Abstract
The Notch1-RBP-Jκ and the transcription factor Runx1 pathways have been independently shown to be indispensable for the establishment of definitive hematopoiesis. Importantly, expression of Runx1 is down-regulated in the para-aortic splanchnopleural (P-Sp) region of Notch1- and Rbpsuh-null mice. Here we demonstrate that Notch1 up-regulates Runx1 expression and that the defective hematopoietic potential of Notch1-null P-Sp cells is successfully rescued in the OP9 culture system by retroviral transfer of Runx1. We also show that Hes1, a known effector of Notch signaling, potentiates Runx1-mediated transactivation. Together with the recent findings in zebrafish, Runx1 is postulated to be a cardinal down-stream mediator of Notch signaling in hematopoietic development throughout vertebrates. Our findings also suggest that Notch signaling may modulate both expression and transcriptional activity of Runx1.
Introduction
Mammalian hematopoietic development is believed to arise from 2 distinct cellular origins. In mice, primitive hematopoiesis arises in the yolk sac (YS) blood island at embryonic day (E) 7.5, while definitive hematopoiesis starts at the ventral region of the aortagonad-mesonephros (AGM) around E10.5, which shifts to the liver, spleen, and bone marrow, in this order. Progenitors for definitive hematopoiesis are first detected in the para-aortic splanchnopleural (P-Sp) region at E7.5 to E9.5,1,2 where the Notch1 gene has a nonredundant role in hematopoietic stem cell (HSC) development.3 Notch1 encodes a 300-kDa heterodimeric single-span transmembrane receptor consisting of a 180-kDa extracellular and a 120-kDa transmembrane subunit. Together with 3 other paralogs, it belongs to the evolutionarily conserved Notch family receptors that mediate cell-fate determination in multiple species. The Notch signaling is initiated by the binding of the Jagged and Delta families of ligands expressed on the neighboring cells, which induces the cleavage of the Notch transmembrane subunit and the release of the Notch intracellular domain. The latter in turn translocates to the nucleus and forms a transactivation complex by interacting with the DNA-binding protein RBP-Jκ and induces the expression of their target genes, such as those for the hairy/enhancer of split (Hes) family of basic helix-loop-helix transcription factors.4 Molecular channels downstream of these, however, are largely unknown.
Mice deficient in Runx1 (also known as AML1, CBFA2, or PEBP2αB), Scl, and Gata2 genes are lethal during the embryonic stage and show failure in the establishment of definitive hematopoiesis.5-7 A connection between Notch signaling and these transcription factors has been shown by the analyses of Notch1- and RBP-Jκ–encoding Rbpsuh-null mice. In the E9.5 P-Sp cells from Notch1-null mice, expression levels of SCL, GATA2, and Runx1 mRNA are significantly reduced.3 Rbpsuh-null mice also show markedly reduced levels of SCL, GATA2, and Runx1 mRNA in the endothelial-cell layer of the E9.5 P-Sp region,8 supporting the notion that the Notch1-RBP-Jκ pathway up-regulates the expression of these key transcription factors. Among these, Runx1, which has close homology to a Drosophila protein, Runt, functions as a transcriptional activator or repressor for its target genes in concert with several specific coactivators or corepressors, depending on the context.9 Importantly, presence of the Notch-Runx pathway has been proposed in Drosophila embryonic hemocytogenesis10 and zebrafish hematopoiesis during both developmental and postnatal periods.11 Similarly reported has been transcriptional regulation by Notch of the Gata2 gene in mouse AGM hematopoiesis8 and of the Gata homolog Serpent gene in Drosophila embryonic hemocytogenesis.12 In mammals, the existence of Notch-Runx pathway has been unclear.
In this study, we show that Notch1 up-regulates Runx1 mRNA expression in NIH3T3 cells. When introduced to the defective prehematopoietic precursor cells derived from the P-Sp region of Notch1-null embryos using retroviruses, Runx1, but neither SCL nor GATA2, restores the definitive hematopoiesis. We also demonstrate that Hes1, one of the Notch signal effectors, augments the transcriptional activity of Runx1 protein. These findings indicate that Runx1 is a key molecule in Notch1-RBP-Jκ–mediated mammalian hematopoiesis.
Materials and methods
Mice and embryos
C57BL/6 mice were purchased from Japan SLC (Hamamatsu, Japan) and Notch1 mutant mice13 were from Jackson Laboratory (Bar Harbor, ME). To generate embryos, timed matings were set up between Notch1+/– mice. The time at midday (12 PM) was taken to be E0.5 for the plugged mice.
In vitro P-Sp culture
P-Sp culture was performed as described previously.14 In brief, isolated P-Sp regions of E9.5 embryos were dissociated by incubation with 250 protease units (PU)/mL dispase (Godo Shusei, Tokyo, Japan) for 20 minutes and cell-dissociation buffer (Gibco BRL, Carlsbad, CA) for 20 minutes at 37°C, followed by vigorous pipetting. Approximately 5 × 104 P-Sp–derived cells were suspended in 300 μL of serum-free StemPro media (Life Technologies, Gaithersburg, MD) supplemented with 50 ng/mL stem-cell factor (SCF), 5 ng/mL interleukin-3 (IL3; gifts from Kirin Brewery, Takasaki, Japan), and 10 ng/mL mouse oncostatin M (R&D Systems, Minneapolis, MN). Single-cell suspensions were seeded on preplated OP9 stromal cells15 in the 24-well plate, followed by incubation at 37°C in a 5% CO2 incubator. Images were visualized with a Nikon Eclipse TE2000-U microscope equipped with 40×/0.60 and 10×/0.30 NA objective lenses (Nikon, Tokyo, Japan), and were captured with a C5810 camera (Hamamatsu Photonics, Hamamatsu, Japan).
Plasmid construction
The cDNA of human Runx1 was subcloned into the EcoRI restriction site of the retrovirus vector pMYs/internal ribosomal entry site–enhanced green fluorescent protein (IRESEGFP; pMYs/IG).16 The cDNAs for FLAG-tagged murine SCL and FLAG-tagged murine GATA2 were inserted into the EcoRI and NotI restriction sites of pMYs/IG. The cDNA for murine Notch1 intracellular domain (NICD)3 was subcloned into the BamHI restriction site of pMYs/IG. To assess the domain functions of Runx1, we used mutants and wild-type Runx1 constructed in pMY/IG.14 The pME18S-HA-Runx1 and pME18S-PEBP2β were described previously.17 The cDNA for FLAG-tagged murine Hes1 was inserted into the EcoRI and NotI restriction sites of the pME18S-expression vector and in-frame into the EcoRI and XbaI restriction sites of the p3xFLAG-myc-CMV-25–expression vector (Sigma, St Louis, MO).
Retroviral transduction
Plat-E packaging cells (2 × 106)16 were transiently transfected with 3 μgof retrovirus vectors, mixed with 9 μL of FuGENE6 (Roche Applied Science, Indianapolis, IN), and incubated at 37°C. Supernatant containing retrovirus was collected 48 hours after transfection and used immediately for infection. Retroviral transduction of the cells derived from Notch1-null P-Sp regions was performed as described previously.14 In brief, the viral supernatant was added to the P-Sp cells seeded on the OP9 stromal-cell layer together with 10 μg/mL polybrene (Sigma). After 72 hours of incubation, virus-containing medium was replaced by the original culture medium. The cells were incubated for another 10 days and processed for analysis. To confirm the expression of proteins, NIH3T3 cells were also infected with the same viral supernatants. The efficiency of infection was evaluated by the positivity of GFP. The proteins were detected by Western blot using anti-Runx1 antibody (PC284L; Oncogene, Cambridge, MA), anti-FLAG monoclonal antibody (M2; Sigma), and anti-FLAG polyclonal antibody (F7425; Sigma) to detect Runx1, GATA2, and SCL, respectively. F7425 antibody was used to exclude the overlap of SCL and nonspecific band by M2 antibody.
CFC assay
The nonadherent or semiadherent cells that emerged from wild-type and Notch1-null P-Sp regions were used for colony-forming–cell (CFC) assays. Cells (6 × 104) were plated into MethoCult GF M3434 medium (StemCell Technologies, Vancouver, BC, Canada) and cultured in a 5% CO2 incubator at 37°C. Colony types were determined at day 7 by morphologic appearance and by Wright-Giemsa staining of each colony. Images were taken with a Nikon Eclipse TE2000-U.
Flow cytometric analysis
Flow cytometric analysis was performed with a BD LSRII (BD Biosciences, San Jose, CA) after addition of 7-amino-actinomycin D (7-AAD) (Via-Probe; BD PharMingen, San Diego, CA) to exclude dead cells. For surface staining, cell suspensions collected from the P-Sp cultures were incubated on ice for 30 minutes in the presence of various mixtures of labeled monoclonal antibodies. The following monoclonal antibodies were purchased from BD PharMingen: phycoerythrin (PE)–conjugated anti–granulocyte 1 (anti–Gr-1), anti–macrophage antigen 1 (anti–Mac-1), anti–stem-cell antigen 1 (anti–Sca-1), anti–Ter-119, allophycocyanin (APC)–conjugated anti-CD45, anti–c-Kit, and biotin-conjugated anti-CD34. Biotinylated antibodies were labeled with PE- or APC-conjugated streptavidin.
Immunoprecipitation and Western blotting
COS7 cells were transfected with expression plasmids (pME-HA-Runx1 and p3xFLAG-myc-CMV-25-Hes1) using the FuGENE6 according to the manufacturer's instruction. The cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) for 48 hours after transfection and were lysed in radioimmunoprecipitation assay (RIPA) buffer.14 These cell lysates were precleared with protein G–sepharose (Amersham Bioscience, Little Chalfont, United Kingdom) and mixed with anti-FLAG antibody (M2; Sigma) or anti-HA antibody (HA.11; Covance Research Products, Berkeley, CA) for 2 hours. The antibody-associated proteins were then recovered on protein G–sepharose beads. The beads were washed 4 times with the RIPA buffer. Whole-cell lysates containing 100 μg of proteins and immunoprecipitates were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon; Millipore, Bedford, MA). The membranes were blocked with 5% skim milk treated with either peroxidase-conjugated anti-FLAG monoclonal antibody (M2; Sigma) or peroxidase-conjugated anti-HA monoclonal antibody (12CA5; Roche Applied Science). The blots were visualized using the enhanced chemiluminescence (ECL) system (Amersham Bioscience).
Transcriptional response assays
Luciferase assays were performed as described previously18 with minor modifications. Briefly, HeLa cells were transfected with 300 ng of reporter (pM-CSF-R-luc),19 and expression plasmids (combinations of 200 ng of pME18S-HA-Runx1 and 160 ng of pME18S-PEBP2β and 60, 200, or 600 ng of pME18S-FLAG-Hes1 or control) using FuGENE6 according to the manufacturer's instructions. As a control of transfection efficiency, a plasmid expressing β-galactosidase was cotransfected. The cells were harvested 48 hours after transfection and assayed for luciferase activity. The data were normalized to β-galactosidase activity.
Quantitative PCR analysis
NIH3T3 cells were infected with NICD or mock retrovirus. The cells were cultured in DMEM medium supplemented with 10% FCS for 48 hours after infection and were selected by the expression of GFP with the FACSAria (BD Biosciences). Total cellular RNA was extracted with RNeasy (QIA-GEN, Hilden, Germany) and converted into cDNAs by reverse transcriptase (Superscript III; Invitrogen, Carisbad, CA). Real-time polymerase chain reaction (PCR) was performed using TaqMan Gene Expression Assays Mm00486762_m1 (Applied Biosystems, Foster City, CA) with the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. Amplification of 18S ribosomal RNA cDNA was used as the endogenous normalization standard.
Results
Retroviral expression of Runx1 rescues hematopoietic defects of Notch1-null P-Sp regions
It has been reported that expression of Runx1 or its homolog, Lozenge, is up-regulated by positive Notch signaling in zebrafish and Drosophila systems, respectively.10,11 We thus first evaluated whether Notch activation results in up-regulation of Runx1 also in the mammalian system. When NIH3T3 cells were transiently transfected with Notch1 intracellular domain (NICD), which represents the constitutive active form of Notch1, the mRNA level of Runx1 increased (Table 1).
Notch activation up-regulates the expression of Runx1
. | RAU . | Mean . | Notch-mock . |
|---|---|---|---|
| Experiment 1 | 2.78* | ||
| NIH3T3-Mock | 0.112634; 0.077514; 0.093663 | 0.094604 | |
| NIH3T3-Notch | 0.263982; 0.241864; 0.282636 | 0.262827 | |
| Experiment 2 | 4.12* | ||
| NIH3T3-Mock | 0.038500; 0.045755; 0.044123 | 0.042792 | |
| NIH3T3-Notch | 0.186016; 0.148638; 0.194443 | 0.176366 |
. | RAU . | Mean . | Notch-mock . |
|---|---|---|---|
| Experiment 1 | 2.78* | ||
| NIH3T3-Mock | 0.112634; 0.077514; 0.093663 | 0.094604 | |
| NIH3T3-Notch | 0.263982; 0.241864; 0.282636 | 0.262827 | |
| Experiment 2 | 4.12* | ||
| NIH3T3-Mock | 0.038500; 0.045755; 0.044123 | 0.042792 | |
| NIH3T3-Notch | 0.186016; 0.148638; 0.194443 | 0.176366 |
Data are from 2 independent experiments in triplicate. RAU, relative arbitrary units; Notch-mock, the ratio of RAU by constitutive active Notch 1 infection and RAU by mock infection.
P < .01 (2-tailed, unequal variance t test).
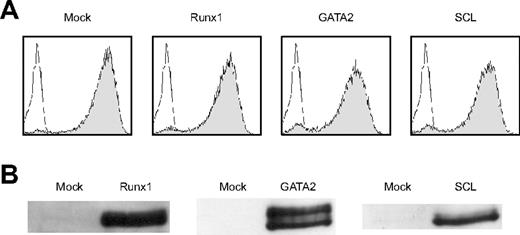
We then examined whether forced expression of Runx1 could rescue the hematopoietic defect of Notch1-null mice. Wild-type P-Sp cells gave rise to round-shaped nonadherent cells when overlayed on the OP9 stromal cells. Flow cytometric analysis of these cells revealed that they were viable (7-AAD negative) CD45-positive cells (top panels in Figure 1A), representing hematopoietic cells. No such cells were generated from Notch1-null P-Sp cells and only background OP9 cells were observed (bottom panels in Figure 1A).3 We retrovirally infected Notch1-null P-Sp cells that were seeded on the OP9 layer with Runx1, SCL, or GATA2, and assessed whether Notch1-null P-Sp cells could generate hematopoietic cells. Titers of the retroviruses containing Runx1, SCL, and GATA2 were similar to each other as evaluated by infecting NIH3T3 cells with these viruses (Figure 2A). Expression of individual proteins was confirmed by a Western blot analysis (Figure 2B). Mock, SCL, and GATA2 transduction did not generate round-shaped nonadherent cells morphologically or viable CD45-positive cells detectable by flow cytometric analysis. In contrast, Runx1-transduced P-Sp cells gave rise to round-shaped nonadherent cells. These cells were shown to be viable CD45-positive cells by flow cytometric analysis (Figure 1B). This pattern was identical to the positive control (Notch1+/+ P-Sp cells; top panels in Figure 1A).
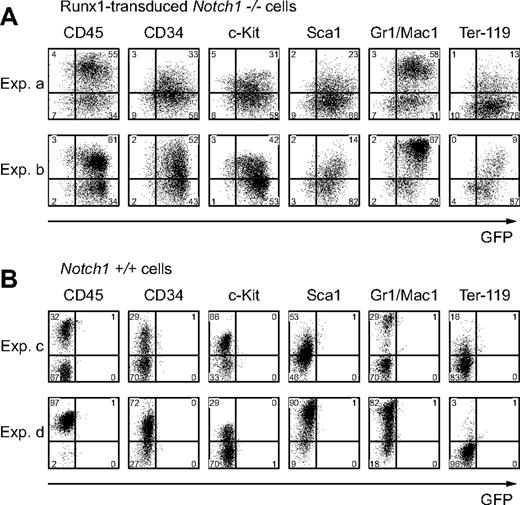
To confirm that the cells developed from Runx1-infected Notch1-null P-Sp cells (hereafter referred to as Runx1-rescued cells) retain the features of hematopoietic cells, we evaluated these cells for surface markers and CFC activities. The flow cytometric analysis of the Runx1-rescued cells at day 12 revealed that they express hematopoietic cell-surface markers such as a panleukocyte marker (CD45), stem-cell markers (c-Kit, CD34, and Sca1), myeloid-cell markers (Gr-1 or Mac-1), and an erythroid-cell marker (Ter-119) (Figure 3A). Their expression profiles were reminiscent of those of hematopoietic cells generated from the wild-type P-Sp cells (Figure 3B). The P-Sp culture system faithfully reproduced the generation of hematopoietic cells, and there were no consistent differences between Runx1-rescued and wild-type P-Sp–derived cells in the surface-marker expression levels, although we observed variable minor differences in individual experiments partly because of the variation in the time required for hematopoietic development (Figure 3A-B).
Retroviral expression of Runx1 rescues hematopoietic defect of Notch1-null P-Sp region. (A) P-Sp cells from wild-type (Notch1+/+) and Notch1-null (Notch1–/–) embryos at E9.5 were cultured for 5 days on OP9 cells. (B) P-Sp cells from Notch1-null embryos at E9.5 were infected with mock retrovirus or retrovirus containing Runx1, SCL, or GATA2, and cultured for 12 days on OP9 cells. Microscopic representation (left column; original magnification, × 100). Only cocultured OP9 cells are shown if hematopoietic cells are not produced. Flow cytometric analyses (center and right columns) of cells generated in the culture. Percentages of cells gated (center columns) and cells in each quadrant (right columns) are indicated.
Retroviral expression of Runx1 rescues hematopoietic defect of Notch1-null P-Sp region. (A) P-Sp cells from wild-type (Notch1+/+) and Notch1-null (Notch1–/–) embryos at E9.5 were cultured for 5 days on OP9 cells. (B) P-Sp cells from Notch1-null embryos at E9.5 were infected with mock retrovirus or retrovirus containing Runx1, SCL, or GATA2, and cultured for 12 days on OP9 cells. Microscopic representation (left column; original magnification, × 100). Only cocultured OP9 cells are shown if hematopoietic cells are not produced. Flow cytometric analyses (center and right columns) of cells generated in the culture. Percentages of cells gated (center columns) and cells in each quadrant (right columns) are indicated.
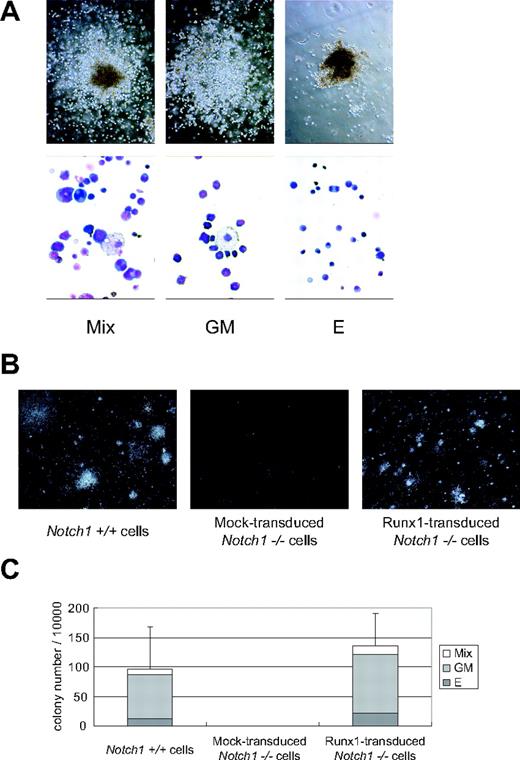
When the Runx1-rescued cells were seeded into semisolid medium at day 12 and cultured for an additional 7 days, they generated mixed, granulocyte/macrophage, and erythroid colonies containing enucleated erythrocytes (Figure 4A) at a frequency comparable to that of wild-type P-Sp–derived cells (Figure 4B-C). There were no statistical differences in the numbers of total (P = .11) and individual (erythroid, P = .20; granulocyte/macrophage, P = .11; mixed, P = .07) colonies generated from Notch1+/+ and Runx1-transduced Notch1–/– P-Sp–derived cells. These observations indicate that the hematopoietic characteristics of Runx1-rescued cells were similar to those of wild-type P-Sp–derived cells.
Retroviruses properly create Runx1, GATA2, and SCL proteins. (A) The efficiency of retrovirus-mediated gene transfer of Runx1, GATA2, or SCL was estimated by infecting NIH3T3 cells. Retrovirus-infected cells were evaluated by the expression of GFP (shaded histograms). Uninfected NIH3T3 cells are also shown as a control (open histograms). (B) Expression of individual proteins was confirmed by a Western blot analysis.
Retroviruses properly create Runx1, GATA2, and SCL proteins. (A) The efficiency of retrovirus-mediated gene transfer of Runx1, GATA2, or SCL was estimated by infecting NIH3T3 cells. Retrovirus-infected cells were evaluated by the expression of GFP (shaded histograms). Uninfected NIH3T3 cells are also shown as a control (open histograms). (B) Expression of individual proteins was confirmed by a Western blot analysis.
Runx1-rescued cells express hematopoietic surface markers. Expression of hematopoietic surface markers of cultured cells at day 12 from Runx1-transduced Notch1-null (Notch1–/–) embryos (A) or wild-type (Notch1+/+) embryos (B) was evaluated by flow cytometric analyses. GFP intensity (marking retrovirus-transduced cells) is plotted on the x-axis and intensity of counterstaining of hematopoietic surface markers is plotted on the y-axis. The results show representative results of independent replicates from 5 experiments. Percentages of cells in each quadrant are indicated.
Runx1-rescued cells express hematopoietic surface markers. Expression of hematopoietic surface markers of cultured cells at day 12 from Runx1-transduced Notch1-null (Notch1–/–) embryos (A) or wild-type (Notch1+/+) embryos (B) was evaluated by flow cytometric analyses. GFP intensity (marking retrovirus-transduced cells) is plotted on the x-axis and intensity of counterstaining of hematopoietic surface markers is plotted on the y-axis. The results show representative results of independent replicates from 5 experiments. Percentages of cells in each quadrant are indicated.
Functional implication of Runx1 at the downstream of Notch-RBP-Jκ pathway
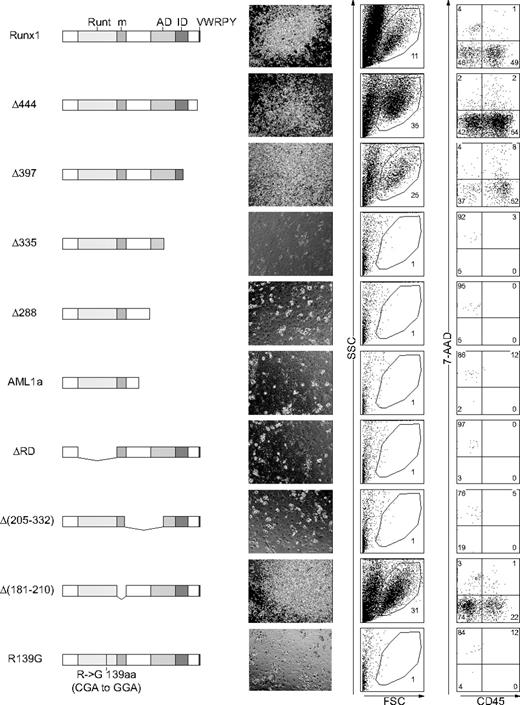
Runx1 has several distinct domains with defined biochemical functions. The Runt domain mediates both binding to DNA and dimerization with a partner protein, CBFβ/PEBP2β, whereas the transactivation domain interacts with transcriptional coactivators. Inhibitory domain counteracts the effect of the transactivation domain. The VWRPY motif located near the C-terminus mediates the interaction with a corepressor, TLE. A domain that interacts with mSin3A corepressor is also identified.9 To assess whether Runx1 functions as an activator or a repressor20 to restore the hematopoietic defect of Notch1-null embryo, we examined a series of Runx1 mutants (Figure 5)14 for hematopoietic rescue.
Infection of retroviruses containing wild-type and several mutants, Δ444, Δ397, and Δ205-332 of Runx1 (Figure 5) resulted in the rescue of the Notch1-null phenotype, giving the same pattern with the culture of wild-type P-Sp cells (Figure 1A, top panels). In contrast, other mutants, Δ335, Δ288, AML1a, ΔRD, Δ205-332, and R139G (Figure 5) could not rescue the Notch1-null phenotype, giving the same pattern with the negative control (Figure 1A, bottom panels). Therefore, wild-type of Runx1 and the mutants that lack the VWRPY domain (Δ444, Δ397) or the mSin3A-binding region (Δ181-210) could restore the production of hematopoietic cells in the Notch1-null P-Sp culture, whereas those mutants that lack transactivation domain (Δ335, Δ288, AML1a, and Δ205-332) or Runt domain (ΔRD) could not rescue hematopoiesis from the Notch1-null P-Sp cells. Since changes in the tertiary structure of the protein could influence the function independent of the role of each domain, we also examined R139G, a mutant isolated from a patient with myelodysplastic syndrome (MDS) that harbors a point mutation causing substitution of Arg139 in the Runt domain with Gly. The DNA-binding ability is severely impaired in R139G, although the ability to heterodimerize with CBFβ/PEBP2β is spared.21 This mutant could not restore hematopoiesis. These results suggest that, in the presence of an intact Runt domain, the transcriptional activating function is necessary and sufficient for Runx1 to rescue the hematopoietic defect of Notch1-null mice in the P-Sp culture system, while the transcriptional repressing function is dispensable.
Notch signaling also regulates transactivating function of Runx1
Hes1 is known to be a canonical Notch-RBP-Jκ target gene in mammals. It is also evident, however, by a number of studies that Hes1 mediates a part of, but not the whole, Notch-RBP-Jκ signaling.22 In adult hematopoiesis, Hes1 maintains HSCs in vitro and expands them in vivo when retrovirally introduced to a highly HSC-enriched population.23 Because Hes1 is expressed in the hematopoietic clusters budding from the dorsal aorta,8 this transcription factor is a candidate as a physiologic target of the Notch-RBP-Jκ pathway in the embryonic hematopoietic development. Hes1 has also been known to mediate cross-talk between Notch and other signaling pathways such as Janus-activating kinase/signal transducer and activator of transcription (JAK/STAT), Wnt, and Ras/mitogen-activated protein kinase (MAPK) pathways.24-26 Furthermore, the transactivating function of Runx2, another Runx family member, is modified by Hes proteins and their relatives Hey proteins. When overexpressed, Hes1 potentiates Runx2-mediated transactivation in the transfected cells,27 while Hey represses Runx2-mediated transactivation.28,29
Runx1-rescued cells generate hematopoietic colonies. Colony formation of the Runx1-rescued cells from Notch1-null embryos. The rescued cells were harvested at day 12 and plated into MethoCult GF M3434 medium. (A) Representative hematopoietic colonies at day 7 are shown. Mix indicates mixed colony; GM, granulocyte/macrophage colony; and E, erythroid colony. Morphology of the colonies (top panels); original magnification, × 100. Wright-Giemsa–stained cytospin preparation of corresponding cell populations (bottom panels); original magnification, × 600. (B) Photographs of representative colonies. Original magnification, × 3. (C) The total number of colonies and the frequencies of different kinds of colonies. The results show the mean values of 5 independent experiments, each in duplicate, with standard deviations for the total colony numbers. Data were statistically analyzed by 2-tailed, unequal-variance t test.
Runx1-rescued cells generate hematopoietic colonies. Colony formation of the Runx1-rescued cells from Notch1-null embryos. The rescued cells were harvested at day 12 and plated into MethoCult GF M3434 medium. (A) Representative hematopoietic colonies at day 7 are shown. Mix indicates mixed colony; GM, granulocyte/macrophage colony; and E, erythroid colony. Morphology of the colonies (top panels); original magnification, × 100. Wright-Giemsa–stained cytospin preparation of corresponding cell populations (bottom panels); original magnification, × 600. (B) Photographs of representative colonies. Original magnification, × 3. (C) The total number of colonies and the frequencies of different kinds of colonies. The results show the mean values of 5 independent experiments, each in duplicate, with standard deviations for the total colony numbers. Data were statistically analyzed by 2-tailed, unequal-variance t test.
The transcriptionally active form of Runx1 is required for hematopoietic rescue. P-Sp cells from Notch1-null embryos at E 9.5 were infected with retroviruses containing Runx1 mutants and cultured on OP9 cells for 12 days. Structures of Runx1 mutants are depicted (left column). Runt indicates the Runt domain; m, a binding region for mSin3A; AD, transactivation domain; ID, inhibitory domain; and VWRPY, VWRPY motif. Microscopic representations (center column; original magnification, × 100) and flow cytometric analyses (right 2 columns) of cells produced in the culture. Percentages of cells gated (center columns) and cells in each quadrant (right columns) are indicated.
The transcriptionally active form of Runx1 is required for hematopoietic rescue. P-Sp cells from Notch1-null embryos at E 9.5 were infected with retroviruses containing Runx1 mutants and cultured on OP9 cells for 12 days. Structures of Runx1 mutants are depicted (left column). Runt indicates the Runt domain; m, a binding region for mSin3A; AD, transactivation domain; ID, inhibitory domain; and VWRPY, VWRPY motif. Microscopic representations (center column; original magnification, × 100) and flow cytometric analyses (right 2 columns) of cells produced in the culture. Percentages of cells gated (center columns) and cells in each quadrant (right columns) are indicated.
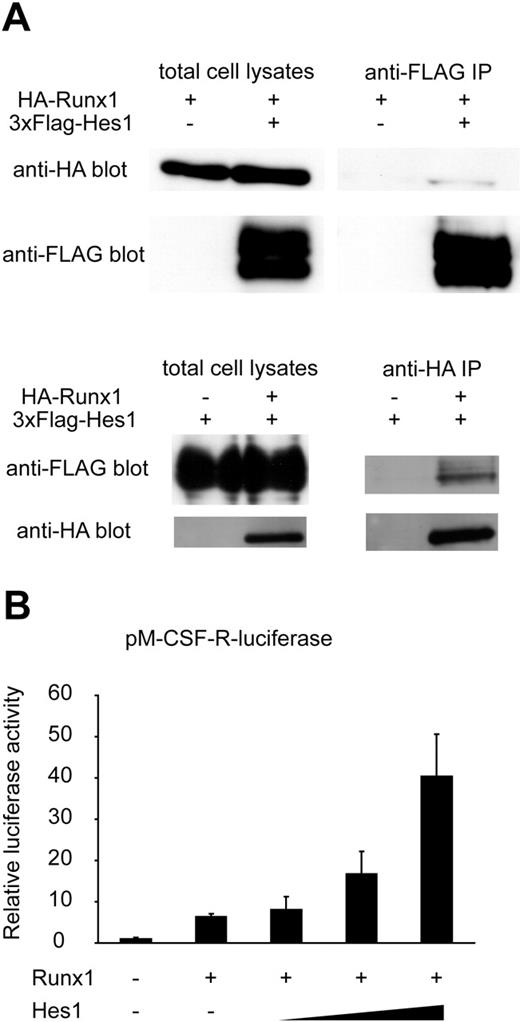
Based on these pieces of information, we assessed whether Hes1 also modulates Runx1-mediated transactivation. Consistent with a previous report in which Hes1 was shown to bind to Runx1 in glutathione S-tranferase (GST) pull-down assays,27 we detected HA-tagged Runx1 protein in the anti-FLAG immunoprecipitant, and reversely, FLAG-tagged Hes1 protein in the anti-HA immunoprecipitant, indicating physical interaction of Hes1 with Runx1 (Figure 6A). Moreover, Hes1 potentiated Runx1-mediated transactivation when expressed in HeLa cells, depending on the expression levels of Hes1 (Figure 6B).
Discussion
In this study, we showed that Runx1 rescues the defective hematopoiesis of Notch1-null mice in the OP9 culture system. The functional relationship between Notch and Runx families during hematopoietic development was first indicated in Drosophila, in which Notch up-regulates the expression of a Runt family gene, Lozenge.10 More recently, it was shown that a zebrafish Notch-signaling mutant mind bomb fails in the specification of definitive HSCs during embryogenesis, and that Runx1 is required for expansion of HSCs in the zebrafish AGM region sufficient to restore the HSC specification in the mind bomb mutant.11 The data shown in the present study strongly indicate that the Notch-Runx pathway is conserved from invertebrates to mammals and that Runx1 locates at a very proximal position in the Notch1 signaling pathway during establishment of definitive hematopoiesis.
GATA2 is also reported to have an important role downstream of Notch signaling in the establishment of definitive hematopoiesis. It was reported that NICD directly binds to the Gata2 promoter and increases its expression level in mouse AGM cells.8 Similarly in Drosophila, Notch up-regulates Serpent and induces emergence of hemocyte progenitors in lymph glands.12 In our retroviral expression system, however, GATA2 could not rescue the hematopoietic defect of Notch1-null P-Sp cells (Figure 1B). It remains unknown whether GATA2 expression in more regulated levels and/or timings could rescue the hematopoietic deficient phenotype of Notch1-knockout P-Sp cells.
We clearly demonstrated that definitive hematopoiesis is rescued by forced expression of Runx1 in the Notch1-null P-Sp cells, but it should be directly shown whether transplantable HSCs are generated from the Notch1-null Runx1-introduced P-Sp cells. Fresh P-Sp cells obtained from wild-type embryos can be engrafted to mouse bone marrow if injected in the preconditioned newborn mice, as described.3,30 It is unknown, however, whether the cultured P-Sp cells are also engraftable with the same method. We were unable to observe engraftment of the cultured P-Sp cells unlike fresh P-Sp cells, when injected to busulfan-pretreated newborn mice (data not shown). Culturing the cells, even for just a short time, is prerequisite for the retroviral gene transfer, which stands as a major technical obstacle to assess the engraftability of the Notch1-null Runx1-introduced P-Sp cells. Transgenic expression of Runx1, under an appropriate promoter, in the Notch1-null background may reveal further that the Notch1-Runx1 pathway represents an essential physiologic channel for the mammalian HSC generation from the P-Sp cells.
Notch signaling regulates transcriptional level of Runx1 and modulates the function of Runx1 protein through the effector protein, Hes1. (A) COS7 cells were transfected with HA-tagged Runx1 and 3xFLAG-tagged Hes1. Whole-cell extracts were immunoprecipitated (IP) with anti-FLAG antibody or anti-HA antibody followed by immunoblotting (blot) using anti-HA antibody or anti-FLAG antibody. (B) Relative luciferase activity in HeLa cells transfected with Runx1 (200 ng) and Runx1-dependent macrophage colony-stimulating factor receptor (pM-CSF-R) luciferase reporter (300 ng) with or without cotransfection of Hes1 (60, 200, or 600 ng). Data are means ± standard errors of duplicate wells in a representative experiment. Reproducible results were obtained in 3 independent experiments.
Notch signaling regulates transcriptional level of Runx1 and modulates the function of Runx1 protein through the effector protein, Hes1. (A) COS7 cells were transfected with HA-tagged Runx1 and 3xFLAG-tagged Hes1. Whole-cell extracts were immunoprecipitated (IP) with anti-FLAG antibody or anti-HA antibody followed by immunoblotting (blot) using anti-HA antibody or anti-FLAG antibody. (B) Relative luciferase activity in HeLa cells transfected with Runx1 (200 ng) and Runx1-dependent macrophage colony-stimulating factor receptor (pM-CSF-R) luciferase reporter (300 ng) with or without cotransfection of Hes1 (60, 200, or 600 ng). Data are means ± standard errors of duplicate wells in a representative experiment. Reproducible results were obtained in 3 independent experiments.
We also showed that Hes1, a known mediator of Notch signaling, cooperatively activates the Runx1-responsive pM-CSF-R luciferase reporter. This observation suggests that the Notch1 pathway modulates expression of Runx1 target genes through multiple mechanisms. There is a possibility that Notch1 directly augments the expression of Runx1 target genes. Although overexpression of Runx1 is sufficient to restore hematopoietic potential in Notch1-null P-Sp cells, both of these mechanisms might cooperatively contribute to HSC generation during normal development.
Prepublished online as Blood First Edition Paper, August 3, 2006; DOI 10.1182/blood-2006-04-019570.
Supported in part by Grant-in-Aid for Scientific Research (KAKENHI no. 17390274) and Grant-in-Aid for Japan Society for the Promotion of Science (JSPS), Fellows from JSPS, Research on Pharmaceutical and Medical Safety, Health and Labour Sciences Research Grants, Ministry of Health, Labour and Welfare of Japan.
The authors declare no competing financial interests.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank M. Ohki for the gift of the human Runx1 cDNA, Y. Ito for the PEBP2β cDNA, D.-E. Zhang for the pM-CSF-R-luc vector, T. Kitamura for the Plat-E packaging cells and the pMYs/IRES-EGFP retrovirus vector, T. Nakano for the OP9 stromal cells, R. Kageyama for the Hes1 cDNA, and Kirin Brewery Pharmaceutical Research Laboratory for the cytokines. We dedicate this paper for the late Prof Hisamaru Hirai, who passed away during the progress of this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal