Abstract
Splenic marginal zone lymphoma (SMZL) is a newly recognized lymphoma type whose precise molecular pathogenesis is still essentially unknown. This hampers differential diagnosis with other small B-cell malignancies. With the aim of characterizing this tumor more comprehensively, and of identifying new diagnostic and prognostic markers, we performed cDNA microarray expression profiling and tissue microarray (TMA) immunohistochemical studies in a relatively large series of 44 SMZLs. The results were related to immunoglobulin heavy chain variable region (IgVH) mutational status and clinical outcome. SMZLs display a largely homogenous signature, implying the existence of a single molecular entity. Of the genes deregulated in SMZLs, special mention may be made of the genes involved in B-cell receptor (BCR) signaling, tumor necrosis factor (TNF) signaling and nuclear factor-κB (NF-κB) activation, such as SYK, BTK, BIRC3, TRAF3, and LTB. Other genes observed were SELL and LPXN, which were highly expressed in spleen, and lymphoma oncogenes, such as ARHH and TCL1. In contrast, the genes CAV1, CAV2, and GNG11 located in 7q31, a commonly deleted area, were down-regulated in the entire series. A comparison with the genes comprising the signature of other small B-cell lymphomas identified 3 genes whose expression distinguishes SMZL, namely ILF1, SENATAXIN, and CD40. Shorter survival was associated with CD38 expression, naive IgVH genes, and the expression of a set of NF-κB pathway genes, including TRAF5, REL, and PKCA. (Blood. 2005;106:1831-1838)
Introduction
Splenic marginal zone lymphoma (SMZL) is a newly described lymphoma type, whose precise diagnosis, patient stratification into risk groups, and appropriate treatment is still in need of further investigation.1-3
The diagnosis of SMZL is often hindered by the lack of conclusive immunophenotypical or molecular data concerning differences from chronic lymphocytic leukemia (CLL), follicular lymphoma (FL), and mantle cell lymphoma (MCL). The expression of CyclinD1 and B-cell lymphoma 6 (Bcl6) can contribute to the diagnosis of MCL or FL, but this is occasionally difficult to measure in samples of peripheral blood or bone marrow from CLL and MCL cases. Consequently, the diagnosis of SMZL is still characterized by strikingly low reproducibility. Even when a correct diagnosis has been established, the clinical course is appreciably variable, with a subset of cases showing frequent and early relapses, and shorter survival. Although patients with this aggressive SMZL form can be recognized by the absence of immunoglobulin heavy chain variable region (IgVH) somatic mutation or the 7q31 deletion,4 this is not accurate enough to allow stratification of patients into different risk groups receiving risk-adjusted specific treatments. Finally, the available treatments for SMZL essentially overlap those of other small B-cell non-Hodgkin lymphomas (NHLs), and the molecular data obtained so far for SMZL are insufficient to form the basis for the proposal of new therapeutic targets.
Lymphoma diagnosis and knowledge has been facilitated in recent years by the development of high-throughput molecular tools, such as expression microarrays aimed at quantifying the expression of RNA or protein.5-7 To characterize this tumor more comprehensively, and to identify new diagnostic and prognostic markers, we performed cDNA microarray expression profiling and tissue microarray (TMA) immunohistochemical studies in a relatively large series of SMZLs. The results were related to IgVH mutational status and clinical outcome and were compared with those obtained for other B-cell NHLs. To ensure the quality of the data produced, the series was restricted to cases diagnosed after splenectomy, because the key features of this entity have been described in splenectomy specimens.
Patients, materials, and methods
Patients and tissue samples
The study involved 44 consecutively diagnosed cases of SMZL, selected from routine and consultation cases of the Centro Nacional de Investigaciones Oncológicas (CNIO), Hospital Virgen de la Salud, Toledo, and the Spanish Tumor Bank Network. We included 5 reactive splenic tissue cases (4 after traumatic rupture and 1 diagnosed with idiopathic thrombocytopenic purpura). The only criterion for inclusion was the availability of frozen tissue from splenectomy specimens for molecular analysis.
Cases were diagnosed on the basis of splenic morphology, immunophenotypical, and molecular findings according to World Health Organization classification criteria.8 Routine diagnosis of these cases was reviewed by M.M. and M.P.
Age, sex, localization and clinical stage at diagnosis, and follow-up data (including overall survival, progression when appropriate, and outcome) were obtained from patient medical records.
Microarray procedures
Total RNA was extracted from 32 frozen blocks (27 tumoral and 5 control spleen) by Trizol (Life Technologies, Grand Island, NY) and RNeasy (Qiagen, Valencia, CA) cleanup. Retrotranscription was carried out by double-strand cDNA synthesis from 4.5 μg good quality total RNA using the Superscript System for cDNA synthesis (Gibco/Invitrogen, Paisley, United Kingdom) and the T7 Megascript in vitro transcription kit (Ambion, Austin, TX) for amplification. The quality of the total and the amplified RNA produced was checked by electrophoresis and their concentration was measured. Cases with low-quality RNA were excluded from further analysis.
The CNIO oncochip was used for all microarray studies.9 Amplified RNA (2.5 μg) was labeled with cyanine 3 (Cy3)-conjugated deoxyuridine triphosphate (dUTP), while 2.5 μg aRNA from the Universal Human Reference RNA (Stratagene, La Jolla, CA) was labeled with cyanine 5 (Cy5)-conjugated dUTP as reference. Hybridizations were performed as described.9 After washing, slides were scanned using the Agilent G2565AA Microarray Scanner System (Agilent, Palo Alto, CA), and images were analyzed with the Genepix 5.0 program (Axon Instruments, Union City, CA).
IgVH study
We extracted DNA from the 44 frozen tissue specimens. Rearranged IgVH genes were amplified and sequenced as described.4 Cases were considered to be mutated when they exhibited less than 98% homology with the closest germ line VH genes. The mutational status of 23 patients in this series has been previously reported.4
Tissue microarray immunostaining
A tissue microarray (TMA) block was constructed using a Tissue Arrayer device (Beecher Instruments, Silver Springs, MD) with two 1-mm diameter cylinders from 2 selected areas of each case and from appropriate controls. Sample cores were obtained from paraffin-embedded splenectomy specimen blocks, corresponding to the initial diagnostic sample of the 44 included in the study. Paraffin sections were examined to select the involved area. The resulting TMA contained 88 cores from cases and 6 from controls. The antibodies Syk, Lyn, p65 (Santa Cruz Biotechnology, Santa Cruz, CA), Bcl2, CD38, IgD, Ki67 (DAKO, Copenhagen, Denmark), ZAP-70, phosphorylated IκBα (p-IκBα) (Cell Signaling, Beverly, MA), and p53 (Novocastra, Newcastle upon Tyne, United Kingdom) were used to determine protein expression. The immunohistochemical protocol used was that described elsewhere.12 The tissues were counterstained with hematoxylin. The specimens were analyzed by microscopy, using an Olympus BX60 (Olympus Optical, Hamburg, Germany). Images were taken with an Olympus DP50 camera at an original magnification of ×100 or ×400, and treated with Viewfinder and StudioLite 1.0 software (Pixera, Los Gatos, CA). All hybridized TMAs were scanned using the BLISS (Bacus Laboratories Incorporated Slide Scanner) system (Bacus Laboratories, Lombard, IL), which uses a 3 CCD RGB sensor optically coupled to a microscope. Tissue microarrays were quantitatively scored with TMAscore v.1.0 image analysis software (Bacus Laboratories), which uses WebSlide v.1.0 (Bacus Laboratories) virtual slides.
Real-time quantitative PCR
To validate microarray experiment data, real-time quantitative reverse-transcription polymerase chain reaction (RT-PCR) was performed. Assays-on-Demand TaqMan MGB probes (Applied Biosystems, Foster City, CA) of 2 of the genes clearly overexpressed in this series (ARHH and SELL) and the endogenous control gene (TFRC) were selected. In all cases a standard curve containing 4 concentrations of a control cDNA (HuT78 cell line)13 was constructed for the endogenous control gene (TFRC) and the genes of interest. Calculations were made from measurements of 3 replicates of each sample. All PCRs were performed using the ABI Prism 7700 (Applied Biosystems) under the conditions recommended by the manufacturers, and results were normalized and analyzed using sequence-detector software (Applied Biosystems). The quantitative PCR control gene TFRC was chosen on the basis of its homogeneous expression in all splenic specimens.
Data and cluster analyses
Cy3-Cy5 ratios were normalized using the Diagnosis and Normalization Array Data (DNMAD) tool (http://dnmad.bioinfo.cnio.es). To obtain the expression profile of each tumor we compared its ratio with the mean ratio of 5 normal splenic specimens. To compare SMZL with MCL, CLL, and FL, we referred SMZL ratios to an internal pool built from the mean of all tumor ratios
Unsupervised cluster analysis was done with the Gene Cluster program.14 To cluster gene expression data into groups with similar genes, we used the K-means algorithm, measuring similarity with Spearman rank correlation as a distance function, with a value of k = 2.
Statistics
To identify genes discriminating between groups established on the basis of the presence or absence of a condition, we used Student t test. Relationships between the expression levels of different markers evaluated in the TMA were explored using Spearman correlation. Associations between categorical variables were tested using Fisher exact test.
Time-to-event analysis, involving Kaplan-Meier, log-rank test, and Cox models, served to identify markers influencing disease-related overall survival (DOS) and progression-free interval (PFI). The estimates of the hazard ratios (HRs) along its 95% confidence interval (95% CI) were computed via the Cox model.
Patients were considered to have progressive disease on the basis of progression to a more advanced stage, or the enlargement or new appearance of lymph nodes, development of systemic symptoms, large B-cell lymphoma progression, or death attributable to the lymphoma.
Statistical analyses were performed using the SPSS (SPSS, Chicago, IL) program, and the random permutation test for multiple testing, available at http://pomelo.bioinfo.cnio.es/cgi-bin/multest.cgi. We present adjusted P values using the FDR (False Discovery Rates) procedure of Benjamini and Hochberg15 assuming an independent FDR less than 0.2.
Definition of functional signatures
Genes were analyzed using the FATIGO gene ontology-based application (http://fatigo.bioinfo.cnio.es). Functional signatures considered were apoptosis (164 genes), metabolism (1479 genes), and cell communication (846 genes). New gene clusters were created for nuclear factor-κB (NF-κB) pathway (152 genes) and B-cell receptor (BCR) signaling (50 genes), as described.10
Results
Clinical features
Forty-four patients were included in this study, of which 19 (43%) were men and 25 (57%) were women. The mean age at diagnosis was 66 years (range, 44-80 years). Forty-one of the cases were diagnosed at clinical stage IV with bone marrow involvement, 1 at clinical stage II, and 2 at clinical stage I. All patients had undergone splenectomy for diagnostic and/or therapeutic purposes. Twelve patients had received chemotherapy (chlorambucil, CHOP [cyclophosphamide, doxorubicin, vincristine, and prednisone], or CHOP-like regimens).
The median actuarial follow-up for DOS was 3 years. At the time of the analysis, 30 (68%) patients were alive, 11 (25%) had died because of the disease, and 3 (7%) had died of unrelated causes. Estimated DOS rate at 3 years was 74%. Progression occurred in 20 (45%) cases with a PFI rate at 3 years of 62%.
SMZL signature
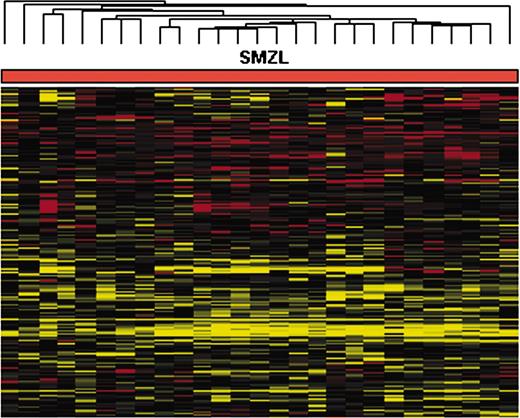
Twenty-seven SMZL samples were studied with cDNA microarrays. The overall expression pattern of this series was first analyzed by hierarchic clustering, using the expression profiles of the 4834 expressed genes that remained after removing duplicates, inconsistent replicates, and genes with a high proportion (> 30%) of missing values. After normalization with a set of 5 normal/reactive spleens, the SMZL analysis revealed a relatively homogeneous signature, with all tumor samples grouped in a single main cluster, as shown in Figure 1.
Molecular signature
From those 4834 genes, a distinctive SMZL signature was obtained, composed of 56 and 88 genes that were found to be more than 2-fold up-regulated or down-regulated, respectively, relative to reactive spleen tissue, in at least 50% of the samples.
A preliminary functional analysis, based on gene ontology, revealed that the largest functional group deregulated in this series was made up of those featuring the cell communication cluster (50%) and those involved in metabolism (46%), followed by cellular physiologic processes, organismal physiologic processes, and response to stimulus (43%, 37%, and 42%, respectively). Distinctive up-regulated SMZL genes and their functions are described in Table 1.
SMZL signature
Gene symbol . | C . | A . | M . | BCR/NFκB . | Median change . | Description . |
|---|---|---|---|---|---|---|
| MIG* | 4.52 | Glucose signal transduction pathway | ||||
| MS4A2 | + | 4.18 | B-cell: Fc (crystallizable fragment) IgE receptor, β chain; found on the surface of mast cells and basophils | |||
| UBD* | + | 3.71 | Ubiquitin D, FAT10, B-cell, and dendritic marker | |||
| HLA-DMA | 3.22 | B-cell: major histocompatibility complex, class II, DM α, expressed in B-cell lysosomes, same as RING6 | ||||
| ARHH* | 3.22 | Ras oncogene superfamily member, involved in translocation t(3;4) with BCL6 | ||||
| SYK* | + | + | + | 3.19 | Positive effector of BCR-stimulated responses; regulation of intracellular calcium ion | |
| PCDH9 | 3.09 | Member of a subfamily of calcium-dependent cell adhesion and recognition proteins of the cadherin superfamily | ||||
| E2F5 | + | 2.75 | Transcriptional regulator: E2F4/E2F5 complex and p 107 act as transducers of transforming growth factor β (TGF-β) receptor signals upstream of cyclin-dependent kinase (CDK) | |||
| TOSO* | + | 2.72 | Regulator of Fas-induced apoptosis in T cells | |||
| SELL* | + | 2.64 | Selectin L (lymphocyte adhesion molecule 1), key for spleen lymphocyte relocalization | |||
| TCL 1A* | 2.54 | Gene for α-chain of human T-cell receptor, Akt coactivator | ||||
| AIM2 | 2.44 | Member of interferon-inducible gene family: expressed in spleen, small intestine, and peripheral blood leukocytes | ||||
| SP140 | + | 2.37 | Lymphocyte-specific nuclear body protein, increased in myeloid precursor cell lines, undetected in nonlymphoid cell lines | |||
| PFTK1 | + | 2.34 | Protein kinase activity, may play a role in meiosis | |||
| LPXN* | + | + | 2.30 | Leupaxin, highly expressed in the spleen | ||
| PIR121 | 2.29 | p53-inducible protein | ||||
| ITM2A | 2.29 | Integral membrane protein 2A: gene expressed in human CD34+ hematopoietic stem/progenitor cells | ||||
| PTPRC | + | + | + | 2.28 | CD45, key role in lymphocyte activation, required for proximal signaling events following triggering of the T - and B-cell antigen (Ag) receptors | |
| NSF | 2.24 | N-ethylmalcimide-sensitive factor required for vesicle-mediated transport | ||||
| PTPN1 | + | + | + | 2.23 | Protein-tyrosine phosphatase: important regulatory component in signal transduction, neoplastic transformation, and control of mitotic cycle | |
| RASSF2 | + | 2.22 | Ras association domain family 2 | |||
| CDW52* | 2.20 | CAMPATH-1 antigen | ||||
| HLA-DMB | 2.06 | B-cell: major histocompatibility complex, class II, DM β | ||||
| BIRC3* | + | + | + | 2.05 | HIAP1 (human inhibitor of apoptosis protein 1), apoptotic suppressor, interacts with tumor necrosis factor (TNF) receptor-associated factor 1 (TRAF1) and TRAF2 | |
| TNFRSF5* | + | + | + | + | 2.05 | CD40, expressed in B cells, essential role for T-cell-dependent Ig class switching, memory B-cell development, and GC formation |
| EIF4B | + | 2.01 | RNA-binding activity and use in the initiation of protein biosynthesis | |||
| BTAF1 | + | 2.01 | Regulates transcription in association with TATA binding protein (TBP) | |||
| TRAF3* | + | + | + | 2.00 | Signal transducer associated with the cytoplasmic domain of tumor necrosis factor type 2 receptor (TNF-R2); also binds to CD40 and lymphotoxin beta receptor (LTB-R) | |
| TRAF5* | + | + | + | 1.99 | Interacts with LTB-R and activates NFκB | |
| AMPD3 | + | 1.96 | Purine metabolism | |||
| POU2AF1 | + | 1.93 | Transcriptional coactivator, essential for the Ag B-cell response and GC formation, involved in t(3;11) in a form of B-cell leukemia with BCL6 | |||
| ADD3 | 1.93 | Binds to calmodulin; a t(10;11), yielding a T-cell acute lymphoblastic leukemia and myeloid markers | ||||
| EGR2 | + | 1.90 | Early growth response 2, zinc finger transcription factor; DNA binding | |||
| WBP1 | 1.85 | Protein-protein interaction; similar function to Src homology 3 (SH3) domain | ||||
| ENPP2 | + | + | 1.80 | Autotaxin (ATX): Purine metabolism; specific target of cell transformation by v-jun | ||
| BTK* | + | + | + | + | 1.71 | BCR signaling: signal transduction in B cells and other tissues, essential for B-cell activation |
| PDE4B | + | 1.66 | Cyclic adenosine monophosphate (cAMP)-specific phosphodiesterase | |||
| ICSBP1 | + | 1.61 | Interferon regulatory factor expressed exclusively in cells of the immune system | |||
| LTB* | + | 1.54 | BCR signaling: lymphotoxin β (TNF superfamily, member 3) |
Gene symbol . | C . | A . | M . | BCR/NFκB . | Median change . | Description . |
|---|---|---|---|---|---|---|
| MIG* | 4.52 | Glucose signal transduction pathway | ||||
| MS4A2 | + | 4.18 | B-cell: Fc (crystallizable fragment) IgE receptor, β chain; found on the surface of mast cells and basophils | |||
| UBD* | + | 3.71 | Ubiquitin D, FAT10, B-cell, and dendritic marker | |||
| HLA-DMA | 3.22 | B-cell: major histocompatibility complex, class II, DM α, expressed in B-cell lysosomes, same as RING6 | ||||
| ARHH* | 3.22 | Ras oncogene superfamily member, involved in translocation t(3;4) with BCL6 | ||||
| SYK* | + | + | + | 3.19 | Positive effector of BCR-stimulated responses; regulation of intracellular calcium ion | |
| PCDH9 | 3.09 | Member of a subfamily of calcium-dependent cell adhesion and recognition proteins of the cadherin superfamily | ||||
| E2F5 | + | 2.75 | Transcriptional regulator: E2F4/E2F5 complex and p 107 act as transducers of transforming growth factor β (TGF-β) receptor signals upstream of cyclin-dependent kinase (CDK) | |||
| TOSO* | + | 2.72 | Regulator of Fas-induced apoptosis in T cells | |||
| SELL* | + | 2.64 | Selectin L (lymphocyte adhesion molecule 1), key for spleen lymphocyte relocalization | |||
| TCL 1A* | 2.54 | Gene for α-chain of human T-cell receptor, Akt coactivator | ||||
| AIM2 | 2.44 | Member of interferon-inducible gene family: expressed in spleen, small intestine, and peripheral blood leukocytes | ||||
| SP140 | + | 2.37 | Lymphocyte-specific nuclear body protein, increased in myeloid precursor cell lines, undetected in nonlymphoid cell lines | |||
| PFTK1 | + | 2.34 | Protein kinase activity, may play a role in meiosis | |||
| LPXN* | + | + | 2.30 | Leupaxin, highly expressed in the spleen | ||
| PIR121 | 2.29 | p53-inducible protein | ||||
| ITM2A | 2.29 | Integral membrane protein 2A: gene expressed in human CD34+ hematopoietic stem/progenitor cells | ||||
| PTPRC | + | + | + | 2.28 | CD45, key role in lymphocyte activation, required for proximal signaling events following triggering of the T - and B-cell antigen (Ag) receptors | |
| NSF | 2.24 | N-ethylmalcimide-sensitive factor required for vesicle-mediated transport | ||||
| PTPN1 | + | + | + | 2.23 | Protein-tyrosine phosphatase: important regulatory component in signal transduction, neoplastic transformation, and control of mitotic cycle | |
| RASSF2 | + | 2.22 | Ras association domain family 2 | |||
| CDW52* | 2.20 | CAMPATH-1 antigen | ||||
| HLA-DMB | 2.06 | B-cell: major histocompatibility complex, class II, DM β | ||||
| BIRC3* | + | + | + | 2.05 | HIAP1 (human inhibitor of apoptosis protein 1), apoptotic suppressor, interacts with tumor necrosis factor (TNF) receptor-associated factor 1 (TRAF1) and TRAF2 | |
| TNFRSF5* | + | + | + | + | 2.05 | CD40, expressed in B cells, essential role for T-cell-dependent Ig class switching, memory B-cell development, and GC formation |
| EIF4B | + | 2.01 | RNA-binding activity and use in the initiation of protein biosynthesis | |||
| BTAF1 | + | 2.01 | Regulates transcription in association with TATA binding protein (TBP) | |||
| TRAF3* | + | + | + | 2.00 | Signal transducer associated with the cytoplasmic domain of tumor necrosis factor type 2 receptor (TNF-R2); also binds to CD40 and lymphotoxin beta receptor (LTB-R) | |
| TRAF5* | + | + | + | 1.99 | Interacts with LTB-R and activates NFκB | |
| AMPD3 | + | 1.96 | Purine metabolism | |||
| POU2AF1 | + | 1.93 | Transcriptional coactivator, essential for the Ag B-cell response and GC formation, involved in t(3;11) in a form of B-cell leukemia with BCL6 | |||
| ADD3 | 1.93 | Binds to calmodulin; a t(10;11), yielding a T-cell acute lymphoblastic leukemia and myeloid markers | ||||
| EGR2 | + | 1.90 | Early growth response 2, zinc finger transcription factor; DNA binding | |||
| WBP1 | 1.85 | Protein-protein interaction; similar function to Src homology 3 (SH3) domain | ||||
| ENPP2 | + | + | 1.80 | Autotaxin (ATX): Purine metabolism; specific target of cell transformation by v-jun | ||
| BTK* | + | + | + | + | 1.71 | BCR signaling: signal transduction in B cells and other tissues, essential for B-cell activation |
| PDE4B | + | 1.66 | Cyclic adenosine monophosphate (cAMP)-specific phosphodiesterase | |||
| ICSBP1 | + | 1.61 | Interferon regulatory factor expressed exclusively in cells of the immune system | |||
| LTB* | + | 1.54 | BCR signaling: lymphotoxin β (TNF superfamily, member 3) |
The most highly expressed genes in the SMZL series are shown. The functional signatures cell communication (C), apoptosis (A), metabolism (M), and BCR/NF-κB activation are indicated with “+”.
The most important genes.
The signature includes notable up-regulated genes involved in apoptosis, such as TOSO and BIRC3, and others involved in BCR signaling, TNF signaling, and NF-κB activation, such as SYK, BTK, BIRC3, TRAF3, TRAF5, CD40, and LTB.
The presence of other genes associated with localization in the spleen, like SELL and LPXN, is of particular note. SELL (L-selectin, CD62L) is a cell-surface protein, a member of a family of adhesion/homing receptors that play important roles in leukocyte-endothelial cell interactions.16 LPXN (Leupaxin) is a leukocyte-specific molecule involved in signaling through association with proline-rich tyrosine kinase 2 (PYK2), a member of the focal adhesion kinase family.17
An interesting group of genes is involved in signal transduction and cell communication, including MIG (CXCL9), a ligand for the CXC chemokine receptor 3 (CXCR3) chemokine receptor, whose expression in response to interferon-γ (IFNγ) seems to require constitutive NF-κB activity, as is often associated with tumor development.18 Protein tyrosine phosphatase, nonreceptor type 1 (PTPN1) is a protein tyrosine phosphatase known to be a signaling molecule that regulates a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation.19
UBD (Ubiquitin D, FAT10) appears up-regulated in SMZL. FAT10 protein has been found to be associated with the human spindle assembly checkpoint protein, mitotic arrest deficient 2 (MAD2), modulating cell cycling during B-cell or dendritic cell development and activation.20 Other genes of particular note are those already known to be associated with hematologic malignancies, such as ARHH (RhoH/TTF) and TCL1. ARHH is an oncogene that encodes a Rho guanosine triphosphate (GTP)-binding protein specifically expressed in hematopoietic tissues,21 while the TCL1 protooncogene is expressed in many small B-cell lymphomas.22
With respect to down-regulated genes, we draw attention to the genes located in the 7q region, an area known to be lost in approximately 40% to 45% of SMZL cases.23 In comparison with the results obtained for genes located in 3p or 7p, the great majority (12 of 13) of genes in the 7q31 region were expressed at a lower level than in normal spleen, and in 3 of the genes (CAV1, CAV2, and GNG11) located in the precise region that is lost in SMZL (7q31-q32), there was a less than 2-fold level of expression. Expression of the genes located in 7q31 has been analyzed, taking into account the presence or absence of 7q31 deletions, as assessed by loss of heterozygosity (LOH) (data not shown). This was restricted to a group of 7 cases, only 2 of which had 7q31 deletions. The level of expression of the genes located in 7q31 was uniformly low, with the sole exception of MET, and independent of the 7q31 LOH.
Validation of results: quantitative RT-PCR
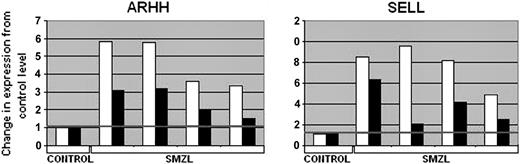
Two genes (ARHH and SELL), belonging to the SMZL signature, were analyzed by quantitative reverse-transcriptase (RT)-PCR in 4 cases, comparing the expression level with a control pool of normal tissues, as shown in Figure 2. The estimates of expression levels of both genes were similar using both techniques.
Tissue microarray
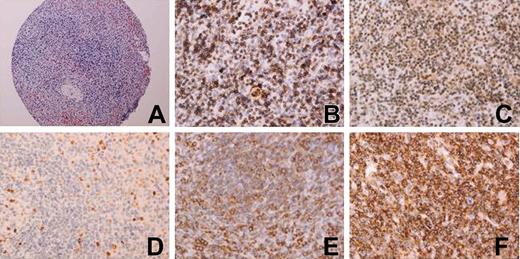
Expression of Syk, Lyn, Bcl2, ZAP-70, p-IκBα, IgD, bcl6, cyclin D1, p65, CD38, Ki67, and p53 was measured in a TMA containing cores from 44 cases. Captured images of slides are shown in Figure 3.
Correlations between markers were remarkably high for Ki67 with CD38, Syk, and p53 and for Syk with CD38 and p-IκBα. Cases were divided into 2 groups with respect to median protein expression in the whole series for survival analysis. This showed that cases with a high level of CD38 expression had shorter survival (P < .05). Likewise, high levels of expression of Ki67 and Syk were shown to be associated with a shorter progression-free interval (P < .05). Details of the results are presented in Tables 2 and 3.
Results of immunohistochemical staining in the TMA
. | . | . | Visual evaluation (expression type) . | Ki67 . | . | SYK . | . | CD38 . | . | p-IκBα . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker . | Median . | Range . | . | rho . | P . | rho . | P . | rho . | P . | rho . | P . | ||||
| Ki67 | 3.19 | 0.04-19.53 | Nuc | — | — | — | — | — | — | — | — | ||||
| p53 | 0.08 | 0.01-20.07 | Nuc | 0.37 | <.05 | — | — | — | — | — | — | ||||
| SYK | 32.78 | 0.54-80.38 | Cyto | 0.36 | <.05 | — | — | — | — | — | — | ||||
| CD38 | 36.4 | 1.42-67.80 | Cyto | 0.69 | <.001 | 0.33 | <.05 | — | — | — | — | ||||
| LYN | 32.51 | 19.95-82.62 | Variable cyto | NS | NS | NS | NS | — | — | — | — | ||||
| p-IκBα | 2.33 | 0.02-67.58 | Cyto and nuc | NS | NS | 0.50 | <.001 | — | — | — | — | ||||
| IgD | 18.46 | 1.44-73.28 | Variable cyto-surf | 0.38 | <.05 | NS | NS | 0.38 | <.05 | — | — | ||||
| p65 | 10.48 | 0.04-73.15 | Cyto and nuc | NS | NS | NS | NS | NS | NS | — | — | ||||
| ZAP-70 | 37.76 | 4.74-71.54 | Cyto | 0.44 | <.01 | 0.38 | <.05 | NS | NS | 0.33 | <.05 | ||||
. | . | . | Visual evaluation (expression type) . | Ki67 . | . | SYK . | . | CD38 . | . | p-IκBα . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker . | Median . | Range . | . | rho . | P . | rho . | P . | rho . | P . | rho . | P . | ||||
| Ki67 | 3.19 | 0.04-19.53 | Nuc | — | — | — | — | — | — | — | — | ||||
| p53 | 0.08 | 0.01-20.07 | Nuc | 0.37 | <.05 | — | — | — | — | — | — | ||||
| SYK | 32.78 | 0.54-80.38 | Cyto | 0.36 | <.05 | — | — | — | — | — | — | ||||
| CD38 | 36.4 | 1.42-67.80 | Cyto | 0.69 | <.001 | 0.33 | <.05 | — | — | — | — | ||||
| LYN | 32.51 | 19.95-82.62 | Variable cyto | NS | NS | NS | NS | — | — | — | — | ||||
| p-IκBα | 2.33 | 0.02-67.58 | Cyto and nuc | NS | NS | 0.50 | <.001 | — | — | — | — | ||||
| IgD | 18.46 | 1.44-73.28 | Variable cyto-surf | 0.38 | <.05 | NS | NS | 0.38 | <.05 | — | — | ||||
| p65 | 10.48 | 0.04-73.15 | Cyto and nuc | NS | NS | NS | NS | NS | NS | — | — | ||||
| ZAP-70 | 37.76 | 4.74-71.54 | Cyto | 0.44 | <.01 | 0.38 | <.05 | NS | NS | 0.33 | <.05 | ||||
Description of the expression and correlation of the markers analyzed in the TMA. Spearman correlation coefficient (rho) and the associated P values are indicated.
Nuc, nuclear; cyto, cytoplasmic; NS, not significant; cyto-surf, cytoplasmic-surface.
Relationships with DOS and PFI
. | DOS . | PFI . | . | . | ||
|---|---|---|---|---|---|---|
. | CD38 . | CD38 . | Ki67 . | SYK . | ||
| n, +/−* | 22/22 | 22/22 | 21/21 | 20/23 | ||
| Events, no., +/− | 2/9 | 6/13 | 7/12 | 7/12 | ||
| HR (% CI) | 5.33 (1.15-24.81) | 3.32 (1.17-9.39) | 3.22 (1.19-8.68) | 0.34 (0.12-0.99) | ||
| P | .033 | .024 | .021 | .048 | ||
. | DOS . | PFI . | . | . | ||
|---|---|---|---|---|---|---|
. | CD38 . | CD38 . | Ki67 . | SYK . | ||
| n, +/−* | 22/22 | 22/22 | 21/21 | 20/23 | ||
| Events, no., +/− | 2/9 | 6/13 | 7/12 | 7/12 | ||
| HR (% CI) | 5.33 (1.15-24.81) | 3.32 (1.17-9.39) | 3.22 (1.19-8.68) | 0.34 (0.12-0.99) | ||
| P | .033 | .024 | .021 | .048 | ||
The number of patients (n) and events observed in each group, estimated HR computed via the univariate Cox model, and the associated P values are indicated.
For CD38, Ki67, and SYK, + indicates high expression and − indicates low expression.
Hierarchical clustering of SMZL series, demonstrating the relative homogeneity of this series, which appears clustered into a single group. Red and green indicate high- and low-level expression, respectively.
Hierarchical clustering of SMZL series, demonstrating the relative homogeneity of this series, which appears clustered into a single group. Red and green indicate high- and low-level expression, respectively.
Given the small number of observations and the strong correlation observed between these markers, only Ki67 remained as statistically significant in the fitted multivariate model.
IgVH mutational status
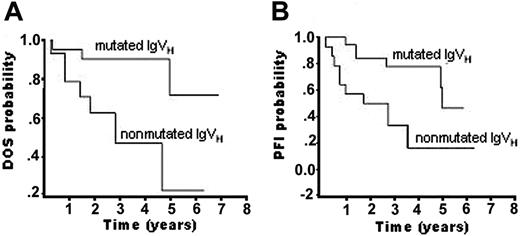
DNA obtained from the 44 cases displayed a monoclonal IgVH rearrangement in 37 patients, with percentages of mutated (59%) and nonmutated (41%) cases overlapping those observed in previous studies.4 Kaplan-Meier analysis confirmed a longer survival in the mutated group (P < .05), whereby DOS rate at 3 years was 41% in the nonmutated group and 90% in the mutated group (Figure 4 and Table 4).
Survival probability in relation to the IgVHmutational status
. | . | DOS . | . | . | PFI . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IgVμstatus . | n . | Events . | HR (95% CI) . | P . | Events . | HR (95% CI) . | P . | ||||
| Nonmutated | 15 | 7 | 0.19 (0.05−0.76) | .01 | 9 | 0.31 (0.11−0.84) | .02 | ||||
| Mutated | 22 | 3 | NA | NA | 8 | NA | NA | ||||
. | . | DOS . | . | . | PFI . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IgVμstatus . | n . | Events . | HR (95% CI) . | P . | Events . | HR (95% CI) . | P . | ||||
| Nonmutated | 15 | 7 | 0.19 (0.05−0.76) | .01 | 9 | 0.31 (0.11−0.84) | .02 | ||||
| Mutated | 22 | 3 | NA | NA | 8 | NA | NA | ||||
The number of patients (n) and events observed in each group, estimated HR computed via the univariate Cox model, and the associated P values are indicated.
Validation of the results achieved using microarray analysis by quantitative RT-PCR. The ratios of the expression level of the genes related to the control pool are represented. □ indicates microarray; ▪, quantitative RT-PCR.
Validation of the results achieved using microarray analysis by quantitative RT-PCR. The ratios of the expression level of the genes related to the control pool are represented. □ indicates microarray; ▪, quantitative RT-PCR.
Images from immunostaining of several markers in the TMA. (A) Hematoxylin and eosin (H&E) of a 1-mm diameter cylinder of an SMZL case; (B) CD38; (C) p-IκBα; (D) Ki67; (E) p65; (F) Syk. Original magnifications and numerical apertures 100 ×/0.30 (A) and 400 ×/0.75 (B-F).
Images from immunostaining of several markers in the TMA. (A) Hematoxylin and eosin (H&E) of a 1-mm diameter cylinder of an SMZL case; (B) CD38; (C) p-IκBα; (D) Ki67; (E) p65; (F) Syk. Original magnifications and numerical apertures 100 ×/0.30 (A) and 400 ×/0.75 (B-F).
The partial overlap with the original series in which this phenomenon was observed needs to be taken into account when interpreting these results.4
The frequency of IgVH mutation was significantly related with CD38 expression (Fisher exact test, P < .05) (Table 5).
Relation between IgVHmutational status and CD38 expression as prognostic factors
. | CD38 expression* . | . | . | ||
|---|---|---|---|---|---|
| IgVHmutational status . | Low . | High . | Median . | ||
| Nonmutated | 4 | 11 | 39.65 | ||
| Mutated | 15 | 7 | 30.42 | ||
. | CD38 expression* . | . | . | ||
|---|---|---|---|---|---|
| IgVHmutational status . | Low . | High . | Median . | ||
| Nonmutated | 4 | 11 | 39.65 | ||
| Mutated | 15 | 7 | 30.42 | ||
Fisher exact test, P < .05.
For CD38 expression, low was defined as less than 36.40%; high, more than 36.40%.
Student t test was used to examine differences in the expression profile associated with mutational status. It revealed that changes in the expression of RAD54L (P < .001; FDR = 0.075) were significantly associated with the mutational index. RAD54L is a gene involved in homologous recombination and DNA repair.24
Differential diagnosis of SMZL with CLL/MCL/FL and normal tissue
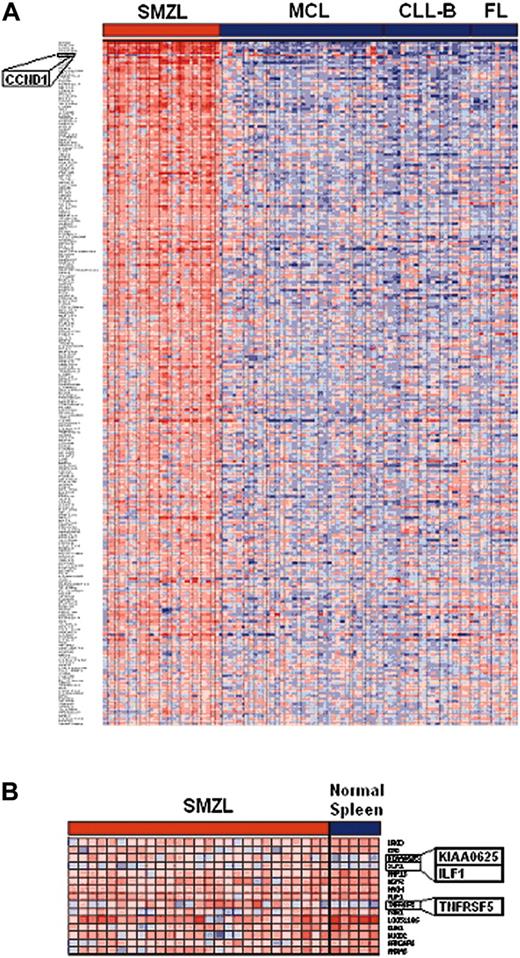
To identify new markers for this entity that would be useful in the differential diagnosis with other low-grade non-Hodgkin lymphomas, we compared the expression profiles of this SMZL series with those from CLL, MCL, and FL series, after hybridization using the same cDNA platform. This was done in 2 steps: first, analysis of variance (ANOVA) identified 266 genes whose expression level was higher in SMZLs than in the other tumor types (Figure 5), using an internal pool of the mean of the ratios of all the small B-cell lymphomas for normalization. As a way of confirming this approach, we should mention that MCL was distinguished by the overexpression of CCND1 (Cyclin D1). Secondly, data from these 266 genes were then examined with Student t test to compare normal spleen and tumoral tissue. This identified a smaller set of SMZL-specific genes that were overexpressed under these conditions. These genes were ILF1 (interleukin enhancer binding factor 1) (P < .01; FDR = 0.172), KIAA0625 (senataxin) (P < .01; FDR = 0.165), and TNFRSF5 (CD40, p50) (P < .01; FDR = 0.172).
Time-to-event analysis
A univariate Cox model was used to determine whether expression profiles of genes were associated with outcome and survival probability. For this only genes included in the following functional clusters were considered: apoptosis, BCR-signaling, cell cycle, NF-κB, and stress response. Each cluster was analyzed independently, revealing that the NF-κB cluster contained the largest group of genes whose expression was associated with changes in survival probability (25 genes for PFI and 5 genes for DOS; FDR < 0.2) (Table 6).
Survival probability in relation to the expression of a set of genes included in the NF-κB cluster
Gene symbol . | HR (95% CI) . | P . | FDR . |
|---|---|---|---|
| DOS | |||
| REL | 0.42 (0.21-0.86) | .003 | 0.16 |
| TRAP1 | 3.20 (1.25-8.19) | .004 | 0.16 |
| ADAM17 | 0.41 (0.20-0.84) | .004 | 0.16 |
| TRAF5 | 0.51 (0.30-0.88) | .005 | 0.16 |
| FZD6 | 0.52 (0.30-0.89) | .005 | 0.16 |
| PFI | |||
| PRKCA | 1.80 (1.27-2.55) | .001 | 0.04 |
| ADAM17 | 0.44 (0.27-0.73) | .001 | 0.04 |
| CCND2 | 2.11 (1.30-3.43) | .001 | 0.04 |
| PAWR | 0.60 (0.42-0.86) | .001 | 0.04 |
| FZD6 | 0.63 (0.45-0.87) | .001 | 0.04 |
| TRAP1 | 2.30 (1.27-4.16) | .003 | 0.07 |
| BAK1 | 2.12 (1.24-3.62) | .003 | 0.07 |
| HSPA8 | 2.45 (1.28-4.69) | .004 | 0.08 |
| MAP3K4 | 0.43 (0.23-0.81) | .005 | 0.08 |
| MAP4K3 | 0.70 (0.53-0.92) | .006 | 0.08 |
| IL2RB | 1.18 (1.04-1.34) | .007 | 0.08 |
| IL 1B | 1.79 (1.14-2.81) | .007 | 0.08 |
| MAP4K4 | 0.62 (0.43-0.89) | .008 | 0.09 |
| PRKCE | 2.40 (1.17-4.93) | .008 | 0.09 |
| PIG7 | 2.02 (1.12-3.62) | .011 | 0.10 |
| TRAF2 | 2.54 (1.17-5.50) | .011 | 0.10 |
| TNFSF13 | 1.56 (1.08-2.27) | .012 | 0.10 |
| HCK | 1.30 (1.05-1.62) | .014 | 0.12 |
| MMP17 | 1.93 (1.11-3.36) | .020 | 0.16 |
| MAP3K14 | 1.30 (1.03-1.64) | .023 | 0.16 |
| CRADD | 2.48 (1.09-5.63) | .024 | 0.16 |
| PSMD2 | 1.68 (1.05-2.70) | .025 | 0.16 |
| PLCG2 | 1.50 (1.03-2.21) | .025 | 0.16 |
| STAT4 | 1.30 (1.02-1.65) | .026 | 0.16 |
| NFKB1E | 1.53 (1.02-2.29) | .027 | 0.16 |
Gene symbol . | HR (95% CI) . | P . | FDR . |
|---|---|---|---|
| DOS | |||
| REL | 0.42 (0.21-0.86) | .003 | 0.16 |
| TRAP1 | 3.20 (1.25-8.19) | .004 | 0.16 |
| ADAM17 | 0.41 (0.20-0.84) | .004 | 0.16 |
| TRAF5 | 0.51 (0.30-0.88) | .005 | 0.16 |
| FZD6 | 0.52 (0.30-0.89) | .005 | 0.16 |
| PFI | |||
| PRKCA | 1.80 (1.27-2.55) | .001 | 0.04 |
| ADAM17 | 0.44 (0.27-0.73) | .001 | 0.04 |
| CCND2 | 2.11 (1.30-3.43) | .001 | 0.04 |
| PAWR | 0.60 (0.42-0.86) | .001 | 0.04 |
| FZD6 | 0.63 (0.45-0.87) | .001 | 0.04 |
| TRAP1 | 2.30 (1.27-4.16) | .003 | 0.07 |
| BAK1 | 2.12 (1.24-3.62) | .003 | 0.07 |
| HSPA8 | 2.45 (1.28-4.69) | .004 | 0.08 |
| MAP3K4 | 0.43 (0.23-0.81) | .005 | 0.08 |
| MAP4K3 | 0.70 (0.53-0.92) | .006 | 0.08 |
| IL2RB | 1.18 (1.04-1.34) | .007 | 0.08 |
| IL 1B | 1.79 (1.14-2.81) | .007 | 0.08 |
| MAP4K4 | 0.62 (0.43-0.89) | .008 | 0.09 |
| PRKCE | 2.40 (1.17-4.93) | .008 | 0.09 |
| PIG7 | 2.02 (1.12-3.62) | .011 | 0.10 |
| TRAF2 | 2.54 (1.17-5.50) | .011 | 0.10 |
| TNFSF13 | 1.56 (1.08-2.27) | .012 | 0.10 |
| HCK | 1.30 (1.05-1.62) | .014 | 0.12 |
| MMP17 | 1.93 (1.11-3.36) | .020 | 0.16 |
| MAP3K14 | 1.30 (1.03-1.64) | .023 | 0.16 |
| CRADD | 2.48 (1.09-5.63) | .024 | 0.16 |
| PSMD2 | 1.68 (1.05-2.70) | .025 | 0.16 |
| PLCG2 | 1.50 (1.03-2.21) | .025 | 0.16 |
| STAT4 | 1.30 (1.02-1.65) | .026 | 0.16 |
| NFKB1E | 1.53 (1.02-2.29) | .027 | 0.16 |
Univariate Cox results for DOS and PFI. All the gene expressions have been rescaled presenting HR associated to a change of 0.25. Adjusted P values using the FDR procedure are indicated.
A subsequent multivariate analysis of the representative genes obtained from K-means clustering of the genes determined by the univariate analysis, chosen on the basis of the lowest P value, identified only PKCA [HR (95% CI): 2.02 (1.27-3.21); P < .001] and ADAM17 [HR (95% CI): 0.37(0.19-0.71); P < .001] as independent prognostic factors for progression-free interval probability.
In summary, Kaplan-Meier analysis confirmed a longer disease-related overall survival and progression-free interval in the IgVH-mutated group (log rank, P < .05).
Likewise, high levels of expression of CD38, Ki67 and Syk were found to be related to poorer survival and progression-free interval, respectively (P < .05).
Thus, time-to-event analysis revealed 3 types of biologic predictor markers: (1) IgVH mutational status, (2) CD38, Syk, and Ki67 protein expression as measured in the TMA, and (3) a set of NF-κB pathway genes.
There were too few cases to allow reliable conclusions to be drawn about whether all these variables are independent prognosis factors.
Discussion
The SMZL series analyzed here has a strongly homogeneous signature, in which all the clinical samples are grouped in a single main cluster. This confirms that SMZL diagnostic criteria, when used according to the World Health Organization (WHO) classification, allow different diseases to be distinguished. The observation is in line with those concerning CLL25,26 and MCL11,27 and confirms previous observations made by other groups.3
Kaplan-Meier survival curves comparing patients with SMZL with mutated and nonmutated IgVH genes. Patients with nonmutated IgVH genes had significantly shorter related-disease overall survival (DOS) (A) and progression-free interval (PFI) (B).
Kaplan-Meier survival curves comparing patients with SMZL with mutated and nonmutated IgVH genes. Patients with nonmutated IgVH genes had significantly shorter related-disease overall survival (DOS) (A) and progression-free interval (PFI) (B).
Normalizing the signature with reactive spleen yields a more reduced set of 144 up-regulated or down-regulated genes that are present in at least 50% of the cases.
Notable up-regulated genes forming part of this SMZL signature are involved in the regulation of apoptosis, cell cycle, and BCR and TNF signaling. Some of these genes were initially described specifically in the spleen, such as the cytoplasmic spleen tyrosine kinase (SYK), a key regulator of BCR signaling, apoptosis, and cell-cycle progression in B-cells.28 A few of these up-regulated SMZL genes have previously been identified in other lymphoproliferative disorders, including MIG, ARHH (RhoH/TTF), and TCL1. Thus, expression of MIG has been described as being associated with a subset of B-cell lymphomas, particularly CLL/SLL (small lymphocytic lymphoma) and SMZL.29 ARHH (RhoH/TTF), whose recurrent chromosomal alterations and aberrant somatic hypermutations have been detected in hematopoietic malignancies (more than 50% of diffuse large B-cell lymphomas, myeloma)30 , is a regulator of the activation of NF-κB and p38 by other Rho GTPases.31 The TCL1 (T-cell leukemia 1) oncogene is highly expressed in many human B-cell lymphomas and leukemias,22 binding to Akt1, and increasing its phosphorylation status and kinase activity.32 The SMZL signature has already been investigated by other groups.6,33 Thieblemont et al6 described the identification of genes associated with intracellular signaling via the AKT1 pathway as characteristic of SMZL. The increased expression of TCL1, as observed in this study, is further evidence in favor of this interpretation. At the same time, this finding could be linked with the increased expression of Syk and Bruton tyrosine kinase (Btk), since BCR-mediated protein kinase B (Akt) activation seems to require Syk and, to some extent, Btk.34
The identification of the down-regulated genes in this series justifies the approach adopted. Thus, SMZLs are distinguished by the loss of the 7q31 to 32 chromosomal region, where the genes CAV1, CAV2, and GNG11 are located. Additionally, the observation that the loss of this chromosomal region is more widespread than expected prompts the identification of the molecular mechanisms leading to this loss.
Interestingly, this SMZL molecular signature mainly includes significant up-regulated genes and proteins related with BCR signaling, apoptosis, and, specifically, NF-κB activation. SYK and BTK genes are overexpressed in this series, implying that BCR signaling contributes to the generation of survival signaling in SMZL cells, which bears some similarities with the situation observed in CLL.10 Of particular interest is our observation that Syk expression is related with the level of expression of CD38 (prognostic marker) and p-IκBα. This thereby links BCR signaling with NF-κB activation and the expression of CD38, which has been shown to constitute a signaling molecule in B-cell chronic lymphocytic leukemia cells.35
Our results coincide with the observations made for other B- and T-cell lymphomas in that the activation of NF-κB may play a relevant role in SMZL. As this is a common phenomenon under different conditions, the shared presence of interesting features by all of them is nevertheless remarkable.10,36 Genes that have been shown to be of particular importance to NF-κB activation in SMZL are CD40, SYK, PKCA, REL, BIRC3, TRAF3, TRAF5, and p-IκBα.
This study also allowed the identification of potential markers for this entity, by which it could be distinguished from other B-cell lymphoproliferative conditions, such as IL1F, SENATAXIN, and CD40. Even though the relationships between ILF1 and senataxin and these types of lymphomas are not well established, CD40 is already known to be down-regulated in follicular lymphomas (from another microarray study37 ), to have a role in all stages of mature B-cell physiology as proliferation or germinal center developments and its pathway effects are known to depend on NF-κB activation,38 probably related to our findings. These genes are different from those described by Tröen et al,33 probably because of the dissimilar criteria for case selection or the smaller sizes of the Tröen et al series.33 Although this needs to be validated in further independent studies, it opens the way for the use of more accurate markers for the recognition of this disorder.
One of the particularly interesting findings of this study is the identification, by time-to-event analysis, of several new prognostic markers, including CD38, IgVH mutational status, and the increased expression of a set of key NF-κB pathway genes. Multiple studies have shown that increased CD38 surface expression may serve as a marker of unfavorable prognosis in CLL,39-42 and our observations indicate that this extends to SMZL cases, thus emphasizing the similarities between the 2 conditions. Here, CD38 appears also to be associated with the use of nonmutated IgVH genes and more aggressive clinical behavior. Likewise, Syk and Ki67 proteins were related to progression-free interval and are also associated with CD38 expression. Since Syk is functionally homologous with the T-cell-associated protein-tyrosine kinase Zap-70, this could be studied as a possible substitute for this CLL prognostic marker in SMZL.
Time-to-event analysis of a cluster of NF-κB pathway genes revealed the expression of c-REL to be associated with a longer disease-related overall survival. Increased expression of c-REL is probably related with NF-κB activation, which again mirrors the observation made in CLL, where the increased expression of p-IκBα, a surrogate of NF-κB activation, was associated with a more favorable clinical course.10 A potential role for TNF and BCR signaling in cell survival in SMZL is implied by the association of PKCA with an unfavorable course. ADAM17 (TACE) plays a prominent role in the activation of the Notch pathway.43
The lack of somatic IgVH mutations and the expression of various markers could reliably identify patients whose disease has a more aggressive behavior. If validated in an independent and larger set of patients, this could allow a more rational approach to be adopted toward the stratification of patients into different risk groups in a neoplastic process where different therapeutic options are available.
Gene-expression profiling using DNA microarrays is providing evidence of new molecular therapeutic target in neoplasias. Our study identified rational molecules, such as CD52, PKCA, and SYK. The demonstration of CD52 overexpression in our series requires prospective evaluation to establish whether the use of anti-CD52 monoclonal therapy (alemtuzumab [Campath]) could be used as therapy in some cases of SMZL. Protein kinase C isozymes have been reported as potential targets for anticancer therapy,44 and PKC modulators directed against PKC-α have shown antitumor activity in human cells. A selective inhibitor of SYK kinase has recently been identified.45
In summary, this study demonstrates that SMZL has a strongly homogeneous signature and identifies several biologic prognostic markers as well as potential therapeutic targets.
Genes differentially expressed in SMZL and other small B-cell lymphomas (MCL, CLL, FL). Red and blue indicate higher and lower expression, respectively. (A) Panel of 266 genes identified by ANOVA to be up-regulated in SMZL versus other NHLs. (B) Overexpressed genes in SMZL, after normalization with nontumor spleen (FDR < 0.2).
Genes differentially expressed in SMZL and other small B-cell lymphomas (MCL, CLL, FL). Red and blue indicate higher and lower expression, respectively. (A) Panel of 266 genes identified by ANOVA to be up-regulated in SMZL versus other NHLs. (B) Overexpressed genes in SMZL, after normalization with nontumor spleen (FDR < 0.2).
Prepublished online as Blood First Edition Paper, May 24, 2005; DOI 10.1182/blood-2004-10-3898.
Supported by grants from the Comunidad Autónoma de Madrid (CAM 08.1/0011/2001.1), Comunidad Autónoma de Castilla La Mancha (02 031-00) (GC2006), the Ministerio de Sanidad y Consumo (G03/179), the Ministerio de Ciencia y Tecnología (SAF 2001-0060, 2004-04 286), Spain, and the Asociación Española Contra el Cáncer (F.I.C. and A.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr P. Domínguez, Madrid; Dr T. Flores, Salamanca; L. Bernardó, Girona; A. Santana, Madrid; A. Alvarez, Barakaldo; E. Sánchez, Talavera; P. Gonzalvo, Jarrió; J. J. Esquivias, Granada; M. F. Fresno, Oviedo; B. J. Blas, Sagunt; R. García-Ligero Algeciras; P. Alemany, Valencia; J. San Juan, Valencia; and the CNIO Tumour Bank for kindly providing the cases included in this series. We also thank Dr L. Sánchez, E. Gómez (CNIO), and C. Granda (HVS) for their excellent technical help.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal