Abstract
The discovery of marker proteins of human blood (BECs) and lymphatic endothelial cells (LECs) has allowed researchers to isolate these cells. So far, efforts to unravel their transcriptional and functional programs made use of cultured cells only. Hence, it is unknown to which extent previously identified LEC- and BEC-specific programs are representative of the in vivo situation. Here, we define the human BEC- and LEC-specific in vivo transcriptomes by comparative genomewide expression profiling of freshly isolated cutaneous EC subsets and of non-EC skin cells (fibroblasts, mast cells, dendritic cells, epithelial cells). Interestingly, the expression of most of the newly identified EC subset-discriminating genes depends strictly on the in vivo tissue environment as revealed by comparative analyses of freshly isolated and cultured EC subsets. The identified environment-dependent, EC subset-restricted gene expression regulates lineage fidelity, fluid exchange, and MHC class II–dependent antigen presentation. As an example for a BEC-restricted in vivo function, we show that non-activated BECs in situ, but not in vitro, assemble and display MHC class II protein complexes loaded with self-peptides. Thus, our data demonstrate the key importance of using precisely defined native ECs for the global identification of in vivo relevant cell functions.
Introduction
The microvasculature is actively involved in metabolism and immune cell trafficking. Although the supply with oxygen and nutrients as well as the attraction of leukocytes is accomplished by blood vessels, the removal of extracellular fluid from the tissues and the guidance of immune cells to lymph nodes are carried out by lymphatic vessels. At the molecular level, these different functional capabilities are likely encoded by the transcriptional repertoires of blood vessel endothelial cells (BECs) and lymphatic ECs (LECs). Recently, the discovery of the LEC-specific marker proteins podoplanin (PDPN) and LYVE-1 allowed researchers to isolate and propagate BECs and LECs in vitro.1,2 Accordingly, several techniques, including genomewide expression profiling coupled to bioinformatic approaches, have been used to identify the spectrum of genes that are expressed in cultured LECs and BECs.3–6 However, probably as a result of technical limitations, genomewide analyses were not performed on freshly isolated ECs. Thus, it is possible that ECs, in their tissue-resident state, have a transcriptional and, thus, functional repertoire that differs from that of cultured ECs. In fact, the specialized ECs of high endothelial venules rapidly lose EC subset-defining antigen expression and morphology when these cells were placed in culture.7 This supports that active signal exchange between ECs and the local tissue environment can also be of critical importance for the maintenance of differentiation and function of LECs and BECs
To address whether and, if so, to which extent tissue-resident LECs or BECs resemble their cultured counterparts, we established for the first time the complete transcriptomes of freshly isolated cutaneous LECs, BECs, and other major skin cell types by combining fluorescence-activated cell sorting (FACS) and microarray technology. The resulting datasets containing cell type-resolved “in vivo” transcriptomes enabled us to identify common vascular gene expression programs as well as the transcriptional signatures that are selective for LECs or for BECs in vivo. Comparative analyses with cultured LECs and BECs revealed a dramatic loss of BEC- and of LEC-specific signatures during culture. These findings allowed us to define functional programs of LECs and BECs that are regulated by the tissue environment and are not adequately recapitulated in conventional in vitro culture systems.
Materials and methods
Antibodies
FITC-conjugated monoclonal antibodies (mAbs) used were directed against CD45 (BD Biosciences [BD], San Jose, CA), CD11b (Immunotech, Marseille, France), CD54 (Immunotech), CD62E (Ancell, Bayport, MN), HLA-DP (BD), CD74 (C-terminus: M-B741; BD; N-terminus: VICY18 ), CLIP (BD), HLA-DR (TÜ36 and L243; BD; UL-5A19 ), and Duffy antigen (BD). Phycoerythrin (PE)–conjugated Abs were CD19 (BD), CD34 (BD), HLA-DR (L243; BD), CD31 (Serotec, Raleigh, NC), CD1a (BD), CD14 (BD), and CD203c (Immunotech). PE 5.1 (PC5)–conjugated mAbs were CD117 (BD) and CD144 (Immunotech). PE-cyanin 7 (PC7)–conjugated CD14 mAbs and allophycocyanin (APC)–CD1a were from Immunotech. CD146 mAb (541/10B2) was a gift from Dr O. Majdic. Biotinylated goat IgG anti–human CD144 was from R&D Systems (Minneapolis, MN). Secondary reagents included Alexa488-conjugated goat anti–rabbit IgG, PE-conjugated goat anti–mouse IgG, Alexa 633-conjugated streptavidin (Molecular Probes, Eugene, OR), and tetramethylrhodamine-isothiocyanate (TRITC)–labeled goat anti–mouse IgG (Jackson ImmunoResearch, West Grove, PA). Abs to PDPN we raised by immunizing rabbits with the recombinant extracellular domain of PDPN (aa104-205).
Cell isolation
Skin was obtained on informed consent, in accordance with the Declaration of Helsinki, from healthy adults undergoing elective surgery (breast reduction and abdominoplasty). Dermal and epidermal sheets were prepared from 0.8-mm split-thickness skin by an incubation with dispase I (3 U/mL; Roche, Basel, Switzerland) for 60 minutes at 37°C. Microvascular ECs were released from dermal sheets by gentle scraping. Cells were pelleted, resuspended in ice-cold EC growth medium MV (PromoCell, Heidelberg, Germany), and subjected to immunostaining and FACS (FACSAria; BD). Dermal cell suspensions containing fibroblasts, mast cells, and dermal dendritic cells (DDCs) were prepared by dissociating collagenase IV (0.5 U/mL, 90 minutes at 37°C; Worthington, Lakewood, NJ)–treated, minced dermal tissue using a cell dissociation sieve (Sigma-Aldrich, St Louis, MO). Cells were pelleted, resuspended in ice-cold RPMI1640/10% FCS, immunostained, and subjected to FACS. Single cells were released from epidermal sheets using 0.25% trypsin/EDTA (30 minutes at 37°C; Invitrogen, San Diego, CA). Cells were sedimented over Ficoll-Paque (Pharmacia, Uppsala, Sweden), and interphase cells were incubated with CD1a mAbs and immunomagnetically separated using rat anti–mouse IgG1 beads (magnetic cell sorting [MACS]; Miltenyi Biotec, Bergisch Gladbach, Germany). Cell populations enriched for and depleted of CD1a-expressing cells were used to isolate, by FACS, Langerhans cells and keratinocytes. Freshly excised cutaneous melanoma metastases were finely minced and dissociated by a brief incubation with collagenase IV. ECs were enriched by CD144-based MACS and purified by FACS. Procedures were approved by the institutional review board of the Medical University of Vienna.
Culture of microvascular ECs
EC-enriched dermal cells were seeded on fibronectin (Invitrogen)–coated dishes and cultured in supplemented EC growth medium MV without hydrocortisone. After having reached confluence, ECs were harvested using a brief incubation with dispase I, and contaminating cells were removed by anti-CD31–based MACS. After this purification step (passage 1) or after the indicated passages, ECs were subjected to immunostaining and FACS-based separation into BECs and LECs.
FACS and RNA isolation
After labeling with appropriate fluorochrome-labeled mAbs, dermal cells were sorted into BECs (VE-cadherin+CD34highPDPN−, n = 5), LECs (VE-cadherin+CD34lowPDPN+, n = 4), fibroblasts (VE-cadherin− CD34highPDPN−, n = 1), mast cells (CD45+CD203c+CD117+, n = 1), CD14+ DDCs (CD11b+CD14+CD1a−, n = 3), and CD1a+ DDCs (CD11b+CD14−CD1a+, n = 3). Cultured BECs (n = 4) and LECs (n = 4) were sorted as VE-cadherin+PDPN− and VE-cadherin+PDPN+ cells. CD11b−CD1a+ Langerhans cells and CD11b−CD1a− keratinocytes were sorted from epidermal cell suspensions. The purity of all cell populations used was at least 98%. Sorted cells were pelleted and lysed in Trizol (Invitrogen), and RNA was isolated according to the manufacturer's recommendations.
RNA amplification and Affymetrix GeneChip hybridization
Total RNA was subjected to 2 rounds of linear amplification as described.10,11 Biotin-labeled ribonucleotides were incorporated using the ENZO Bio-Array High-Yield RNA Transcript Labeling Kit (Affymetrix, Santa Clara, CA) during the second round of in vitro transcription. Fragmented cRNA (10 μg) was hybridized to Human Genome U133 Plus 2.0 Array (Affymetrix).
Data normalization and bioinformatic analyses
Microarray data were normalized using the Robust Multi-Array Analysis as implemented in Bioconductor.12,13 All analyses were performed with log2-transformed data. The volcano plot analysis was used to provide a summary of test statistics for differential expression of genes. BEC and LEC samples were compared with the remaining non-EC cell types either together or alone. Hypothesis tests were performed using a modified t statistics with an empirical Bayes approach as implemented in Bioconductor LIMMA package.14 Unsupervised hierarchical clustering was performed using the Unweighted Pair Group Method with Arithmetic Mean method after sorting all probe set results by their coefficient of variation using the Pearson correlation coefficient distance measure. For unsupervised network analyses, the Ingenuity Pathway Analysis System (Ingenuity Systems, Redwood City, CA) was used.
Quantitative PCR
Quantitative polymerase chain reaction (qPCR) was performed as described previously11 using the Taqman real-time reverse transcriptase (RT)–PCR assay and commercial primers and probe sets (Applied Biosystems, Foster City, CA). Briefly, cDNA was synthesized from total RNA of freshly isolated BECs and LECs using Superscript III (Invitrogen). EC-derived cDNAs or serial dilutions of a reference cDNA pool were used as templates for PCR reactions containing primers for target genes or an internal control (B2M). Each reaction was performed in duplicate using a MX 4000 thermal cycler (Stratagene, La Jolla, CA). Conditions included polymerase activation at 95°C for 10 minutes and 40 PCR cycles at 95°C for 15 seconds and 60°C for 60 seconds. Target gene expression was calculated by plotting their Ct values onto the standard curve obtained with the reference cDNA pool and data normalization for B2M expression.
Flow cytometry analyses
Freshly prepared and confluent, cultured ECs were incubated with fluorochrome-labeled anti–VE-cadherin and anti-PDPN Abs to discriminate BECs and LECs. Cells were further incubated with the indicated fluorochrome-labeled mAbs or isotype control mAbs. Nonspecific Ab binding was blocked with 5% rat serum (Sigma). For the detection of intracellular antigens, cells were fixed and permeabilized before the immunostaining procedure. Cellular fluorescence was analyzed on a FACScan or FACSAria flow cytometer.
Immunofluorescence microscopy
Cryostat sections from fresh normal human skin (n = 3) or cutaneous melanoma metastases (n = 5) were mounted on slides and fixed for 10 minutes in acetone at −20°C. Sections were incubated for 30 minutes in 1% BSA/PBS before simultaneous exposure to goat anti–CD144-biotin, rabbit anti-PDPN, and the indicated mouse mAbs. Binding of primary Abs was revealed by an incubation with Alexa 633–conjugated streptavidin, Alexa 488–conjugated goat anti–rabbit IgG, and TRITC-labeled goat anti–mouse IgG. Samples were analyzed by confocal laser scanning microscopy. The microscopy system included an inverted Axiovert 200M microscope (Zeiss, Oberkochen, Germany) equipped with the laser scanning module LSM510 (Zeiss). For all image acquisitions, we used a 40×/1.3 NA oil Plan-Neofluar objective (Zeiss) and the LSM 510 image acquisition software version 3.0-Sp2 (Zeiss). Fluoprep (BioMerieux, Marcy l'Etoile, France) was used as imaging medium.
Results
Genomewide expression profiling of freshly isolated human skin cell types
To analyze freshly isolated EC subsets, we established optimized protocols for tissue (skin) dissociation, cell purification, and extraction as well as linear amplification of total RNAs for genomewide expression profiling. Accordingly, dermal cells were dissociated from human split-thickness skin of healthy donors, and ECs were identified by their homogeneous expression of VE-cadherin (Figure 1A). VE-cadherin+ cells were subgrouped and sorted into BECs (PDPN−CD34high) and LECs (PDPNhighCD34intermediate)1 (Figure 1A). Typically, 1 to 3 × 105 LECs and BECs were recovered at a purity of greater than 98%, and 100 ng RNA/1 × 105 cells was obtained. After linear amplification, cRNAs were hybridized to Affymetrix HG U133 Plus 2.0 microarrays. To complete a unique transcriptome data bank of essential primary human skin cell types, fibroblasts (FBs), mast cells (MCs), CD1a+ and CD14+ dermal dendritic cells (DDCs), epidermal Langerhans cells (LCs), and keratinocytes (KCs) were sorted and subjected to expression profiling. The purity of cell samples and the fidelity of hybridization were assessed by analysis of lineage marker gene expression (Figure 1B). LECs but not BECs expressed PROX-1.15 whereas both cell types expressed similar amounts of VE-cadherin. FBs, MCs, and KCs displayed exclusive expression of collagen I, tryptase, and cytokeratin 14, respectively. CD14+ DDCs, CD1a+ DDCs, and epidermal LCs were clearly discriminated by the expression of CD14, CD1b, and the LC-specific marker langerin.16 Thus, our strategy provided high-quality datasets containing the genomewide transcriptomes of important human skin cell types in their fresh, noncultured state.
Purification and GeneChip analysis of fresh BECs, LECs, and other major skin cell types. (A) Identification and isolation of fresh BECs and LECs by triple immunolabeling and FACS. Microvascular ECs in freshly prepared dermal cell suspensions were identified by their homogeneous VE-cadherin expression (y-axis, left). The sort gate was set to actively exclude VE-cadherin− non-ECs and VE-cadherin+ dead ECs that are characterized by their low forward scatter (FSC) signals (x-axis). Within the vital EC population, nonoverlapping subpopulations of BECs (red dots) and LECs (green dots) were resolved by anti–podoplanin-FITC (x-axis) anti–CD34-PE immunolabeling (y-axis, right). BECs and LECs were purified to at least 98% purity by FACS. (B) Purity assessment of isolated BECs, LECs, and other major skin cell types by analysis of marker gene expression. BECs and LECs as well as fibroblasts (FBs), mast cells (MCs), CD14+ dermal dendritic cells (DDCs), CD1a+ DDCs, epidermal Langerhans cells (LCs), and keratinocytes (KCs), isolated according to the criteria described in “Materials and methods” by FACS, were subjected to RNA isolation. After 2 rounds of linear amplification, cRNAs were hybridized to Affymetrix HG-U133 plus 2.0 GeneChips. Hybridization intensities with probe sets corresponding to cell lineage-associated marker genes (VE-cadherin, PROX-1, collagen 1 [COL1], tryptase, CD14, CD1b, langerin, keratin 14) are given for all cell types (x-axis). If more than one sample was available from a defined cell type, mean values and standard deviations (SDs) are shown.
Purification and GeneChip analysis of fresh BECs, LECs, and other major skin cell types. (A) Identification and isolation of fresh BECs and LECs by triple immunolabeling and FACS. Microvascular ECs in freshly prepared dermal cell suspensions were identified by their homogeneous VE-cadherin expression (y-axis, left). The sort gate was set to actively exclude VE-cadherin− non-ECs and VE-cadherin+ dead ECs that are characterized by their low forward scatter (FSC) signals (x-axis). Within the vital EC population, nonoverlapping subpopulations of BECs (red dots) and LECs (green dots) were resolved by anti–podoplanin-FITC (x-axis) anti–CD34-PE immunolabeling (y-axis, right). BECs and LECs were purified to at least 98% purity by FACS. (B) Purity assessment of isolated BECs, LECs, and other major skin cell types by analysis of marker gene expression. BECs and LECs as well as fibroblasts (FBs), mast cells (MCs), CD14+ dermal dendritic cells (DDCs), CD1a+ DDCs, epidermal Langerhans cells (LCs), and keratinocytes (KCs), isolated according to the criteria described in “Materials and methods” by FACS, were subjected to RNA isolation. After 2 rounds of linear amplification, cRNAs were hybridized to Affymetrix HG-U133 plus 2.0 GeneChips. Hybridization intensities with probe sets corresponding to cell lineage-associated marker genes (VE-cadherin, PROX-1, collagen 1 [COL1], tryptase, CD14, CD1b, langerin, keratin 14) are given for all cell types (x-axis). If more than one sample was available from a defined cell type, mean values and standard deviations (SDs) are shown.
Set of genes defining EC identity in vivo
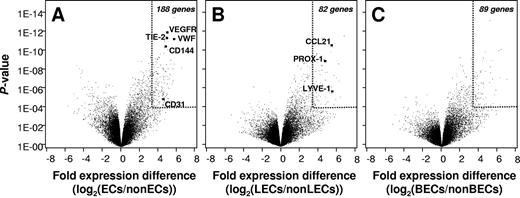
To identify the genes that are expressed in ECs in vivo but not in other skin cell types (referred to as pan-EC genes), we displayed the pool of genes that are expressed by both BECs and LECs against the pooled transcriptomes of all non-EC cell types. The comparison of primary ECs and pooled primary control cells revealed the average difference in expression and the corresponding P value for each expressed gene (Figure 2A). Selective pan-EC genes were defined by an expression in ECs that is greater than 10-fold higher than in control cell types and by a P value of less than 10−4. Using this criterion, 188 genes were found to define EC identity. Twenty-eight pan-EC genes remain when the required expression difference is raised to more than 20 and the P value threshold is lowered to less than 10−7 (genes listed in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Confirming the validity and quality of the results, several identified genes code for proteins with established vascular functions (VE-cadherin, von Willebrand factor, VEGFR2/KDR, angiopoietin receptor 2/TIE-2). Our analysis also revealed additional human pan-EC–defining genes like APOLD1, previously identified as a vessel-specific transcript in rats,17 as well as ECSM2 and Hs.638786, coding for proteins of still unknown functions. Interestingly, the homeobox gene HOXD10 was expressed in both LECs and BECs abundantly but not in other skin cell types. Moreover, several yet poorly investigated GTPase-related molecules (Rho GAP29, IMAP6, and IMAP8), G protein–coupled receptors (EDG1), tyrosine phosphatases (PTPRB), and mucin-type receptors (endomucin, podocalyxin-like/PODXL) define EC identity in vivo.
Identification of in vivo–expressed EC-specific and BEC and LEC sublineage-specific genes. (A) Pan-EC–specific gene expression. Genes that are differentially expressed in ECs (pooled data of BECs and LECs) and non-ECs (pooled data of FBs, MCs, CD14+ DDCs, CD1a+ DDCs, LCs, and KCs) were identified by variance analysis as described in “Materials and methods.” In the Volcano presentation of data, the mean logarithmic expression difference (x-axis) and the statistical confidence of differential expression (P value, y-axis) are shown for each individual identified gene. The identification of several established pan-EC gene products (VEGFR2/KDR, angiopoietin receptor 2/TIE2, von Willebrand factor/VWF, VE-caherin/CD144) in the group of gene products whose expression is highly overrepresented in ECs validates the quality of cell samples and the statistical approach. (B-C) Identification of in vivo expressed, LEC- and BEC-specific genes. Genes that are differentially expressed in LECs and non-LECs (pooled data of BECs and the other skin cell types) (B), or in BECs and in non-BECs (pooled data of LECs and the other skin cell types) (C), were extracted and the mean logarithmic expression difference (x-axis) and P values of differential expression (y-axis) for each identified gene are shown. Arbitrary cut offs for statistically significant EC-specific and EC subtype-specific gene expression (> 10-fold expression difference, P < 10−4) are marked by dotted lines in all volcano plots.
Identification of in vivo–expressed EC-specific and BEC and LEC sublineage-specific genes. (A) Pan-EC–specific gene expression. Genes that are differentially expressed in ECs (pooled data of BECs and LECs) and non-ECs (pooled data of FBs, MCs, CD14+ DDCs, CD1a+ DDCs, LCs, and KCs) were identified by variance analysis as described in “Materials and methods.” In the Volcano presentation of data, the mean logarithmic expression difference (x-axis) and the statistical confidence of differential expression (P value, y-axis) are shown for each individual identified gene. The identification of several established pan-EC gene products (VEGFR2/KDR, angiopoietin receptor 2/TIE2, von Willebrand factor/VWF, VE-caherin/CD144) in the group of gene products whose expression is highly overrepresented in ECs validates the quality of cell samples and the statistical approach. (B-C) Identification of in vivo expressed, LEC- and BEC-specific genes. Genes that are differentially expressed in LECs and non-LECs (pooled data of BECs and the other skin cell types) (B), or in BECs and in non-BECs (pooled data of LECs and the other skin cell types) (C), were extracted and the mean logarithmic expression difference (x-axis) and P values of differential expression (y-axis) for each identified gene are shown. Arbitrary cut offs for statistically significant EC-specific and EC subtype-specific gene expression (> 10-fold expression difference, P < 10−4) are marked by dotted lines in all volcano plots.
Sets of genes defining EC subset identity in vivo
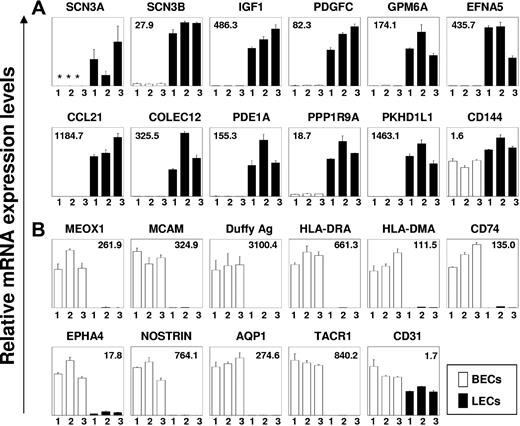
As shown in Figure 2B, 82 genes were found that define LEC identity in vivo (> 10-fold higher expression in LECs than in control cells, P < .001). Twenty-one genes remain when we superimposed the further criterion of greater than 10-fold higher expression in LECs than in BECs and a corresponding P value of less than 10−6 (bold in Table S2). The identified set of gene products validates LEC-specific expression of CC chemokine ligand 21 (CCL21) and PROX-1 in vivo. Three other previously suggested LEC-specific molecules (ie, PDPN, LYVE-1, and FOXC2) were clearly enriched in LECs in vivo but are also dimly expressed in other skin cells (Table S2). Three novel LEC-specific genes or gene regions were identified (KIAA0711, FLJ23728, LOC572558). Interestingly, LECs in vivo selectively display several neuronally expressed molecules, including SLC24A1, a potassium-dependent sodium/calcium exchanger, a voltage-gated sodium channel (SCN3A/SCN3B), neuronal glycoprotein (GP) M6A, neurabin I (protein phosphatase I, regulatory subunit 9A/PPP1R9A), neuralin as well as neurotensin. Besides these newly discovered LEC-specific molecules, some transcripts that were previously identified in cultured LECs fulfilled the stringent criteria of selective in vivo expression (reelin, intestinal trefoil factor 3).5,6 Expression of 11 selected genes that showed LEC-restricted expression in profiling experiments was analyzed in 3 prospectively prepared pairs of BECs and LECs by quantitative qRT-PCR (Figure 3A). Eleven of 11 genes were drastically overexpressed in LECs as compared with BECs with a range of mean expression difference from 18.7 to 1463. Thus, data obtained by qRT-PCR verify unequivocally the expression profiling results of LEC-specific gene expression. Expectedly, the dynamic range of differential gene expression was even higher in qRT-PCR than in the microarray analysis.12
Validation of LEC- and BEC-specific gene expression by quantitative RT-PCR. mRNA expression of LEC- (A) and BEC-specific genes (B), as identified by gene chip analysis, was analyzed in 3 pairs of prospectively prepared samples of LECs and BECs by qRT-PCR. qPCR was performed for the expression of voltage-gated sodium channel type III α (SCN3A) and β chains (SCN3B), insulin-like growth factor 1 (IGF1), platelet-derived growth factor C (PDGFC), glycoprotein M6A (GPM6A), ephrin-A5 (EFNA5), CC chemokine ligand 21 (CCL21, SLC), collectin subfamily member 12 (COLEC12), phosphodiesterase 1A (PDE1A), protein phosphatase 1, regulatory (inhibitor) subunit 9A (PPP1R9A), polycystic kidney and hepatic disease 1-like 1 (PKHD1L1), mesenchyme homeobox 1 (MEOX1), melanoma cell adhesion molecule (MCAM), Duffy blood group antigen (FY), α chain of HLA-DR (HLA-DRA), α chain of HLA-DM (HLA-DMA), MHC class II invariant chain (CD74), EPH receptor A4 (EPHA4), nitric oxide synthase trafficker (NOSTRIN), aquaporin 1 (AQP1), tachykinin receptor 1 (TACR1), VE-cadherin (CD144), and platelet/endothelial cell adhesion molecule (CD31). Data shown are mean mRNA expression values and standard error of means from 2 independent PCR measurements normalized to the expression of β2-microglobulin (B2M). Asterisks indicate samples in which mRNA levels were below the detection limit of RT-PCR. In each diagram, the mean fold difference of mRNA expression of the 3 paired BEC and LEC samples is given.
Validation of LEC- and BEC-specific gene expression by quantitative RT-PCR. mRNA expression of LEC- (A) and BEC-specific genes (B), as identified by gene chip analysis, was analyzed in 3 pairs of prospectively prepared samples of LECs and BECs by qRT-PCR. qPCR was performed for the expression of voltage-gated sodium channel type III α (SCN3A) and β chains (SCN3B), insulin-like growth factor 1 (IGF1), platelet-derived growth factor C (PDGFC), glycoprotein M6A (GPM6A), ephrin-A5 (EFNA5), CC chemokine ligand 21 (CCL21, SLC), collectin subfamily member 12 (COLEC12), phosphodiesterase 1A (PDE1A), protein phosphatase 1, regulatory (inhibitor) subunit 9A (PPP1R9A), polycystic kidney and hepatic disease 1-like 1 (PKHD1L1), mesenchyme homeobox 1 (MEOX1), melanoma cell adhesion molecule (MCAM), Duffy blood group antigen (FY), α chain of HLA-DR (HLA-DRA), α chain of HLA-DM (HLA-DMA), MHC class II invariant chain (CD74), EPH receptor A4 (EPHA4), nitric oxide synthase trafficker (NOSTRIN), aquaporin 1 (AQP1), tachykinin receptor 1 (TACR1), VE-cadherin (CD144), and platelet/endothelial cell adhesion molecule (CD31). Data shown are mean mRNA expression values and standard error of means from 2 independent PCR measurements normalized to the expression of β2-microglobulin (B2M). Asterisks indicate samples in which mRNA levels were below the detection limit of RT-PCR. In each diagram, the mean fold difference of mRNA expression of the 3 paired BEC and LEC samples is given.
Few genes with BEC-specific expression patterns have been described so far. Our analysis reveals that 89 genes define BEC identity in vivo using the criterion of greater than 10-fold higher expression in BECs than in the pooled reference cell group and a P value of less than .0001 (Figure 2C). Twenty-three genes remain when the further criterion of greater than 10-fold higher expression in BECs than in LECs and a corresponding P value of less than 10−6 is superimposed (bold in Table S3). Several of these most explicitly BEC-specific gene products are of importance for blood vessel integrity/cell adhesion (JAM2, melanoma cell adhesion molecule/MCAM/CD146). Newly identified BEC-specific molecules are involved in substrate transport across vessel walls (aquaporin 1, glutamate transporter SLC1A1), or in nitric oxide (nitric oxide synthase trafficker/NOSTRIN) and neurotrophic signaling (neurokinin/substance P receptor/TACR1). Although the homeobox genes PROX1 and FOXC2 are of established importance for LEC differentiation,18,19 no master regulator of BEC-specific gene transcription has been identified so far. We find that the mesenchyme homeobox gene MEOX1 is the superior BEC-defining gene in vivo. Also interesting was that BECs, but not other skin cells, even in their nonactivated in vivo state express transcripts coding for molecules with importance for leukocyte adhesion (E- and P-selectin) and migration (CCL23, Duffy antigen).
Expression of 10 selected genes that were found overexpressed in BECs in the microarray studies was also analyzed in the 3 prospective pairs of BECs and LECs (Figure 3B). By qRT-PCR, 10 of 10 genes were found drastically overexpressed in BECs as compared with LECs with a range of mean expression difference from 17.8 to 3100. As a control, the pan-EC gene products CD144 and CD31 were expressed in BECs and LECs almost equally (mean expression difference 1.6 and 1.7, respectively; Figure 3). Thus, data obtained by qRT-PCR also verify the identification of BEC-specific gene expression by expression profiling.
MHC class II expression is a hallmark feature of the in vivo differentiation program of BECs
In an unbiased approach, lists of genes that are overexpressed in BEC as compared with LECs and vice versa were subjected to bioinformatic data mining to identify functional programs. With BECs, transcripts coding for the full complement of MHC class II (HLA-DR, HLA-DP, HLA-DQ) and MHC class II–related molecules (HLA-DM, invariant chain/CD74) formed the prominent part of the interaction plot (Figure S1). Within the pan-EC gene list, networks were identified that are important for vessel wall formation (TIE-1, TIE-2), the formation of cell junctions (CD31, VE-cadherin), angiogenesis (VEGFR2/KDR), and the establishment of the basement membrane (COLIV).
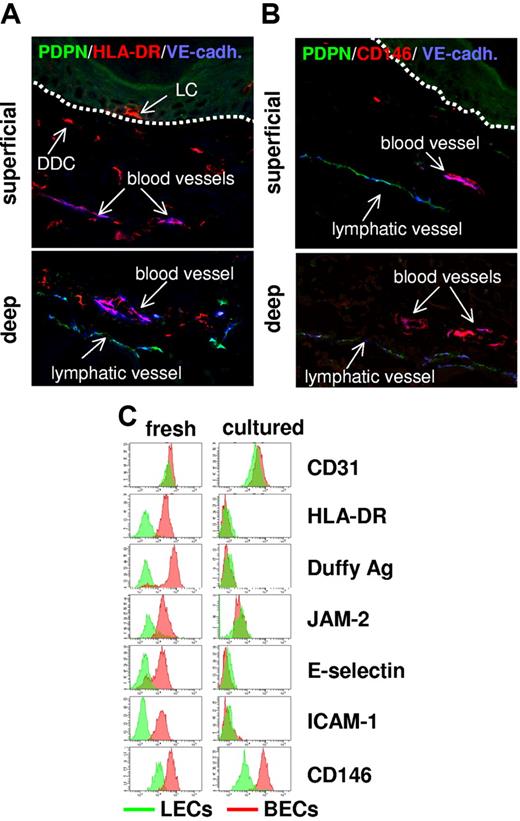
To corroborate the surprising observation of constitutive MHC class II expression by normal BECs at the protein level, we performed confocal microscopy on snap-frozen normal human skin and flow cytometry on freshly isolated BECs and LECs. Substantial MHC class II protein expression was detected in the ECs lining blood vessels of both the superficial and deep vascular plexus of the skin (Figure 4A). Lymphatic vessels in situ were devoid of anti–MHC class II immunoreactivity which is in agreement with the results of our transcriptional analysis. The striking difference in MHC class II protein expression between BECs and LECs was also evident in flow cytometry analysis of freshly isolated EC subsets (Figure 4C).
Expression profiling-defined BEC-specific proteins are expressed in vivo but lost in vitro. (A-B) In normal human skin, BECs but not LECs express MHC class II and melanoma cell adhesion molecule (MCAM)/CD146. Sections (7 μm) of snap-frozen normal human skin were subjected to triple immunolabeling using Abs specific for VE-cadherin (blue), PDPN (green), and MHC class II (red) (A) or CD146 (red) (B) and confocal laser scanning microscopy. Uniform MHC class II (A) and CD146 expression (B) by VE-cadherin+PDPN− blood vessel ECs is shown by purple coloration of these vessels in the overlay exposures. In contrast, VE-cadherin+PDPN+ lymph vessel ECs fail to express MHC class II (A) or CD146 (B). Images shown in the upper and lower panels were taken from areas containing the superficial and the deep vascular plexus of the skin, respectively. Dotted lines in the upper panels denote the epidermodermal junction. MHC class II+VE-cadherin− cells in panel A are epidermal LCs and DDCs. BEC-restricted protein expression in normal human skin was also verified for Duffy antigen, junctional adhesion molecule (JAM2), C1q receptor protein 1 (C1QRp1), and E- and P-selectin (data not shown). (C) Most identified BEC-restricted antigens are expressed by freshly isolated cells but are lost in cell culture. BECs (red histograms) and LECs (green histograms) either freshly isolated (left) or cultured (right) were immunostained with mAbs recognizing the indicated pan-EC (CD31) or BEC-restricted antigens (HLA-DR, Duffy antigen, JAM2, E-selectin, ICAM-1, CD146) and analyzed by flow cytometry. Cultured, nonproliferative BECs and LECs were harvested from confluent cultures at passage 4. Note that, although BEC-restricted antigen expression clearly separated freshly isolated BECs from freshly isolated LECs, this divergent expression was lost in cultured ECs mostly because of a dramatic down-regulation of antigen expression in BECs. The only notable exception is the preferential expression CD146 in BECs that is maintained in culture.
Expression profiling-defined BEC-specific proteins are expressed in vivo but lost in vitro. (A-B) In normal human skin, BECs but not LECs express MHC class II and melanoma cell adhesion molecule (MCAM)/CD146. Sections (7 μm) of snap-frozen normal human skin were subjected to triple immunolabeling using Abs specific for VE-cadherin (blue), PDPN (green), and MHC class II (red) (A) or CD146 (red) (B) and confocal laser scanning microscopy. Uniform MHC class II (A) and CD146 expression (B) by VE-cadherin+PDPN− blood vessel ECs is shown by purple coloration of these vessels in the overlay exposures. In contrast, VE-cadherin+PDPN+ lymph vessel ECs fail to express MHC class II (A) or CD146 (B). Images shown in the upper and lower panels were taken from areas containing the superficial and the deep vascular plexus of the skin, respectively. Dotted lines in the upper panels denote the epidermodermal junction. MHC class II+VE-cadherin− cells in panel A are epidermal LCs and DDCs. BEC-restricted protein expression in normal human skin was also verified for Duffy antigen, junctional adhesion molecule (JAM2), C1q receptor protein 1 (C1QRp1), and E- and P-selectin (data not shown). (C) Most identified BEC-restricted antigens are expressed by freshly isolated cells but are lost in cell culture. BECs (red histograms) and LECs (green histograms) either freshly isolated (left) or cultured (right) were immunostained with mAbs recognizing the indicated pan-EC (CD31) or BEC-restricted antigens (HLA-DR, Duffy antigen, JAM2, E-selectin, ICAM-1, CD146) and analyzed by flow cytometry. Cultured, nonproliferative BECs and LECs were harvested from confluent cultures at passage 4. Note that, although BEC-restricted antigen expression clearly separated freshly isolated BECs from freshly isolated LECs, this divergent expression was lost in cultured ECs mostly because of a dramatic down-regulation of antigen expression in BECs. The only notable exception is the preferential expression CD146 in BECs that is maintained in culture.
MHC class II expression is part of a strictly environmentally regulated BEC differentiation program
Although few older reports noted anti–MHC class II immunoreactivity on some microvessels in normal human skin,20,21 resting ECs in vitro lack MHC class II expression unless stimulated with inflammatory cytokines.22–24 Thus, MHC class II expression may be part of a distinctive BEC differentiation program that is regulated by the tissue environment but lost under in vitro conditions. To identify the pool of genes that are under stringent control of the physiologic tissue environment, we comparatively analyzed gene expression by freshly isolated and in vitro propagated EC subsets.
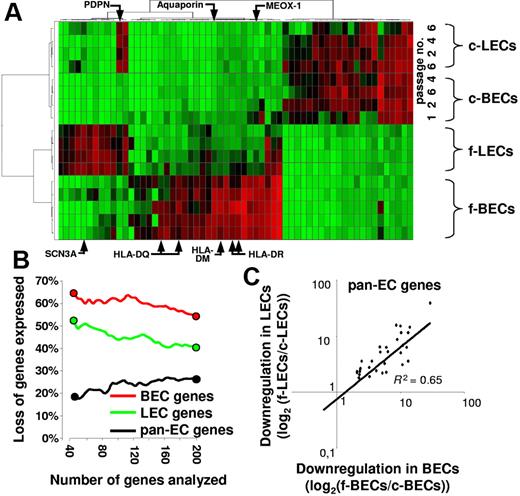
Unsupervised hierarchical clustering of genes expressed by primary and cultured LECs and BECs clearly separated primary ECs from their cultured counterparts (Figure 5A). Intriguingly, the BEC- and the LEC-specific in vivo gene expression programs were largely lost on propagation of the EC subsets in vitro (Figure 5A). The BEC- and LEC-defining gene expression profiles were lost until the earliest possible harvest of the in vitro propagated cells (passage 1; Figure 5A). Only a subset of down-regulated BEC-specific transcripts was expressed at detectable levels until passage 1 (MEOX1, E-selectin, ELOVL family member 7, SPARC-like-1) or passage 2 (COL15A1; Figure 5A). In fact, greater than 65% of the 40 most selectively in vivo–expressed BEC genes were silenced in vitro (Figure 5B). Similarly, greater than 50% of the 40 transcripts that best define LECs in vivo were down-regulated in vitro. Comparatively less pronounced was the loss of pan-EC–defining gene expression (only 20% of the 40 most selective pan-EC transcripts are lost in vitro) with a strictly coordinated loss of expression in BECs and LECs (Figure 5C). Thus, our analysis shows a loss of EC subset identity and a striking assimilation of LEC and BEC transcriptomes in vitro.
BEC- and LEC-defining gene expression is lost in vitro. (A) BEC and LEC transcriptomes are most divergent in vivo but assimilate in vitro. The 60 genes showing the highest coefficient of variation of expression in normalized datasets from freshly isolated (f)–BECs, f-LECs, cultured (c)–BECs, and c-LECs harvested at the indicated passages (passage 1 to 6) were subjected to unsupervised hierarchical clustering. Each column shows the expression of one gene in the various samples analyzed and color ranges indicate the relative expression level (red, high; green, low). Note that 2 distinctive clusters of expressed genes define f-BECs and f-LECs. With few exceptions (eg, PDPN), both gene expression patterns are lost in vitro. (B-C) Preferential loss of in vivo BEC- and LEC-defining gene expression but maintenance of pan-EC gene expression in vitro. Pan-EC–specific genes (black symbols), BEC-specific genes (red symbols), and LEC-specific genes (green symbols) were ranked according to their power to discriminate ECs from non-ECs, BECs from LECs, and LECs from BECs. The percentage of genes that were down-regulated at least 5-fold in the comparison of fresh and cultured ECs was plotted against the number of ranked genes analyzed. Note that a high percentage of the 40 genes that best discriminate BECs or LECs are down-regulated in vitro. The next 160 ranked genes are lost with much lower prevalence. In contrast, few pan-EC genes are down-regulated in vitro. (C) The alteration of expression of down-regulated pan-EC genes is of similar magnitude in cultured BECs and in cultured LECs. Shown is the expression of genes that are at least 5-fold down-regulated on culture of LECs and BECs. Log2-transformed ratios of mean expression values obtained with fresh and cultured EC subsets are shown. The degree of down-regulation of gene expression is similar in LECs (y-axis) and BECs (x-axis); R2 = 0.65.
BEC- and LEC-defining gene expression is lost in vitro. (A) BEC and LEC transcriptomes are most divergent in vivo but assimilate in vitro. The 60 genes showing the highest coefficient of variation of expression in normalized datasets from freshly isolated (f)–BECs, f-LECs, cultured (c)–BECs, and c-LECs harvested at the indicated passages (passage 1 to 6) were subjected to unsupervised hierarchical clustering. Each column shows the expression of one gene in the various samples analyzed and color ranges indicate the relative expression level (red, high; green, low). Note that 2 distinctive clusters of expressed genes define f-BECs and f-LECs. With few exceptions (eg, PDPN), both gene expression patterns are lost in vitro. (B-C) Preferential loss of in vivo BEC- and LEC-defining gene expression but maintenance of pan-EC gene expression in vitro. Pan-EC–specific genes (black symbols), BEC-specific genes (red symbols), and LEC-specific genes (green symbols) were ranked according to their power to discriminate ECs from non-ECs, BECs from LECs, and LECs from BECs. The percentage of genes that were down-regulated at least 5-fold in the comparison of fresh and cultured ECs was plotted against the number of ranked genes analyzed. Note that a high percentage of the 40 genes that best discriminate BECs or LECs are down-regulated in vitro. The next 160 ranked genes are lost with much lower prevalence. In contrast, few pan-EC genes are down-regulated in vitro. (C) The alteration of expression of down-regulated pan-EC genes is of similar magnitude in cultured BECs and in cultured LECs. Shown is the expression of genes that are at least 5-fold down-regulated on culture of LECs and BECs. Log2-transformed ratios of mean expression values obtained with fresh and cultured EC subsets are shown. The degree of down-regulation of gene expression is similar in LECs (y-axis) and BECs (x-axis); R2 = 0.65.
Expression of MHC class II and MHC class II–related proteins was also completely lost on EC culture in vitro (Figures 4C and 5A), contrasting the maintained expression of the pan-EC gene cluster of VEGF-R/VE-cadherin signaling (Table S3; Figure S2). Like MHC class II, other molecules that were identified as part of the prototypic in vivo BEC differentiation program were rapidly lost during culture as well (Duffy antigen, JAM2, E-selectin, ICAM-1, complement subcomponent receptor 1/CD93; Figure 4C). A remarkable exception is the maintained expression of the BEC- and LEC-specific molecules CD146 and PDPN (Figure 4B-C, Figure 5A), qualifying them as reliable markers for the discrimination of LECs and BECs in culture.
Resting microvascular BECs in vivo express cell-surface–bound MHC class II complexes loaded with self-peptides
To characterize the in vivo status of MHC class II molecules on BECs, freshly isolated BECs, LECs, and dermal DCs were gated and analyzed simultaneously for their reactivity with mAbs recognizing MHC class II framework epitopes or reacting with peptide-bound HLA-DRαβ dimers selectively (Figure S3). This analysis revealed that BECs in normal human skin, but not LECs, express peptide-bound HLA-DRαβ uniformly (Figure S3). Thus, nonactivated BECs in normal human skin appear to display a functional pathway of MHC class II processing and presentation.
The repertoire of peptides presented by MHC class II–expressing ECs in vivo is still unknown but may include moieties that are derived from self-antigens. To address this, we used a mAb (UL-5A1) that recognizes a distinctive MHC class II:self-peptide complex containing HLA-DRB1*0101 and a peptide derived from HLA-A2 (HLA-A2103-117).9 Accordingly, this reagent stained B cells of HLA-DRB1*0101+HLA-A2+ donors but not B cells from donors lacking one of these or both alleles (Figure 6A). To see whether ECs express defined self-peptide:MHC class II complexes in vivo, dermal cells from HLA-genotyped, healthy volunteers were subjected to 5-color flow cytometry. As shown in Figure 6B, freshly isolated BECs from DRB1*0101+HLA-A2+ donors displayed substantial levels of these self-peptide:MHC complexes. As a control, no mAb reactivity was seen on BECs from DRB1*0101−HLA-A2+ donors while total peptide:MHC class II complexes were expressed equally in the 2 BEC preparations. Interestingly, BECs expressed comparable levels of self-peptide:MHC II complexes as tissue DCs, whereas the latter displayed higher total amounts of total peptide:MHC class II complexes (Figure 6B). Thus, the relative level of endogenous self-peptide display in MHC class II is higher in BECs than in DCs (Figure 6C). This indicates that BECs in vivo are able and, perhaps even, specialized to present self-antigens efficiently.
BECs constitutively present defined self-antigens in the context of MHC class II. (A) Specific detection of self-peptide:MHC class II complexes on antigen-presenting cells. B cells from HLA-DRB1*0101+HLA-A*0201+ donors (top) but not B cells from HLA-DRB1*0101+HLA-A*0201− donors (bottom) react with mAb UL-5A1 specific for a MHC class II complex composed of HLA-DRB1 and a peptide of HLA-A2 [amino acids 103-117; DR1:A2(aa103-117); x-axis]. Peripheral blood B cells were identified by anti-CD19 immunoreactivity in mononuclear cell preparations from healthy donors (y-axis). (B) Comparable levels of self-peptide-MHC complexes are constitutively displayed by BECs and DCs in normal human skin. Dermal single-cell suspensions from HLA-DRB1*0101+HLA-A*0201+ (left panels) and HLA-DRB1*0101−HLA-A*0201+ donors (right panels) were labeled with mAb UL-5A1-FITC (top) or L243-FITC (bottom) together with anti–VE-cadherin-PC5, anti–PDPN-PE, anti–CD1a-APC, and anti–CD14-PC7. BECs (red histograms) and CD1a+ DDCs (blue histograms) were gated and analyzed for surface expression of DR1:A2(aa103-117) complexes (x-axis, upper panels) and total HLA-DR–peptide complexes (x-axis, lower panels). Gray histograms show the reactivity of BECs and CD1a+ DDCs with FITC-labeled control Abs. (C) Efficient presentation of self-peptide–MHC complexes by BECs. The relation between DR1:A2(aa103-117) and total DR:peptide complex display was calculated by forming the ratio between the respective measured fluorescence intensities for BECs, CD1a+ DDCs, and CD14+ DDCs (y-axis). Data are expressed as mean values (+ SDs) obtained in experiments with dermal cells from 2 unrelated HLA-DRB1*0101+HLA-A*0201+ donors.
BECs constitutively present defined self-antigens in the context of MHC class II. (A) Specific detection of self-peptide:MHC class II complexes on antigen-presenting cells. B cells from HLA-DRB1*0101+HLA-A*0201+ donors (top) but not B cells from HLA-DRB1*0101+HLA-A*0201− donors (bottom) react with mAb UL-5A1 specific for a MHC class II complex composed of HLA-DRB1 and a peptide of HLA-A2 [amino acids 103-117; DR1:A2(aa103-117); x-axis]. Peripheral blood B cells were identified by anti-CD19 immunoreactivity in mononuclear cell preparations from healthy donors (y-axis). (B) Comparable levels of self-peptide-MHC complexes are constitutively displayed by BECs and DCs in normal human skin. Dermal single-cell suspensions from HLA-DRB1*0101+HLA-A*0201+ (left panels) and HLA-DRB1*0101−HLA-A*0201+ donors (right panels) were labeled with mAb UL-5A1-FITC (top) or L243-FITC (bottom) together with anti–VE-cadherin-PC5, anti–PDPN-PE, anti–CD1a-APC, and anti–CD14-PC7. BECs (red histograms) and CD1a+ DDCs (blue histograms) were gated and analyzed for surface expression of DR1:A2(aa103-117) complexes (x-axis, upper panels) and total HLA-DR–peptide complexes (x-axis, lower panels). Gray histograms show the reactivity of BECs and CD1a+ DDCs with FITC-labeled control Abs. (C) Efficient presentation of self-peptide–MHC complexes by BECs. The relation between DR1:A2(aa103-117) and total DR:peptide complex display was calculated by forming the ratio between the respective measured fluorescence intensities for BECs, CD1a+ DDCs, and CD14+ DDCs (y-axis). Data are expressed as mean values (+ SDs) obtained in experiments with dermal cells from 2 unrelated HLA-DRB1*0101+HLA-A*0201+ donors.
ECs of tumor blood vessels, as normal BECs, can express peptide:MHC II complexes along with T-cell activation–inhibiting members of the B7 family of costimulatory molecules
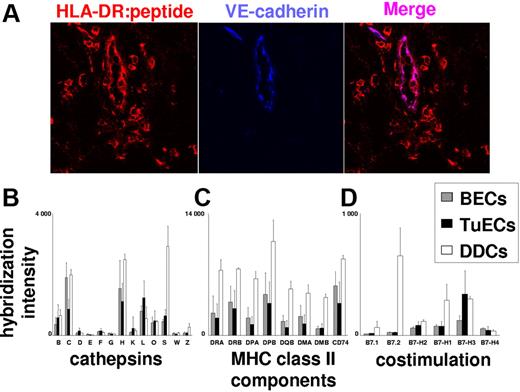
To see whether constitutive MHC class II presentation by BECs is of relevance in human cancer, we also analyzed the transcriptomes of tumor-derived BECs (TuECs) for the expression of MHC class II and costimulatory molecules. TuECs were purified from melanoma skin metastases, and RNA was subjected to expression profiling. As shown in Figure 7C, TuECs and normal BECs expressed comparable levels of transcripts encoding MHC class II, HLA-DM, and invariant chain. At the protein level, TuECs expressed peptide:MHC class II complexes at levels comparable to VE-cadherin− melanoma-infiltrating macrophages (Figure 7A). The functionality of MHC class II processing and presentation in TuECs is further supported by the detection of transcripts encoding lysosomal proteases that are essential for antigen and Ii processing (cathepsins B, H, L, and S)25 at levels comparable to those in normal ECs. As shown in Figure 7D, normal BECs and TuECs, unlike DDCs, did not express transcripts encoding the stimulatory B7 family members B7.1 and B7.2. In contrast, BECs and TuECs expressed transcripts for the T-cell function–inhibitory/–modulatory B7 molecules B7-H1, B7-H2, B7-H3, and B7-H4. Among those, the negative costimulatory molecule B7-H3 was most prominently expressed in TuECs. In aggregate, these data show that MHC class II biosynthesis, processing, and peptide display are part of a strictly environmentally regulated BEC program that is operative in normal as well as in neoplastic tissues.
ECs in metastatic melanoma, as normal BECs, express the MHC class II:peptide complexes along with T-cell activation–inhibiting costimulatory molecules. (A) Detection of MHC class II:peptide complexes on tumor ECs. Immunofluorescence double labeling using Abs to MHC class II:peptide complexes (red) and VE-cadherin (blue) were performed on 7-μm snap-frozen sections from freshly excised cutaneous melanoma metastases. Expression of MHC class II:peptide complexes by VE-cadherin+ tumor vessel ECs is shown by purple coloration of these vessels in the overlay exposure. Results are representative for experiments with metastases from 5 donors. (A-C) Normal and tumor ECs express similar amounts of mRNAs coding for proteases relevant for antigen processing and peptide-loading in MHC class II (A), MHC class II and MHC class II-associated molecules (B), as well as T-cell activation–inhibiting costimulatory molecules (C). Normalized expression data were derived from purified TuECs (n = 8), normal BECs (n = 5), and CD1a+ DDCs (n = 3) as obtained by GeneChip analysis. Mean mRNA expression values and standard errors of means are shown for cathepsins B, C, D, E, F, G, H, K, L, O, S, W, and Z (B); HLA-DR, HLA-DP, HLA-DQ, HLA-DM, and invariant chain/CD74 (C); and the costimulatory molecules B7.1, B7.2, B7-H1, B7-H2, B7-H3, and B7-H4 (D).
ECs in metastatic melanoma, as normal BECs, express the MHC class II:peptide complexes along with T-cell activation–inhibiting costimulatory molecules. (A) Detection of MHC class II:peptide complexes on tumor ECs. Immunofluorescence double labeling using Abs to MHC class II:peptide complexes (red) and VE-cadherin (blue) were performed on 7-μm snap-frozen sections from freshly excised cutaneous melanoma metastases. Expression of MHC class II:peptide complexes by VE-cadherin+ tumor vessel ECs is shown by purple coloration of these vessels in the overlay exposure. Results are representative for experiments with metastases from 5 donors. (A-C) Normal and tumor ECs express similar amounts of mRNAs coding for proteases relevant for antigen processing and peptide-loading in MHC class II (A), MHC class II and MHC class II-associated molecules (B), as well as T-cell activation–inhibiting costimulatory molecules (C). Normalized expression data were derived from purified TuECs (n = 8), normal BECs (n = 5), and CD1a+ DDCs (n = 3) as obtained by GeneChip analysis. Mean mRNA expression values and standard errors of means are shown for cathepsins B, C, D, E, F, G, H, K, L, O, S, W, and Z (B); HLA-DR, HLA-DP, HLA-DQ, HLA-DM, and invariant chain/CD74 (C); and the costimulatory molecules B7.1, B7.2, B7-H1, B7-H2, B7-H3, and B7-H4 (D).
Discussion
In this study, we established the transcriptomes of freshly ex vivo isolated human cutaneous BECs and LECs. This became possible by the combined use of high-capacity multiparametric cell sorting and efficient linear RNA amplification for reliable and reproducible genomewide expression profiling. Several other skin cell types were analyzed in parallel. This allowed the establishment of the “private” in vivo transcriptomes of BECs and LECs that do not overlap with transcriptomes of other skin cells. The obtained “in vivo” BEC and LEC transcriptomes revealed striking differences to transcriptomes that were previously obtained using cultured EC subsets obtained after 4 or 5 passages in vitro.5,6 This difference is explained by our observation that the transcriptional programs which define BECs and LECs in vivo were lost rapidly in vitro as revealed by unsupervised comparison of gene expression of freshly isolated ECs and resting confluent ECs harvested after various in vitro passages. Dedifferentiation in vitro occurred to an unexpected extent because we detected a strikingly greater relatedness of freshly isolated LECs with freshly isolated BECs than with cultured LECs. Moreover, cultured LECs and BECs displayed a much higher degree of similarity than did LECs and BECs in vivo. Thus, the transcriptional repertoires of LECs and BECs differ in vivo but undergo dramatic assimilation during cell culture. In contrast, the pan-EC–restricted transcriptome is rather stable in culture. Accordingly, stably expressed were pan-EC genes involved in vessel growth and cell-cell adhesion, emphasizing their comparable role in vascular biology in vitro and in vivo. Thus, it appears that EC sublineage differentiation and functionality is largely directed by environmental cues but is not irreversibly determined even when ECs have reached terminal differentiation in vivo.
These findings urged for a detailed analysis of the prototypic in vivo transcriptomes of LECs and BECs and, in particular, for the definition of those programs which are strictly environmentally regulated. Of interest, gene expression that best defined BECs and LECs in vivo was lost preferentially in vitro. Selectively expressed in vivo but down-regulated in vitro were molecules that are involved in the regulation of fluid exchange between blood and tissues. One most prominent example is the channel-forming integral protein aquaporin-1 which is expressed by BECs but not by any other skin cell type. Aquaporin-1 is an essential water channel, but may be also involved in angiogenesis and cell migration.26 However, LECs in vivo but not in vitro express the voltage-gated sodium channel type III α chain (SCN3A along with the β chain SCN3B) as their most discriminative feature in normal human skin. Voltage-sensitive sodium channels play a fundamental role in excitable cells, transiently increasing the sodium permeability of the plasma membrane in response to changes in membrane potential.27 It is thus tempting to speculate that LEC-expressed SCN3 functions as critical sensor of osmolarity and regulator of osmotic fluid balance in the tissues.
The comparison of ex vivo–isolated LECs and BECs revealed LEC-restricted expression of almost the complete spectrum of gene products that are of known importance for lymphangiogenesis (ie, PROX-1, podoplanin, FOXC2) and LEC function (LYVE-1, SLC). Interestingly, expression of the 2 essential LEC differentiation-promoting transcription factors, PROX-118 and FOXC2,19 was maintained in vitro, in line with our previous observation of partial preservation of LEC differentiation in vitro.1 In contrast to LECs, BECs have not been assigned an EC sublineage-defining homeobox gene. We find that the mesenchyme homeobox gene MEOX1 is expressed abundantly and selectively in BECs. In fact, we find that MEOX-1 expression is the best defining criterion of BEC differentiation in normal human skin. Although a possible functional role of MEOX-1 in BECs still has to be established, its structural relative MEOX-2, which is expressed in both BECs and LECs (Figure S2), is a positive regulator of angiogenesis in vivo.28 Thus, it is tempting to speculate that a concerted action of the 2 MEOX gene products is critical for BEC differentiation and function.
The homeobox gene HOXD10 was identified as an EC-defining gene in our study. In contrast to MEOX-1, we find HOXD10 to be stably expressed in culture. Functionally, HOXD10 expression is essential to maintain a nonangiogenic state in the endothelium.29 This confirms that the ECs investigated, both ex vivo and in vitro, were in a bona fide resting, nonproliferative state. It was interesting to note that HOXD10 was among the genes the expression of which is most prominently down-regulated in tumor ECs as compared with normal BECs (data not shown). Thus, it appears that in vivo HOXD10 can be the critical repressor of both angiogenesis and lymphangiogenesis.
The in vivo mechanisms of MHC class II-dependent antigen presentation by ECs and its consequences for T-cell activation are still incompletely understood. ECs in vivo are commonly thought to express MHC class II only after encounter of proinflammatory cytokines.21–24 As a major example for environment-dependent gene expression, we find that nonactivated BECs in vivo, but not LECs, express prominently the full repertoire of gene products that are required for MHC class II–dependent antigen presentation. It is highly unlikely that MHC class II expression on BECs is due to the procedure of EC isolation because immunohistochemistry on fresh, snap-frozen human skin revealed substantial MHC class II expression on BECs. Moreover, transcriptional and protein expression analyses revealed a dramatic down-regulation but never an up-regulation of MHC class II expression following the isolation of BECs from the tissue.
In vivo, the level of constitutive peptide:MHC class II expression by BECs is substantial because tissue DCs expressed only moderately higher amounts. The availability of unique mAbs and of MHC-genotyped, fresh human skin samples allowed us to measure surface expression of defined species of peptide:MHC class II complexes on ECs. These analyses clearly showed that BECs in vivo display self-protein–derived peptides in the context of MHC class II. The fraction of MHC class II complexes bearing peptides derived from our test self-antigen was even higher in BECs than in DCs. This suggests that the repertoire of peptide display differs in BECs and DCs what may have important implications for the selection of T-cell specificities in vivo.
This together with our observation that BECs in normal human skin constitutively express B7 family members that are essential for the down-regulation of T-cell responses30,31 suggests that in vivo BECs negatively regulate CD4+ T-cell reactivity to self-proteins. The physical interaction of T cells and microvascular BECs in vivo may be promoted under slow flow conditions by the observed low level constitutive expression of E- and P-selectins, chemokines, and ICAM-1, all features that we found to be strictly environment-regulated and lost on EC culture (Figure 4C; Table S3).32 BECs in metastatic melanoma, to a similar extent as in normal skin, were found to express peptide:MHC class II complexes and display negative costimulatory molecules for T-cell activation. This indicates that BECs could also contribute to T-cell tolerance to tumor antigens. Because of the demonstrated rapid dedifferentiation of BECs in vitro, the resolution of the biologic impact of MHC class II on ECs for T-cell activation clearly requires novel in vivo models that will allow for controlled expression of defined MHC class II complexes on BECs in their natural tissue surroundings.
In summary, the “in vivo” transcriptomes of BECs and LECs reveal novel EC subtype-restricted functionalities that are determined by the environment in which these cells reside. The further understanding of the environment-derived signals regulating these functions will be instrumental for the full understanding of lymphatic and blood vessel physiology and the ability to manipulate these cells in therapeutic settings.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Marion Gröger for critical reading of the manuscript and Roland Varecka for technical help.
This work was supported by the GENome Research in AUstria (GEN-AU) program of the Austrian Ministry of Science, CeMM–Research Center for Molecular Medicine of the Austrian Academy of Sciences, and the Austrian Science Foundation (FWF; SFB F2308) (D.M.).
Authorship
Contribution: S.A. and D.M. designed the study and wrote the manuscript; S.A., E.K., W.B., B.R., P.M., and N.S. performed the experiments; A.W. contributed essential reagents; C.H. performed bioinformatic analyses; and G.S. provided general support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dieter Maurer, Department of Dermatology, Medical University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail: Dieter.Maurer@meduniwien.ac.at.

![Figure 1. Purification and GeneChip analysis of fresh BECs, LECs, and other major skin cell types. (A) Identification and isolation of fresh BECs and LECs by triple immunolabeling and FACS. Microvascular ECs in freshly prepared dermal cell suspensions were identified by their homogeneous VE-cadherin expression (y-axis, left). The sort gate was set to actively exclude VE-cadherin− non-ECs and VE-cadherin+ dead ECs that are characterized by their low forward scatter (FSC) signals (x-axis). Within the vital EC population, nonoverlapping subpopulations of BECs (red dots) and LECs (green dots) were resolved by anti–podoplanin-FITC (x-axis) anti–CD34-PE immunolabeling (y-axis, right). BECs and LECs were purified to at least 98% purity by FACS. (B) Purity assessment of isolated BECs, LECs, and other major skin cell types by analysis of marker gene expression. BECs and LECs as well as fibroblasts (FBs), mast cells (MCs), CD14+ dermal dendritic cells (DDCs), CD1a+ DDCs, epidermal Langerhans cells (LCs), and keratinocytes (KCs), isolated according to the criteria described in “Materials and methods” by FACS, were subjected to RNA isolation. After 2 rounds of linear amplification, cRNAs were hybridized to Affymetrix HG-U133 plus 2.0 GeneChips. Hybridization intensities with probe sets corresponding to cell lineage-associated marker genes (VE-cadherin, PROX-1, collagen 1 [COL1], tryptase, CD14, CD1b, langerin, keratin 14) are given for all cell types (x-axis). If more than one sample was available from a defined cell type, mean values and standard deviations (SDs) are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/11/10.1182_blood-2006-10-053280/4/m_zh80110703800001.jpeg?Expires=1767778917&Signature=F4GalMp9U37WZhOC8Z1t2KKEFp1MmS7K1wKE8EkkwhuDaPX~mFdqu-18kqb1KnOWv8PkQ2BbCZm3KMPHdMcV2BTcadA8FquK3tK3JVV6grMfd~VLauvMbUdS43PcvMK892E7sL8d6AV-Zi2nhnYvl-Yh-24KadGMblnw6kon6480PByhwob75l2wq8lGgpeIKqTfQdbKdpFCchpqVI19qKg0DMfp5plbyB0RshviCOjG7wVAa8f3OFeGogNM6lBRui54k~1UfE~xlzG7ErKGk509Ezq9qEhXkK6oUxkeXgrjSQBuDP8IvRXJHo4wPQyZfc38USnaDIUKvdadUYdGjA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. BECs constitutively present defined self-antigens in the context of MHC class II. (A) Specific detection of self-peptide:MHC class II complexes on antigen-presenting cells. B cells from HLA-DRB1*0101+HLA-A*0201+ donors (top) but not B cells from HLA-DRB1*0101+HLA-A*0201− donors (bottom) react with mAb UL-5A1 specific for a MHC class II complex composed of HLA-DRB1 and a peptide of HLA-A2 [amino acids 103-117; DR1:A2(aa103-117); x-axis]. Peripheral blood B cells were identified by anti-CD19 immunoreactivity in mononuclear cell preparations from healthy donors (y-axis). (B) Comparable levels of self-peptide-MHC complexes are constitutively displayed by BECs and DCs in normal human skin. Dermal single-cell suspensions from HLA-DRB1*0101+HLA-A*0201+ (left panels) and HLA-DRB1*0101−HLA-A*0201+ donors (right panels) were labeled with mAb UL-5A1-FITC (top) or L243-FITC (bottom) together with anti–VE-cadherin-PC5, anti–PDPN-PE, anti–CD1a-APC, and anti–CD14-PC7. BECs (red histograms) and CD1a+ DDCs (blue histograms) were gated and analyzed for surface expression of DR1:A2(aa103-117) complexes (x-axis, upper panels) and total HLA-DR–peptide complexes (x-axis, lower panels). Gray histograms show the reactivity of BECs and CD1a+ DDCs with FITC-labeled control Abs. (C) Efficient presentation of self-peptide–MHC complexes by BECs. The relation between DR1:A2(aa103-117) and total DR:peptide complex display was calculated by forming the ratio between the respective measured fluorescence intensities for BECs, CD1a+ DDCs, and CD14+ DDCs (y-axis). Data are expressed as mean values (+ SDs) obtained in experiments with dermal cells from 2 unrelated HLA-DRB1*0101+HLA-A*0201+ donors.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/11/10.1182_blood-2006-10-053280/4/m_zh80110703800006.jpeg?Expires=1767778917&Signature=TT1zbLS4x7ud-ih1lF9tUkPQ4bXIcp-smAHjd2gzCT4ap~K2D88uLllb6vqeAyHj1lDmdy33y3Lk~-TQFWXUNDr1No3g02d~xIxJ4bg4lhBYRD6CnW7sTdTNFFZQJbXgU~48AePcI06kTH~hsAa7vTTKE4~879KUWyLh-U6qGa0PKYexNLhsqXLLrdvTEycg~LySdk90uRG3NT5AcTmXnFjNB5niuuGhq1U1KhCIR42j1KJ0Nifa~Ruz6ozm~TnpLzoXc19U6KjZ5qwE6aLgdX-0nBl4GTpNoOWCXoNdEWd~3iyepdyg2ajOqN8LnOHlkXzFk5NWY7C1Zx4f9zNpgg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)