Abstract
Osteoclasts differentiate from hematopoietic precursors under systemic and local controls. Chemokines and receptors direct leukocyte traffic throughout the body and may help regulate site-specific bone resorption. We investigated bone gene expression in vivo during rapid osteoclast differentiation induced by colony-stimulating factor 1 (CSF-1) in Csf1-null toothless (tl/tl) rats. Long-bone RNA from CSF-1–treated tl/tl rats was analyzed by high-density microarray over a time course. TRAP (tartrate-resistant acid phosphatase)–positive osteoclasts appeared on day 2, peaked on day 4, and decreased slightly on day 6, as marrow space was expanding. TRAP and cathepsin K mRNA paralleled the cell counts. We examined all chemokine and receptor mRNAs on the arrays. CCL9 was strongly induced and peaked on day 2, as did its receptor, CCR1, and regulatory receptors c-Fms (CSF-1 receptor) and RANK (receptor activator of nuclear factor κB). Other chemokines and receptors showed little or no significant changes. In situ hybridization and immunohistochemistry revealed CCL9 in small, immature osteoclasts on day 2 and in mature cells at later times. Anti-CCL9 antibody inhibited osteoclast differentiation in culture and significantly suppressed the osteoclast response in CSF-1–treated tl/tl rats. While various chemokines have been implicated in osteoclastogenesis in vitro, this first systematic analysis of chemokines and receptors during osteoclast differentiation in vivo highlights the key role of CCL9 in this process.
Introduction
Osteoclasts are specialized, multinucleated bone-resorbing cells that differentiate by fusion of mononuclear hematopoietic precursor cells of the myeloid lineage. This complex process is closely regulated and requires numerous systemic and local factors, including growth factors, hormones, transcription factors, enzymes, and transporters for resorption to occur at the necessary levels and appropriate skeletal sites. The anabolic and catabolic activities of osteoblasts, osteocytes, and osteoclasts are normally exquisitely balanced to maintain normal bone mass.1 Excessive resorptive activity causes bone deficiency, for example in osteoporosis, arthritis, or other bone-loss syndromes, whereas defective resorptive activity can cause osteopetrosis, characterized by dense, brittle bones, lack of marrow cavities, and lack of teeth due to blockage of eruption.2
There are many known mutations in human patients and in animal models that inhibit osteoclastogenesis and/or osteoclast activity.3,4 Each of these osteopetrotic mutations has provided insights into mechanisms that regulate the differentiation and activity of osteoclasts. Two growth factors essential for early steps in the commitment of F4/80-positive, mononuclear leukocytes to differentiate along the osteoclast pathway are colony stimulating factor-1 (CSF-1, or M-CSF) and the tumor necrosis factor (TNF) family member, RANKL (also called TRANCE, OPGL, or TNFSF11), which are supplied in the skeleton by osteoblasts. Following CSF-1 and RANKL binding, precursor cells fuse and become multinucleated, attach to bone, differentiate, and become active, bone-resorbing cells. One naturally occurring osteopetrotic model is the toothless (tl/tl) rat,5,6 an autosomal recessive mutation in which a frameshift in the CSF-1 gene causes severe osteopetrosis due a virtually complete lack of osteoclasts.7,8 Injections of soluble, recombinant CSF-1 rapidly restore osteoclast populations and rescue many, but not all, aspects of the phenotype.9-11
While cell and organ culture models are essential experimental tools, no ex vivo system can replicate the complex interactions that occur in living bone. The CSF-1-treated tlCsf1/tlCsf rat thus provides a powerful model to study molecular and cellular events that accompany osteoclastogenesis in vivo. We used high-density microarrays to investigate gene expression changes that occur in the tibia during the burst of osteoclastogenesis caused by CSF-1 injections in the tl/tl rat. One gene that was very highly up-regulated was the chemokine CCL9 (also called MIP-1γ, CCF18, Scya9, or MRP-2).12 Recent cell culture studies by several groups13-17 suggest that chemokines and their receptors could play roles in extravasation, migration, and/or survival of osteoclastic cells in vivo. In addition, CCL3 (MIP-1α) has been implicated in local osteolysis due to myeloma18 and prosthesis wear particles.19
Chemokines are a large family of small, secreted (with very few exceptions) proteins that regulate the migration and interactions of white blood cells of all types.20-22 Their receptors are also numerous: some can bind more than one chemokine, and vice versa. This produces some redundancies and overlap of functions. Chemokines are categorized structurally based on the arrangement of N-terminal region Cys residues. CC denotes adjacent cysteines and CX(N)C denotes a residue (or the number of residues) between them. They are also characterized as “homeostatic” or “inflammatory,” depending on whether they participate in the normal regulation of leukocyte differentiation and targeting or are mainly involved in inflammatory responses. Some are known as “dualfunction” chemokines, participating in both processes.20 The chemokine receptors are named by the structural category of their ligands, followed by a number. For example, CCR1 is the first member of the family of receptors that bind ligands of the CC subfamily. When ligand binds, receptors activate cellular responses via coupled G-proteins, inducing chemotactic and other cellular responses.
Recent investigations have surveyed chemokine and receptor expression in vitro in a culture model in which mouse bone marrow cells were induced to differentiate under the influence of CSF-1 and RANKL.14 CCL9 mRNA expression increased markedly in this system, suggesting a role in osteoclast differentiation. During the course of our investigations, another study of CCL9 in differentiating mouse osteoclast cultures showed similar in vitro up-regulation of CCL9, that CCL9 promoted survival of mouse osteoclast-lineage cells, and that anti-CCL9 antibody could inhibit mouse osteoclast differentiation in cultures.13 How these model systems correspond to the complexities of in vivo osteoclastogenesis has not been established. Here we report our findings of strong up-regulation of CCL9 and its receptor, CCR1, during rapid osteoclast differentiation in CSF-1–treated tl/tl rats and that anti-CCL9 antibody inhibited osteoclast differentiation not only in cultures, but also in vivo, demonstrating a genuine physiologic role for CCL9 in osteoclastogenesis.
The evolutionary conservation of the chemokines and their receptors is looser than many other gene families. Given their biological importance, the complexity of their ligand/receptor cross reactivities, and the dependence of many lines of investigation on animal models, we also present results of comparative sequence analysis of chemokines and their receptors in rat versus mouse and discuss comparisons with the human system.
Materials and methods
Animals and tissue samples
Procedures involving animals were carried out under University of Massachusetts Medical School Institution of Animal Care and Use Committees–approved protocols. Three-week-old tlCSF1/tlCSF1 rats and +/+ littermates were obtained from the inbred colony maintained under specific pathogen–free (SPF) conditions at the University of Massachusetts Medical School. Mutants and wild-type littermates were identified by neonatal X-ray and by polymerase chain reaction (PCR) genotyping.8 Beginning at 21 days after birth, some tl/tl animals and wild-type (+/+) littermates received daily injections of 106 U of recombinant human CSF-1 (generously provided by Chiron, Emeryville, CA) as described,9,11 a dose that produces roughly normal circulating levels of CSF-1. Preliminary experiments established the timing of the osteoclast response. At appropriate time points, tibiae and femora were harvested. For RNA extraction, bones were carefully dissected to remove extraneous connective tissue and muscle, and joint cartilage and epiphyseal growth plates were also removed, leaving the metaphysis and diaphysis. Bones were split longitudinally and flushed with cold phosphate-buffered saline (PBS) to remove as much marrow as possible, although in untreated and 2-day-treated mutants, no discernible marrow spaces had formed. Dissected bones were flash-frozen in liquid nitrogen for later RNA extraction. For studies of in vivo inhibition of osteoclast differentiation by anti-CCL9, tl/tl rats were injected at 20 days after birth with 50 μg affinity-purified rabbit anti-murine CCL9 (MIP-1γ) antibody (Peprotech, Rocky Hill, NJ) in 100 μL PBS, and again on days 21 and 23, with 50 μg anti-CCL9 plus 106 U CSF-1 in 100 μL PBS. On alternate days, they received only CSF-1, as described.
Osteoclast counts
Contralateral tibiae were dissected from animals used in the CSF-1 time course, immersion-fixed in glutaraldehyde and paraformaldehyde, demineralized in EDTA, embedded in glycol methacrylate, and processed for TRAP (tartrate-resistant acid phosphatase) histochemistry to visualize osteoclasts as described previously.23,24 TRAP-positive cells were counted as follows. A histologic microscope with an attached digitizing tablet was used to project a fixed rectangle into the field of view. With a × 20 objective, the size of the rectangle seen in the field of view was 755 × 545 μm, and all the TRAP-positive cells in the rectangle were counted. The rectangle was positioned in the metaphysis just below the chondro-osseous junction of the proximal tibia or the distal femur for Area A, and it was moved toward the diaphysis by 1.5 times its height for counting Area B. Counts were done by 2 individuals on each section, at least 6 sections were counted per animal, and at least 3 individual animals were evaluated for each time point and condition. Alternate sections were briefly counterstained with toluidine blue (0.1%) to visualize the tissue context of the osteoclasts.
RNA extraction and microarray analysis
Each time point was measured in each of 4 animals using RNA from cleaned limb bones (1 tibia and 1 femur). Frozen bones were pulverized in a Bessler mortar and pestle cooled with dry ice, and RNA was extracted from the bone powder using Trizol (Invitrogen, Carlsbad, CA). After extraction, RNA was processed for microarray analysis according to Affymetrix's instructions (Santa Clara, CA). Briefly, RNA was cleaned using RNeasy columns (Qiagen, Valencia, CA), reverse transcribed (Superscript; Invitrogen), and transcribed in vitro to make cRNA with biotin incorporated (ENZO RNA in vitro transcript kit; Enzo Biochem, Farmingdale, NY). The cRNA was then subjected to limited alkaline hydrolysis for hybridization with the DNA microarrays. High-density microarray analysis was done using the rat RAE230 rat chipset (Affymetrix) according to manufacturer's instructions. This set represents some 30 200 different cDNA sequences on 2 chips. For each chip set, we tested RNA from a single animal. Chip image files were obtained through Affymetrix GeneChip software (MAS5). Subsequently, D-Chip (http://www.biostat.harvard.edu/complab/dchip/) analysis was performed to analyze differentially expressed genes, and further analyses were done using Excel (Microsoft, Redmond, WA). Expression levels are means for 4 individual animals, given in relative units, normalized to GAPDH mRNA.
Real-time PCR
RNA was isolated and reverse-transcribed, and real-time reverse transcriptase (RT)–PCR was carried out as described.12 Briefly, the LightCycler System (Roche Diagnostics, Indianapolis, IN) was used according to the manufacturer's instructions. Reactions were set up in micro capillary tubes using 0.5 μL cDNA with 9.5 μL of a LightCycler FastStart DNA Master SYBR Green I mix (Roche Diagnostics) to which gene-specific upstream and downstream PCR primers had been added. The final concentrations of the reaction components were 1.0 mM of each primer and 2.5 mM MgCl2. The primers for CCL9 were 5′-GATGAAGCCCTTTCATACTGC -3′ (forward) and 5′-GTGGTTGTGAGTTTTGCTCCAATC -3′ (reverse). Primers for RANK were 5′-CTGCTCCTCTTCATCTCTGTG -3′ (forward) and 5′-CTTCTGGAACCATCTTCTCCTC-3′ (reverse). The cycling conditions were 95°C preincubation for 10 minutes; 95°C for 10 seconds; 55°C for 10 seconds; and 72°C for 5 seconds; for 45 cycles. A dilution series of a test sample was set up for the standard curve. Parallel running of GAPDH was used for normalization, and results are given in relative units.
Osteoclast differentiation in vitro
Osteoclasts were differentiated in culture according to standard procedures.25 Bone marrow was harvested from tibiae and femora of 2-week-old wild-type rats by flushing with cold PBS, and the resulting cell suspension was centrifuged on Histopaque-1077 (Sigma, St Louis, MO) to isolate the mononuclear cell fraction. After washing in PBS, the cells were suspended in complete α–modified Eagle medium (MEM) with 10% fetal bovine serum, plated in 75-cm2 flasks at roughly 5 × 106 cells per flask with 5 ng/mL CSF-1 added to the medium, and incubated for 24 hours at 37°C in 5% CO2 in a humidified incubator to allow stromal cells to adhere, after which the nonadherent cells were collected, suspended in complete medium containing 75 ng/mL CSF-1 and plated at a density of at least 5 × 105 cells per well in 12-well plates. After 2 days, the medium was changed, this time containing 75 ng/mL CSF-1 and 30 ng/mL RANKL. Subsequently, medium was replaced every 2 to 3 days. In some wells, antichemokine antibodies (anti–mouse -CCL9, -CCL3, or -CCL5; all affinity-purified immunoglobulin G [IgG] fraction rabbit antibodies; Peprotech) were added to the medium at the time of RANKL addition at concentrations of 0.5 and 1.5 μg/mL. Following 1 week of differentiation, cells were fixed in 4% paraformaldehyde, processed for TRAP histochemical staining as described, and TRAP-positive cells with 3 or more nuclei were counted in at least 6 wells in each of at least 3 replicate experiments.
In situ hybridization
Long bones were harvested, fixed in 4% paraformaldehyde, demineralized with EDTA, paraffin-embedded, and subjected to in situ hybridization as previously described.26 Briefly, digoxigenin-UTP (Roche Diagnostics) was incorporated into a riboprobe for rat CCL9 using the T7 Ampliscribe kit27 (Epicentre, Madison, WI) with the rat CCL9–containing plasmid pCDNA3-rCCL9 as a template.12 Following blocking and hybridization at 42°C, probe was detected with alkaline phosphatase–conjugated antidigoxigenin, and signal was developed with NBT/BCIP substrate.
Immunohistochemistry
Bones were fixed, demineralized, and paraffin-embedded as described, and 7-μm-thick sections were cut. Peroxidase activity was blocked with 3% H2O2 and the sections were then blocked with 5% BSA and 5% normal goat serum for 2 hours. After blocking, they were incubated with rabbit polyclonal anti-CCL9 IgG fraction (Peprotech) overnight at 4°C. Following 4 washes in PBS with 5% normal goat serum, biotinylated secondary goat anti-rabbit antibody (Dako, Carpinteria, CA) was added and incubated for 1 hour at room temperature. After additional rinsing, ABC signal amplification reagent (Dako) was added, followed by avidin horseradish peroxidase (HRP) complex (Dako), and finally by DAB substrate. The reaction was stopped with H2O rinses and slides were treated with AquaPerm (Thermo Shandon, Pittsburgh, PA) reagent before coverslipping.
Microscopic imaging
Images were obtained using a Zeiss Axioskop 2 Plus with a brightfield condenser and equipped with a Zeiss Axiocam HRc color digital camera and Zeiss Axiovision software (4.0; Zeiss, Oberkochen, Germany)
Statistical analysis
Routine statistical comparisons were performed using Microsoft Excel. Following F tests to determine equality of sample variances, 2-tailed t tests were done to compare sample means obtained from replicates of microarray data, real-time PCR, and osteoclast counts. All data are reported as mean plus or minus 1 standard deviation.
Results
CSF-1–induced osteoclastogenesis
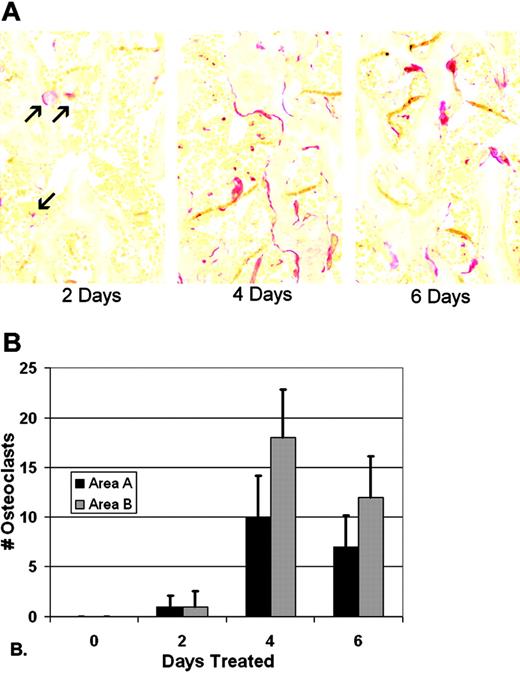
Preliminary experiments were done to determine the time course over which CSF-1 injections would induce osteoclastogenesis in 3-week-old tl/tl rats. No osteoclasts were seen in multiple sections of tibia from untreated tl/tl rats at any time point, and treatment for 2, 4, 6, and 24 hours produced no TRAP-positive cells (not shown). Later time points were analyzed, and the results are shown in Figure 1. TRAP-positive osteoclasts first appeared after 2 days of CSF-1 treatments, with about 1 osteoclast per slide in areas A (near the chondroosseous junction) and B (further down in the metaphysis). The counts reached a maximum on day 4, averaging approximately 10 and 18 cells per slide in the 2 areas. By day 6, counts declined slightly to about 7 and 12 cells per slide in areas A and B, respectively. This decline coincided with the reduction in bone area that resulted from the rapid, ongoing bone resorption, leaving less bone surface for osteoclast attachment. Using these results as a guide, we established a time course of 2, 4, and 6 days to measure gene expression changes by microarray from the first appearance of differentiated osteoclasts through and just past its peak.
Osteoclast gene expression
We ruled out any naturally occurring changes in osteoclast-associated genes by following untreated tl/tl and +/+ rats over the same time course. Microarray analysis showed no significant changes in the mRNA levels for the osteoclast markers TRAP or cathepsin K in either tl/tl mutants or +/+ littermates, nor did CSF-1 injections induce significant changes in osteoclast-associated genes in +/+ littermates. The average value for wild-type rats, treated or untreated, was 7490 RU for TRAP and 11 485 for cathepsin K. Table 1 summarizes array results for osteoclast-associated genes in the CSF-1–treated tl/tl mutants over the time course. TRAP mRNA levels increased significantly in a manner that paralleled the osteoclast counts, rising 5-fold on day 2, doubling again on day 4, and slightly decreasing on day 6 (P values compared with day 0: day 2, P < .01; day 4, P < .001; and day 6, P < .001). The pattern for cathepsin K mRNA in treated tl/tl mutants was nearly identical to TRAP, as was its statistical significance. The CSF-1 receptor (c-Fms) had a different pattern, increasing more than 5-fold over the day 0 to a peak on day 2, then slightly decreasing on days 4 and 6 (P < .001 for days 2, 4, and 6 compared with day 0). RANK, the receptor for RANKL, which is expressed in committed preosteoclasts and mature osteoclasts and is induced by CSF-1 in vitro, was not present on the microarrays. Because of its importance in osteoclast differentiation, we analyzed RANK mRNA by real-time PCR. RANK expression was strongly induced by CSF-1 and paralleled CSF-1 receptor mRNA, increasing sharply on day 2, then subsiding gradually on days 4 and 6. RANKL and its “decoy” receptor OPG, which, like CSF-1, are supplied by osteoblasts in bone, showed modest changes. The decrease in RANKL seen in Table 1 was significant only on day 6 (P < .05), whereas the slight increase in OPG was not statistically significant (P > .05).
TRAP-positive osteoclasts in tl rats in response to CSF-1 injections. (A) TRAP staining of proximal tibial metaphysis reveals few osteoclasts by day 2 (red, arrows), and well-established populations at days 4 and 6 (images were obtained with a Zeiss Axioskop 2 Plus microscope with a 20 ×/0.5 NA brightfield objective). None were seen at day 0 (not shown). (B) Cell counts were performed in a window of defined size in 2 areas, area A near the chondro-osseous junction, and area B, in the lower metaphysis, in replicate slides of multiple animals (n = 24-36; bars show mean + 1 SD).
TRAP-positive osteoclasts in tl rats in response to CSF-1 injections. (A) TRAP staining of proximal tibial metaphysis reveals few osteoclasts by day 2 (red, arrows), and well-established populations at days 4 and 6 (images were obtained with a Zeiss Axioskop 2 Plus microscope with a 20 ×/0.5 NA brightfield objective). None were seen at day 0 (not shown). (B) Cell counts were performed in a window of defined size in 2 areas, area A near the chondro-osseous junction, and area B, in the lower metaphysis, in replicate slides of multiple animals (n = 24-36; bars show mean + 1 SD).
Relative gene expression for osteoclast-associated genes, chemokines, and chemokine receptors
. | . | . | . | Expression, microarray relative units ± 1 SD . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene name . | Alternative names . | GenBank accession no. . | RAE230 array probe set . | Day 0 . | Day 2 . | Day 4 . | Day 6 . | |||
| Control/osteoclast genes | ||||||||||
| GAPDH | — | NM_017008 | 1367557_s_at | 9750 ± 2181 | 10 737 ± 1 446 | 11 282 ± 2 216 | 11 111 ± 1 496 | |||
| Acid phosphatase 5 | TRAP | NM_019144 | 1367942_at | 1212 ± 378 | 6 677 ± 2 294 | 12 001 ± 739 | 10 454 ± 265 | |||
| Cathepsin K | — | NM_031560 | 1369947_at | 1789 ± 487 | 8 906 ± 2 818 | 14 564 ± 994 | 12 657 ± 1 475 | |||
| RANKL | TRANCE, TNF superfamily member 11 | NM_057149 | 1369282_at | 679 ± 209 | 981 ± 263 | 484 ± 117 | 385 ± 59 | |||
| OPG | Osteoprotegerin, TNF receptor superfamily, member 11b | NM_012870 | 1369407_at | 325 ± 54 | 331 ± 15 | 452 ± 75 | 422 ± 63 | |||
| CSF-1 receptor | c-Fms | BI285793 | 1388784_at | 770 ± 99 | 4 310 ± 1 337 | 3 127 ± 644 | 2 462 ± 327 | |||
| Chemokines/Receptors | ||||||||||
| CC chemokines | ||||||||||
| CCL1 (NRS) | — | — | — | — | — | — | — | |||
| CCL2 | MCP-1 | NM_031530 | 1367973_at | 189 ± 80 | 456 ± 184 | 534 ± 361 | 257 ± 77 | |||
| CCL3 | MIP-1α | U22414 | 1369815_at | 420 ± 94 | 566 ± 46 | 425 ± 70 | 429 ± 63 | |||
| CCL4 | MIP-1β | NM_053858 | 1370832_at | 385 ± 103 | 386 ± 34 | 323 ± 42 | 321 ± 36 | |||
| CCL5 | RANTES | NM_031116 | 1369983_at | 565 ± 83 | 450 ± 74 | 532 ± 112 | 545 ± 120 | |||
| CCL6 | MRP-1α | BE095824 | 1389123_at | 1131 ± 378 | 2 244 ± 720 | 919 ± 247 | 1 358 ± 367 | |||
| CCL7 | MCP-3 | BF419899 | 1379935_at | 117 ± 20 | 113 ± 10 | 128 ± 24 | 93 ± 26 | |||
| CCL8 (NRS) | MCP-2 | — | — | — | — | — | — | |||
| CCL9 | MIP-1γ | AI169984 | 1392172_at | 100 ± 94 | 7 832 ± 2 260 | 1 898 ± 783 | 858 ± 385 | |||
| CCL11 | Eotaxin | NM_019205 | 1387319_at | 257 ± 20 | 243 ± 26 | 218 ± 29 | 213 ± 18 | |||
| CCL12 (NP) | MCP-5 | XM_213425 | — | — | — | — | — | |||
| CCL13 (NRS) | MCP-4 | — | — | — | — | — | — | |||
| CCL15 (NRS) | HCC-2 | — | — | — | — | — | — | |||
| CCL17 | TARC | NM_057151 | 1370118_at | 187 ± 11 | 175 ± 8 | 166 ± 12 | 185 ± 26 | |||
| CCL19 | ELC, MIP-3β | AA996885 | 1391925_at | 622 ± 195 | 478 ± 144 | 517 ± 108 | 517 ± 196 | |||
| CCL20 | MIP-3α | AF053312 | 1369814_at | 229 ± 23 | 196 ± 22 | 223 ± 13 | 274 ± 50 | |||
| CCL21 | 6CKINE | BI282920 | 1378015_at | 320 ± 26 | 417 ± 56 | 374 ± 104 | 431 ± 49 | |||
| CCL22 | MDC | AF432871 | 1370619_at | 207 ± 12 | 208 ± 23 | 178 ± 34 | 169 ± 44 | |||
| CCL25 (NP) | TECK | — | — | — | — | — | — | |||
| CCL26 | Eotaxin-3 | BI294084 | 1385309_at | 662 ± 181 | 742 ± 67 | 804 ± 145 | 636 ± 64 | |||
| CCL27 | CTACK | XM_342823 | 1376850_a_at | 463 ± 27 | 390 ± 40 | 356 ± 66 | 386 ± 84 | |||
| CXC chemokines | ||||||||||
| CXCL1 | GROα | NM_030845 | 1387316_at | 96 ± 12 | 115 ± 6 | 123 ± 21 | 108 ± 34 | |||
| CXCL2 | GROβ, MIP-2α | NM_053647 | 1368760_at | 498 ± 124 | 406 ± 72 | 408 ± 47 | 362 ± 172 | |||
| CXCL4 | Platelet factor 4, PF-4 | AI169104 | 1371 250_at | 5004 ± 632 | 5 172 ± 297 | 7 687 ± 566 | 10 210 ± 538 | |||
| CXCL5 | CXC chemokine LIX, ENA-78 | NM_022214 | 1387648_at | 233 ± 100 | 148 ± 148 | 156 ± 41 | 119 ± 72 | |||
| CXCL8 (NRS) | IL-8 | — | — | — | — | — | — | |||
| CXCL9 | MIG | AI044222 | 1382454_at | 155 ± 19 | 137 ± 12 | 142 ± 30 | 137 ± 15 | |||
| CXCL10 | CRG-2, IP-10 | U22520 | 1387969_at | 173 ± 20 | 152 ± 29 | 171 ± 18 | 190 ± 29 | |||
| CXCL11 | I-TAC | BF281987 | 1379365_at | 90 ± 21 | 90 ± 15 | 86 ± 12 | 94 ± 19 | |||
| CXCL12 | SDF-1 | AI171777 | 1369633_at | 4661 ± 1007 | 5 563 ± 1 201 | 5 371 ± 917 | 5 893 ± 1 062 | |||
| CXCL13 | BCA-1 | AA892854 | 1398390_at | 353 ± 54 | 351 ± 54 | 359 ± 66 | 421 ± 129 | |||
| CXCL14 | BRAK, BOLOKINE | AI412979 | 1396129_at | 505 ± 69 | 838 ± 440 | 493 ± 106 | 675 ± 101 | |||
| XCL1 | Lymphotactin | NM_134361 | 1387831_at | 53 ± 27 | 36 ± 44 | 63 ± 62 | 64 ± 30 | |||
| CX3CL1 | Fraktalkine | NM_134455 | 1368200_at | 228 ± 112 | 257 ± 29 | 215 ± 69 | 239 ± 53 | |||
| Receptors | ||||||||||
| CCR1 | MIP1α-R, RANTES-R | NM_020542 | 1370083_at | 237 ± 74 | 1 122 ± 310 | 829 ± 306 | 523 ± 117 | |||
| CCR2 | MCP-1 R | NM_021866 | 1387742_at | 135 ± 45 | 155 ± 27 | 125 ± 9 | 115 ± 38 | |||
| CCR3 | Eotaxin R | NM_053958 | 1369808_at | 297 ± 97 | 244 ± 67 | 224 ± 21 | 223 ± 63 | |||
| CCR4 | — | NM_133532 | 1369555_at | 189 ± 49 | 187 ± 39 | 156 ± 19 | 185 ± 68 | |||
| CCR5 | — | NM_053960 | 1369290_at | 115 ± 18 | 126 ± 23 | 107 ± 17 | 90 ± 14 | |||
| CCR6 | GRO/MGSA-R, STRL22 | AI045155 | 1393929_at | 1148 ± 126 | 1 075 ± 142 | 1 180 ± 122 | 1 031 ± 168 | |||
| CCR7 (NP) | — | — | — | — | — | — | — | |||
| CCR8 (NP) | — | — | — | — | — | — | — | |||
| CCR9 (NP) | — | — | — | — | — | — | — | |||
| CCR10 (NP) | — | — | — | — | — | — | — | |||
| CXCR1 | IL-8 RA | NM_019310 | 1369875_at | 288 ± 33 | 259 ± 52 | 272 ± 30 | 270 ± 29 | |||
| CXCR2 | IL-8 RB | NM_017183 | 1369697_at | 241 ± 37 | 223 ± 20 | 260 ± 31 | 242 ± 50 | |||
| CXCR3 | MIG-R, IP-10 | NM_053415 | 1368192_at | 312 ± 46 | 312 ± 56 | 356 ± 69 | 318 ± 56 | |||
| CXCR4 | LCR1, SDF1-R, Fusin | U54791 | 1389244_x_at | 4083 ± 521 | 4 902 ± 110 | 3 203 ± 614 | 4 156 ± 619 | |||
| CXCR5 | Burkitt lymphoma receptor 1 | NM_053303 | 1369911_at | 669 ± 104 | 513 ± 65 | 524 ± 106 | 507 ± 63 | |||
| CXCR6 (NRS) | — | — | — | — | — | — | — | |||
| CX3CR1 | V28 | NM_133534 | 1369527_at | 520 ± 96 | 411 ± 43 | 468 ± 96 | 508 ± 94 | |||
| XCR1 (NP) | GPR-5 | — | — | — | — | — | — | |||
. | . | . | . | Expression, microarray relative units ± 1 SD . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene name . | Alternative names . | GenBank accession no. . | RAE230 array probe set . | Day 0 . | Day 2 . | Day 4 . | Day 6 . | |||
| Control/osteoclast genes | ||||||||||
| GAPDH | — | NM_017008 | 1367557_s_at | 9750 ± 2181 | 10 737 ± 1 446 | 11 282 ± 2 216 | 11 111 ± 1 496 | |||
| Acid phosphatase 5 | TRAP | NM_019144 | 1367942_at | 1212 ± 378 | 6 677 ± 2 294 | 12 001 ± 739 | 10 454 ± 265 | |||
| Cathepsin K | — | NM_031560 | 1369947_at | 1789 ± 487 | 8 906 ± 2 818 | 14 564 ± 994 | 12 657 ± 1 475 | |||
| RANKL | TRANCE, TNF superfamily member 11 | NM_057149 | 1369282_at | 679 ± 209 | 981 ± 263 | 484 ± 117 | 385 ± 59 | |||
| OPG | Osteoprotegerin, TNF receptor superfamily, member 11b | NM_012870 | 1369407_at | 325 ± 54 | 331 ± 15 | 452 ± 75 | 422 ± 63 | |||
| CSF-1 receptor | c-Fms | BI285793 | 1388784_at | 770 ± 99 | 4 310 ± 1 337 | 3 127 ± 644 | 2 462 ± 327 | |||
| Chemokines/Receptors | ||||||||||
| CC chemokines | ||||||||||
| CCL1 (NRS) | — | — | — | — | — | — | — | |||
| CCL2 | MCP-1 | NM_031530 | 1367973_at | 189 ± 80 | 456 ± 184 | 534 ± 361 | 257 ± 77 | |||
| CCL3 | MIP-1α | U22414 | 1369815_at | 420 ± 94 | 566 ± 46 | 425 ± 70 | 429 ± 63 | |||
| CCL4 | MIP-1β | NM_053858 | 1370832_at | 385 ± 103 | 386 ± 34 | 323 ± 42 | 321 ± 36 | |||
| CCL5 | RANTES | NM_031116 | 1369983_at | 565 ± 83 | 450 ± 74 | 532 ± 112 | 545 ± 120 | |||
| CCL6 | MRP-1α | BE095824 | 1389123_at | 1131 ± 378 | 2 244 ± 720 | 919 ± 247 | 1 358 ± 367 | |||
| CCL7 | MCP-3 | BF419899 | 1379935_at | 117 ± 20 | 113 ± 10 | 128 ± 24 | 93 ± 26 | |||
| CCL8 (NRS) | MCP-2 | — | — | — | — | — | — | |||
| CCL9 | MIP-1γ | AI169984 | 1392172_at | 100 ± 94 | 7 832 ± 2 260 | 1 898 ± 783 | 858 ± 385 | |||
| CCL11 | Eotaxin | NM_019205 | 1387319_at | 257 ± 20 | 243 ± 26 | 218 ± 29 | 213 ± 18 | |||
| CCL12 (NP) | MCP-5 | XM_213425 | — | — | — | — | — | |||
| CCL13 (NRS) | MCP-4 | — | — | — | — | — | — | |||
| CCL15 (NRS) | HCC-2 | — | — | — | — | — | — | |||
| CCL17 | TARC | NM_057151 | 1370118_at | 187 ± 11 | 175 ± 8 | 166 ± 12 | 185 ± 26 | |||
| CCL19 | ELC, MIP-3β | AA996885 | 1391925_at | 622 ± 195 | 478 ± 144 | 517 ± 108 | 517 ± 196 | |||
| CCL20 | MIP-3α | AF053312 | 1369814_at | 229 ± 23 | 196 ± 22 | 223 ± 13 | 274 ± 50 | |||
| CCL21 | 6CKINE | BI282920 | 1378015_at | 320 ± 26 | 417 ± 56 | 374 ± 104 | 431 ± 49 | |||
| CCL22 | MDC | AF432871 | 1370619_at | 207 ± 12 | 208 ± 23 | 178 ± 34 | 169 ± 44 | |||
| CCL25 (NP) | TECK | — | — | — | — | — | — | |||
| CCL26 | Eotaxin-3 | BI294084 | 1385309_at | 662 ± 181 | 742 ± 67 | 804 ± 145 | 636 ± 64 | |||
| CCL27 | CTACK | XM_342823 | 1376850_a_at | 463 ± 27 | 390 ± 40 | 356 ± 66 | 386 ± 84 | |||
| CXC chemokines | ||||||||||
| CXCL1 | GROα | NM_030845 | 1387316_at | 96 ± 12 | 115 ± 6 | 123 ± 21 | 108 ± 34 | |||
| CXCL2 | GROβ, MIP-2α | NM_053647 | 1368760_at | 498 ± 124 | 406 ± 72 | 408 ± 47 | 362 ± 172 | |||
| CXCL4 | Platelet factor 4, PF-4 | AI169104 | 1371 250_at | 5004 ± 632 | 5 172 ± 297 | 7 687 ± 566 | 10 210 ± 538 | |||
| CXCL5 | CXC chemokine LIX, ENA-78 | NM_022214 | 1387648_at | 233 ± 100 | 148 ± 148 | 156 ± 41 | 119 ± 72 | |||
| CXCL8 (NRS) | IL-8 | — | — | — | — | — | — | |||
| CXCL9 | MIG | AI044222 | 1382454_at | 155 ± 19 | 137 ± 12 | 142 ± 30 | 137 ± 15 | |||
| CXCL10 | CRG-2, IP-10 | U22520 | 1387969_at | 173 ± 20 | 152 ± 29 | 171 ± 18 | 190 ± 29 | |||
| CXCL11 | I-TAC | BF281987 | 1379365_at | 90 ± 21 | 90 ± 15 | 86 ± 12 | 94 ± 19 | |||
| CXCL12 | SDF-1 | AI171777 | 1369633_at | 4661 ± 1007 | 5 563 ± 1 201 | 5 371 ± 917 | 5 893 ± 1 062 | |||
| CXCL13 | BCA-1 | AA892854 | 1398390_at | 353 ± 54 | 351 ± 54 | 359 ± 66 | 421 ± 129 | |||
| CXCL14 | BRAK, BOLOKINE | AI412979 | 1396129_at | 505 ± 69 | 838 ± 440 | 493 ± 106 | 675 ± 101 | |||
| XCL1 | Lymphotactin | NM_134361 | 1387831_at | 53 ± 27 | 36 ± 44 | 63 ± 62 | 64 ± 30 | |||
| CX3CL1 | Fraktalkine | NM_134455 | 1368200_at | 228 ± 112 | 257 ± 29 | 215 ± 69 | 239 ± 53 | |||
| Receptors | ||||||||||
| CCR1 | MIP1α-R, RANTES-R | NM_020542 | 1370083_at | 237 ± 74 | 1 122 ± 310 | 829 ± 306 | 523 ± 117 | |||
| CCR2 | MCP-1 R | NM_021866 | 1387742_at | 135 ± 45 | 155 ± 27 | 125 ± 9 | 115 ± 38 | |||
| CCR3 | Eotaxin R | NM_053958 | 1369808_at | 297 ± 97 | 244 ± 67 | 224 ± 21 | 223 ± 63 | |||
| CCR4 | — | NM_133532 | 1369555_at | 189 ± 49 | 187 ± 39 | 156 ± 19 | 185 ± 68 | |||
| CCR5 | — | NM_053960 | 1369290_at | 115 ± 18 | 126 ± 23 | 107 ± 17 | 90 ± 14 | |||
| CCR6 | GRO/MGSA-R, STRL22 | AI045155 | 1393929_at | 1148 ± 126 | 1 075 ± 142 | 1 180 ± 122 | 1 031 ± 168 | |||
| CCR7 (NP) | — | — | — | — | — | — | — | |||
| CCR8 (NP) | — | — | — | — | — | — | — | |||
| CCR9 (NP) | — | — | — | — | — | — | — | |||
| CCR10 (NP) | — | — | — | — | — | — | — | |||
| CXCR1 | IL-8 RA | NM_019310 | 1369875_at | 288 ± 33 | 259 ± 52 | 272 ± 30 | 270 ± 29 | |||
| CXCR2 | IL-8 RB | NM_017183 | 1369697_at | 241 ± 37 | 223 ± 20 | 260 ± 31 | 242 ± 50 | |||
| CXCR3 | MIG-R, IP-10 | NM_053415 | 1368192_at | 312 ± 46 | 312 ± 56 | 356 ± 69 | 318 ± 56 | |||
| CXCR4 | LCR1, SDF1-R, Fusin | U54791 | 1389244_x_at | 4083 ± 521 | 4 902 ± 110 | 3 203 ± 614 | 4 156 ± 619 | |||
| CXCR5 | Burkitt lymphoma receptor 1 | NM_053303 | 1369911_at | 669 ± 104 | 513 ± 65 | 524 ± 106 | 507 ± 63 | |||
| CXCR6 (NRS) | — | — | — | — | — | — | — | |||
| CX3CR1 | V28 | NM_133534 | 1369527_at | 520 ± 96 | 411 ± 43 | 468 ± 96 | 508 ± 94 | |||
| XCR1 (NP) | GPR-5 | — | — | — | — | — | — | |||
NRS indicates no sequence for rat in GenBank with sufficient homology to assign identity; NP, no sequence present on the RAE 230 chipset used. mRNA levels were determined by DNA microarray analysis of tibial RNA isolated from toothless (tl/tl) rats on day 0 (3 weeks old), or following 2, 4, and 6 days of CSF-1 injections to induce osteoclast differentiation. Values shown are the mean ± 1 SD of 4 individual animals. GAPDH served as an internal standard. Osteoclast-specific genes include TRAP, cathepsin K, and the CSF-1 receptor (c-Fms). We also show results for the osteoblast-derived osteoclast-inducing factor RANKL and its decoy receptor OPG, which underwent little or no change in this experimental system. TRAP and cathepsin K mRNA levels follow the same time course as osteoclast counts in bone sections. CCL9 and its receptor CCR1 showed by far the greatest responses of all chemokines and receptors and peaked a full 2 days before osteoclast counts, as did CSF-1 receptor levels. Chemokine and receptor identities were assigned based on highest homology to mouse, determined as described in “Results.”
In summary, osteoclast-specific gene expression in total bone RNA from treated mutants rose and fell with the same temporal pattern as the osteoclast counts in bone tissue, and TRAP and cathepsin K expression levels were of the same order as for wild-type bone. Receptors required for osteoclastogenesis (c-Fms and RANK) peaked early, on day 2, whereas markers of mature, active osteoclasts (TRAP and cathepsin K) peaked later, simultaneously with osteoclast populations, roughly 4 days after commencing treatments with CSF-1. There was evidence that the rapid wave of osteoclast induction and bone resorption did not feed back quickly to affect the levels of either OPG or RANKL mRNA expressed in bone tissue.
CCL9 and CCR1 expression
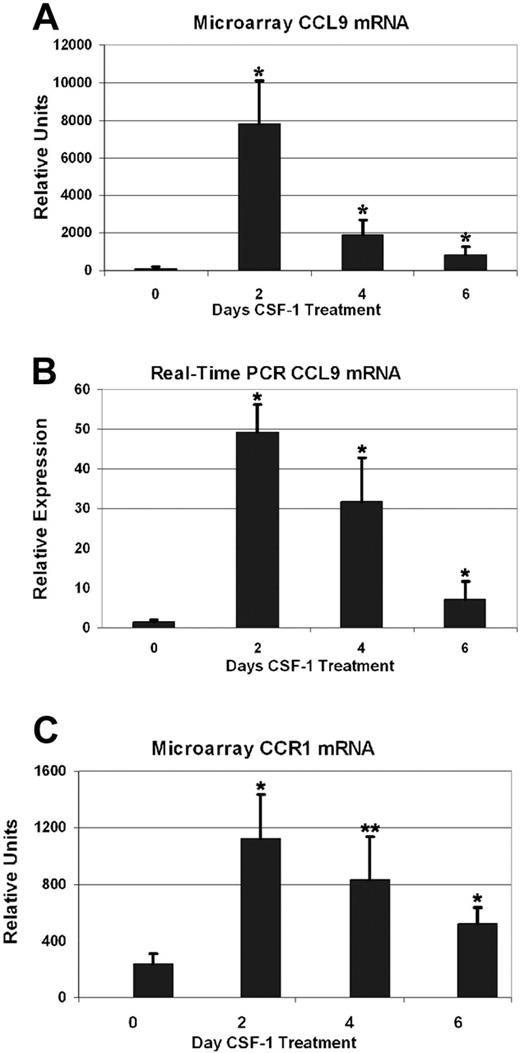
Among all genes induced in long-bone RNA from CSF-1-treated tl/tl rats, 1 sequence, highly similar to the mouse CC chemokine CCL9, stood out as being particularly strongly responsive. We therefore cloned the complete cDNA and characterized it in greater detail (GenBank accession no. AY863011), confirming it as the rat ortholog of mouse CCL9, and demonstrating its potent chemotactic effects in bone marrow mononuclear cell cultures.12 Figure 2 shows that the expression profile for CCL9 mRNA as determined by microarray (2A) was in good agreement with real-time PCR results (2B). All 3 postinjection time points had highly elevated CCL9 mRNA compared with uninjected tl/tl mutants. Microarray results were 78-fold higher on day 2 than day 0 (2260 vs 94 RU), 19-fold higher on day 4, and 9-fold higher on day 6. All these differences from the baseline value were statistically significant (P < .01 for all postinjection time points). The real-time PCR results confirm a large increase by day 2, up 32-fold (49 vs 1.5 RU), 21-fold on day 4, and 5-fold on day 6. We also tested samples 1 day after injection, and the results showed that CCL9 had already risen to 2280 RU, but also confirmed that the 2-day time point is the genuine peak of CCL9 mRNA. In comparison, CCL9 mRNA did not change significantly in untreated or CSF-1–treated +/+ littermates over the time course, averaging 2003 RU. The receptor for CCL9 is CCR1, or MIP-1α receptor. It changed in a similar pattern to its ligand (Table 1 and Figure 2C). The level rose to roughly 5 times the baseline value on day 2, then declined gradually over the next 4 days. The differences between day 0 and subsequent days were significant (P < .005 on days 2 and 6, and P < .02 on day 4). CCR1 mRNA did not change significantly in untreated or treated +/+ littermates over the time course.
Sequence identification and expression of other chemokines and receptors
Because the interactions among chemokines and their receptors are complex, we closely examined expression of all the chemokines and receptors represented on the rat microarrays. The rat chemokine and receptor repertoire is not as thoroughly established as that for human or mouse. We therefore did BLAST searches of potential chemokine and receptor sequences represented on the gene arrays against the rat genome to obtain full-length cDNA sequences, where available. These in turn we could use to BLAST mouse and human sequences to identify homologous genes using the GenBank UniGene resource. We also did the reverse, querying the chip sequences with known chemokine and receptor sequences. This allowed us to assign identities to the sequences present on the rat arrays. The identities given in Table 1 are either established in the rat or have the highest homology to known mouse chemokines/receptors (> 70% identical and > 80% similar amino acids). If no sequence on the chips met this level of homology, we report it as “not present” on the chip. Likewise, if there was no rat sequence in GenBank with sufficient homology to known chemokines or receptors, we report it as such. The expression levels for all chemokines and receptors on the microarrays over the time course are shown in Table 1.
In addition to the dramatic changes in CCL9 and CCR1 described, several other genes were of interest. For example, CXCL4 (platelet factor 4 [PF-4]) was reported to inhibit parathyroid hormone (PTH)–stimulated bone resorption,28 to promote monocyte survival and differentiation into macrophages,29 and to inhibit angiogenesis.30 Its expression level doubled over the time course from approximately 5000 to just over 10 000 RU. The slight increase on day 2 was not significant (P > .1), but it was significant on day 4 (P < .05), and highly significant on day 6 (P < .001). Whether this increase is related to its potential regulatory effect on monocytes or to other activities (eg, regulating vascularization of the nascent marrow space) remains to be investigated. A recent report31 suggested that CXCR3 exists in 2 isoforms, A and B, and that CXCR3-B may be the receptor that mediates the actions of CXCL4.31 We did not detect significant changes in this receptor over the time course.
Yu et al15 demonstrated a potential role for CXCL12 (SDF-1) and its receptor, CXCR4, in the migration of preosteoclasts to sites of bone resorption, based on their expression in endothelial and stromal cells and chemotactic effect on the murine preosteoclast-like cell line, RAW 264.7. In our microarray time course, the mRNA level for neither CXCL12 nor CXCR4 varied significantly (P > .05), except for a slight increase in the receptor seen on day 2 (P < .05). Thus, the levels in vivo were fairly constant, despite the extraordinary increases taking place in osteoclasts and resorptive activity.
The modest increase in CCL6 on day 2 was significant (P < .05), but not for the later time points (P > .1). Similarly, for CCL2 (monocyte chemoattractant protein-1 [MCP-1]), there was a small increase on day 2 that was significant (P < .05), but not on subsequent days. CCL3 (MIP-1α) did not change significantly, and its expression was at low levels throughout the time course. All other chemokines and receptors showed little or no change in RNA levels.
In situ hybridization
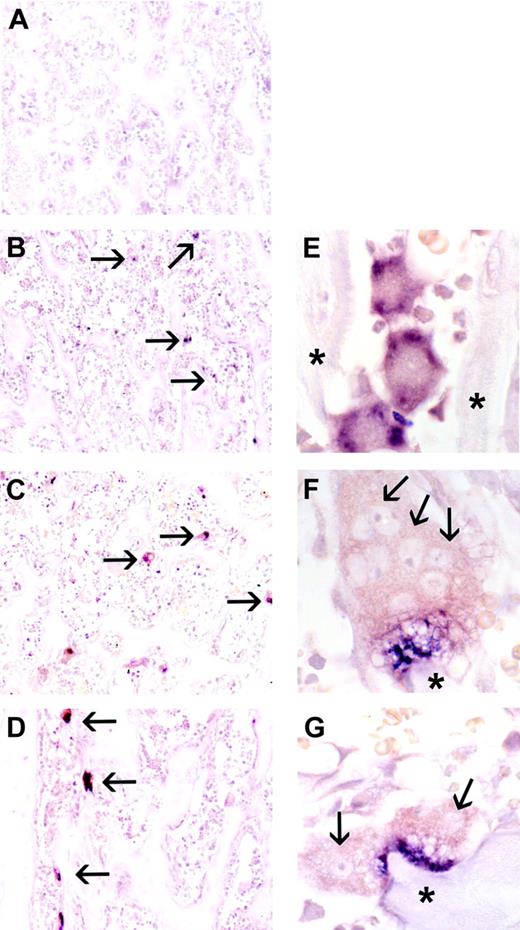
Because of the complexity of bone tissue, with osteoblasts, osteoclasts, endothelium, and stromal and hematopoietic cells all in close proximity, we sought to determine whether the cellular source of CCL9 mRNA in vivo could be unambiguously attributed to osteoclasts and/or their precursors. In situ hybridization using a CCL9 riboprobe was therefore performed on the contralateral tibiae of the animals used for RNA isolation, and the results are shown in Figure 3 (negative controls using sense probe gave no reactivity; not shown). CCL9 mRNA was abundant in immature osteoclasts (Figure 3B-C) at day 2 and in mature osteoclasts at days 4 and 6 (Figure 3D-G). In immature cells, prior to fusion and attainment of the mature differentiated state, there was strong signal for CCL9 mRNA dispersed throughout the cytoplasm. In mature osteoclasts, however, the message often had a very unusual distribution, appearing highly concentrated in the area of the ruffled border, adjacent to the bone surface, and very little was detected in the remainder of the cell. This was a consistent finding and is very different from the distribution of TRAP mRNA in similar experiments (not shown) and from the CCL9 protein (Figure 4), although its significance remains to be explored.
CCL-9 and CCR1 mRNA expression in long bones determined by microarray and real-time PCR.tl rats were treated for the days indicated and total bone RNA was analyzed. CCL9 and CCR1 mRNA were highest after 2 days of treatment, after which the levels subsided. Means + 1 SD are shown. Compared with untreated animals (day 0), all time points were significantly elevated. (A-B) *Difference from day 0, P < .01. (C) *Difference from day 0, P < .005; **P < .02.
CCL-9 and CCR1 mRNA expression in long bones determined by microarray and real-time PCR.tl rats were treated for the days indicated and total bone RNA was analyzed. CCL9 and CCR1 mRNA were highest after 2 days of treatment, after which the levels subsided. Means + 1 SD are shown. Compared with untreated animals (day 0), all time points were significantly elevated. (A-B) *Difference from day 0, P < .01. (C) *Difference from day 0, P < .005; **P < .02.
In situ hybridization for CCL-9 mRNA during CSF-1 time course, proximal tibia of tl rat. No CCL-9-expressing cells were visible at day 0 (A). CCL-9-positive cells are indicated in panels B-D with arrows. Many small cells were detected at day 2 (B,E), and larger osteoclasts were seen at days 4 (C,F) and 6 (D,G). Higher magnification revealed cytological features of the CCL-9-expressing cells. Arrows in panels F and G indicate some individual nuclei. The cells seen at day 2 were typically smaller, mononuclear, and less differentiated, while at days 4 and 6, osteoclasts generally contained multiple nuclei and were better differentiated. The CCL-9 mRNA is diffusely distributed throughout the cytoplasm in the differentiating mononuclear pre-osteoclasts in panel E, but appears highly concentrated in the ruffled border area in days 4 (F) and 6 (G). *Bone trabeculae. Original magnifications: × 100 (A-D); × 630 (E-G). No counterstain was used.
In situ hybridization for CCL-9 mRNA during CSF-1 time course, proximal tibia of tl rat. No CCL-9-expressing cells were visible at day 0 (A). CCL-9-positive cells are indicated in panels B-D with arrows. Many small cells were detected at day 2 (B,E), and larger osteoclasts were seen at days 4 (C,F) and 6 (D,G). Higher magnification revealed cytological features of the CCL-9-expressing cells. Arrows in panels F and G indicate some individual nuclei. The cells seen at day 2 were typically smaller, mononuclear, and less differentiated, while at days 4 and 6, osteoclasts generally contained multiple nuclei and were better differentiated. The CCL-9 mRNA is diffusely distributed throughout the cytoplasm in the differentiating mononuclear pre-osteoclasts in panel E, but appears highly concentrated in the ruffled border area in days 4 (F) and 6 (G). *Bone trabeculae. Original magnifications: × 100 (A-D); × 630 (E-G). No counterstain was used.
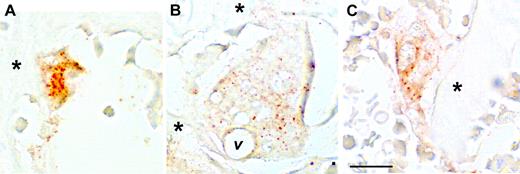
Immunohistochemistry for CCL-9 in the proximal tibia of tl rats. Immunohistochemistry after 2 (A), 4 (B), or 6 (C) days of CSF-1 injections is shown. Orange-brown color indicates CCL-9 protein. At day 2, the staining was most evident in small, newly formed osteoclasts, while at days 4 and 6, large, mature osteoclasts were present with much weaker, more diffuse labeling. *Bone trabeculae; v indicates a prominent vacuole in the large osteoclast in panel B. Scale bar in panel C equals 21 μm. Shown are 10-μm paraffin sections without counterstain. Objective magnification: 63 ×/1.25 NA.
Immunohistochemistry for CCL-9 in the proximal tibia of tl rats. Immunohistochemistry after 2 (A), 4 (B), or 6 (C) days of CSF-1 injections is shown. Orange-brown color indicates CCL-9 protein. At day 2, the staining was most evident in small, newly formed osteoclasts, while at days 4 and 6, large, mature osteoclasts were present with much weaker, more diffuse labeling. *Bone trabeculae; v indicates a prominent vacuole in the large osteoclast in panel B. Scale bar in panel C equals 21 μm. Shown are 10-μm paraffin sections without counterstain. Objective magnification: 63 ×/1.25 NA.
Immunohistochemistry
Immunohistochemistry was performed on sections from the same tissues examined by in situ hybridization to determine whether the CCL9 mRNA is translated into protein and to establish its distribution in the bone tissues (Figure 4). CCL9 was not detected in tl/tl day-0 samples (not shown), but was seen in day-2, -4, and -6 bone sections. Close inspection confirmed that CCL9-positive cells were typically small on day 2 (Figure 4A), and were relatively strongly labeled. Lower intensity of anti-CCL9 labeling was seen at the later time points, and tended to occur in large, multinucleated osteoclasts that often contained vacuoles (Figure 4B), indicative of active ingestion of bone matrix. This confirms that both the mRNA and its translated product are expressed by osteoclasts in vivo beginning early in the differentiation process.
Osteoclast differentiation in bone marrow cell cultures
CSF-1 and RANKL were added to normal rat bone marrow cell cultures to induce osteoclast differentiation in the presence or absence of the same anti-CCL9 polyclonal antibody used for immunostaining. Multinucleated TRAP-positive cells were counted (Figure 5). In control wells, without anti-CCL9 added, there were on average 379 ± 147 (mean ± SD; n = 19) osteoclasts per well. With 0.5 μg antibody per well added, that decreased to 106 ± 62 (n = 9), and in the presence of 1.5 μg antibody, the average was 83 ± 77 (n = 9). The decreases were highly significant for both concentrations of anti-CCL9 antibody (P < .01). In control experiments, the addition of anti-CCL3 or anti-CCL5 (both of these chemokines bind CCR1, the same receptor as CCL9) had no significant effect on differentiation at either concentration (data not shown).
Inhibition of CCL9 blocks osteoclast differentiation in vitro. Rat BMCs were induced to differentiate into osteoclasts using CSF-1 and RANKL. Anti-CCL9 antibody blocked the formation of TRAP-positive, multinucleated cells. Means + 1 SD are shown. For both antibody concentrations tested, the decrease was highly significant. *P < .001.
Inhibition of CCL9 blocks osteoclast differentiation in vitro. Rat BMCs were induced to differentiate into osteoclasts using CSF-1 and RANKL. Anti-CCL9 antibody blocked the formation of TRAP-positive, multinucleated cells. Means + 1 SD are shown. For both antibody concentrations tested, the decrease was highly significant. *P < .001.
Anti-CCL9 inhibition of osteoclastogenesis in vivo
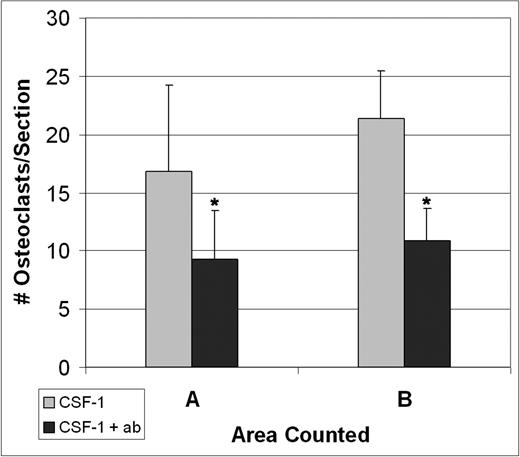
Three tl/tl rats were treated with CSF-1, but were also given anti-CCL9 antibody injections prior to and during their treatment course to determine whether it would interfere with CSF-1–induced osteoclastogenesis. Tissues were collected at 4 days, the time of the osteoclast peak counts, and TRAP-positive osteoclasts were counted (Figure 6). The anti-CCL9 treatment reduced the osteoclast counts in both area A and area B in comparison to tl/tl mutants treated with CSF-1 alone. The differences were highly significant: in area A, 9.3 ± 4.1 versus 16.8 ± 2.8 (P < .001); for area B, 10.8 ± 7.4 versus 21.5 ± 4.0 (P < .001) (mean ± SD).
Discussion
Together, the gene expression profiles for TRAP, cathepsin K, CCL9, and CCR1, and the osteoclast counts in bone over the time course suggest that CCL9 acts in vivo early in osteoclast differentiation. Given CCL9's chemotactic effects on mononuclear precursors in mouse13,14 and rat,12 it presumably is guiding them to sites where they will fuse and fully differentiate. The results also suggest that the average time required for CSF-1–induced osteoclast differentiation from precursors in vivo was roughly 4 days.
The fact that anti-CCL9 antibody suppressed the appearance of TRAP-positive osteoclasts when injected in vivo provides strong evidence for its importance in early events in osteoclastogenesis. Because the antibody was in limited supply, we have not determined whether the suppression of osteoclast differentiation would have been greater if higher doses of antibody were used, or was partially compensated by other chemokines. CCL-9 was recently shown to be highly expressed during mouse osteoclast differentiation in vitro, and inhibiting it with anti-CCL9 antibody interfered with osteoclast differentiation in primary mouse cell cultures.13,14 The extremely strong induction of CCL9, and not of other chemokines, in long bones of CSF-1–treated tl/tl rats establishes several things. First, it demonstrates that the increases seen in earlier studies of cultured cells also take place within the context of living bone. Second, it establishes that CCL9, among all the chemokines we studied, plays the decisive role in promoting osteoclastogenesis in response to CSF-1 and RANKL. Third, it shows that CCL9 is playing a similar role in the rat as in the mouse, which is important to confirm since the chemokines are such a large and diverse family, with multiple ligand/receptor interactions, and substantial variation from species to species.
Injections of anti-CCL9 suppress osteoclast differentiation in vivo.tl/tl rats were injected either with CSF-1 alone or with anti-CCL9 antibody and CSF-1, and TRAP+ osteoclasts were counted on multiple sections of the proximal femur from 3 animals for each condition after 4 days of injections. Anti-CCL9 significantly reduced the osteoclastogenic response to CSF-1 injections. Means + 1 SD are shown. In both areas, differences were highly significant (*P < .001).
Injections of anti-CCL9 suppress osteoclast differentiation in vivo.tl/tl rats were injected either with CSF-1 alone or with anti-CCL9 antibody and CSF-1, and TRAP+ osteoclasts were counted on multiple sections of the proximal femur from 3 animals for each condition after 4 days of injections. Anti-CCL9 significantly reduced the osteoclastogenic response to CSF-1 injections. Means + 1 SD are shown. In both areas, differences were highly significant (*P < .001).
The lack of marked changes in other chemokines and their receptors is consistent with the work of Lean et al,14 in which a survey of all known chemokine and receptor mRNAs was carried out during the RANKL-induced differentiation of normal mouse bone marrow mononuclear cells. They found CCL9 and its receptor, CCR1, to be those most highly expressed after 5 days of treatment with CSF-1 and RANKL, but this did not occur if only CSF-1 was used. The next most highly expressed chemokine mRNAs in their analysis were CCL12 and CCL25. Unfortunately, neither of those ligands was represented on the rat arrays, so more detailed investigations in rat bones have not been done by us at present. Given the dependence on CSF-1 and RANKL that those authors found for CCL9 and CCR1 expression, we infer that the increases in our test system result not from the actions of CSF-1 alone, but rather from the combined effects of the injected CSF-1 and normal RANKL expressed in the tl/tl rat bone.
Another ligand for CCR1, CCL3 (MIP-1α), was shown several years ago to promote osteoclast differentiation in vitro.32 A report showed that CCL3 promoted tumor invasiveness in mouse models and that its inhibition by anti–MIP-1α antibody reduced myeloma-induced osteolysis.18 CCL3 and the other ligands that bind CCR1—CCL5 and 7—were examined in RAW264.7 cells, a mouse cell line that becomes osteoclast-like in response to RANKL, and in primary mouse marrow-derived cells.17 These authors found that CCL3, CCL5, and CCL7 all exerted chemotactic effects. Because those chemokines are all normally expressed by osteoblastic or other cell types in the bone environment, the authors correctly point out that their misexpression could lead to increased fusion and differentiation of osteoclasts at inappropriate sites and could contribute to pathologic bone loss in vivo, as CCL3 has been shown to do. During our time course, however, expression of CCL3, CCL5, and CCL7 did not change. Additionally, neither anti-CCL3 nor anti-CCL5 inhibited osteoclast differentiation in bone marrow cell culture experiments, whereas anti-CCL9 had a highly significant inhibitory effect (Figure 5). We conclude that although the other CCR1 ligands are potential mediators of pathological bone resorption, at least in rodent models, CCL9 normally plays the decisive role.
One question that remains open is whether, if the CCL9 gene were knocked out, other chemokines would be able to compensate. The CCR1 receptor has been knocked out, but those authors did not note a skeletal phenotype,33-35 implying that compensation has occurred. A knockout mouse model for CCL9 deficiency would shed light on this question and provide further information on whether chemokine pathways overlap to regulate bone mass.
The lack of change in several other chemokines and receptors also deserves consideration. CXCL12 (SDF-1) has been shown to exert significant chemotactic activity on RAW cells, which express high levels of its putative receptor, CXCR4.15 Further work in RAW cells showed an induction of the tissue remodeling enzyme MMP9 by CXCL12,16 suggesting a role for CXCL12 in promoting the extravasation of osteoclast precursors and guiding their migration to appropriate bone resorption sites. Our findings are not inconsistent with those results, in that the near-constant levels of CXCL12 and CXCR4 in our time-course may be sufficient for these activities without necessitating additional transcription. It is clear, however, that the levels we saw in vivo were not responsive to the sudden appearance of normal CSF-1 levels and were independent of the resulting rapid, large-scale osteoclast differentiation.
The validity of rodent models to study the skeletal biology of higher vertebrates is fully supported by the high conservation of many signaling pathways, growth factors, and receptors between them and humans. The chemokines, however, are not so closely conserved across species. In our own investigations, we found many examples of extremely high conservation between rat and human, but also abundant cases of comparatively large divergence or even missing family members. Thus, precisely how our results in the rat and others' findings in the mouse correspond to human skeletal biology remains to be firmly established, and has not been thoroughly addressed to date. This is a significant challenge, given the complex interactions among chemokines and their receptors. In human osteoclast cultures, a recent report showed that the CXCL10 and CXCL12 were up-regulated specifically when cells were cultured on calcium phosphate–coated surfaces, but not when grown on plastic.36,37 In our experimental system, however, we did not observe changes in those ligands during osteoclast differentiation in vivo, although CXCL12 was indeed present at fairly high levels throughout the time course. Neither did we observe significant changes in several other chemokines and receptors those workers found in human osteoclast cultures. Another example is CXCL8 (IL-8), which was shown to have chemotactic and activating effects on human peripheral blood mononuclear cell–derived osteoclasts, and also to stimulate RANKL synthesis by MC3T3 cells.38 To date, however, no highly homologous gene to CXCL8 has been identified in either the rat or mouse genome, leaving the question open as to how the equivalent response would be triggered in rodent osteoclasts.
CCL9 clearly plays a key role in nonpathological osteoclast induction in vivo in the rat, and almost certainly in the mouse, but which chemokine plays the analogous role in human osteoclast recruitment and differentiation remains an important unanswered question. The nearest matches at the amino acid sequence level are CCL15, or A15 (GenBank no. NP_116740.2), and CCL23, or A23 (GenBank accession no. NP_005055.2), both of which are CCR1 ligands and can also bind to CCR3. Both are only moderately similar to rat CCL9. CCL15 has 42% identity and 59% total similarity, and CCL23 has 38% identity and 58% similarity. With human osteoclastoma cells, CCL15 was found to have chemoattractant activity.39 The degree of similarity of these human chemokines to rat CCL9 is not especially noteworthy. For example, rat CCL3 has a much closer human homolog, also designated CCL3 (GenBank accession no. AAH27888). The rat and human CCL3 proteins share 74% amino acid identities and 84% total similarity, which is fairly typical. It would therefore appear that evolution has selected a different chemokine to modulate osteoclast ontogeny in humans versus the 2 rodent species. Future work focused on the role of CCL15 and/or other, less homologous candidates should establish the identity of the key human osteoclastogenic chemokine(s) in nonpathological osteoclast induction.
Prepublished online as Blood First Edition Paper, November 22, 2005; DOI 10.1182/blood-2005-08-3365.
Supported by grants DE 07444 and DE13961 from the National Institute of Dental and Craniofacial Research (NIDCR) to P.R.O. and by core facilities support from the Diabetes Endocrinology Research Center grant DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDKD).
M.Y. performed research, designed project, designed experiments, and wrote the paper; G.M., C.A.M., A.M.-S., and J.A. performed research; and P.R.O. performed research, designed the project, designed research, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This work is solely the responsibility of the authors and does not necessarily represent official views of the NIDCR or the National Institutes of Health (NIH).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal