Abstract
Modulation of the proteins released during activation is one mechanism whereby aspirin may influence platelet-mediated human disease. We investigated the effect of aspirin on the platelet releasate using mass spectrometry and found that different agonists evoked different releasate profiles, with aspirin having a general moderating effect on the amount of protein released regardless of the agonist. These observations were confirmed for several cytokines using an antibody array approach.
Introduction
Platelets are small anucleate cells rapidly deployed to sites of injury to maintain hemostasis. Recently, however, activated platelets have not only been implicated in thrombosis but also in inflammatory reactions and in distinct aspects of atherosclerosis.1 Activated platelets can release cytokines on vascular surfaces, triggering the recruitment of inflammatory cells, and several platelet-released proteins have been identified in atherosclerotic lesions such as platelet factor 4 whose deposition has been correlated with lesion severity.2 Early lesion development can, however, be prevented using cyclooxygenase (COX) inhibitors.3 Aspirin, a potent COX inhibitor, retards platelet function by inhibiting prostaglandin and thromboxane formation, and has been shown at low doses to prevent arterial thrombosis under certain conditions.4
Aspirin can exert its effects at least indirectly by reducing the release of inflammatory cytokines and growth factors at sites of vascular injury by, for example, decreasing the levels of platelet-released IL-7.5 Additionally, it has been shown to reduce levels of the platelet secretory marker P-selectin in many clinical studies, including a recent report where the use of aspirin in high-risk hypertensive patients led to a reduction in both P-selectin levels and the platelet angiogenic growth factor angiopoieten.6–8 The effect of aspirin on the wider repertoire of secreted proteins remains to be determined. Here we use 2 complementary proteomic approaches to examine the effect of aspirin on the protein composition of the platelet releasate.
Materials and methods
Platelet aggregation and isolation of the platelet releasate
Washed platelets were prepared and aggregations performed as previously described,9 with the following modifications. Platelets were preincubated with 20 μM aspirin or vehicle control for 30 minutes at 37°C. The platelets (2.5 × 108) were stimulated with either ADP (5 μM), collagen (0.05 mg/mL), or TRAP (SFLLRN; 5 μM) for 3 minutes in an aggregometer. The platelet releasate fraction was isolated as previously described.10 For the time course experiment, platelets (2.5 × 108) were stimulated for 30 seconds, 3 minutes, and 20 minutes with collagen (0.05 mg/mL) and their releasate fractions isolated as above.
SDS-PAGE and immunoblotting
The releasate fractions were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as previously described.9 High-molecular-weight proteins were resolved on 4% to 20% gradient gels (Pierce, Rockford, IL). Low-molecular-weight proteins were resolved on 16% tris-tricine gels (Invitrogen, Carlsbad, CA). Western blotting was performed as previously described.10 The primary monoclonal antibody to thrombospondin clone p10 (1:1000 dilution) was purchased from Chemicon International (Hampshire, United Kingdom) and anti–mouse horseradish peroxidase (HRP) antibody was obtained from Pierce. Fluorescence was detected using West Pico Supersignal chemiluminescent substrate (Pierce).
EIA for the measurement of TXB2
Measurement of thromboxane B2 (TXB2) was performed on all platelet releasate fractions (diluted 1:50) using an enzyme-linked immunoassay (EIA) kit (Metachem Diagnostics, Northhampton, United Kingdom) as per the manufacturer's instructions.
Mass spectrometry
SDS-PAGE gels were cut into bands and digested in gel with trypsin according to the method of Shevchenko et al.11 The resulting peptide mixtures were resuspended in 1% formic acid and analyzed by nano-electrospray liquid chromatography mass spectrometry (Nano-LC MS/MS). An HPLC instrument (Dionex, Surrey, United Kingdom) was interfaced with an LTQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA). Chromatography buffer solutions (Buffer A, 5% acetonitrile and 0.1% formic acid; Buffer B, 80% acetonitrile and 0.1% formic acid) were used to deliver a 60-minute gradient (35 minutes to 45% Buffer B, 10 minutes to 90%, hold 10 minutes, 3 minutes to 5%, hold for 15 minutes). A flow rate of 2 μL/min was used at the electrospray source. Spectra were searched using the SEQUEST algorithm as described previously,12 against the International Protein Index (IPI) database.13 The probability-based evaluation program, Protein Prophet (Institute for Systems Biology [ISB], Seattle, WA), was used for filtering identifications.14 Hierarchical clustering of log-transformed spectral count values was performed with the Cluster 3.0 software package (Eisen Lab, Berkeley, CA) using the Spearman distance metric.
Detection of platelet-released cytokines using antibody arrays
Human Cytokine Antibody Arrays III (Raybiotech, Norcross, GA) were used according to the manufacturer's instructions. Briefly, the arrays were blocked, incubated with 100 μL of releasate overnight, incubated with biotin-conjugated antibodies (1/250) for 2 hours and with HRP antibodies (1/1000) overnight. The membranes were incubated with chemiluminescent substrate and exposed to x-ray film for 15 minutes before development. Quantitative array analysis was performed using Array Vision Evaluation 8.0 (GE Healthcare Life Science, Buckinghamshire, United Kingdom).
Results
To investigate the effects of aspirin on platelet secretion, platelets were stimulated through 3 different pathways in the presence and absence of aspirin. Equal volumes of platelets were stimulated with ADP, TRAP, and collagen following preincubation with low-dose aspirin (20 μM) or with a vehicle control. A significant decrease in platelet aggregation was observed in the presence of the drug with all agonists: 92%, 83%, and 72% reduced for ADP, collagen, and TRAP, respectively (P < .03; Figure 1A). Platelet releasate fractions were isolated and separated on coomassie-stained 4% to 20% mid- to high-molecular-weight gels and 16% silver-stained low-molecular-weight gels (Figure 1B). Different protein profiles were observed in the releasate fractions following activation by the different agonists. Moreover, at least for the case of proteins visible on stained gels, aspirin appeared to evoke a general decrease in the amount of protein released for a given agonist, rather than result in a change to the profile of proteins released.
Modulation of platelet aggregation and protein release by aspirin. Equal volumes of platelets (2.5 × 108) were stimulated with ADP, TRAP, and collagen in the presence or absence of low-dose aspirin (20 μM). The aggregation response was plotted (mean ± standard deviation [SD] of 3 independent experiments using different donors) (A). Platelet releasate from a single representative donor was visualized using coomassie-stained (4%-20% acrylamide) and silver-stained (16% acrylamide) gels (B). Thromboxane levels in the platelet releasate (mean ± SD of 3 experiments) were plotted for each fraction (C).
Modulation of platelet aggregation and protein release by aspirin. Equal volumes of platelets (2.5 × 108) were stimulated with ADP, TRAP, and collagen in the presence or absence of low-dose aspirin (20 μM). The aggregation response was plotted (mean ± standard deviation [SD] of 3 independent experiments using different donors) (A). Platelet releasate from a single representative donor was visualized using coomassie-stained (4%-20% acrylamide) and silver-stained (16% acrylamide) gels (B). Thromboxane levels in the platelet releasate (mean ± SD of 3 experiments) were plotted for each fraction (C).
In order to confirm that this effect was largely independent of the donor, the experiment was repeated with 2 additional different donors. Very similar platelet releasate profiles were observed for all donors (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Similarly, to investigate whether the profiles observed were reflective of agonist dosing (in particular for ADP, which evokes a weaker response than collagen and TRAP at low dose levels15 ), the experiment was repeated with 2 doses of ADP (5 μM, 20 μM). The releasate profile observed when platelets were stimulated with higher doses of ADP was identical to that seen in the lower dose experiment (Figure S2), although the effect of aspirin was more pronounced in platelets stimulated with the lower dose of ADP. While the gel reflects the gross pattern of ADP-induced protein release, mass spectrometry analysis also confirmed this observation with many common proteins being identified at both doses (data not shown). Finally, to ensure that aspirin was indeed suppressing the formation of thromboxane, this compound was assayed in the releasate of platelets stimulated with the different agonists. Thromboxane levels decreased significantly for all agonists with a reduction of 99% for collagen and TRAP and 50% for ADP. Total levels of thromboxane released from platelets stimulated with ADP were much lower than the amounts released from collagen- or TRAP-activated platelets (Figure 1C).
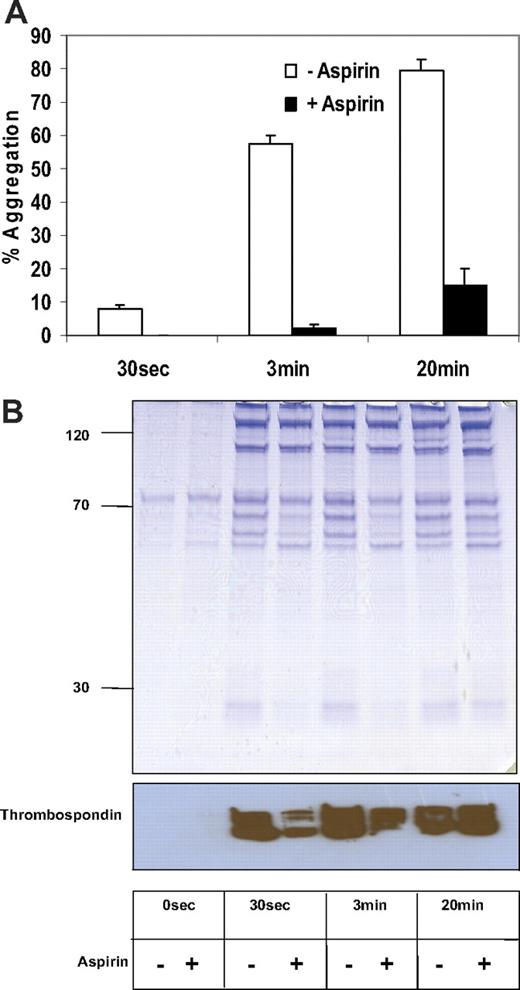
A time course of platelet activation was carried out to examine the levels of platelet secretion in the presence of aspirin at different timepoints. Platelets were stimulated with collagen for 30 seconds, 3 minutes, and 20 minutes. Platelet aggregation was 13%, 58%, and 80%, respectively, at these time points (Figure 2A). Aspirin inhibited platelet aggregation almost completely at 30 seconds and 3 minutes, while more than 20% of platelets remained aggregated after 20 minutes following aspirin addition. By 20 minutes, additional inside-out pathways will contribute to aggregation, thus bypassing the aspirin effect. As previously noted, a decrease in protein was observed by SDS-PAGE following aspirin addition that corresponded to the aggregation results (Figure 2B). Immunoblot analysis for the platelet secretory marker thrombospondin was performed on these time course fractions to confirm that secretion was reduced by aspirin at least up to 3 minutes following addition of agonist (Figure 2B).
Time course of collagen-stimulated platelet releasate. Equal volumes of platelets (2.5 × 108) were stimulated with collagen in the presence or absence of low-dose aspirin (20 μM) at different time points (30 seconds, 3 minutes, and 30 minutes). The aggregation response was plotted (mean ± SD of 3 independent experiments) using different donors (A). Platelet releasate from a single representative donor was visualized using coomassie-stained (4%-20%) acrylamide (B, upper panel). An immunoblot for thrombospondin was performed on these fractions (B, lower panel).
Time course of collagen-stimulated platelet releasate. Equal volumes of platelets (2.5 × 108) were stimulated with collagen in the presence or absence of low-dose aspirin (20 μM) at different time points (30 seconds, 3 minutes, and 30 minutes). The aggregation response was plotted (mean ± SD of 3 independent experiments) using different donors (A). Platelet releasate from a single representative donor was visualized using coomassie-stained (4%-20%) acrylamide (B, upper panel). An immunoblot for thrombospondin was performed on these fractions (B, lower panel).
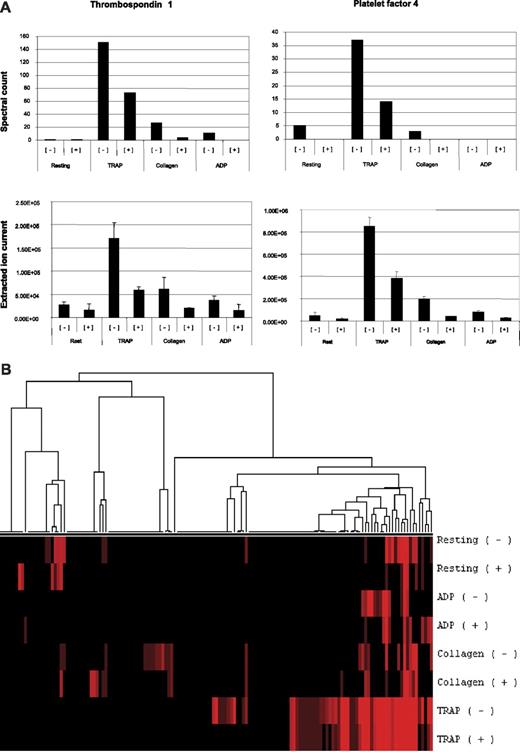
In order to more fully characterize the effect of aspirin on platelet secretion, we used proteomics to identify the proteins released in response to different agonists in the presence and absence of aspirin. Proteins from the experiment illustrated in Figure 1 were digested and subjected to peptide tandem mass spectrometry, and 146 proteins were identified. The fractions mostly contained platelet secretory proteins as previously reported10 (Figure 3A, Table S1). Spectral counts (the number of peptide mass spectra collected per sample leading to a protein identification) and extracted ion counts (a measure of the electronic signal representing an identified ion detected by the mass spectrometer), were used as a semiquantitative tool to define the pattern of proteins released in response to agonist-drug combinations.12 Notably, the levels of 2 platelet secretory markers, thrombospondin and platelet factor 4, were decreased in the aspirin-treated samples (P < .03; Figure 3A). Pretreatment with aspirin resulted in lower numbers of proteins identified and lower spectral counts except in the case of ADP, where fibrinogen is added exogenously and contributes to a disproportionate spectral count total for that agonist (Tables S2 and S3). Hierarchical clustering of log-transformed spectral count values was performed and showed that secretion of a common set of more abundant proteins is evoked by all agonists, whereas lower abundance proteins could be detected in the TRAP-stimulated fractions (Figure 3B).
Mass spectrometry analysis of aspirin-treated releasate. The complete number of proteins identified and the number of spectral counts for each condition is available in Table S1. Spectral counts (top) and extracted ion currents (XICs; bottom) for representative ions from thrombospondin (FASAEAEEDGDLQCLCVK, GKEESLDSDLYAELR, EAEEDGDLQCLCVK) and platelet factor 4 (FTGSQPFGQGVEHATANK, RPPLCYHNGVQYR, TIVTTLQDSIR) (errors bars are SD for mean of peptide XICs) indicated that the abundance of these proteins was decreased in the aspirin-treated fractions (A). Hierarchical clustering of agonist-induced platelet releasate protein profiles in the presence or absence of aspirin was perfomed. Spectral counts for each fraction were tabulated and null values were replaced with the value 0.01. The data were normalized across samples, log-transformed, and clustered using the Spearman rank correlation coefficient (average linkage) using Cluster 3.0 (B).
Mass spectrometry analysis of aspirin-treated releasate. The complete number of proteins identified and the number of spectral counts for each condition is available in Table S1. Spectral counts (top) and extracted ion currents (XICs; bottom) for representative ions from thrombospondin (FASAEAEEDGDLQCLCVK, GKEESLDSDLYAELR, EAEEDGDLQCLCVK) and platelet factor 4 (FTGSQPFGQGVEHATANK, RPPLCYHNGVQYR, TIVTTLQDSIR) (errors bars are SD for mean of peptide XICs) indicated that the abundance of these proteins was decreased in the aspirin-treated fractions (A). Hierarchical clustering of agonist-induced platelet releasate protein profiles in the presence or absence of aspirin was perfomed. Spectral counts for each fraction were tabulated and null values were replaced with the value 0.01. The data were normalized across samples, log-transformed, and clustered using the Spearman rank correlation coefficient (average linkage) using Cluster 3.0 (B).
Proteins identified in all samples
| IPI no. . | Resting . | TRAP . | Collagen . | ADP . | Protein . | ||||
|---|---|---|---|---|---|---|---|---|---|
| − . | + . | − . | + . | − . | + . | − . | + . | ||
| IPI00022434.2 | — | 28 | 844 | 489 | 15 | 20 | 12 | 81 | Alb protein |
| IPI00029717.1 | — | 2 | 187 | 117 | 7 | 2 | 75 | 251 | Fibrinogen; alpha chain; isoform alpha preproprotein |
| IPI00298497.3 | — | 8 | 167 | 93 | 2 | 26 | 13 | 25 | Fibrinogen; beta chain; preproprotein |
| IPI00296099.5 | — | 1 | 151 | 73 | 27 | 4 | 11 | — | Thrombospondin-1 precursor |
| IPI00219713.1 | 1 | — | 145 | 80 | — | — | — | — | Fibrinogen; gamma polypeptide |
| IPI00021439.1 | 1 | 3 | 60 | 22 | 12 | 10 | 11 | 7 | Hypothetical protein dkfzp459a127 |
| IPI00022445.1 | — | — | 37 | 11 | 20 | 3 | — | 1 | Platelet basic protein precursor (pbp) (small inducible cytokine b7) (cxcl7) (leukocyte-derived growth factor) (ldgf) (macrophage-derived growth factor) (mdgf) |
| IPI00022446.1 | — | — | 48 | 18 | 4 | — | — | — | Hypothetical protein pf4 (platelet factor 4) (chemokine (c-x-c motif) ligand 4) (fragment) |
| IPI00027547.2 | — | 2 | 26 | 19 | 1 | 2 | 2 | 5 | Proteolysis-inducing factor |
| IPI00411626.2 | — | 7 | 1 | — | 2 | 5 | 14 | 31 | Hypothetical protein dkfzp779n0926 |
| IPI00411506.1 | — | — | 6 | 6 | — | — | — | 14 | 48-kDa protein |
| IPI00553177.1 | — | — | 25 | 16 | 6 | 6 | 1 | 4 | Full-length cDNA clone cs0de007yp21 of placenta of Homo sapiens (human) (full-length cDNA clone cs0di028yp15 of placenta of Homo sapiens) |
| IPI00022295.1 | — | — | 37 | 14 | 3 | — | — | — | Platelet factor 4 variant precursor (pf4var1) (pf4alt) (cxcl411) [contains: platelet factor 4 variant(4-74); platelet factor 4 variant(5-74); platelet factor 4 variant(6-74)] |
| IPI00022463.1 | — | — | 22 | 15 | 2 | 5 | 4 | 1 | Serotransferrin precursor (transferrin) (siderophilin) (beta-1-metal-binding globulin) |
| IPI00515047.1 | — | — | 24 | 21 | 5 | — | — | — | Actin; alpha 1; skeletal muscle |
| IPI00021440.1 | — | — | 25 | 20 | — | — | — | — | Actin; gamma 1 |
| IPI00373937.2 | — | 1 | 16 | 9 | 1 | — | 1 | 1 | Hlar698 |
| IPI00430839.1 | — | — | 13 | 13 | — | 1 | — | 5 | Hypothetical protein |
| IPI00550315.1 | — | — | 19 | 10 | — | 1 | — | — | Ig kappa chain c region |
| IPI00240503.5 | — | 25 | — | — | — | — | — | — | 48-kDa protein |
| IPI00551005.1 | 7 | 2 | 13 | 2 | — | — | — | 5 | Ig lambda chain c regions |
| IPI00014572.1 | — | — | 14 | 5 | 3 | — | — | — | Sparc protein |
| IPI00549654.2 | — | — | 13 | 9 | — | — | — | — | Ig gamma-1 chain c region |
| IPI00302592.2 | 2 | — | 11 | 11 | — | — | — | — | Filamin a; alpha (actin binding protein 280) |
| IPI00307162.2 | — | — | 9 | 9 | — | 2 | — | — | Vinculin isoform meta-vcl |
| IPI00013508.4 | 5 | 1 | 8 | 7 | — | — | 3 | — | Alpha-actinin-1 |
| IPI no. . | Resting . | TRAP . | Collagen . | ADP . | Protein . | ||||
|---|---|---|---|---|---|---|---|---|---|
| − . | + . | − . | + . | − . | + . | − . | + . | ||
| IPI00022434.2 | — | 28 | 844 | 489 | 15 | 20 | 12 | 81 | Alb protein |
| IPI00029717.1 | — | 2 | 187 | 117 | 7 | 2 | 75 | 251 | Fibrinogen; alpha chain; isoform alpha preproprotein |
| IPI00298497.3 | — | 8 | 167 | 93 | 2 | 26 | 13 | 25 | Fibrinogen; beta chain; preproprotein |
| IPI00296099.5 | — | 1 | 151 | 73 | 27 | 4 | 11 | — | Thrombospondin-1 precursor |
| IPI00219713.1 | 1 | — | 145 | 80 | — | — | — | — | Fibrinogen; gamma polypeptide |
| IPI00021439.1 | 1 | 3 | 60 | 22 | 12 | 10 | 11 | 7 | Hypothetical protein dkfzp459a127 |
| IPI00022445.1 | — | — | 37 | 11 | 20 | 3 | — | 1 | Platelet basic protein precursor (pbp) (small inducible cytokine b7) (cxcl7) (leukocyte-derived growth factor) (ldgf) (macrophage-derived growth factor) (mdgf) |
| IPI00022446.1 | — | — | 48 | 18 | 4 | — | — | — | Hypothetical protein pf4 (platelet factor 4) (chemokine (c-x-c motif) ligand 4) (fragment) |
| IPI00027547.2 | — | 2 | 26 | 19 | 1 | 2 | 2 | 5 | Proteolysis-inducing factor |
| IPI00411626.2 | — | 7 | 1 | — | 2 | 5 | 14 | 31 | Hypothetical protein dkfzp779n0926 |
| IPI00411506.1 | — | — | 6 | 6 | — | — | — | 14 | 48-kDa protein |
| IPI00553177.1 | — | — | 25 | 16 | 6 | 6 | 1 | 4 | Full-length cDNA clone cs0de007yp21 of placenta of Homo sapiens (human) (full-length cDNA clone cs0di028yp15 of placenta of Homo sapiens) |
| IPI00022295.1 | — | — | 37 | 14 | 3 | — | — | — | Platelet factor 4 variant precursor (pf4var1) (pf4alt) (cxcl411) [contains: platelet factor 4 variant(4-74); platelet factor 4 variant(5-74); platelet factor 4 variant(6-74)] |
| IPI00022463.1 | — | — | 22 | 15 | 2 | 5 | 4 | 1 | Serotransferrin precursor (transferrin) (siderophilin) (beta-1-metal-binding globulin) |
| IPI00515047.1 | — | — | 24 | 21 | 5 | — | — | — | Actin; alpha 1; skeletal muscle |
| IPI00021440.1 | — | — | 25 | 20 | — | — | — | — | Actin; gamma 1 |
| IPI00373937.2 | — | 1 | 16 | 9 | 1 | — | 1 | 1 | Hlar698 |
| IPI00430839.1 | — | — | 13 | 13 | — | 1 | — | 5 | Hypothetical protein |
| IPI00550315.1 | — | — | 19 | 10 | — | 1 | — | — | Ig kappa chain c region |
| IPI00240503.5 | — | 25 | — | — | — | — | — | — | 48-kDa protein |
| IPI00551005.1 | 7 | 2 | 13 | 2 | — | — | — | 5 | Ig lambda chain c regions |
| IPI00014572.1 | — | — | 14 | 5 | 3 | — | — | — | Sparc protein |
| IPI00549654.2 | — | — | 13 | 9 | — | — | — | — | Ig gamma-1 chain c region |
| IPI00302592.2 | 2 | — | 11 | 11 | — | — | — | — | Filamin a; alpha (actin binding protein 280) |
| IPI00307162.2 | — | — | 9 | 9 | — | 2 | — | — | Vinculin isoform meta-vcl |
| IPI00013508.4 | 5 | 1 | 8 | 7 | — | — | 3 | — | Alpha-actinin-1 |
A sample of the 146 proteins released from platelets and identified by LC-MS/MS, listed by IPI number, and ranked by number of spectral counts (descending) are highlighted.
− indicates absence of aspirin; +, presence of aspirin; —, no spectral counts detected.
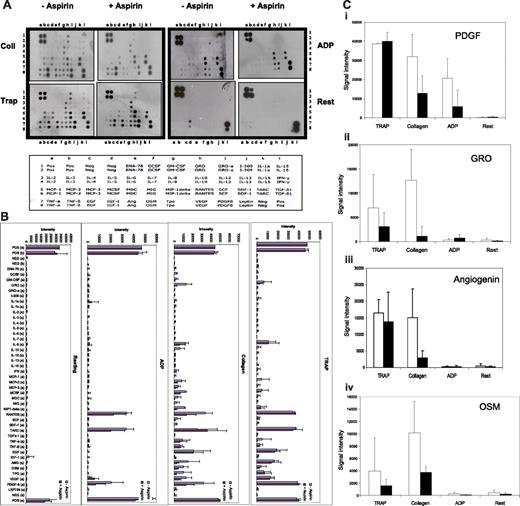
The sensitivity of mass spectrometry limits protein detection to relatively abundant proteins. We therefore employed human antibody arrays to examine the expression of 42 different cytokines in the platelet releasate (Figure 4A). Following quantitative array analysis, similar but not identical cytokine profiles were observed between 3 different donors with the expression of several cytokines lower in the aspirin-treated than in untreated fractions, confirming the results of the mass spectrometry experiments (Figure 4B). High levels of abundant platelet cytokines, such as PDGF, were seen for all releasate fractions together with cytokines not previously reported to be present in platelets, including OSM, ANG, GRO (Figure 4C), and MCP-2.
Aspirin decreases agonist-induced release of platelet cytokines. Equal volumes of platelets (2.5 × 108) were stimulated with ADP, TRAP, and collagen in the presence and absence of low-dose aspirin (20 μM) and the releasate was incubated with human cytokine protein antibody arrays (A). The intensity of each cytokine on the array in the presence (■) and absence (□) of low-dose aspirin with each agonist is compared (B). Signals for platelet-derived growth factor (PDGF) (i), growth regulating growth factor (GRO) (ii), angiogenin (Ang) (iii), and oncostatin M (OSM) (iv) are shown (mean ± SD of 3 independent experiments with different donors). Signal intensities for different donors were normalized to the positive control spots (C).
Aspirin decreases agonist-induced release of platelet cytokines. Equal volumes of platelets (2.5 × 108) were stimulated with ADP, TRAP, and collagen in the presence and absence of low-dose aspirin (20 μM) and the releasate was incubated with human cytokine protein antibody arrays (A). The intensity of each cytokine on the array in the presence (■) and absence (□) of low-dose aspirin with each agonist is compared (B). Signals for platelet-derived growth factor (PDGF) (i), growth regulating growth factor (GRO) (ii), angiogenin (Ang) (iii), and oncostatin M (OSM) (iv) are shown (mean ± SD of 3 independent experiments with different donors). Signal intensities for different donors were normalized to the positive control spots (C).
Discussion
While previous reports focusing on individual proteins indicated that their release from platelets was reduced by aspirin,5–7 we used mass spectrometry to monitor the secretion of over 100 proteins. Using this more comprehensive approach, we examined the platelet releasate upon stimulation with ADP, collagen, and TRAP in the presence or absence of the cyclooxygenase inhibitor, aspirin. Hierarchical cluster analysis and other experiments revealed 3 potential trends. First, the strength of agonist appears to be important for achieving maximal secretion. Stimulation with ADP resulted in fewer protein identifications and lower spectral count values than stimulation with either TRAP or collagen. Second, an overall decrease in protein expression was observed in the presence of aspirin. This can be seen in the in-gel analysis, particularly for the collagen- and ADP-stimulated fractions. Li et al16 reported 2 waves of platelet secretion, the first of which occurs even when platelet aggregation is inhibited, indicating that it does not require integrin outside-in signaling. Our findings support this 2-wave model; an initial wave upon agonist stimulation that leads to release of thromboxane, which upon binding to its receptor amplifies the platelet response, causing aggregation and another wave of platelet secretion. Inhibition of thromboxane production and aggregation by aspirin appear to inhibit this second wave, leading to a decrease in protein observed. This observation was less pronounced in the TRAP releasate fraction, possibly because signaling arising from low-dose collagen and ADP stimulation is more thromboxane-dependant than signaling arising from TRAP stimulation, and therefore, aspirin-treated platelets stimulated with the former release fewer granules.17–19 Increasing the stimulation time to 20 minutes for collagen did not appear to result in additional secretion, although more quantitative study would be required to examine this. The third observation was that the secretion profile differs depending on the agonist used to stimulate the platelets. This suggests the presence of different populations of α-granules under different exocytosis regulatory mechanisms, as has been reported for neuronal secretion.14
Platelet-derived cytokines have been shown to play significant roles during various stages of atherosclerosis. For example, activated platelets deposit the chemokines ‘regulated upon activation, normal T-cell expressed and secreted' (RANTES) and CXCL4 directly on the surface of monocytes and the endothelium of atherosclerotic arteries.1 Additionally, a small number of studies have shown a decrease in the levels of certain cytokines released from platelets (IL-7 and CTGF) in the presence of aspirin.5,20 Using quantitative antibody microarray technology we determined the levels of 42 different cytokines in the platelet releasate when platelets were stimulated with different agonists in the presence and absence of aspirin. High levels of abundant platelet cytokines, RANTES, PDGF, and EGF were detected. The interleukin family was generally underrepresented with the exception of interleukin 8 (IL-8), a potent chemoattractant for neutrophils. The presence of IL-8 in platelets has been suggested previously,21–23 but additional experiments are required for confirmation, due to potential leukocyte contamination. We detected the presence of several novel platelet cytokines including MCP2, a functional C-C chemokine inhibitor of inflammation,24 and oncostatin M, which promotes fibrinogen biosynthesis and smooth muscle proliferation.25 Furthermore, we found angiogenin, an inducer of angiogenesis, to be present in platelets.26
We show here that proteomics technologies can be used to provide new insights into the pharmacology of platelet activation. Specifically, the composition of the platelet releasate in response to different agonists and in the presence and absence of aspirin was characterized with greater resolution than previously. Moreover, this work resulted in the identification of novel platelet proteins that merit further functional studies, as well as raising the hypothesis that stimulation of different pathways leads to the release of proteins from different, as-yet-uncharacterized subcompartments housed within the platelet.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by a fellowship from Enterprise Ireland (R.O'C.), research grants from the Health Research Board of Ireland (P.M., D.F.), the Higher Education Authority of Ireland (D.F., G.C.), and the Science Foundation Ireland award (K.W., M.S., M.F., P.M, and G.C.; grant no. 02/IN.1/B117).
Authorship
Contribution: G.C., P.B.M., and D.J.F. designed the research; J.A.C. and R.O'C. carried out the experiments; K.W. and M.F. did the mass spectrometry; and M.S. did the bioinformatics analyses. J.A.C., R.O'C., and G.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
J.A.C. and R.O'C. contributed equally to this report.
Correspondence: Gerard Cagney, School of Biomolecular & Biomedical Science, Conway Institute, University College Dublin, Dublin 4, Ireland; e-mail: gerard.cagney@ucd.ie.

![Figure 1. Modulation of platelet aggregation and protein release by aspirin. Equal volumes of platelets (2.5 × 108) were stimulated with ADP, TRAP, and collagen in the presence or absence of low-dose aspirin (20 μM). The aggregation response was plotted (mean ± standard deviation [SD] of 3 independent experiments using different donors) (A). Platelet releasate from a single representative donor was visualized using coomassie-stained (4%-20% acrylamide) and silver-stained (16% acrylamide) gels (B). Thromboxane levels in the platelet releasate (mean ± SD of 3 experiments) were plotted for each fraction (C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/11/10.1182_blood-2006-07-038539/4/m_zh80110704300001.jpeg?Expires=1767775527&Signature=uy8hWnw8Da3oKySp2Ycg-9trYQ4jk8Utnp7yEFRGLjrLK4NjGGQ6017uVZUBmd~ECi9m2AVe8vajuIkJgz1gtVSxA8Xk0C1Kxz7bCydIfeHWWKvc5wPhktvyh8uP9Wmi1NTwaXTziNH~GJJG4vNaKIgOHUm77RkiM4PDBI7do~COLvMTEixD5WJbOMeFhyweAW8N9OBC1p8opAVnsZd5c77DjK~t9BGJM5rpHi0W7X8HtfNnvDnTVYMeARfFqqTki8WGPUGGRpoCRzA9DlZ39erWW1jB2J-WlVZFKPnyXSe~NthhUjxv4DWARXDV6k~h~u9qZF2uzwFo7sBtloKddg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)