Abstract
In T-cell–dependent antibody responses, antigen-specific B cells undergo a phase of secondary antibody diversification in germinal centers (GCs). Somatic hypermutation (SHM) introduces mutations into the rearranged immunoglobulin (Ig) variable (V) region genes, and class-switch recombination (CSR) alters the Ig heavy (H) chain constant region. Aberrant SHM or CSR is thought to contribute to the development of GC-derived B-cell malignancies. Diffuse large B-cell lymphomas (DLBCLs) are a heterogeneous group of such GC-derived tumors. Based on their gene expression profile, DLBCLs can be divided into activated B-cell–like and GC-like subgroups. The human gene HGAL is predominantly expressed in GCs. It is also part of the gene expression signature of GC-like DLBCL, and its high expression in DLBCL has been associated with a better clinical prognosis. We have generated mice deficient of the HGAL homologue M17 in order to investigate its functional significance. The mutant animals form normal GCs, undergo efficient CSR and SHM, and mount T-cell–dependent antibody responses similar to wild-type controls. Thus, M17 is dispensable for the GC reaction, and its potential function in the pathogenesis of DLBCL remains elusive.
Introduction
Germinal centers (GCs) are distinct histologic structures in secondary lymphoid organs that form upon activation of B cells in T-cell–dependent immune responses.1-3 Here, activated B cells undergo a phase of rapid expansion during which 2 major genetic changes occur: somatic hypermutation (SHM) and class-switch recombination (CSR). CSR alters the constant (C) region of the immunoglobulin (Ig) heavy chain by switching from Cμ to the C region of another isotype.4,5 SHM modifies the affinity of the B-cell receptor (BCR) for the cognate antigen by introducing predominantly point mutations into the variable region of the Ig genes.6-8 B cells carrying unfavorable mutations undergo apoptosis, while B cells expressing high-affinity BCRs are selected and differentiate into memory B cells or antibody-secreting plasma cells.3
GCs are thought to be the origin of most B-cell malignancies as a result of aberrant CSR or SHM, which promote chromosome translocations and mutations of proto-oncogenes, respectively.9-11 Diffuse large B-cell lymphomas (DLBCLs) are a heterogeneous group of B-cell–derived tumors. They usually carry mutated Ig genes indicating their GC or post-GC origin and harbor SHM-associated mutations in proto-oncogenes such as BCL6 and c-MYC.10 Based on their gene-expression profiles, DLBCLs can be divided into distinct subsets that represent separate stages of B-cell differentiation and show a different clinical outcome.12-16 Activated B-cell–like DLBCLs express genes characteristic of activated B cells and plasma cells and have a poor clinical prognosis.12 In contrast, GC-like DLBCLs show a gene-expression profile typical of GC B cells, display ongoing SHM,17 and are associated with a favorable clinical prognosis.12 The expression levels of some, but not all, GC-specific genes can predict the clinical outcome of DLBCL.18,19 The human germinal center–associated lymphoma (HGAL) gene encodes a cytosolic protein that is expressed mainly in the GCs.19-21 HGAL is also expressed in many GC-derived cancers, including follicular lymphomas, Burkitt lymphomas, and DLBCLs.12,21 In DLBCL, HGAL gene expression is heterogeneous and part of the gene expression signature of GC-like DLBCL.12 HGAL can serve as a marker for the clinical prognosis of DLBCL, since its high expression correlates with a better survival rate of patients suffering from this malignancy.12,19 However, little is known about the biologic role of HGAL in DLBCL.
In an attempt to identify genes that are specifically involved in the GC reaction in mice, we had previously isolated a gene termed M17 by subtraction of cDNA libraries derived from GC B cells and LPS-stimulated splenocytes.22 M17 is the mouse homologue of HGAL.19,20 Like its human counterpart, it is a protein of unknown function that is predominantly expressed in the GCs. M17 is encoded by 5 exons and contains several potential phosphorylation sites, including a putative immune tyrosine activation motif (ITAM), suggesting a role in cell signaling.19,22 In order to elucidate the biologic function of M17 in the GC reaction and thus to obtain clues about the potential function of HGAL in DLBCL pathogenesis, we have generated M17-deficient mice by gene targeting. Here, we report that M17-deficient B cells are able to mount normal T-cell–dependent immune responses, undergo efficient CSR and mutate their Ig genes indistinguishably from wild-type animals. Thus, M17 is dispensable for the GC reaction.
Materials and methods
Targeting vector
The construction of the targeting vector has been described elsewhere in detail.23 Briefly, DNA fragments containing parts of the M17 locus were obtained by screening a phage library containing C57BL/6 mouse genomic DNA. The resulting cloning vectors contained a fragment of 6.9 kb encoding M17 exons 3, 4, and a part of exon 5 and a fragment of 7.8 kb encoding M17 exon 5, respectively. A 3.2-kb XbaI-ScaI fragment was used as the left arm and a 3.4-kb XbaI-SalI fragment was used as the right arm of homology. The loxP-flanked region consisted of a 4.5-kb ScaI-XbaI fragment containing exons 4 and 5 of the M17 gene. The frt-flanked SAS-IRES-BAP-31N-EGFP cassette contained a splice acceptor site (SAS), followed by an internal ribosome entry site (IRES), and the BAP31N-EGFP fragment encoding a membrane-bound form of EGFP. This cassette was inserted immediately after the third loxP site. Finally, a DNA fragment encoding a thymidine kinase gene under the control of a phosphoglycerol kinase promoter was introduced into the vector. The final targeting vector was named pBS-M17-TV and linearized by digestion with SalI for the transfection of embryonic stem cells.
Generation of mutant mice
Embryonic stem (ES) cell culture has been described previously.24 Briefly, 1 × 107 C57BL/6-derived Bruce 4 ES cells were transfected with 30 μg linearized targeting vector by electroporation and subsequently subjected to G418 (170 μg/mL) and gancyclovir (2 μM) selection. Double-resistant colonies were screened for homologous recombination by Southern hybridization of EcoRI-digested genomic DNA with probe A located 5′ of the targeted region. Probe A was derived from plasmid pDS10 by double digestion with the HincII and XbaI restriction enzymes. Cointegration of the third loxP site was confirmed by EcoRI digestion of genomic DNA using probe B located at the 3′ end of the targeted region. To obtain probe B, plasmid pDS9 was cut with SalI, and the ligated plasmid was cut again with HinDIII and PstI to obtain a 1.3-kb fragment, which was used as probe. Cre-mediated deletion was confirmed with the internal probe C. Probe C was generated by polymerase chain reaction (PCR) with the primer pair MP57 and MP26 using plasmid pDS10 as template. Chimeric mice were derived from 2 correctly targeted ES cell clones that had been injected into blastocysts of CB.20 mice and transplanted into the uteri of pseudopregnant CB.20 female mice.
Mice
Following germ-line transmission, mice containing the targeted M17 allele were intercrossed with deleter mice.25 Mice carrying a deleted M17 allele were crossed to homozygosity. All mice were kept on the C57BL/6 background and housed in conventional animal facilities at the Institute for Genetics in Cologne, Germany, or at the Brigham & Woman's Hospital and the Harvard School of Public Health in Boston, MA. Mice were used for analysis at the age of 8 to 12 weeks.
RT-PCR
RNA was isolated from B cells with Trizol (Invitrogen, Frederick, MD) according to the manufacturer's instructions. Reverse transcription (RT) was performed with an oligo-T primer using the Thermoscript kit (Invitrogen). Subsequently, the cDNA was amplified by PCR using the primer pairs M17Seq1 (5′-ATGGGGAACTGTTTGCAGAGGACAACCAG) and M17Seq2 (5′-GGGAGCTGAAGTCATCCCTTCA), M17Seq1 and M17Seq3 (5′-CTTTGGAGACTCTTGTCTGGC), or M17Seq1 and M17Seq4 (5′-GCTGTTGAAAGGCATGTGGAG). Equal loading of cDNA was controlled by the amplification of β-actin cDNA using the primers m-b-actinB (5′-TCTTCATGGTGCTAGGAGCCA) and m-b-actinT (5′-CCTAAGGCCAACCGTGAAAAG). All PCR reactions involved the denaturation of the DNA for 30 seconds at 94°C, primer annealing for 45 seconds at 57°C, and elongation for 60 seconds at 72°C and used for 30 to 34 cycles.
Flow cytometry
Single-cell suspensions prepared from primary or secondary lymphoid organs were stained with the following monoclonal antibodies conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), CyChrome (CyC), allophycocyanin (APC), or biotin: anti-CD3 (145-2C11), anti-CD4 (L3T4), anti-CD5 (53-7.3), anti-CD8 (53-6.7), anti-CD19 (ID3), anti-CD21, anti-CD23 (B3B4), anti-CD25, anti-CD43 (S7), anti-CD69 (H1.2F3), anti-IgG1, anti-IgG3, anti–c-kit, and anti-Fas (Jo2) (all from BD Pharmingen, San Diego, CA). Additionally, monoclonal antibodies against IgM (R33-2412), IgD (1.3-5), B220 (RA3-6B2), and MHC-II (M5/114) were prepared and conjugated in our laboratory. PNA conjugates were PNA-FITC or PNA-biotin (Vector Laboratories, Burlingame, CA). Biotin conjugates were visualized with CyChrome- or allophycocyanin-streptavidin (BD Pharmingen). All samples were acquired on a FACSCalibur (BD Biosciences, San Jose, CA) and analyzed using CellQuest software.
Immunizations
Mice were immunized via intraperitoneal injection with 50 μg or 10 μg alum-precipitated NP36-CG ((4-hydroxy-3-nitrophenyl)acetyl coupled to chicken γ-globulin; Biosearch Technologies, Novato, CA). To determine NP-specific Ig serum titers, mice were bled every 7 days from the tail vain.
ELISA
The determination of Ig isotype concentrations and of NP-specific antibody concentrations in the serum has been published elsewhere.26,27 Briefly, microtiter plates (Costar, Cambridge, MA) were coated with NP-BSA or antibodies of known isotype in PBS at 4°C overnight, and subsequently blocked at room temperature for 30 minutes with PBS, 0.5% BSA, 0.01% N3 (pH 7.2). Serially diluted serum samples were applied to the wells and incubated at 4°C overnight. The plates were then incubated with a secondary biotinylated anti-Ig antibody at 37°C for 1 hour, followed by the incubation with SA-conjugated alkaline phosphatase (AP; Roche, Indianapolis, IN) at room temperature for 30 minutes. The amount of bound AP was detected by incubation with p-nitrophenylphosphate as substrate (Roche). Following each incubation step, unbound antibodies or SA-conjugated AP was removed by 3 washes with tap water. The OD405 was measured with an enzyme-linked immunosorbent assay (ELISA)–photometer (Spectramax 340; Molecular Devices, Sunnyvale, CA), and antibody concentrations were determined by comparison with a standard curve.
Immunohistology
Mice were immunized with 50 μg NP-CG. At 14 days after immunization, spleens or Peyer patches were embedded in Tissue-Tek OTC compound (Sekura-Finetek, Torrance, CA) and frozen in methyl butane that was cooled in liquid nitrogen. Frozen sections were fixed in cold acetone and air-dried. After rehydration in PBS, the sections were first incubated in blocking buffer (PBS containing 1% BSA and 5% goat serum) for 30 minutes at room temperature, followed by a mixture of either rat anti–mouse CD19 (Pharmingen) and biotinylated PNA (Biosearch) or rat anti–mouse FDCM1 (Pharmingen) and biotinylated PNA in PBS for 30 minutes at room temperature. The sections were washed 3 times in PBS and subsequently stained with a mixture of goat anti–rat IgG1-FITC and streptavidin-PE for 30 minutes at room temperature. Finally, the sections were washed in PBS, mounted with Fluorotec medium, and examined by fluorescence microscopy. The images were acquired on a Zeiss Axiovert 200M microscope (10 ×/0.3 PH.1 objective, 20 ×/0.25 PH.1 objective; 10 ×/18 ocular; Carl Zeiss, Thornwood, NJ) using a Hamamatsu C47-42-95 camera (Hamamatsu Photonics, Hamamatsu City, Japan) and the Openlab 3.1.2 software (Improvision, Lexington, MA) and further processed using Photoshop and Illustrator software (Adobe Systems, San Jose, CA). Alternatively, the frozen sections were incubated with anti-BCL6 or anti-B220 antibodies, followed by HRP-conjugated goat anti–rabbit IgG, as published previously.28
B-cell stimulation and class-switch analysis
B cells were purified from splenic single-cell suspensions by magnetic-activated cell sorting (MACS) depletion using anti-CD43 microbeads (Miltenyi Biotech, Auburn, CA). Subsequently, the cells were cultured at a concentration of 106 cells/mL and stimulated with 20 μg LPS/mL alone (Sigma, St Louis, MO), 20 μg LPS/mL and 25 ng IL-4/mL (R&D Systems, Minneapolis, MN), 20 μg LPS/mL and 2 ng IFN-γ/mL (R&D Systems), 20 μg LPS/mL and 2 ng TGFγ/mL (R&D Systems), 0.5 μg anti-CD40 mAb/mL (clone HM40-3; Pharmingen) and 25 ng IL-4/mL, or 25 ng IL-4/mL alone. Cells were cultured for 5 days, during which the number of cells per volume was kept constant by addition of fresh medium. The percentage of class-switched cells was determined on day 4 or day 5 by flow cytometry.
Isolation of GC B cells and analysis of SHM
The analysis of SHM in GC B cells has been published previously.27,29 Mice were immunized with 50 μg NP-CG. At 14 days after immunization, GC B cells from spleen or Peyer patches were purified directly following incubation with PNA-FITC, anti–Fas-PE, and anti–B220-APC mAb on a FACSVantage cell sorter (Becton Dickinson, Mountain View, CA) and sorted into a naive fraction (B200+PNAlowFaslow) and a GC fraction (B220+PNAhighFashigh). PCR fragments from isolated DNA of sorted cell populations were amplified using primer J558Fr3, which anneals in the framework 3 region of most VHJ558 genes, and primer JHCHint, which hybridizes in the intron 3′ of exon JH4.30 The PCR fragments were subcloned into the T/A cloning vector pGEM-Teasy (Promega, Madison, WI), and the resulting plasmids were sequenced using the primer JHCHint. A stretch of 500 bp intron sequence immediately downstream of the JH4 element was analyzed for somatic mutations using the Genejockey II (Biosoft, Ferguson, MO) or Lasergene (DNASTAR, Madison, WI) software. A similar analysis was performed for the determination of the frequency of the high-affinity W33L mutation in the V186.2 gene using primer pairs that are specific for the V186.2 and JH2 genes.29
Results
Generation of M17-deficient mice
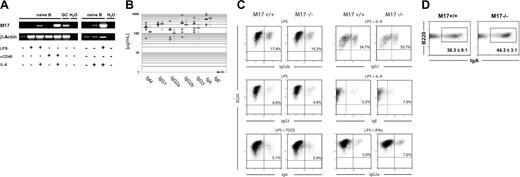
To investigate the in vivo function of M17, we modified the genomic M17 locus in the mouse by conditional gene targeting. Exons 4 and 5 of the published cDNA contain most of the coding sequence of the M17 gene,22 including the putative ITAM motif and the 3′ UTR, and their deletion likely results in a nonfunctional M17 protein. We introduced loxP sites 5′ of exon 4 and 3′ of exon 5, thus rendering these exons susceptible to Cre-mediated recombination. In addition, we introduced a cassette containing a splice-acceptor site, an internal ribosome entry site (IRES), and a green fluorescence protein gene 3′ of exon 5 to identify cells that underwent Cre-mediated deletion. The wild-type M17 locus, the targeting vector, the modified locus after homologous recombination, and the targeted locus after Cre-mediated recombination are depicted in Figure 1A. Homologous recombination in double-resistant embryonic stem (ES) cell colonies was confirmed by Southern blotting of EcoRI-digested genomic DNA using probe A (Figure 1B) or probe C (not shown). We injected 2 independently targeted ES cell clones into blastocysts to generate chimeric animals and crossed the chimeras to the deleter strain25 in order to transmit the targeted allele into the germ line and to delete the neomycin resistance (neor) gene and exons 4 and 5 in vivo. The successful deletion of the loxP-flanked exons and the neor gene was confirmed by Southern blotting of BglII-digested tail DNA with probe C (Figure 1B). Mice homozygous for the deletion of M17 gene were viable, born at mendelian ratios, and displayed no obvious abnormalities.
Generation of M17–/– mice. (A) Schematic representation of the gene targeting in the M17 locus by homologous recombination. C57BL/6-derived ES cells were targeted with a vector containing the loxP-flanked exons 4 and 5, a frt-flanked IRES-gfp cassette, and a neomycin resistance cassette for positive selection. Negative selection of clones harboring randomly integrated vectors was mediated by a thymidine kinase gene. Only exons 3 to 5 are shown. Rectangles indicate coding DNA (black, translated region; gray, untranslated region); filled triangles, loxP sites; ovals, frt sites; RI, EcoRI; and BII, BglII. Bold lines indicate regions of homology and Southern probes are shown as thin black lines under the wild-type locus. The map is not drawn to scale. (B) Successful homologous recombination was identified by Southern blot of EcoRI-digested genomic ES cell DNA and probe A located 5′ of exon 3. The wild-type fragment migrates at 6.9 kb, while the fragment from the targeted locus migrates at 4.6 kb. Cre-mediated deletion of M17 exons 4 and 5 was confirmed by Southern blot of BglII-digested genomic DNA using probe C located 3′ of exon 3. The wild-type fragment migrates at 2.1 kb and the fragment of the deleted locus migrates at 1.0 kb. (C) Confirmation of the successful inactivation of the M17 gene by RT-PCR. RNA was isolated from sorted CD19+PNA–Fas– naive B cells (N) or CD19+PNA+Fas+ GC B cells from either M17+/+ or M17–/– mice and reverse transcribed using an oligoT primer. PCR products were amplified with primers annealing either in exons 1 and 4 (M17 1/4) or in exons 1 and 3 (M17 1/3). Intron-spanning primers annealing in the β-actin gene were used to control for equal amounts of RNA. The PCR products were consistent with the expected sizes for the 2 alternative transcripts of M17.
Generation of M17–/– mice. (A) Schematic representation of the gene targeting in the M17 locus by homologous recombination. C57BL/6-derived ES cells were targeted with a vector containing the loxP-flanked exons 4 and 5, a frt-flanked IRES-gfp cassette, and a neomycin resistance cassette for positive selection. Negative selection of clones harboring randomly integrated vectors was mediated by a thymidine kinase gene. Only exons 3 to 5 are shown. Rectangles indicate coding DNA (black, translated region; gray, untranslated region); filled triangles, loxP sites; ovals, frt sites; RI, EcoRI; and BII, BglII. Bold lines indicate regions of homology and Southern probes are shown as thin black lines under the wild-type locus. The map is not drawn to scale. (B) Successful homologous recombination was identified by Southern blot of EcoRI-digested genomic ES cell DNA and probe A located 5′ of exon 3. The wild-type fragment migrates at 6.9 kb, while the fragment from the targeted locus migrates at 4.6 kb. Cre-mediated deletion of M17 exons 4 and 5 was confirmed by Southern blot of BglII-digested genomic DNA using probe C located 3′ of exon 3. The wild-type fragment migrates at 2.1 kb and the fragment of the deleted locus migrates at 1.0 kb. (C) Confirmation of the successful inactivation of the M17 gene by RT-PCR. RNA was isolated from sorted CD19+PNA–Fas– naive B cells (N) or CD19+PNA+Fas+ GC B cells from either M17+/+ or M17–/– mice and reverse transcribed using an oligoT primer. PCR products were amplified with primers annealing either in exons 1 and 4 (M17 1/4) or in exons 1 and 3 (M17 1/3). Intron-spanning primers annealing in the β-actin gene were used to control for equal amounts of RNA. The PCR products were consistent with the expected sizes for the 2 alternative transcripts of M17.
The inactivation of the M17 gene was confirmed by RNA expression analysis. Reverse-transcription PCR (RT-PCR) of RNA derived from naive and GC B cells using a primer pair that anneals in exons 1 and 4 led to PCR products from RNA derived of GC B cells but not of naive B cells of M17+/+ mice. In contrast, amplification of equal amounts of GC B-cell–derived cDNA from M17–/– mice did not yield a specific PCR product (Figure 1C). A similar RT-PCR with a reverse primer that anneals to exon 3 also failed to amplify a specific PCR product in M17–/– mice, suggesting a destabilized mRNA of the remaining M17 gene and hence a nonfunctional GFP cassette. The amplification of cDNA from M17+/+ mice revealed the presence of 2 products, which were consistent with the expected sizes for the 2 predicted alternative transcripts of M17.22,31
B-cell compartments in M17-deficient mice
While M17 is predominantly expressed in GCs, low levels of M17 transcripts are also detected in the bone marrow.22 However, only a very small population of lymphocytes in the bone marrow expresses M17 (around 0.4%).22,32 Plasma cells residing in the bone marrow do not express M17.22,32 We therefore isolated B220– cells, B220+ IgM– c-kit+ pro-B cells, B220+ IgM– CD25+ pre-B cells, and B220+ IgM+ B cells by cell sorting in order to determine the expression of M17 mRNA transcripts in these cell populations by RT-PCR. The expression of M17 was confined to the fraction of B220+ IgM+ B cells (Figure 2A). Consistent with this finding, the analysis of bone marrow–derived cells of M17-deficient mice by flow cytometry did not reveal differences in absolute cell numbers or proportions of major subsets of developing B cells, indicating that M17 is dispensable for B-cell development (data not shown). In spleen and mesenteric lymph nodes, the number of total cells was equivalent in M17–/– mice and wild-type controls. The proportions of B- and T-cell subsets (including CD19+CD21– CD23– transitional, CD19+CD21+CD23+ follicular, and CD19+CD21+CD23– marginal zone B cells) were also not affected (data not shown). In contrast, the number of total cells in Peyer patches was 2- to 3-fold reduced (Figure 2B). This reduction reflected both a reduced number of Peyer patches per mouse (5.1 ± 1.3 versus 7.8 ± 1.1 in M17–/– and M17+/+ mice, respectively) and a reduced size of some but not all Peyer patches of M17–/– mice. However, the ratio of B and T cells in the Peyer patches remained unchanged (data not shown).
M17 may be required for the recruitment of B cells into the GCs. To analyze the proportions of CD19+Fas+PNAhigh GC B cells in M17–/– mice and wild-type controls, we immunized the mice with 50 μg of the T-cell–dependent antigen NP-CG and determined the percentages of GC B cells 14 days later. No significant differences between mutants and controls were observed (Figure 2C). In addition, despite the general reduction of total cells in the Peyer patches, we still observed spontaneously arising GC B cells in this lymphoid compartment in unimmunized mice (data not shown). Hence, M17 is dispensable for the generation and maintenance of the major B- and T-cell populations in bone marrow, spleen, and mesenteric lymph nodes, but affects the number and size of Peyer patches.
Cell numbers and GC formation. (A) Expression of M17 cDNA in developing B cells. Bone marrow–derived lymphocytes were sorted into B220– cells, B220+ IgM– c-kit+ pro-B cells, B220+ IgM– CD25+ pre-B cells, and B220+ IgM+ B cells by fluorescent-activated cell sorting (FACS), and the expression of M17 mRNA transcripts in these cell populations was determined by RT-PCR. (B) The total number of cells in peripheral lymphoid organs. SP indicates spleen; MLN, mesenteric lymph nodes; and PP, Peyer patches. (C) Generation of GC B cells. Mice were immunized with 50 μg NP-CG and analyzed for the presence of CD19+PNA+Fas+ GC B cells 14 days after immunization. Only CD19+ cells are shown. Numbers represent the mean in percent plus standard deviation. SP indicates spleen; MLN, mesenteric lymph nodes; and PP, Peyer patches. (D-E) GC architecture in M17–/– mice. Mice were immunized with 50 μg NP-CG. Frozen splenic sections were prepared on day 14 after immunization and analyzed by immunofluorescence. (D) Sections were incubated with αCD19 mAb (green) and PNA (red) to visualize B-cell follicles and GCs, respectively. (E) Sections were incubated with αBCL-6 mAb (brown) and counterstained with hematoxylin (blue). Representative pictures are shown. SP indicates spleen; PP, Peyer patches. Numbers below pictures indicate the magnification.
Cell numbers and GC formation. (A) Expression of M17 cDNA in developing B cells. Bone marrow–derived lymphocytes were sorted into B220– cells, B220+ IgM– c-kit+ pro-B cells, B220+ IgM– CD25+ pre-B cells, and B220+ IgM+ B cells by fluorescent-activated cell sorting (FACS), and the expression of M17 mRNA transcripts in these cell populations was determined by RT-PCR. (B) The total number of cells in peripheral lymphoid organs. SP indicates spleen; MLN, mesenteric lymph nodes; and PP, Peyer patches. (C) Generation of GC B cells. Mice were immunized with 50 μg NP-CG and analyzed for the presence of CD19+PNA+Fas+ GC B cells 14 days after immunization. Only CD19+ cells are shown. Numbers represent the mean in percent plus standard deviation. SP indicates spleen; MLN, mesenteric lymph nodes; and PP, Peyer patches. (D-E) GC architecture in M17–/– mice. Mice were immunized with 50 μg NP-CG. Frozen splenic sections were prepared on day 14 after immunization and analyzed by immunofluorescence. (D) Sections were incubated with αCD19 mAb (green) and PNA (red) to visualize B-cell follicles and GCs, respectively. (E) Sections were incubated with αBCL-6 mAb (brown) and counterstained with hematoxylin (blue). Representative pictures are shown. SP indicates spleen; PP, Peyer patches. Numbers below pictures indicate the magnification.
We then isolated spleens and Peyer patches of M17–/– mice and wild-type controls 14 days after immunization with NP-CG and prepared frozen sections in order to evaluate the impact of M17 deficiency on GC architecture. The sections were stained with peanut agglutinin (PNA) plus mAbs against CD19 or FDCM1 coupled to fluorescent conjugates to visualize GCs, B-cell follicles, and the network of follicular dendritic cells (FDCs), respectively. Alternatively, the sections were stained with mAbs against BCL6 and counterstained with hematoxylin. M17–/– mice developed GCs (PNA+ cells, red) and B-cell follicles (CD19+ cells, green) comparable in size and shape with wild-type controls (Figure 2D). GCs of M17–/– mice contained a polarized network of FDCs (data not shown), and the architecture of GCs remained unchanged in these tissues. The expression of BCL6, which is essential for GC formation, also did not change in M17–/– mice (Figure 2E). Hence, M17 is not an essential gene required for GC formation.
M17 mRNA is up-regulated by IL-4
Interleukin-4 (IL-4) is an essential cytokine for the induction of CSR to IgG1 and IgE. HGAL expression is induced by IL-4 in peripheral blood B cells.19 IL-4 signal transduction is mediated by STAT6. The M17 gene contains 2 putative STAT6 binding sites near the start of transcription, thus suggesting a similar response to IL-4 stimulation as in the case of the HGAL gene. To test this possibility, we stimulated purified splenic B cells with either LPS or an αCD40 mAb in the presence or absence of IL-4. After 48 hours, we isolated total RNA from equal numbers of cells and assessed M17 mRNA expression by RT-PCR using intron-spanning primers specific for M17 and housekeeping gene β-actin. Unstimulated cells or cells stimulated with either LPS or αCD40 mAb alone did not express M17 mRNA. In contrast, B cells stimulated with IL-4 and LPS or IL-4 and αCD40 mAb up-regulated the expression of M17-specific transcripts (Figure 3A). We conclude that the M17 gene, as HGAL, is the target of IL-4 stimulation in activated B cells.
Antibody titers and class-switch recombination. (A) M17 is up-regulated by IL-4. Isolated splenocytes were MACS depleted of CD43+ cells and subsequently activated with the indicated stimuli. Following the isolation of total RNA 48 hours later, RT-PCR was performed using intron-spanning primers for the M17 and β-actin genes. Naive B indicates naive B cells in vitro; GC, GC B cells; and H20, water control. A representative experiment is shown. (B) Antibody titers in the serum of unimmunized wild-type and M17–/– mice were determined in an ELISA assay. Each circle represents one mouse. Black bars indicate the geometric means. Closed circles indicate wild-type mice; open circles, M17-deficient mice. (C) In vitro stimulation of isolated B cells of M17–/– mice and wild-type controls. B cells were induced to undergo CSR with the indicated stimuli. The percentage of class-switched cells was determined 4 days later by flow cytometry. Numbers in the graphs represent the percentages of switched cells. A representative experiment is shown. (D) Percentage of IgA+ GC B cells in the Peyer patches of M17–/– mice and wild-type controls. The differences in percentages of IgA+ B cells were not statistically significant (P = .15).
Antibody titers and class-switch recombination. (A) M17 is up-regulated by IL-4. Isolated splenocytes were MACS depleted of CD43+ cells and subsequently activated with the indicated stimuli. Following the isolation of total RNA 48 hours later, RT-PCR was performed using intron-spanning primers for the M17 and β-actin genes. Naive B indicates naive B cells in vitro; GC, GC B cells; and H20, water control. A representative experiment is shown. (B) Antibody titers in the serum of unimmunized wild-type and M17–/– mice were determined in an ELISA assay. Each circle represents one mouse. Black bars indicate the geometric means. Closed circles indicate wild-type mice; open circles, M17-deficient mice. (C) In vitro stimulation of isolated B cells of M17–/– mice and wild-type controls. B cells were induced to undergo CSR with the indicated stimuli. The percentage of class-switched cells was determined 4 days later by flow cytometry. Numbers in the graphs represent the percentages of switched cells. A representative experiment is shown. (D) Percentage of IgA+ GC B cells in the Peyer patches of M17–/– mice and wild-type controls. The differences in percentages of IgA+ B cells were not statistically significant (P = .15).
Normal Ig serum titers and efficient class switching in M17-deficient mice
To analyze the ability of M17–/– mice to undergo CSR, we first measured Ig antibody isotype titers in the serum of unimmunized mice by ELISA. M17–/– mice were able to produce antibodies of all isotypes and their titers were comparable with wild-type controls (Figure 3B). In order to assess whether M17 modulates CSR in more subtle ways, we then determined the ability of M17–/– B cells to undergo CSR in vitro. We induced splenic B cells from M17–/– mice and wild-type controls with the indicated stimuli and measured the percentage of class-switched cells by flow cytometry 4 days later (Figure 3C). Stimulation of the cells by LPS, which does not induce M17 expression, was able to induce efficient CSR to IgG2b and IgG3 in B cells of M17–/– mice. The activation of M17–/– B cells with LPS plus TGF-β or LPS plus IFN-β led to similar results. More importantly, M17–/– B cells switched efficiently to IgG1 and IgE isotypes after stimulation by αCD40 mAb plus IL-4, a condition in which M17 is up-regulated in wild-type B cells. In addition, CFSE-labeled M17–/– B cells proliferated upon stimulation with LPS or with αCD40 mAb plus IL-4 at comparable levels with wild-type controls (data not shown). Since we had observed a reduction of lymphocytes in the Peyer patches, we determined whether CSR to IgA, the predominant isotype in the Peyer patches, is affected in this cellular compartment in vivo (Figure 3D). GCs in the Peyer patches of M17–/– mice contained similar percentages of IgA+ B cells as wild-type controls (44.3% ± 3.1% versus 36.1% ± 8.1%, P = .15). In summary, we conclude that M17 is not required for B-cell proliferation or CSR.
GC B cells of M17-deficient mice mutate their Ig genes efficiently
To elucidate whether M17 is involved in SHM of the Ig V genes, we monitored the accumulation of somatic mutations in the intron downstream of the rearranged V gene in the IgH locus by immunizing 2 sets of wild-type and M17-deficient mice with 50 μg NP-CG and isolating GC B cells and naive B cells 14 days after immunization.30 All mice used for this analysis responded to NP-CG, as demonstrated by the elevated titers of NP-CG–specific IgG1 antibodies in the blood (data not shown). The PCR reaction was performed using a primer pair that anneals in the framework region 3 of most VHJ558 genes and in the intron downstream of the JH4 gene, thus amplifying the intron sequence downstream of a large proportion of rearranged V genes.30 The PCR products obtained from VDJ rearrangements involving a JH4 element were selected for further analysis. The products were subcloned and sequenced. Clonally related sequences were excluded from the analysis and a stretch of 500 bp of intron sequence was analyzed for the presence of somatic mutations. As expected from previous analyses, sequences derived from naive B cells were largely unmutated27,29 (Table 1). In contrast, the majority of sequences derived from GC B cells of both wild-type and M17–/– mice contained somatic mutations. Sequences harboring one or more mutations were considered for the evaluation of the frequency of somatic mutations and the analysis of mutational patterns. Both wild-type and M17–/– mice mutated their Ig genes with a similar mutation frequency (0.83% versus 0.75% for GC B cells in the spleen and 1.57% versus 1.71% for GC B cells in the Peyer patches; Table 1) and range of mutations per sequence (1-12 versus 1-14 in the GC B cell in the spleen and 1-20 versus 1-25 in the GC B cells in the Peyer patches). The analysis of the mutational patterns did not reveal significant differences between M17–/– mice and wild-type controls (Table 2). There was no difference in the number of deletions or duplications (data not shown), and the ratio of transitions versus transversions did not change significantly when compared with control animals. Hence, M17 is not required for somatic hypermutation.
Frequency and range of mutations in a 500-bp-long region in the intron downstream of the rearranged VHDHJH4 joints of GC B cells derived from pairs of M17+/+ and M17–/– mice
Organ and genotype . | No. clones . | No. mutated clones . | Range, mutations/clone . | No. mutations/no. bp (%) . |
|---|---|---|---|---|
| Spleen | ||||
| M17+/+ | 35 | 25 | 1-14 | 102/12 248 (0.83) |
| M17-/- | 56 | 31 | 1-12 | 117/15 500 (0.75) |
| Peyer patches | ||||
| M17+/+ | 26 | 25 | 1-20 | 196/12 500 (1.57) |
| M17-/- | 30 | 25 | 1-25 | 213/12 447 (1.71) |
Organ and genotype . | No. clones . | No. mutated clones . | Range, mutations/clone . | No. mutations/no. bp (%) . |
|---|---|---|---|---|
| Spleen | ||||
| M17+/+ | 35 | 25 | 1-14 | 102/12 248 (0.83) |
| M17-/- | 56 | 31 | 1-12 | 117/15 500 (0.75) |
| Peyer patches | ||||
| M17+/+ | 26 | 25 | 1-20 | 196/12 500 (1.57) |
| M17-/- | 30 | 25 | 1-25 | 213/12 447 (1.71) |
Naive M17-/- B cells had a 0.03% mutation frequency.
GC B cells were isolated 14 days after immunization with NP-CG. Following cell lysis, a 600-bp fragment was PCR amplified from 40 000 cell equivalents using a primer pair that anneals in the framework 3 region of most J558 V genes and in the intron downstream of JH4 gene segment.
Patterns of mutations in the intron downstream of ther rearranged VHDHJH4 joints of GC B cells
. | M17+/+, % mutations . | . | . | . | . | M17-/-, % mutations . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ and base from which mutation occurred . | Base to which mutation occurred . | . | . | . | . | Base to which mutation occurred . | . | . | . | . | ||||||
| . | A . | G . | C . | T . | Total . | A . | G . | C . | T . | Total . | ||||||
| Spleen | ||||||||||||||||
| A | NA | 25 | 8 | 12 | 45 | NA | 14 | 10 | 6 | 30 | ||||||
| G | 14 | NA | 9 | 1 | 24 | 13 | NA | 3 | 3 | 19 | ||||||
| C | 1 | 5 | NA | 5 | 11 | 1 | 2 | NA | 12 | 15 | ||||||
| T | 3 | 2 | 16 | NA | 21 | 9 | 9 | 19 | NA | 37 | ||||||
| Peyer patches | ||||||||||||||||
| A | NA | 25 | 7 | 11 | 43 | NA | 20 | 11 | 15 | 46 | ||||||
| G | 16 | NA | 6 | 3 | 25 | 12 | NA | 6 | 5 | 23 | ||||||
| C | 1 | 1 | NA | 6 | 8 | 1 | 1 | NA | 11 | 13 | ||||||
| T | 5 | 9 | 9 | NA | 23 | 6 | 3 | 10 | NA | 19 | ||||||
. | M17+/+, % mutations . | . | . | . | . | M17-/-, % mutations . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ and base from which mutation occurred . | Base to which mutation occurred . | . | . | . | . | Base to which mutation occurred . | . | . | . | . | ||||||
| . | A . | G . | C . | T . | Total . | A . | G . | C . | T . | Total . | ||||||
| Spleen | ||||||||||||||||
| A | NA | 25 | 8 | 12 | 45 | NA | 14 | 10 | 6 | 30 | ||||||
| G | 14 | NA | 9 | 1 | 24 | 13 | NA | 3 | 3 | 19 | ||||||
| C | 1 | 5 | NA | 5 | 11 | 1 | 2 | NA | 12 | 15 | ||||||
| T | 3 | 2 | 16 | NA | 21 | 9 | 9 | 19 | NA | 37 | ||||||
| Peyer patches | ||||||||||||||||
| A | NA | 25 | 7 | 11 | 43 | NA | 20 | 11 | 15 | 46 | ||||||
| G | 16 | NA | 6 | 3 | 25 | 12 | NA | 6 | 5 | 23 | ||||||
| C | 1 | 1 | NA | 6 | 8 | 1 | 1 | NA | 11 | 13 | ||||||
| T | 5 | 9 | 9 | NA | 23 | 6 | 3 | 10 | NA | 19 | ||||||
For M17+/+ spleens, number of mutations (n) = 102 and the ratio of transitions to transversions (Tr/Tv) = 1.5; for M17-/- spleens, n = 117 and Tr/Tv = 1.3; for M17+/+ Peyer patches, n = 196 and Tr/Tv = 1.3; for M17-/- Peyer patches, n = 213 and Tr/Tv = 1.1. NA indicates not applicable.
While M17 is not essential for SHM, it may affect positive selection and affinity maturation during the GC reaction. B cells expressing a V186.2 gene–containing heavy chain in conjunction with a λ1 light chain dominate the immune response to the hapten NP in C57BL/6 mice.33-35 Mutations leading to a tryptophan to leucin exchange in codon 33 of the V186.2 gene result in a 10-fold increase of the affinity of the anti-NP antibody and can serve as a measure of affinity maturation.36,37 We therefore amplified and sequenced the V(D)J rearrangements involving the V186.2 and JH2 genes of splenic GC B cells 14 days after immunization and scored the frequency of the W33L mutation.29 Both wild-type and M17–/– mice accumulated the W33L mutation with similar frequency (44% versus 56%; Table 3). Thus, antibody affinity maturation does not depend on M17.
Frequency of the high-affinity tryptophan to leucin (W33L) mutation in codon 33 of the V186.2 gene using primer pairs that anneal in the V186.2 and JH2 gene
Genotype . | No. clones . | No. mutated clones . | Clones with high affinity position 33 W/L mutation, % . |
|---|---|---|---|
| M17+/+ | 18 | 18 | 44 |
| M17-/- | 18 | 16 | 56 |
Genotype . | No. clones . | No. mutated clones . | Clones with high affinity position 33 W/L mutation, % . |
|---|---|---|---|
| M17+/+ | 18 | 18 | 44 |
| M17-/- | 18 | 16 | 56 |
Antibody response and generation of memory B cells in M17-deficient mice
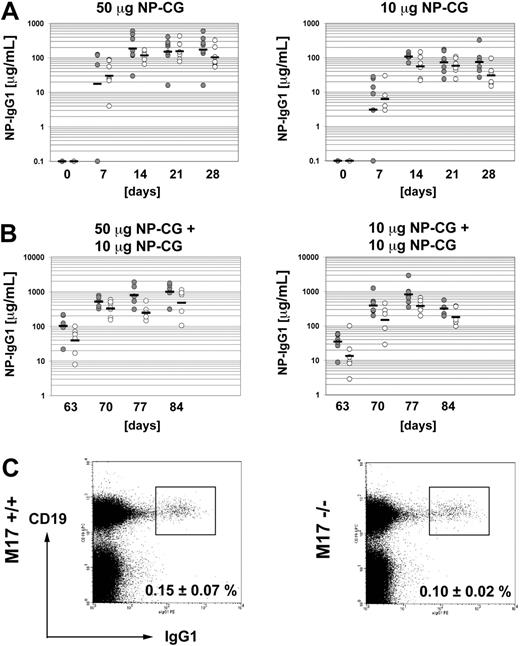
Finally, we tested the primary antibody response of M17–/– mice against the T-cell–dependent antigen NP-CG. To this end, we immunized groups of 7 wild-type or M17-deficient mice with either 50 μg or 10 μg NP-CG in alum per mouse and measured the serum titers of NP-specific IgG1 antibodies every 7 days for 4 weeks. The M17-deficient cohort immunized with 50 μg NP-CG yielded a robust and consistent antibody response with a geometric mean of 117 μg/μL NP-specific IgG1 at the peak of the response (Figure 4A). The response was comparable with that of wild-type controls. M17–/– mice immunized with 10 μg NP-CG gave rise to a similar antibody response against NP-CG, albeit at lower levels: The response peaked at 56 μg/mL NP-specific IgG1 14 days after immunization and was indistinguishable from that in the wild-type. HGAL has been implicated in the generation of memory B cells.19 Therefore, we evaluated the ability of M17–/– mice to mount a secondary antibody response. We immunized the same mice used for the analysis of the primary immune response with 10 μg NP-CG without alum 63 days after primary immunization. M17–/– mice that had been primed with 50 μg NP-CG for the primary immune response mounted a secondary response, which was only moderately weaker in M17–/– mice than that of the wild-type controls (Figure 4B). Wild-type mice reached a geometric mean of 1004 μg/mL at the peak of the response and M17–/– mice peaked at 482 μg/mL. Immunization of mice that had been primed with 10 μg NP-CG gave a similar result (821 μg/mL versus 381 μg/mL). The differences between wild-type and M17–/– mice were not statistically significant (P = .12 and P = .15, respectively). In addition, we measured the fraction of splenic IgG1+ CD19+ B cells, which consists predominantly of memory B cells, in the 6-month-old immunized animals 9 weeks after the second immunization. Equal numbers of IgG1+ B cells were present in the M17–/– mice and the wild-type controls (Figure 4C). While only a fraction of the memory B cells detected in the animals may result from the intentional immunization, they clearly accumulated over time, as the fraction of IgG1+ B cells was smaller in 6-week-old unimmunized M17–/– mice (≤ 0.05%), but again similar to age-matched wild-type mice (data not shown). The IgG1+ B cells in these animals had the typical phenotype of memory B cells, in that they expressed the CD38 surface marker (data not shown). We conclude that M17–/– mice are able to undergo a robust T-cell–dependent antibody response and generate memory B cells.
T-cell–dependent immune response of M17–/– mice and generation of memory B cells. (A) Primary immunization with 50 μg (left panel) or 10 μg (right panel) NP-CG in alum. Each circle represents one mouse. Bars indicate the geometric means. Closed circle indicates M17+/+ mice; open circle, M17–/– mice. (B) Secondary immunization with 10 μg NP-CG without alum of mice previously immunized with 50 μg (left panel) or 10 μg (right panel) NP-CG. (C) Amount of splenic IgG1+ memory B cells 9 weeks after the secondary immunization. Numbers are in percent plus standard deviation of total B cells (n = 5). The differences were not statistically significant.
T-cell–dependent immune response of M17–/– mice and generation of memory B cells. (A) Primary immunization with 50 μg (left panel) or 10 μg (right panel) NP-CG in alum. Each circle represents one mouse. Bars indicate the geometric means. Closed circle indicates M17+/+ mice; open circle, M17–/– mice. (B) Secondary immunization with 10 μg NP-CG without alum of mice previously immunized with 50 μg (left panel) or 10 μg (right panel) NP-CG. (C) Amount of splenic IgG1+ memory B cells 9 weeks after the secondary immunization. Numbers are in percent plus standard deviation of total B cells (n = 5). The differences were not statistically significant.
Discussion
The GC-specific expression of M17 suggested a function of this gene in GC-associated processes such as somatic hypermutation, class-switch recombination, or B-cell differentiation into memory B cells or plasma cells. The IL-4–dependent expression of M17 was consistent with such a function, since IL-4 contributes to B-cell activation and CSR to the IgG1 and IgE isotypes.38 Here, we show that M17-deficient mice were able to generate GC B cells during the course of a T-cell–dependent antibody response and formed GCs with an intact architecture. M17-deficient mice underwent a normal T-cell–dependent antibody response even after immunization with limiting amounts of antigen. Moreover, M17 was dispensable for SHM or CSR, and was not required for affinity maturation. HGAL is expressed at low levels in memory B cells, suggesting a participation of this molecule in the differentiation or function of memory B cells.12,19 However, we did not observe evidence for the involvement of M17 in memory B-cell formation. M17-deficient mice produced a robust NP-specific IgG1 antibody response upon secondary antigen challenge and formed wild-type levels of IgG1+ memory B cells. As M17 is not expressed by plasma cells in the bone marrow,32 we did not suspect a function in this cell type. Nonetheless, a very small fraction of lymphocytes (around 0.4%) in the bone marrow expresses M17.22,32 Our studies showed that these cells are part of the much larger IgM+ B220+ B-cell population. The nature and function of these cells is currently unknown. However, it was recently shown that mature lymphocytes in the bone marrow occupy a functionally distinct niche where they can mount T-independent immune responses.39 It is possible that M17 is expressed during the activation of such cells by blood-borne antigens.
We noticed a moderate reduction of the number of B cells residing in the Peyer patches. While the reasons for this effect are unknown, we may speculate that M17 modulates signaling pathways, which guide the development of Peyer patches or the homing of lymphocytes to this cellular compartment. HGAL is a cytosolic protein and M17 is most likely present in the same cellular compartment.21,22 The presence of a putative lipid-binding domain and an ITAM motif in M17 is consistent with a function as a cytosolic signaling molecule.22 It should be noted, however, that M17 contains an ITAM motif that deviates from the classical consensus sequence by a longer spacer between the YXXL motifs and would be, to our knowledge, the only cytosolic ITAM-containing protein.
HGAL is found in many lymphomas of GC origin. Most Burkitt lymphomas and follicular lymphomas express HGAL. In DLBCL, the expression pattern of HGAL is more heterogeneous. Gene expression profiling separates DLBCLs into distinct subtypes that resemble the profiles of activated B cells or GC B cells and are associated with different clinical outcomes.12,16 The GC-specific expression pattern of individual genes is not a universal marker for the identification of GC-like DLBCL. Some GC-specific genes such as CD10 do not possess a predictive value,18 while others, such as BCL6 and HGAL, have been identified as predictive markers for the survival of DLBCL patients.18,19 It is unclear at the present time whether the high expression of the IL-4 target genes BCL6 or HGAL in DLBCL incidentally identifies particular stages during GC B-cell differentiation whose associated tumors have a better prognosis, or is indeed a reflection of the gene-specific effect in DLBCL. Moreover, the up-regulation of IL-4 target genes may just indicate a strong immune response against the transformed cells that involves the secretion of IL-4, which can exhibit antitumor activity.40-42 Alternatively, different effects of IL-4 signaling have been suggested to contribute to the diverging clinical outcome of the DLBCL subsets.43 IL-4 signaling induces the expression of target genes such as HGAL and BCL6 in the GC B-cell–like subset, but fails to do so in the activated B-cell–like subset.43 Therefore, IL-4 target genes may be involved in the pathogenesis of GC-like DLBCL. For example, the transcriptional repressor BCL6, which is essential for GC formation and suppression of plasma cell differentiation, was shown to promote the development of GC-derived B-cell tumors in mice.44-47 Our studies on M17 failed to establish a role of this protein in the GC reaction. Thus, its high expression in the GC-like subset of DLBCL can serve as a marker for GC origin or for the activation of the IL-4 signaling pathway, but the relationship between the expression of M17 and a gene-specific effect in the pathogenesis of DLBCL remains elusive.
Prepublished online as Blood First Edition Paper, February 21, 2006; DOI 10.1182/blood-2005-10-4154.
Supported by the Deutsche Forschungsgemeinschaft through SFB 243 and the National Institutes of Health through grant AI 054636. D.S. was supported by the Boehringer Ingelheim Fonds.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Jeffrey L. Kutok, Victoria Smith, and Simone Wills for technical assistance and Philipp Oberdoerffer, Marc Schmidt-Supprian, and Stefano Casola for advice, discussion, and critical reading of the paper.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal