 different vertebrate TERC RNA sequences (adapted from Chen et al21 ). The predicted paired (P1 and P4.1) and junctional regions (J4/4.1) are indicated. (D-E) Telomere-length measurements of the granulocytes (D) and lymphocytes (E) from the individual family members are shown relative to those (curves) of individuals in the general population to have the expected telomere lengths at certain ages (curves are based on a study of 392 control individuals; G. Baerlocher and P.M.L., unpublished data). (F) Diagram showing wild-type and mutant TERC RNA vector sequences used in this study. Note the adenine to guanine substitution at position 37 should still allow for “wobble” base-pairing interaction with its uracil base at position 187. Deletion of nucleotides 216 to 229 in the unpaired region of the RNA is shown by the dashed line. Replacements of primary sequences with complementary sequences are shown in bold.
Molecular analysis and telomere length in a family with hTERC mutations. (A) Sequence and PCR analysis of the hTERC gene in patient F17 and family members F43, M44, M14, and F10. The 37A>G substitution is shown in yellow and the nucleotide (nt) 216 to 229 deletions are shown in blue. The presence of both abnormalities is represented by the black symbol. (B) Family pedigree showing inheritance of variant TERC genes (color codes as in panel A). Numbers indicate the ages of the individuals at the time of enrollment into the study. Question marks indicate the absence of clinical information for these individuals. (C) Cartoon illustration of the 2 sequence variants detected in this study (37A>G and 216_229del) relative to the predicted secondary structure of the human telomerase RNA (TERC), based on the phylogenetic analysis of more than 30 different vertebrate TERC RNA sequences (adapted from Chen et al21 ). The predicted paired (P1 and P4.1) and junctional regions (J4/4.1) are indicated. (D-E) Telomere-length measurements of the granulocytes (D) and lymphocytes (E) from the individual family members are shown relative to those (curves) of individuals in the general population to have the expected telomere lengths at certain ages (curves are based on a study of 392 control individuals; G. Baerlocher and P.M.L., unpublished data). (F) Diagram showing wild-type and mutant TERC RNA vector sequences used in this study. Note the adenine to guanine substitution at position 37 should still allow for “wobble” base-pairing interaction with its uracil base at position 187. Deletion of nucleotides 216 to 229 in the unpaired region of the RNA is shown by the dashed line. Replacements of primary sequences with complementary sequences are shown in bold.
Telomere lengths of the affected family members are shorter than age-matched healthy individuals
The telomere length in nucleated blood cells from the patient and her relatives was measured using the flow FISH technique (Figure 2D-E). The telomere length in both granulocytes and lymphocytes from the patient and her siblings was well below that seen in the first (lowest) percentile of age-matched control individuals. The telomere length of the parents was at the 1% percentile of age-matched control individuals, whereas cells from the patient's 67-year-old maternal grandmother (who carried the 216_229del allele) showed a more modestly reduced telomere length. Taken together, these data are consistent with the progressive telomere shortening observed in sequential generations of individuals with an abnormal telomerase RNA gene resulting in “disease anticipation.”22
Telomerase reconstitution assays show reduced enzymatic activities
Comparative phylogenetic sequence analysis of more than 30 different vertebrate telomerase RNAs suggests a shared highly conserved secondary structure composed of 4 major domains: the pseudoknot, CR4-5, box H/ACA, and CR7.21 Experimental data indicate that all 4 domains contribute to telomerase function in vivo.10,11,23 The DKC-associated mutations reported here (ie, 37A>G and 216_229del) map to the P1 helix and to a highly variable region that consists of the P4.1 stem and the adjacent J4/4.1 junction, respectively (Figure 2C).
To study the functional effects of these novel TERC abnormalities, the mutant alleles were used to reconstitute telomerase activity in human cells. For these studies, TERC expression vectors carrying each of the mutant alleles were used to transfect telomerase-deficient human VA13 cells that had been engineered to constitutively express the TERT protein. Cell extracts from such VA13-TERT cells collected approximately 48 hours after transfection were then used in a PCR-based TRAP assay to measure telomerase activity.
As shown in Figure 3A, the DKC-associated 37A>G mutation did not show a noticeable reduction in telomerase enzymatic activity (Figure 3A, lanes 4-6) as compared with the wild-type gene (lanes 1-3). In contrast, a significant reduction in telomerase function was observed for the 216_229del mutation (lanes 7-9). However, when both mutations were coexpressed, telomerase enzymatic activity was only moderately reduced (by approximately 50%) as compared with samples in which either 2 wild-type copies of the TERC gene or a wild-type and the 37A>G TERC gene were coexpressed from a single vector (lanes 10-12, 15-17, and 21-23). A modest reduction in telomerase activity was also observed in cells expressing both the wild-type and the 216_229del genes (lanes 18-20). These results indicated that neither mutant allele had a dominant-negative phenotype. Surprisingly, both mutant alleles appeared equally capable to reconstitute telomerase activity in a rabbit reticulocyte lysate in vitro telomerase reconstitution assay (Figure 3B).
Telomerase activity in telomerase reconstitution assays and in primary cells. (A) Wild-type (WT) or the different indicated abnormal TERC sequence variants were expressed either individually (lanes 1-9) or simultaneously in different combinations (lanes 10-12, 15-23) from either the cellular elongation factor 1α (EF1α) or the viral cytomegalovirus (CMV) promoter, which are located within the pBud-CE 4.3 vector (Strategene) in the transfected VA13+ TERT cell line. Serial 5-fold dilutions of the transfected cell lysates were used to ensure the linearity of the PCR reactions. (–) is the result of the heat-inactivated sample similar to the one shown in lane 1, whereas (+) shows the PCR products amplified from the positive TSR8 DNA template provided in the TRAPeze kit (Chemicon International). IC shows PCR products amplified from an internal control to ensure equal loading of the samples. (B) TRAP results of the in vitro reconstitutions of the telomerase complexes using the rabbit reticulocyte lysate (RRL; Promega) to translate a wild-type copy of the TERT protein in the presence of the indicated synthetic TERC RNA variants individually or in combinations. Various dilutions of the TERC RNAs (100 ng, 10 ng, and 1 ng) were used for each RNA sample. No TERC indicates similar RRL sample without the TERC RNA; –, heat-inactivated sample containing wild-type TERC; +, PCR products amplified from the positive TSR8 DNA template provided in the TRAPeze kit (Chemicon International). (C) TRAP results of the VA13+ TERT cells that were transfected with plasmid DNAs that contain nucleotide substitutions shown in Figure 2F, lines 5 to 9, including the junctional J4/4.1 sequence and sequences predicted to either abolish the formation of the P4.1 helical stem [P4.1(lt) or P4.1(rt)] or restore it [P4.1(ltrt)]. (D) TRAP results of the primary T lymphocytes obtained from either the patient F17 (lanes 1-3) with 2 different TERC abnormalities or an unrelated healthy donor control F19 (lanes 4-6). Lanes 7 to 9 show the PCR products obtained from the positive control K562 cell lysate, indicated as (+). IC shows PCR products amplified from an internal control to ensure equal loading of the samples. (E) Northern blotting analysis of total cellular RNA extracted from VA13+ TERT cells that were transfected with a similar set of DNA constructs as shown in panel A. TERC indicates telomerase RNA template transcripts; β-actin, housekeeping β-actin message. The amounts of TERC RNAs and those of the β-actin in all samples were quantified using the ImageQuant analysis software (Molecular Dynamics). The expression levels of the TERC RNA were normalized to those of the housekeeping β-actin gene by dividing the TERC values in each of the lanes with the amounts of the β-actin in the respective lanes. The ratios of TERC gene expression for each of the samples (lanes 1-5) to that of the wild-type TERC sample (lane 6) are shown in the bottom of the gel.
Telomerase activity in telomerase reconstitution assays and in primary cells. (A) Wild-type (WT) or the different indicated abnormal TERC sequence variants were expressed either individually (lanes 1-9) or simultaneously in different combinations (lanes 10-12, 15-23) from either the cellular elongation factor 1α (EF1α) or the viral cytomegalovirus (CMV) promoter, which are located within the pBud-CE 4.3 vector (Strategene) in the transfected VA13+ TERT cell line. Serial 5-fold dilutions of the transfected cell lysates were used to ensure the linearity of the PCR reactions. (–) is the result of the heat-inactivated sample similar to the one shown in lane 1, whereas (+) shows the PCR products amplified from the positive TSR8 DNA template provided in the TRAPeze kit (Chemicon International). IC shows PCR products amplified from an internal control to ensure equal loading of the samples. (B) TRAP results of the in vitro reconstitutions of the telomerase complexes using the rabbit reticulocyte lysate (RRL; Promega) to translate a wild-type copy of the TERT protein in the presence of the indicated synthetic TERC RNA variants individually or in combinations. Various dilutions of the TERC RNAs (100 ng, 10 ng, and 1 ng) were used for each RNA sample. No TERC indicates similar RRL sample without the TERC RNA; –, heat-inactivated sample containing wild-type TERC; +, PCR products amplified from the positive TSR8 DNA template provided in the TRAPeze kit (Chemicon International). (C) TRAP results of the VA13+ TERT cells that were transfected with plasmid DNAs that contain nucleotide substitutions shown in Figure 2F, lines 5 to 9, including the junctional J4/4.1 sequence and sequences predicted to either abolish the formation of the P4.1 helical stem [P4.1(lt) or P4.1(rt)] or restore it [P4.1(ltrt)]. (D) TRAP results of the primary T lymphocytes obtained from either the patient F17 (lanes 1-3) with 2 different TERC abnormalities or an unrelated healthy donor control F19 (lanes 4-6). Lanes 7 to 9 show the PCR products obtained from the positive control K562 cell lysate, indicated as (+). IC shows PCR products amplified from an internal control to ensure equal loading of the samples. (E) Northern blotting analysis of total cellular RNA extracted from VA13+ TERT cells that were transfected with a similar set of DNA constructs as shown in panel A. TERC indicates telomerase RNA template transcripts; β-actin, housekeeping β-actin message. The amounts of TERC RNAs and those of the β-actin in all samples were quantified using the ImageQuant analysis software (Molecular Dynamics). The expression levels of the TERC RNA were normalized to those of the housekeeping β-actin gene by dividing the TERC values in each of the lanes with the amounts of the β-actin in the respective lanes. The ratios of TERC gene expression for each of the samples (lanes 1-5) to that of the wild-type TERC sample (lane 6) are shown in the bottom of the gel.
The naturally occurring mutations abolish telomerase function by perturbing the proper secondary structural folding of the RNA
Since the 216_229del deletion spans 2 structural regions of the TERC RNA (ie, the P4.1 stem and the J4/4.1 junction) (Figure 2C), we also examined each of the regions separately. Replacing 7 bases of the J4/4.1 junction from positions 217 to 223 by their complementary bases (Figure 2F, line 5) abolished telomerase enzymatic activity (Figure 3C, J4/4.1). The 216_229del mutation also spans 6 of the 8 nucleotides that make up the left strand of the P4.1 stem (Figure 2C). We also substituted these 6 bases from positions 224 to 229 with their complements (Figure 2F, line 7), which were designed to disrupt the secondary structure of this stem. Likewise, the corresponding bases located on the opposite strand of the helix at positions 343 to 349 were also mutated to their complementary bases (Figure 2F, line 8). Each of the latter mutations significantly reduced telomerase activity [Figure 3C, P4.1(lt) and P4.1(rt)]. Interestingly, combining these complementary mutations in a single TERC molecule to restore the helical structure of this stem (Figure 2F, line 9) did not fully restore the optimal telomerase enzymatic activity [Figure 3C, P4.1(ltrt)]. Together, these data indicate that both the primary sequences and the predicted secondary structures of the P4.1 stem and the J4/4.1 junction may be required for optimal telomerase function.
Reduced telomerase activity observed in primary lymphocyte culture
Lymphocytes collected from patient F17 and an unrelated closely age-matched healthy individual (F19) were expanded in tissue culture. Cell lysates from exponentially growing cultures were used for semiquantitative measurements of telomerase activity using the PCR-based TRAP assay. As shown in Figure 3D, telomerase levels appeared modestly reduced in the F17 patient sample (lanes 1-3) as compared with either the F19 control (lanes 4-6) or cell lysate prepared from the K562 leukemia cells (lanes 7-9).
To study the possibility that the 216_229del mutation alters the stability or expression level of TERC RNA, we performed Northern blot analysis on total cellular RNA extracted from VA13+ TERT cells transfected with a similar set of plasmids as shown in Figure 3A. Relative to TERC RNA levels of either the wild-type sequence or the 37A>G variant (Figure 3E, lanes 1, 3, 4, and 6) a noticeable reduction in the level of the 216_229del RNA level was observed (lane 2). Because telomerase activity was recovered in rabbit reticulocyte lysate reconstituted with the synthetic 216_229del RNA (Figure 3B) we speculate that the 216_229del RNA is unstable when expressed in cells.
Discussion
Telomerase levels in human hematopoietic stem cells are critically important to sustain hematopoiesis over a life time. Patients with modest telomerase deficiency resulting from inherited mutations in one of the copies of the genes encoding components of the telomerase enzyme such as the telomerase RNA template (TERC) gene2,6,11 (this report) or the telomerase reverse transcriptase (hTERT) gene24 often suffer from what appears to be an acquired marrow failure syndrome. Patients with mutations in TERC often present with physical signs of dyskeratosis congenita,2 which were not observed in patients with abnormalities in the TERT gene.24 The requirement for adequate telomerase levels in human hematopoietic stem cells is strikingly different from the situation in the laboratory mouse where complete lack of telomerase RNA gene is tolerated for several generations.13 The nature of this species difference in the role of telomerase is currently poorly understood. Telomerase levels appear to limit the growth of normal human lymphocytes25 and leukemic cells,26 and further studies on the role of telomeres and telomerase in the biology of normal and malignant (stem) cells are urgently needed.
In this report, we describe a patient who inherited mutations in both alleles of the TERC gene (37A>G and 216_229del). She presented with typical features of dyskeratosis congenita: abnormal skin pigmentation, nail dystrophy, and oral leucoplakia (Figure 1). Of note, the expression of these features varied markedly between tissues and sites. For example, nail dystrophy was evident on the toenails but the fingernails only showed signs of ridging. It is possible that variable penetrance of the phenotypic features in individual patients with DKC reflects heterogeneity at the level of stem cells more or less compromised as a consequence of overall telomere shortening. This notion is supported by the observation that disease anticipation is observed in families with inherited mutations in the TERC gene which is associated with progressive telomere shortening22 (and this report).
The 2 TERC mutations observed in this family were characterized in various functional assays. On the basis of the collective evidence, we conclude that the 37A>G mutation results in a minor telomerase deficiency that is not readily evident in telomerase reconstitution assays. In contrast, the 216_229del mutation causes a marked reduction in telomerase activity. We extrapolate that the presence of both mutations in the cells of our patient disabled telomerase activity to slightly below 50% of normal levels. The extremely short telomeres and hypoplastic marrow in this patient underscores the importance of telomerase levels in the biology of human hematopoietic stem cells.
The 37A>G mutation is located in the lower strand of a 19-bp bulged P1 stem that is not highly conserved.21 Nevertheless, compensatory mutational analysis reported elsewhere10 has indicated that some base pairing in the P1 stem occurs and is required for optimal telomerase function. Recently, Ly et al10,11 and Chen and Greider27 reported that the base pairings of this P1 stem serve to define the template boundary for telomeric DNA repeat synthesis of the human telomerase RNP complex. Nevertheless, we found no major deleterious effect of the 37A>G mutation in our in vitro assays. Perhaps, this single nucleotide change is not sufficient to disrupt the P1 helix, especially since this guanine base substitution should still allow hydrogen bonding to occur via “wobble” base-pairing interaction with its uracil partner at position 187 (Figure 2F, line 2). It is therefore possible that the effect of the 37A>G substitution on telomerase function is minor, at a level that is undetectable by our in vitro assays, yet may still cause defective telomere maintenance in vivo. Notably, the telomere lengths in cells from family members who carry this allele in addition to a wild-type copy (Figure 2A-B, M44 and M14) fell within the extreme lower end of the normal range observed in a reference population (Figure 2D-E).
The deletion of nucleotides 216 to 229 (216_229del) present in the patient F17 and her relatives F67, F43, and F10 spans 1 of the 3 hypothetical helices that have been predicted by aligning various vertebrate telomerase RNA species.21 The deletion spans part of the P4.1 stem and the J4/4.1 junctional sequence (Figure 2C, F) that constitute the most variable region in the predicted vertebrate telomerase RNA secondary structure, the “hypervariable paired region.”21 The variation in sequence and in length of this hypervariable region between the distantly related vertebrate species made the structural prediction less reliable, and it was suggested that this region does not play an important role in telomerase biologic function.21 However, the results presented here (Figure 3A,D) suggest that at least part of the hypervariable region is required for optimal telomerase enzymatic function and that the primary sequences of both the J4/4.1 junction and the P4.1 stem appear to be indispensable for optimal telomerase function (Figure 3C). While our data do not exclude the possible formation of alternative structures involving these sequences, our results assign for the first time a functional role for this part of the telomerase RNA. We therefore infer from these data that telomerase function in family members F67, F43, and F10 who carry the 216_229del TERC variant may also be compromised.
While TERC RNA was expressed at essentially normal levels from the 37A>G variant (Figure 3E, lanes 1, 3, 4, and 6), expression of the 216_229del RNA appeared somewhat reduced (lane 2). As shown in Figure 3C, maintaining both the primary sequence and/or secondary structure of this region are also needed for optimal telomerase enzymatic function. As a result, a decrease in 216_229del RNA integrity coupled with an alteration of TERC RNA secondary structural folding because of the deletion might effectively compromise both the assembly and function of the telomerase RNP complex in cells. However, when this form of RNA was expressed together with either the 37A>G variant or the wild-type TERC, only moderate reductions in telomerase activity were observed (Figure 3A,D). These observations are consistent with deficient production of stable, functional telomerase RNA in these cells. The striking clinical phenotype produced by the modest telomerase deficiency in this patient supports the notion that telomerase levels are very tightly controlled in human hematopoietic stem cells. Patients who inherit both telomerase deficiency and short telomeres appear to be particularly at risk for the development of DKC and marrow failure.
Prepublished online as Blood First Edition Paper, May 10, 2005; DOI 10.1182/blood-2005-01-0247.
Supported by the special fellowship of the Leukemia-Lymphoma Society of America and the American Cancer Society-Emory Winship Cancer Institute grant (H.L.) and the National Institutes of Health (NIH) (AI36636 and AI40317) (T.G.P.). Work in the laboratory of P.M.L. is supported by NIH (grant AI029524), the Canadian Institute of Health Research (MOP38075), and the Terry Fox Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Irma Vulto, Elizabeth Chavez, and Paulette Allard for technical help with, respectively, telomere length analysis, cytogenetic procedures, and telomerase enzymatic assays.


![Figure 3. Telomerase activity in telomerase reconstitution assays and in primary cells. (A) Wild-type (WT) or the different indicated abnormal TERC sequence variants were expressed either individually (lanes 1-9) or simultaneously in different combinations (lanes 10-12, 15-23) from either the cellular elongation factor 1α (EF1α) or the viral cytomegalovirus (CMV) promoter, which are located within the pBud-CE 4.3 vector (Strategene) in the transfected VA13+ TERT cell line. Serial 5-fold dilutions of the transfected cell lysates were used to ensure the linearity of the PCR reactions. (–) is the result of the heat-inactivated sample similar to the one shown in lane 1, whereas (+) shows the PCR products amplified from the positive TSR8 DNA template provided in the TRAPeze kit (Chemicon International). IC shows PCR products amplified from an internal control to ensure equal loading of the samples. (B) TRAP results of the in vitro reconstitutions of the telomerase complexes using the rabbit reticulocyte lysate (RRL; Promega) to translate a wild-type copy of the TERT protein in the presence of the indicated synthetic TERC RNA variants individually or in combinations. Various dilutions of the TERC RNAs (100 ng, 10 ng, and 1 ng) were used for each RNA sample. No TERC indicates similar RRL sample without the TERC RNA; –, heat-inactivated sample containing wild-type TERC; +, PCR products amplified from the positive TSR8 DNA template provided in the TRAPeze kit (Chemicon International). (C) TRAP results of the VA13+ TERT cells that were transfected with plasmid DNAs that contain nucleotide substitutions shown in Figure 2F, lines 5 to 9, including the junctional J4/4.1 sequence and sequences predicted to either abolish the formation of the P4.1 helical stem [P4.1(lt) or P4.1(rt)] or restore it [P4.1(ltrt)]. (D) TRAP results of the primary T lymphocytes obtained from either the patient F17 (lanes 1-3) with 2 different TERC abnormalities or an unrelated healthy donor control F19 (lanes 4-6). Lanes 7 to 9 show the PCR products obtained from the positive control K562 cell lysate, indicated as (+). IC shows PCR products amplified from an internal control to ensure equal loading of the samples. (E) Northern blotting analysis of total cellular RNA extracted from VA13+ TERT cells that were transfected with a similar set of DNA constructs as shown in panel A. TERC indicates telomerase RNA template transcripts; β-actin, housekeeping β-actin message. The amounts of TERC RNAs and those of the β-actin in all samples were quantified using the ImageQuant analysis software (Molecular Dynamics). The expression levels of the TERC RNA were normalized to those of the housekeeping β-actin gene by dividing the TERC values in each of the lanes with the amounts of the β-actin in the respective lanes. The ratios of TERC gene expression for each of the samples (lanes 1-5) to that of the wild-type TERC sample (lane 6) are shown in the bottom of the gel.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/4/10.1182_blood-2005-01-0247/7/m_zh80160582740003.jpeg?Expires=1769092099&Signature=5OzpzOyXR6xH0JHjVwHw0WzKjFvePcet42mFRETRPUXiroEA36R8yx8XsW~WPuBBxbLJaAVhhD6rKaDohb7htQoX2JMVPe3Y9FEryQA2UKqadTy~lWDZ9Wjvb1svO0fpbqL0dyUYrBRUxv3PaU70lLIS1~Ly0rI83PnNTHFVwuXnCqyP5i159suzXRFd4shtlv1iwlfpxdDn89wkSx8u11jerKi62uyK8sz3nD-DAWCs5pOelhTxyUE-4ys1zchQfVu3DsVZbyegYc4Xf2XOWtwmxzdumedY~e~3E3MiSdNGgfeAzwahsmkdkDUOrSSTuWYQ-Bc5y3RaP3AEiMrGUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal