Abstract
Early growth-response factor 1 (Egr-1) is a zinc-finger transcription factor that plays a regulatory role in the expression of many genes important for inflammation. Whether Egr-1 is involved in IgE-dependent mast-cell activation was investigated. We demonstrated that IgE and antigen (TNP) stimulation induced a rapid expression of Egr-1 mRNA in mouse bone marrow–derived mast cells (BMMCs). As early as 15 to 20 minutes after IgE + TNP stimulation, Egr-1 protein was detectable in the nucleus of BMMCs by immunofluorescence or electrophoretic mobility shift assay. To examine a role for Egr-1 in IgE-dependent cytokine production by mast cells, Egr-1–deficient (Egr-1–/–) BMMCs were developed from the bone marrow cells of Egr-1 knockout mice. Egr-1–/– BMMCs express similar levels of surface c-kit and IgE receptor as compared with those on Egr-1+/+ BMMCs. Importantly, IgE + TNP-induced TNF and IL-13 expression was significantly reduced at both mRNA and protein levels in Egr-1–/– BMMCs as compared with those in Egr-1+/+ BMMCs. Thus, our results suggest that de novo synthesis of Egr-1 represents a novel mechanism in FcϵRI signaling and is required for the full responsiveness of IgE-dependent TNF and IL-13 production by mast cells.
Introduction
Mast cells are critical effector cells in allergic disorders through secretion of mast-cell mediators.1 Antigen-mediated aggregation of IgE bound to its high-affinity IgE receptor (FcϵRI) on the mast-cell surface initiates a complex series of biochemical events, leading to the exocytosis of granule-associated mediators and the generation of leukotrienes and cytokines.2,3 FcϵRI aggregation induces rapid tyrosine phosphorylation of its β and γ subunits and activation of cytoplasmic protein tyrosine kinase Syk and Fyn.2,4 This initial event is followed by activation of multiple signaling pathways that consist of a complex network of enzymes and adaptor molecules.2-4 Mast-cell degranulation can occur in seconds after FcϵRI aggregation,5 suggesting a rapid interaction between signaling molecules that control the exocytosis machinery. Contrary to the exocytosis of granule-associated mediators, IgE-dependent production of cytokines normally occurs in hours.6,7 Production of cytokines and chemokines requires activation of transcription factors. Although diverse cytokines and chemokines, including TNF and IL-13, that are important in inflammation have been described to be produced by mast cells,8 the number of the transcription factors that have been associated with IgE-dependent mast-cell activation is limited. Moreover, individual cytokines are likely regulated by different transcription factors. Transcription factors involved in TNF and IL-13 production by mast cells are not entirely clear.
The early growth response factor-1 (Egr-1),9 also known as zif268,10 krox-24,11 NGFI-A,12 TIS8,13 ETR103,14 and ZENK,15 is an 80- to 82-kDa nuclear protein consisting of 533 amino acids.16 The DNA-binding domain of Egr-1 consists of 3 zinc finger motifs, through which Egr-1 preferentially binds to GC-rich DNA sequences.16 Once bound to DNA, Egr-1 is capable of activating or repressing gene transcription.16 Cell type and specific stimulus determine whether Egr-1 acts to activate or inhibit promoter activity of target genes.16-18 This is likely due to the interaction between Egr-1 and tissue-specific factors and/or transcription factors that associate with proximal promoter elements such as Sp-1.16-18 The biologic activities of Egr-1 include activation or inhibition of gene expression and differentiation of macrophages.16,19 Egr-1 has been implicated in allergic responses because Egr-1–deficient mice showed diminished TNF mRNA and protein in the lung in response to ovalbumin sensitization and airway challenge.20 Egr-1–deficient mice had elevated IgE levels and were hyporesponsiveness to airway challenge with methacholine.20 Although Egr-1 has been reported to be expressed by mast cells,21,22 a role for Egr-1 in mast-cell development or in IgE-dependent cytokine production is unclear.
In this study, we demonstrated that aggregation of FcϵRI by antigen stimulation induced rapid Egr-1 expression at mRNA (15 minutes) and protein (30-60 minutes) levels. IgE-mediated Egr-1 expression precedes TNF and IL-13 production at both mRNA and protein levels. Using bone marrow–derived Egr-1–deficient mast cells, we showed that IgE-dependent TNF and IL-13 production at mRNA and protein levels were both reduced because of Egr-1 deficiency as compared with wild-type mast cells. Thus, Egr-1 is required for the full responsiveness of mast cells to produce TNF and IL-13 by IgE and antigen stimulation. The need of de novo synthesis of Egr-1 in IgE-mediated mast-cell activation likely contributes to the delayed cytokine response and represents a novel mechanism in FcϵRI-mediated TNF and IL-13 production.
Materials and methods
Animals
Egr-1–deficient mice and control C57BL/6NTac mice were purchased from Taconic Farms (Germantown, NY). The protocols were approved by the University Committee on Laboratory Animals, Dalhousie University, in accordance with the guidelines of the Canadian Council on Animal Care.
Antibodies
Antibodies to Egr-1 (sc-189 for immunofluorescence study and sc-189X for the blockade of DNA–protein complex formation), Egr-2 (sc-190X), Egr-3 (sc-22801X), and Egr-4 (sc-19868X) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa 594–conjugated goat anti–rabbit IgG F(ab′)2 fragment was purchased from Molecular Probes (Eugene, OR). FITC-conjugated rat anti–mouse CD117 (c-kit) mAb (IgG2a, CL8936F) and FITC-rat IgG2a (CLCR2A01) were purchased from Cedarlane Laboratories (Hornby, ON, Canada). FITC-conjugated rat anti–mouse IgE (IgG1; catalog no. 553415) and FITC-rat IgG1 (catalog no. 553924) were purchased from BD Biosciences (San Jose, CA).
Mast-cell culture and sensitization
Murine primary cultured bone marrow–derived mast cells (BMMCs) were cultured as previously described.23 Following 4 to 5 weeks of culture, mast-cell purity of greater than 98% was achieved as assessed by toluidine blue staining (pH = 1.0) of fixed cytocentrifuged preparations. BMMCs were passively sensitized with IgE from TIB-141 cells (ATCC no. TIB-141; American Type Culture Collection, Manassas, VA).24 Briefly, cells were resuspended in fresh complete medium supplemented with TIB-141–conditioned medium enriched in IgE directed against trinitrophenyl (TNP) at a ratio of 3:1. Following sensitization, experimental groups were washed extensively and resuspended at the density of 1 million/mL. Cells were activated by the addition of 10 ng/mL TNP-BSA.
Nuclear extract preparation and electrophoretic mobility shift assay (EMSA)
Nuclear protein extracts were obtained using a nuclear extract kit (catalog no. 40010; Active Motif, Carlsbad, CA), according to the manufacturer's protocol. All preparation procedures were carried out at 4°C. Total protein concentration was determined using the Bio-Rad protein assay (catalo no. 500-0006; Bio-Rad Laboratories, Hercules, CA).
EMSA was carried out as previously described.25 The following double-stranded oligonucleotides were used: wild-type Egr-1 probe, 5′-GGA TCC AGC GGG GGC GAG CGG GGG CGA ACG-3′ (catalog no. 1200011; Geneka Biotechnology, Montreal, QC, Canada. Note, this probe recognizes all 4 egr family members, although it is indicated as an Egr-1 probe by the manufacturer); mutant Egr-1 probe, 5′-GGA TCC AGC GGG GTA GAG CGG GTA CGA ACG-3′ (catalog no. 1400011; Geneka Biotechnology); and Sp-1 probe, 5′-ATT CGA TCG GGG CGG GGC GAG C-3′ (catalog no. E3231; Promega, Madison, WI). Probe-labeling was accomplished by treatment with T4 poly kinase (catalog no. M410A; Promega) in the presence of 32P-adenosine 5′-triphosphate (catalog no. PB10218; Amersham Pharmacia Biotech, Piscataway, NJ). Labeled oligonucleotides were purified using a MicroSpin G-25 column (catalog no. 275325; Amersham). Nuclear protein (8 μg) was added to a total volume of 10 μL binding reaction with 1 μL (0.75 μg/μL) poly (dI-dC) (catalog no. 27-7880-02; Amersham) and incubated at room temperature for 15 minutes. Labeled oligonucleotides were added to each reaction mixture and incubated at room temperature for 30 minutes. Samples were then separated by electrophoresis on a 6% polyacrylamide gel in 0.5 × Tris-boric acids–EDTA buffer at room temperature for 1 hour at 300 V. Gels were prerun for 30 minutes at 100 V before samples were loaded. Gels were vacuum-dried and subjected to autoradiography overnight at –80 °C.
For competition assays, 1 μL nonradiolabeled wild-type Egr-1, mutant Egr-1, or Sp1 oligonucleotides (50-fold excess of radiolabeled probe) were added and incubated for 15 minutes before the addition of the radiolabeled probe. For assays using antibodies to block the DNA–protein complex formation, nuclear extracts were incubated in a total volume of 20 μL with 6 μL appropriate polyclonal antibodies to Egr-1 (sc-189X), Egr-2 (sc-190X), Egr-3 (sc-22 801X), or Egr-4 (sc-19868X), respectively, for 2 hours on ice before the addition of the radiolabeled probe.
Real-time quantitative PCR
Total RNA was isolated from BMMCs using TRIZOL Reagent (catalog no. 15596-026; Invitrogen, Carlsbad, CA) and reverse transcribed using SuperScript II RNase H-Reverse Transcriptase (catalog no. 18064-014; Invitrogen) according to the manufacturer's instruction. Real-time quantitative polymerase chain reaction (PCR) was performed using a 7000 Sequence detector (PE Applied Biosystems, Foster City, CA). Specific quantitative assays for Egr-1, TNF, and IL-13 were performed using Assays-on-Demand reagents containing 6-FAM dye-labeled TaqMan MGB probes (Applied Biosystems) according to the manufacturer's protocol.26 GAPDH was used as an endogenous reference. Data were analyzed using relative standard curve method according to the manufacturer's protocol. An average value of each gene after GAPDH normalization at the time point showing highest expression was used as a calibrator to determine the relative levels of Egr-1, TNF, or IL-13 at different conditions.
In addition, PCR products of Egr-1 and GAPDH were separated on 2% agarose gel and stained with ethidium bromide.
Cytokine assays
The levels of TNF and IL-13 were measured by enzyme-linked immunosorbent assay (ELISA) using antibody (Ab) pairs from R&D Systems (Minneapolis, MN) according to the manufacturer's protocol.
Immunofluorescence study
BMMCs (3 × 106 cells/sample) were treated with TNP-BSA (10 ng/mL) for various times. After stimulation, BMMCs were fixed and permeabilized using Cytofix/Cytoperm solution (catalog no. 554714; BD Biosciences Pharmingen, San Diego, CA) for 20 minutes at 4°C. Cells were then blocked with 5% normal goat serum (catalog no. CL1200; Cedarlane Laboratories.) for 1 hour at 4°C. Then, cells were incubated with rabbit polyclonal anti–Egr-1 antibody (catalog no. sc-189; Santa Cruz Biotechnology) for 3 hours at 4°C. Cells were then stained for 2 hours with Alexa Fluor-594–conjugated goat anti–rabbit IgG (F(ab)2) (Molecular Probes). Fluorescence-labeled mast cells were cytocentrifuged (Cytospin 3, Shandon, United Kingdom) onto slides at 4.5g (200 rpm) for 5 minute. To visualize cell nuclei, slides were mounted with DAPI, a fluorescent groove-binding probe for DNA, before cover slip attachment. Cells were examined using a fluorescence microscope (Nikon Eclipse E600; Nikon, Tokyo, Japan).
Histology
Tongue, skin, and trachea tissues from Egr-1+/+ and Egr-1–/– mice were fixed in Carnoy fluid overnight and then in 100% ethanol for paraffin embedding and sectioning. Slides were deparaffinized with Citrisolv (Fisher, Fair Lawn, NJ) and rehydrated through decreasing concentrations of ethanol. Slides were stained with Alcian blue to visualize mast cells.
Fluorescence-activated cell sorting (FACS) analysis
For analysis of c-kit expression, BMMCs were stained with the FITC-conjugated rat anti–mouse CD117 (c-kit) monoclonal Ab (mAb) (IgG2a) for 1 hour at 4°C. FITC-rat IgG2a was used as control. For analysis of FcϵRI expression, BMMCs were sensitized with IgE and then stained with FITC-conjugated rat anti–mouse IgE mAb for 1 hour at 4°C. FITC-rat IgG1 was used as a control. Cells were analyzed by a FACScaliber flow cytometer (BD Biosciences).
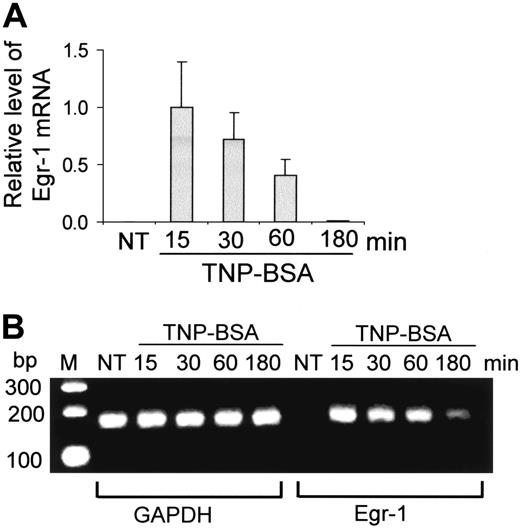
Expression of Egr-1 by mast cells after FcϵRI aggregation. (A-B) Mouse bone marrow–derived mast cells (BMMCs) were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL) for 15, 30, 60, or 180 minutes. RNA isolated from these cells was reverse transcribed to cDNA and subjected to real-time quantitative PCR. Egr-1 expression was normalized to endogenous control GAPDH. The data are expressed as relative mRNA levels compared with the average expression level in BMMCs treated with TNP-BSA for 15 minutes (= 1) because at this time point Egr-1 showed the highest expression level. The PCR products were also separated by agarose gel and stained with ethidium bromide. Untreated BMMCs (NT) showed no Egr-1 expression, whereas TNP induced a rapid and transient expression of Egr-1. Error bars represent standard errors from 3 independent experiments.
Expression of Egr-1 by mast cells after FcϵRI aggregation. (A-B) Mouse bone marrow–derived mast cells (BMMCs) were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL) for 15, 30, 60, or 180 minutes. RNA isolated from these cells was reverse transcribed to cDNA and subjected to real-time quantitative PCR. Egr-1 expression was normalized to endogenous control GAPDH. The data are expressed as relative mRNA levels compared with the average expression level in BMMCs treated with TNP-BSA for 15 minutes (= 1) because at this time point Egr-1 showed the highest expression level. The PCR products were also separated by agarose gel and stained with ethidium bromide. Untreated BMMCs (NT) showed no Egr-1 expression, whereas TNP induced a rapid and transient expression of Egr-1. Error bars represent standard errors from 3 independent experiments.
Results
Transient and early expression of Egr-1 by mast cells after IgE + TNP stimulation
To determine whether Egr-1 is expressed in mast cells after Ag stimulation, mouse BMMCs were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL) for various times (15, 30, 60, 180 minutes). Egr-1 mRNA expression was evaluated by real-time quantitative PCR. Egr-1 level was normalized to GAPDH in each sample. Egr-1 level in the unstimulated mast cells was undetectable. An average value of Egr-1 after GAPDH normalization at the time point of 15 minutes (the highest Egr-1 expression level) was used as a calibrator to determine the relative levels of Egr-1 expression at various conditions (Figure 1A). TNP stimulation induced a transient and rapid Egr-1 expression. A strong Egr-1 expression can be seen at 15 minutes after TNP stimulation. Subsequently, the level of Egr-1 began to return toward basal conditions. After 180 minutes of TNP stimulation, little Egr-1 mRNA can be detected (Figure 1A). PCR-amplified Egr-1 products were also separated in agarose gels and visualized by ethidium bromide staining. A representative gel was presented in Figure 1B. A rapid and strong Egr-1 expression can be seen at 15 minutes after TNP stimulation. Egr-1 levels decreased after this time point.
Nuclear localization of TNP-induced Egr-1 protein in mast cells
To examine TNP-induced Egr-1 expression at protein level and its cellular localization, mouse BMMCs were treated with TNP for various times (15-360 minutes), permeabilized, and stained with anti–Egr-1 Ab. Nucleus was visualized by DAPI staining. Expression of Egr-1 protein can be seen as early as 15 minutes after TNP stimulation. Strong Egr-1 expression was observed at 30 minutes to 1 hour after TNP stimulation. Interestingly, Egr-1 proteins were exclusively found in the nuclei of TNP-stimulated mast cells (Figure 2).
To evaluate the time course relation between Egr-1 expression and TNF and IL-13 production, BMMCs were treated with TNP for various times (15 minutes to 24 hours), and cell-free supernatants were used to determine TNF and IL-13 production by ELISA. Unlike Egr-1, which is detectable at 15 minutes and reaches its peak expression at 1 hour after TNP stimulation (Figure 2), production of TNF and IL-13 were undetectable at 30 minutes (data now shown). TNP-induced TNF and IL-13 production peaked at 3 to 6 hours (TNF, 3 hours at 799.3 ± 114.4 pg/mL; TNF, 6 hours at 787.1 ± 134.9 pg/mL; IL-13, 3 hours at 490.1 ± 63.5 pg/mL; IL-13, 6 hours at 417.6 ± 55.5 pg/mL). Thus, TNP-induced Egr-1 expression precedes TNF and IL-13 production.
Egr-1 activation induced by TNP stimulation in mast cells
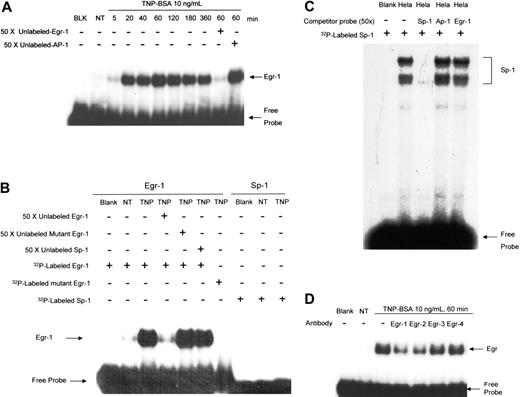
To determine the specific DNA binding activity of TNP-induced Egr-1 in mast cells, nuclear proteins from TNP-stimulated BMMCs were isolated and examined by EMSA using a DNA probe that can be recognized by the Egr family. A strong DNA binding activity of nuclear proteins from mast cells treated with TNP can be observed (Figure 3A). Consistent with the immunofluorescence experiment, Egr binding was not observed in unstimulated mast cells and became apparent as early as 5 to 20 minutes after TNP stimulation. The binding specificity of nuclear proteins to Egr DNA sequence was verified through the competitive binding by nonradioisotope-labeled Egr probes (Figure 3A-B) but not by AP-1 (Figure 3A, last lane) or Sp-1 probes (Figure 3B). Interestingly, although Sp-1 has been shown to be closely associated with Egr-1 function because of similarity in their DNA binding sequences, no Sp-1 activation can be observed in TNP-stimulated mast cells (Figure 3B). As a control, the specific binding of Sp-1 was shown using nuclear extracts from HeLa cells (Figure 3C).
There are 4 members of Egr family members, including Egr-1, -2, -3, and -4. To further examine the binding specificity, nuclear proteins from TNP-treated BMMCs were preincubated with anti–Egr-1 Ab, anti–Egr-2 Ab, anti–Egr-3 Ab, or anti–Egr-4 Ab, respectively. Then Ab-treated samples were used for binding with 32P-labeled Egr probe. Treatment of nuclear proteins with anti–Egr-1 Ab, but not anti–Egr-3 or anti–Egr-4 Ab, reduced nuclear protein binding to Egr probe, suggesting the specificity of Egr-1 binding (Figure 3D). Interestingly, anti–Egr-2 Ab also reduced nuclear protein binding to 32P-labeled Egr probe, suggesting that TNP stimulation may also induce Egr-2 activation (Figure 3D).
Development of mast cells in the absence of Egr-1
A number of cytokines that are important in allergy, such as TNF, contain an Egr-1–binding sequence in their promoters.27,28 TNP-induced expression of Egr-1 precedes TNF and IL-13 production. Accordingly, Egr-1 may play a role in TNP-induced TNF and IL-13 production by mast cells. To test this hypothesis, we attempted to use Egr-1–deficient BMMCs. Because Egr-1 has been shown to be important for the differentiation of hematopoietic precursor cells into cells of the myeloid lineage19 and is involved in apoptosis,29 we determined whether mature mast cells can be developed from Egr-1–deficient mouse bone marrow progenitor cells.
Bone marrow cells from Egr-1–deficient and wild-type mice were cultured in conditioned medium for 4 to 6 weeks. Cells were analyzed by flow cytometry for c-kit and IgE receptor expression at the end of each week during the culturing process. No difference of c-kit and IgE receptor expression pattern between wild-type and Egr-1–deficient cells was observed during mast-cell maturation (week 1to week 6; data not shown). The c-kit and IgE receptor expression after 5 weeks of culture was presented in Figure 4A. Cells after 5 weeks of culture were also used for toluidine blue staining. Morphologically, no difference was observed between Egr-1–deficient and wild-type mast cells (Figure 4B). In addition, various tissues from Egr-1–deficient mice were used to examine the presence of mast cells. Similar number and morphology of mast cells were observed in tongue (Figure 4C), skin and trachea (data not shown), in both Egr-1–deficient and wild-type mice.
Immunofluorescence staining of Egr-1. After IgE sensitization, mouse BMMCs from wild-type mice or Egr-1–deficient mice were either left untreated (no treatment) or stimulated with TNP-BSA (10 ng/mL) for various times. Cells were fixed, permeabilized, and then stained with anti–Egr-1 Abs or control rabbit serum. Alexa 594–conjugated goat anti–rabbit IgG F(ab′)2 was used as a secondary Ab. DAPI staining was used to visualize the nucleus of the cells. TNP stimulation induced a transient expression of Egr-1, which is localized in the nucleus of the cells. As a control, no Egr-1 staining was observed in BMMCs from Egr-1–deficient mice. Original magnification, × 40. Immunolabeled specimens were mounted in DAPI containing Vectashield (Vector Laboratories, Burlingame, CA). Cells were examined using a fluorescence microscope (Nikon E600; Nikon, Tokyo, Japan) equipped with a DMX1200 camera and a CFI Plan-Fluor DDL 40 ×/0.75 objective lens. Images were processed using Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA).
Immunofluorescence staining of Egr-1. After IgE sensitization, mouse BMMCs from wild-type mice or Egr-1–deficient mice were either left untreated (no treatment) or stimulated with TNP-BSA (10 ng/mL) for various times. Cells were fixed, permeabilized, and then stained with anti–Egr-1 Abs or control rabbit serum. Alexa 594–conjugated goat anti–rabbit IgG F(ab′)2 was used as a secondary Ab. DAPI staining was used to visualize the nucleus of the cells. TNP stimulation induced a transient expression of Egr-1, which is localized in the nucleus of the cells. As a control, no Egr-1 staining was observed in BMMCs from Egr-1–deficient mice. Original magnification, × 40. Immunolabeled specimens were mounted in DAPI containing Vectashield (Vector Laboratories, Burlingame, CA). Cells were examined using a fluorescence microscope (Nikon E600; Nikon, Tokyo, Japan) equipped with a DMX1200 camera and a CFI Plan-Fluor DDL 40 ×/0.75 objective lens. Images were processed using Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA).
Reduced IgE-dependent TNF and IL-13 production resulting from Egr-1 deficiency
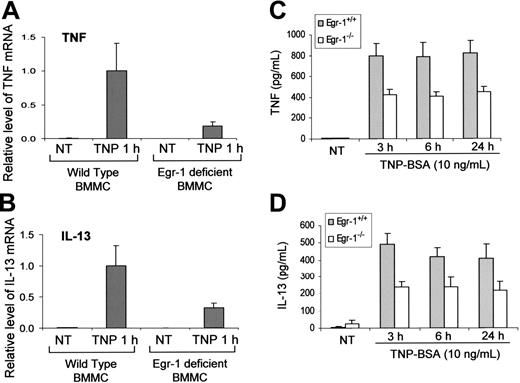
Egr-1–deficient mature mast cells have been obtained by culturing bone marrow cells from Egr-1–deficient mice. These Egr-1–deficient BMMCs as well as control BMMCs cultured from wild-type mice were used to determine the specific contribution of Egr-1 in IgE-dependent TNF and IL-13 production. To examine the TNF and IL-13 expression at the mRNA level, Egr-1–deficient and wild-type BMMCs were stimulated with TNP-BSA for 1 hour and used for RNA isolation. Real-time quantitative PCR was performed to determine the relative levels of TNF and IL-13 mRNA. TNF and IL-13 levels were normalized to GAPDH. An average value after GAPDH normalization at the time point of 1 hour after TNP stimulation from wild-type BMMCs, which showed the highest level of TNF and IL-13 expression, was used as a calibrator to determine the relative levels of TNF and IL-13 at different conditions. There were 70% to 80% reductions of IgE-dependent TNF and IL-13 mRNA expression in Egr-1–deficient BMMCs as compared with that in wild-type BMMCs (Figure 5A-B).
Similarly, TNP-induced production of TNF and IL-13 at the protein levels by Egr-1–deficient BMMCs was significantly reduced compared with that by wild-type BMMCs (Figure 5C-D). Interestingly, Egr-1 deficiency only had a minor effect on TNP-induced IL-6 production by BMMCs (data not shown), suggesting that Egr-1 may selectively regulate specific genes in mast cells.
Discussion
An earlier study showed that treatment of mast cells with cycloheximide (a protein synthesis inhibitor) inhibits IgE-dependent mediator release, suggesting a need of de novo synthesis of proteins in FcϵRI-mediated mast-cell activation.30 In this study, we demonstrated a clear role for the newly synthesized immediate early gene Egr1 in FcϵRI-induced TNF and IL-13 production by mast cells.
Both Egr-1 mRNA and protein are undetectable in resting mast cells. Expression of Egr-1 mRNA and protein was rapidly and transiently induced in mast cells by FcϵRI aggregation. The level of Egr-1 mRNA was found to be the highest at 15 minutes after TNP stimulation, whereas Egr-1 protein reached a peak at 60 minutes. FcϵRI-induced Egr-1 expression precedes TNF and IL-13 production at both the mRNA and protein levels. Importantly, FcϵRI-induced TNF and IL-13 expression was reduced at both mRNA and protein levels in Egr-1–deficient mast cells when compared with that in wild-type mast cells. These data suggest that FcϵRI aggregation induces the new synthesis of Egr-1, which in turn amplifies Tnf and Il13 gene expression by mast cells. Thus, the de novo synthesis of Egr-1 represents an indirect mechanism through which FcϵRI aggregation induces the full strength of gene expression by mast cells.
Stimulation of mast cells by FcϵRI aggregation induces Egr-1 but not Sp-1 activation. (A) Mouse BMMCs were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL) for various times. Nuclear proteins were isolated and subjected to EMSA analysis (see “Materials and methods”) using 32P-labeled Egr-1– or Sp-1–specific probes. BLK indicates blank, no nuclear proteins were added; NT, no treatment, nuclear proteins were isolated from BMMCs without TNP stimulation. Numbers indicated are minutes after TNP stimulation; nuclear proteins were isolated from BMMCs after the indicated times following TNP stimulation. Fifty times concentrated unlabeled Egr-1 oligonucleotide was used to compete with 32P-labeled Egr-1 oligonucleotide, whereas 50 × concentrated unlabeled AP-1 oligonucleotide was used as a nonspecific control probe. (B) Nuclear proteins from TNP-BSA (10 ng/mL, 60 minutes) treated BMMCs (TNP) or from untreated BMMCs (NT) were subjected to DNA probe competition experiment using unlabeled probes or 32P-labeled mutant probes to demonstrate specific Egr-1 binding. No Sp-1 binding was observed in TNP-stimulated BMMCs. (C) As a control, nuclear extracts from HeLa cells (Promega) were subjected to EMSA using 32P-labeled Sp-1 oligonucleotide. 32P-labeled Sp-1 binding was blocked by unlabeled Sp-1 probe, but not by unlabeled Egr-1 or AP-1 probes (50 ×). (D) Antibody blockade of the DNA-protein complex formation. Nuclear proteins from TNP-BSA (10 ng/mL, 60 minutes) treated BMMCs or from untreated BMMCs (NT) were incubated with or without specific antibodies to Egr-1, Egr-2, Egr-3, or Egr-4 for 2 hours on ice before EMSA experiment using 32P-labeled Egr-1 oligonucleotide.
Stimulation of mast cells by FcϵRI aggregation induces Egr-1 but not Sp-1 activation. (A) Mouse BMMCs were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL) for various times. Nuclear proteins were isolated and subjected to EMSA analysis (see “Materials and methods”) using 32P-labeled Egr-1– or Sp-1–specific probes. BLK indicates blank, no nuclear proteins were added; NT, no treatment, nuclear proteins were isolated from BMMCs without TNP stimulation. Numbers indicated are minutes after TNP stimulation; nuclear proteins were isolated from BMMCs after the indicated times following TNP stimulation. Fifty times concentrated unlabeled Egr-1 oligonucleotide was used to compete with 32P-labeled Egr-1 oligonucleotide, whereas 50 × concentrated unlabeled AP-1 oligonucleotide was used as a nonspecific control probe. (B) Nuclear proteins from TNP-BSA (10 ng/mL, 60 minutes) treated BMMCs (TNP) or from untreated BMMCs (NT) were subjected to DNA probe competition experiment using unlabeled probes or 32P-labeled mutant probes to demonstrate specific Egr-1 binding. No Sp-1 binding was observed in TNP-stimulated BMMCs. (C) As a control, nuclear extracts from HeLa cells (Promega) were subjected to EMSA using 32P-labeled Sp-1 oligonucleotide. 32P-labeled Sp-1 binding was blocked by unlabeled Sp-1 probe, but not by unlabeled Egr-1 or AP-1 probes (50 ×). (D) Antibody blockade of the DNA-protein complex formation. Nuclear proteins from TNP-BSA (10 ng/mL, 60 minutes) treated BMMCs or from untreated BMMCs (NT) were incubated with or without specific antibodies to Egr-1, Egr-2, Egr-3, or Egr-4 for 2 hours on ice before EMSA experiment using 32P-labeled Egr-1 oligonucleotide.
Several studies using differentiation-inducible myeloid cell lines as well as normal bone marrow cells have demonstrated a role of Egr-1 in the monocytic differentiation of myeloid cells.19,31-33 Egr-1 stimulates development along the macrophage lineage at the expense of development along the granulocyte or erythroid lineages.32 In addition, Egr-1 also has a role in T-cell, B-cell, and neuronal-cell development.28,34,35 We used bone marrow cells from Egr-1–deficient mice as well as wild-type mice and examined their differentiation into mast cells. IgE receptor and c-kit were used as cell-surface markers. Levels of these 2 receptors were examined every week during the differentiation process. No difference of IgE receptor or c-kit was observed between Egr-1–deficient cells and wild-type cells during the entire course of differentiation. Metachromatic staining (toluidine blue) showed normal morphology and granulation of Egr-1–deficient mast cells. In addition, toluidine blue staining showed a similar number of mast cells in various tissues (skin, tongue, and trachea) between Egr-1–deficient mice and wild-type mice. Thus, Egr-1 appears to have no effects on mast-cell granulation and c-kit and IgE receptor expression during mast-cell development. This effect is distinct from that in macrophages, T cells, or B cells. This further supports the notion that the function of Egr-1 is cell-type specific.
TNF and IL-13 are important inflammatory mediators. The Tnf gene is activated in response to multiple signals of stress and inflammation. A distinct set of transcription factors are recruited to the TNF promoter dependent on a specific stimulus.36-38 Although, it has been well characterized that FcϵRI aggregation induces TNF production by mast cells, the transcription factors involved in this process are less clear. Our finding of a role for Egr-1 in the regulation of IgE-dependent TNF and IL-13 production by mast cells is consistent with an in vivo study, demonstrating that Egr-1 modulates TNF and airway responsiveness in mice.20
The TNF promoter contains an Egr-1 responsive element.27 Binding of Egr-1 to the TNF promoter can either activate or inhibit TNF production.36,38 For example, LPS-induced Egr-1 binding activates TNF expression.39 In contrast, PGE2-induced Egr-1 binding inhibits TNF expression.17 This is likely due to the distinct transcription factors that may interact with Egr-1 under different stimulation conditions. Because Egr-1 shares similar consensus binding sites with transcription factor Sp-1 (-GGGCGG-)16 and forms a complex with Sp-1,40 we examined whether FcϵRI aggregation also induces Sp-1 activation. Interestingly, no Sp-1 activation can be observed in TNP-stimulated mast cells by EMSA analysis. Interaction between Egr-1 and NF-κB family members has been described to be involved in TNF production.41 We reported that FcϵRI aggregation-induced TNF production requires IKK-IκB-NF-κB pathway activation.42 Thus, it is likely that FcϵRI aggregation-induced Egr-1 may interact with NF-κB family members in the regulation of TNF production.
No effect of Egr-1 deficiency on mast-cell maturation was observed. Bone marrow cells from wild-type mice or Egr-1–deficient mice were cultured in conditioned media in vitro for 5 weeks and examined by flow cytometry for c-kit and IgE receptor expression (A) or by toluidine blue staining (B) Original magnification, × 100. Egr-1–deficient or wild-type mast cells express similar levels of c-kit and IgE receptor and show similar levels of granulation. (C) Tongue tissues from wild-type mice or Egr-1–deficient mice were fixed and stained by Alcian blue for mast cells. Similar metachromatic staining between wild-type and Egr-1–deficient tissues was observed. Original magnification, × 40. Specimens were viewed on a Nikon Eclipse E600 microscope equipped with a DMX1200 camera, a CFI Plan-Fluor DDL 100 ×/1.30 oil-immersion objective lens (B), and a DDL 40 ×/0.75 objective lens (C). Acquisition was performed with Nikon ACT-1 software version 2.20. Images were processed using Adobe Photoshop 5.0.
No effect of Egr-1 deficiency on mast-cell maturation was observed. Bone marrow cells from wild-type mice or Egr-1–deficient mice were cultured in conditioned media in vitro for 5 weeks and examined by flow cytometry for c-kit and IgE receptor expression (A) or by toluidine blue staining (B) Original magnification, × 100. Egr-1–deficient or wild-type mast cells express similar levels of c-kit and IgE receptor and show similar levels of granulation. (C) Tongue tissues from wild-type mice or Egr-1–deficient mice were fixed and stained by Alcian blue for mast cells. Similar metachromatic staining between wild-type and Egr-1–deficient tissues was observed. Original magnification, × 40. Specimens were viewed on a Nikon Eclipse E600 microscope equipped with a DMX1200 camera, a CFI Plan-Fluor DDL 100 ×/1.30 oil-immersion objective lens (B), and a DDL 40 ×/0.75 objective lens (C). Acquisition was performed with Nikon ACT-1 software version 2.20. Images were processed using Adobe Photoshop 5.0.
The role of Egr-1 in the regulation of IL-13 production has not been reported previously. IL-13 plays a central role in allergic inflammation.43 Mast cells are ready to produce and release IL-13 protein on stimulation.44 Impaired IL-13 production by Egr-1–deficient mast cells following FcϵRI aggregation suggests a role for Egr-1 in the regulation of this important cytokine in allergic response. Detailed analysis of the IL-13 promoter may shed new light onto the mechanisms involved.
Egr-1 is required for the full responsiveness of TNF and IL-13 production by mast cells in response to FcϵRI aggregation. Mouse BMMCs from wild-type mice or Egr-1–deficient mice were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL). (A-B) Cell pellets were used for RNA isolation. Real-time quantitative PCR was performed to determine TNF and IL-13 expression. TNF and IL-13 expression was normalized to endogenous control GAPDH. The data are expressed as relative mRNA level compared with the average expression level in wild-type BMMCs treated with TNP-BSA for 1 hour (= 1) because at this time point TNF and IL-13 showed the highest expression level. (C-D) Cell-free supernatants were used for examining TNF and IL-13 production by ELISA. Error bars represent standard error (A-B, n = 3; C-D, n = 5).
Egr-1 is required for the full responsiveness of TNF and IL-13 production by mast cells in response to FcϵRI aggregation. Mouse BMMCs from wild-type mice or Egr-1–deficient mice were sensitized with anti-TNP IgE and stimulated with TNP-BSA (10 ng/mL). (A-B) Cell pellets were used for RNA isolation. Real-time quantitative PCR was performed to determine TNF and IL-13 expression. TNF and IL-13 expression was normalized to endogenous control GAPDH. The data are expressed as relative mRNA level compared with the average expression level in wild-type BMMCs treated with TNP-BSA for 1 hour (= 1) because at this time point TNF and IL-13 showed the highest expression level. (C-D) Cell-free supernatants were used for examining TNF and IL-13 production by ELISA. Error bars represent standard error (A-B, n = 3; C-D, n = 5).
It is noticeable that Egr-1 deficiency only partially inhibited FcϵRI aggregation-induced TNF and IL-13 production by mast cells. Others also found that, although substantial evidence supports a role for Egr-1 in macrophage differentiation,19,31-33 little defect of macrophage lineage was observed in Egr-1–deficient mice.45 It is possible that other transcription factors such as Egr-2 may compensate for the effect of Egr-1 under the circumstance of Egr-1 deficiency. All 4 Egr family members, Egr-1, Egr-2, Egr-3, and Egr-4, share highly homologous DNA-binding domains, and all bind to the sequence GCGGGGGCG.46 Our EMSA experiment revealed that FcϵRI aggregation-induced protein binding to the Egr recognition sequence was blocked by anti–Egr-1 or anti–Egr-2 Abs, but not by anti–Egr-3, or anti–Egr-4 Abs, suggesting that Egr-2 may also be induced in TNP-stimulated mast cells. Further studies are needed to clarify the role of Egr-2 in the regulation of mast-cell function.
In an effort to examine potential mechanisms involved in FcϵRI-induced Egr-1 expression, we used various kinase and phosphatase inhibitors, including protein tyrosine kinase inhibitor PP2, PKC inhibitor Ro 31-8220, Erk1/2 inhibitor PD 98059, PI3K inhibitor wortmannin, p38 MAP kinase inhibitor SB 203580, mTOR inhibitor rapamycin, and PP2A inhibitor okadaic acid. Although, the MAPK pathway47 and MEK-Erk1/248 were reported to be required for Egr-1 activation and PD 98059 blocked Egr-1 activation in monocytes,48 no effects of PD 98059, SB 203580, wortmannin, or rapamycin were observed on TNP-induced Egr-1 activation using EMSA analysis (data not shown). Interestingly, tyrosine kinase inhibitor PP2 and PKC inhibitor Ro 31-8220 individually partially inhibited TNP-induced Egr-1 activation. When these 2 inhibitors were combined, an additive inhibitory effect on Egr-1 activation was observed (data not shown). Thus, the early tyrosine kinase phosphorylation following FcϵRI aggregation and subsequent PKC activation are likely upstream signaling events required for Egr-1 activation. The mechanism involved in Egr-1 activation in mast cells is different from that in other cells and requires further study.
In summary, we demonstrated that FcϵRI aggregation induced a rapid and transient de novo synthesis of Egr-1 and subsequent DNA binding. Egr-1 deficiency resulted in impaired IgE-dependent TNF and IL-13 production. The identification of Egr-1 in this context represents a novel mechanism involved in FcϵRI aggregation-induced TNF and IL-13 production by mast cells.
Prepublished online as Blood First Edition Paper, November 29, 2005; DOI 10.1182/blood-2005-09-3610.
Supported by grants from the Canadian Institutes of Health Research, Nova Scotia Health Research Foundation, and Izaak Walton Killam Health Center; by a New Investigator Award from the Canadian Institutes of Health Research (T.-J.L.); and by an Investigatorship from Izaak Walton Killam Health Center (T.-J.L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal