Abstract
Heat shock protein 90 (Hsp90) regulates diverse signaling pathways. Emerging evidence suggests that Hsp90 inhibitors, such as 17-allylamino-17-demethoxygeldanamycin (17-AAG), enhance DNA damage-induced cell death, suggesting that Hsp90 may regulate cellular responses to genotoxic stress. However, the underlying mechanisms are poorly understood. Here, we show that the Fanconi anemia (FA) pathway is involved in the Hsp90-mediated regulation of genotoxic stress response. In the FA pathway, assembly of 8 FA proteins including FANCA into a nuclear multiprotein complex, and the complex-dependent activation of FANCD2 are critical events for cellular tolerance against DNA cross-linkers. Hsp90 associates with FANCA, in vivo and in vitro, in a 17-AAG–sensitive manner. Disruption of the FANCA/Hsp90 association by cellular treatment with 17-AAG induces rapid proteasomal degradation and cytoplasmic relocalization of FANCA, leading to impaired activation of FANCD2. Furthermore, 17-AAG promotes DNA cross-linker–induced cytotoxicity, but this effect is much less pronounced in FA pathway-defective cells. Notably, 17-AAG enhances DNA cross-linker–induced chromosome aberrations. In conclusion, our results identify FANCA as a novel client of Hsp90, suggesting that Hsp90 promotes activation of the FA pathway through regulation of intracellular turnover and trafficking of FANCA, which is critical for cellular tolerance against genotoxic stress.
Introduction
Hsp90 is an abundant, evolutionarily conserved molecular chaperone whose function depends on its ability to bind and hydrolyze ATP. Through an ATPase cycle, Hsp90 facilitates proper folding of “client” proteins, thereby regulating their stability, protein interactions, intracellular trafficking, and functions.1,2 To fulfill these functions, Hsp90 interacts with its cofactors and cochaperones including Hsp70, immunophilins, and p23, to form the Hsp90-based chaperone complex.1,2 Natural compounds such as geldanamycin and radicicol bind the ATP-binding pocket of Hsp90 and disrupt its chaperone function.3,4 Hsp90 is required for function and stability of diverse signal transduction proteins including oncogenic proteins such as ErbB2 and Raf-1.2–7 Hence, the chaperone is an attractive target for cancer therapeutics. Indeed, Hsp90 inhibitors show antitumor activities in preclinical models, and geldanamycin analogues such as 17-allylamino-17-demethoxygeldanamycin (17-AAG) are currently undergoing clinical trials.3,4 Importantly, Hsp90 inhibitors sensitize tumor cells to various genotoxic agents used for standard cancer therapeutics, including DNA cross-linkers,8,9 ionizing radiation,10 and replication inhibitors.11,12 In line with these observations, it is suggested that Hsp90 regulates cell cycle checkpoints and DNA repair,11–13 but the underlying mechanisms are poorly understood.
Fanconi anemia (FA) is a genetically heterogeneous inherited disorder characterized by progressive bone marrow failure, cancer susceptibility, and cellular hypersensitivity to DNA cross-linkers such as mitomycin C (MMC).14–16 Multiple FA proteins cooperate in a common biochemical pathway, termed the FA pathway, which is involved in cellular response to DNA damage. At least 8 FA proteins, specifically, FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL/PHF9, and FANCM, form a nuclear multiprotein complex (FA core complex), which is required for FANCD2 monoubiquitination in response to DNA damage.17–32 DNA cross-linkers and replication inhibitors such as hydroxyurea (HU) are potent inducers of FANCD2 monoubiquitination.14–16 The monoubiquitinated form of FANCD2 is targeted to the chromatin and participates in maintenance of genomic stability interacting with BRCA1 and BRCA2/FANCD1, at least in part, through homology-directed repair.25,33–35 FANCJ/BRIP1, previously identified as a BRCA1-interacting helicase, may function downstream of FANCD2 activation or independently of the FA pathway.36–38 Previous studies suggested that nuclear levels of FANCA have profound effects on FA core complex formation.17–20,24,25,27 Nuclear levels of FANCA are determined by protein synthesis, degradation, and nucleocytoplasmic shuttling mediated by a bipartite nuclear localization signal (NLS) and 3 leucine-rich nuclear export signals.18,39,40 However, little is known about the regulatory mechanisms for intracellular turnover and trafficking of FANCA.
In an attempt to elucidate the molecular mechanisms of the FA pathway, we found that Hsp90 was included in the FANCA-containing protein complex. The goal of the present study was to elucidate the molecular basis and functional significance of this interaction. We here provide evidence indicating that Hsp90 functions as a chaperone of FANCA and that Hsp90 promotes activation of the FA pathway through regulation of FANCA, which is critical for cellular tolerance to DNA cross-linkers. These findings reveal that the FA pathway is involved in the Hsp90-mediated regulation of DNA damage response.
Materials and methods
Plasmids
Human FANCA cDNA with a FLAG-HA tandem-tag at the N-terminus (FH-FANCA) was inserted into a retroviral expression vector pLPCX (Clontech, Palo Alto, CA). An expression vector for HA-tagged ubiquitin was previously described.25 Human FANCA cDNA and FANCG cDNA were cloned into pcDNA3 Myc vector41 or pcDNA3 (Invitrogen, Carlsbad, CA).
Cell culture and transfections
FANCA-null SV40-immortalized fibroblasts GM6914, MCF7, 293T, NIH3T3, and HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum. K562, Jurkat, and RPMI8226 cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum. HeLa cells stably expressing FH-FANCA (HeLa/FH-FANCA cells) were obtained using a recombinant retrovirus as described previously.27 A clone with appropriate expression of FH-FANCA (∼5-fold of endogenous FANCA) was used for the present study. GM6914 cells stably expressing FANCA proteins were generated as described previously.20,27 For transient expression, subconfluent HeLa cells were transfected with FuGENE 6 (Roche Diagnostics, Pleasanton, CA) according to the manufacturer's protocol. Hsp90 small interfering RNA (siRNA) was designed and transfected into HeLa cells using Lipofectamine 2000 (Invitrogen), as described.42
Antibodies
The antibodies used in this study were as follows: mouse monoclonal anti-Hsp90, which recognizes both Hsp90 α and Hsp90 β (F-8, Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-Hsp90 β (SPA-843, StressGen, Victoria, BC, Canada); rabbit polyclonal anti-Hsp90 α (SPS-771, StressGen); mouse monoclonal anti-Hsp70 (SPA-810, StressGen); rabbit polyclonal anti-FANCD2 (Novus Biologicals, Littleton, CO); rabbit polyclonal antiubiquitin (SPA-200, StressGen); mouse monoclonal anti-HA (6E2, Cell Signaling Technology, Beverly, MA); mouse monoclonal anti-FLAG (M2, Sigma, St Louis, MO); mouse monoclonal anti-p23 (JJ3, Affinity Bioreagents, Golden, CO); mouse monoclonal anti–tubulin-β (TBN06, NeoMarkers, Fremont, CA); mouse monoclonal anti–γ-H2AX (JBW301, Upstate Cell Signaling Solutions, Charlottesville, VA); rabbit polyclonal anti-H2AX (Upstate Cell Signaling Solutions); mouse monoclonal anti-Myc (9E10, Santa Cruz Biotechnology); rabbit polyclonal anti-CHIP (Calbiochem, San Diego, CA); and mouse monoclonal anti–topoisomerase II (Transduction Laboratories, Lexington, KY). Rabbit polyclonal anti-FANCG (a gift from Dr J. P. de Winter, VU Medical Center, Amsterdam, The Netherlands),22 rabbit polyclonal anti-FANCF (a gift from Dr M. E. Hoatlin, Oregon Health and Science University, Portland, OR),43 rabbit polyclonal anti-FANCL and rabbit polyclonal anti-FANCM (gifts from Dr W. Wang, National Institutes of Health, Bethesda, MD),29,31 rabbit polyclonal anti-FANCC,19 and rabbit polyclonal anti-FANCA19 antibodies were previously described.
Protein identification by liquid chromatography-tandem mass spectrometry analysis
HeLa/FH-FANCA cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 5 mM Na3VO4, 5 mM NaF) containing a protease inhibitor cocktail (Roche Diagnostics) for 1 hour. After centrifugation at 16 000g for 30 minutes at 4°C, the supernatants were incubated with anti-FLAG M2-agarose at 4°C for 6 hours. After extensive washing, the bound complexes were eluted with Tris-buffered saline containing 0.1 mg/mL of the FLAG peptide (Sigma) and digested with endoproteinase Lys-C. The resulting peptides were analyzed by a liquid chromatography-tandem mass spectrometry (LC-MS/MS) system at the femtomole level as described.44,45
Immunoprecipitation and immunoblot analysis
Immunoprecipitation and immunoblotting were performed as described previously.27 For detection of polyubiquitinated FANCA, cells were lysed in ubiquitin lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.2% Sarkosyl) supplemented with 5 mM N-ethylmaleimide, 1 mM dithiothreitol, 2 mM Na3VO4, 5 mM NaF, and a protease inhibitor cocktail. Immunoprecipitated proteins and cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Quantification of protein signals was performed by densitometry using National Institutes of Health Image software.
Immunofluorescence microscopy
Subcellular fractionation
Cytoplasmic and nuclear fractions were prepared from HeLa cells using CelLytic NuCLEAR extraction kit (Sigma) according to the manufacturer's protocol. Cytoplasmic and nuclear extracts were prepared as described.27
In vitro binding assay
Myc-FANCA, Myc-FANCG, and FANCA proteins were expressed in rabbit reticulocyte lysates (RRLs) using the TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI) according to the supplier's instructions. In vitro transcription/translation reactions were performed in a total volume of 50 μL for 90 minutes at 30°C, using pcDNA3 plasmids encoding the proteins. Reaction mixture was diluted with 500 μL HEPES buffer (10 mM HEPES, pH. 7.6, 150 mM KCl, 10 mM MgCl2, 5 mM NaF, 20 mM Na2MoO4, 0.1% NP40) and the synthesized Myc-FANCA and Myc-FANCG were immunoprecipitated with an anti-Myc antibody, followed by immunoblot analyses. FLAG-tagged wild-type and truncated polypeptides of FANCA were synthesized in vitro, using DNA templates containing the T7 promoter and corresponding nucleotide sequences, and immunoprecipitated from the reaction using anti-FLAG antibody, followed by immunoblot analyses.
Cell survival assay
Cells were seeded in 6-well tissue culture plates at a density of 2 × 105 cells/well and allowed to grow overnight, and then treated with drugs, as indicated. Cells were replated in 96-well tissue culture plates at a density of 5 × 103 cells/well, and then incubated with drug-free culture medium to complete a total 72 hours from the time of initial drug application. Cell survival was colorimetrically measured using 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate assay kit (Dojindo Molecular Technologies, Gaithersburg, MD) as described.27
TUNEL staining
Drug-treated cells were replated into 8-well chamber slides (Electron Microscopy Science, Fort Washington, PA) at a density of 2 × 104 cells/well and incubated in drug-free culture medium, as indicated. Cells were fixed in fresh 4% paraformaldehyde for 25 minutes followed by permeabilization with 0.3% Triton X-100 in PBS for 5 minutes. TdT-mediated dUTP nick-end labeling (TUNEL) staining was performed with a detection kit (Roche Diagnostics).
Chromosome breakage analysis
HeLa cells were treated with drugs for 24 hours and Colcemid (Gibco BRL, Carlsbad, CA) for the last 3 hours. Cells were swollen using 0.075 M KCl and fixed with methanol-acetic acid (3:1). Slides were stained with Giemsa and 50 to 100 metaphases were examined.
Results
Hsp90 associates with FANCA in vivo
To elucidate regulatory mechanisms of the FA core complex, we conducted LC-MS/MS analysis of FANCA-containing protein complexes immunopurified from HeLa/FH-FANCA cells. We identified 96 potential FANCA-associated proteins after subtracting proteins in the immunoprecipitated fraction from mock-transfected cells. This experiment was validated by detection of known FANCA-associated proteins—that is, FANCG,20,22 FANCB,30 FANCL,29 and BLM.46 Among the potential FANCA-associated proteins, Hsp90 α and Hsp90 β were the major components. The association of Hsp90 with FANCA was confirmed by immunoblotting analysis and reciprocal immunoprecipitation followed by immunoblotting (Figure 1A, lanes 1-4). In similar studies, the association of FANCA with both isoforms of Hsp90 was confirmed (data not shown). Moreover, in parental HeLa cells (Figure 1A, lanes 5-8) and K562 human myeloid leukemic cells (data not shown), endogenous FANCA associated with Hsp90 and its cochaperone p23, which preferentially binds to an ATP-bound form of Hsp90, stabilizing the interaction between Hsp90 and its client proteins.2 In addition, Hsp70 coimmunoprecipitated with endogenous FANCA in these cells (data not shown). These data suggest that the Hsp90-based chaperone complex1,2 associates with FANCA.
Hsp90 specifically associates with FANCA in vivo. (A) Coimmunoprecipitation of Hsp90 with FANCA. Lysates from control HeLa cells and HeLa/FH-FANCA cells were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with anti-FLAG and anti-Hsp90 antibodies (lanes 1 and 2). Reciprocally, lysates from HeLa/FH-FANCA cells were immunoprecipitated with either control mouse IgG or mouse anti-Hsp90 monoclonal antibody and immunoblotted with anti-FLAG and anti-Hsp90 antibodies (lanes 3 and 4). Lysates from HeLa cells were immunoprecipitated with either control rabbit IgG or anti-FANCA and immunoblotted with the indicated antibodies (lanes 5 and 6), or immunoprecipitated with either control mouse IgG or anti-Hsp90 antibody and immunoblotted with the indicated antibodies (lanes 7 and 8). (B) Inhibition of the interaction between FANCA and Hsp90 by 17-AAG. HeLa/FH-FANCA cells were treated with vehicle (−) or 250 nM 17-AAG (+) for 15 minutes (lanes 2 and 3). Vehicle-treated parental HeLa cells were used as control (lane 1). Lysates from these cells were immunoprecipitated with anti-FLAG antibody and immunoblotted with the indicated antibodies (upper panels), or were immunoprecipitated with anti-Hsp90 antibody and immunoblotted with the indicated antibodies (middle panels). Lysates were directly immunoblotted with the indicated antibodies to confirm that FH-FANCA and Hsp90 protein levels were constant after 17-AAG treatment (lower panels). (C) Stabilization of the interaction between FANCA and Hsp90 by molybdate. HeLa/FH-FANCA cells were lysed in a buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2, 150 mM KCl, 0.2% Tween 20) in the presence (+) or absence (−) of 20 mM molybdate. Lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with the indicated antibodies (upper panels), or immunoprecipitated with anti-Hsp90 antibody and immunoblotted with the indicated antibodies (lower panels).
Hsp90 specifically associates with FANCA in vivo. (A) Coimmunoprecipitation of Hsp90 with FANCA. Lysates from control HeLa cells and HeLa/FH-FANCA cells were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with anti-FLAG and anti-Hsp90 antibodies (lanes 1 and 2). Reciprocally, lysates from HeLa/FH-FANCA cells were immunoprecipitated with either control mouse IgG or mouse anti-Hsp90 monoclonal antibody and immunoblotted with anti-FLAG and anti-Hsp90 antibodies (lanes 3 and 4). Lysates from HeLa cells were immunoprecipitated with either control rabbit IgG or anti-FANCA and immunoblotted with the indicated antibodies (lanes 5 and 6), or immunoprecipitated with either control mouse IgG or anti-Hsp90 antibody and immunoblotted with the indicated antibodies (lanes 7 and 8). (B) Inhibition of the interaction between FANCA and Hsp90 by 17-AAG. HeLa/FH-FANCA cells were treated with vehicle (−) or 250 nM 17-AAG (+) for 15 minutes (lanes 2 and 3). Vehicle-treated parental HeLa cells were used as control (lane 1). Lysates from these cells were immunoprecipitated with anti-FLAG antibody and immunoblotted with the indicated antibodies (upper panels), or were immunoprecipitated with anti-Hsp90 antibody and immunoblotted with the indicated antibodies (middle panels). Lysates were directly immunoblotted with the indicated antibodies to confirm that FH-FANCA and Hsp90 protein levels were constant after 17-AAG treatment (lower panels). (C) Stabilization of the interaction between FANCA and Hsp90 by molybdate. HeLa/FH-FANCA cells were lysed in a buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2, 150 mM KCl, 0.2% Tween 20) in the presence (+) or absence (−) of 20 mM molybdate. Lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with the indicated antibodies (upper panels), or immunoprecipitated with anti-Hsp90 antibody and immunoblotted with the indicated antibodies (lower panels).
To test whether the FANCA/Hsp90 interaction was specific, we examined effects of 17-AAG and molybdate. 17-AAG disrupts the interaction between Hsp90 and its clients,2–4 whereas molybdate stabilizes the interaction.47,48 Our results showed that cellular treatment with 17-AAG induced a rapid dissociation between Hsp90 and FANCA (Figure 1B). As previously described,49 17-AAG disrupted the interaction between Hsp90 and p23 under the same conditions. Figure 1C shows that the FANCA/Hsp90 interaction was labile in a buffer without molybdate, whereas the interaction was prominently stabilized in the presence of molybdate (Figure 1C). Since vanadate has similar function,48 our standard lysis buffer containing vanadate presumably made it easier to detect the Hsp90/FANCA complex. Collectively these results strongly suggest that the FANCA/Hsp90 interaction is associated with the chaperone activity of Hsp90.
Hsp90 inhibition induces proteasomal degradationand cytoplasmic redistribution of FANCA
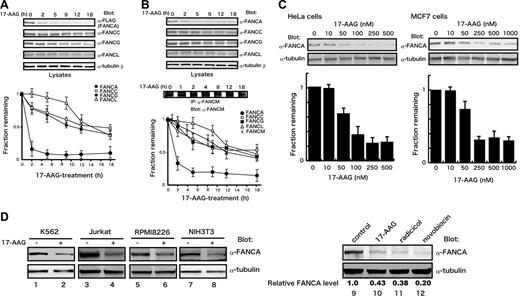
To understand the functional significance of the association of Hsp90 with FANCA, we examined effects of 17-AAG on FANCA and other FA proteins included in the FA core complex in HeLa/FH-FANCA (Figure 2A) and parental HeLa (Figure 2B) cells. In both cells, FANCA was markedly down-regulated within 1 to 2 hours after cellular treatment with 17-AAG, whereas other FA proteins decreased gradually following rapid reduction in FANCA levels. The different kinetics of FANCA and FANCG decreases seemed to conflict with previous observations that the 2 proteins bind and stabilize each other, and that deficiency of either causes accelerated degradation of the other.24 However, it should be noted that our experimental conditions were different from previous studies.24 Most importantly, FANCA- and FANCG-deficient cells were used in the previous studies,24 whereas effects of the drug-induced acute depletions of FANCA and FANCG were analyzed in the present study. Figure 2C shows that 17-AAG induced decreases of FANCA protein levels in HeLa cells and MCF7 human breast cancer cells, in a dose-dependent manner within the range of 50 to 500 nM, which was consistent with known effective doses of the drug.50 The dramatic effect of 17-AAG on FANCA protein levels suggested that FANCA might be a client of Hsp90. To confirm that the effect of 17-AAG was not cell-type specific, we examined the effect of the drug in other cells. Following drug treatment, FANCA was markedly decreased in K562 cells, Jurkat human T-cell leukemia/lymphoma cells, RPMI8226 human myeloma cells, and NIH3T3 murine fibroblasts (Figure 2D, lanes 1-8). In addition, the structurally unrelated Hsp90 inhibitors radicicol and novobiocin,4 showed similar effects on FANCA protein levels (Figure 2D, lanes 9-12), excluding nonspecific effects of 17-AAG. To confirm the effects of Hsp90 inhibitors, we tried to deplete Hsp90 by transient transfection of siRNA, but Hsp90 levels were only mildly reduced and FANCA protein levels were not affected (data not shown).
17-AAG induces a rapid down-regulation of FANCA. HeLa/FH-FANCA (A) and parental HeLa (B) cells were treated with 250 nM 17-AAG for the indicated times. Lysates were immunoblotted with the indicated antibodies. To assess total cellular levels of FANCM, whole-cell lysates were immunoprecipitated (IP) and immunoblotted with anti-FANCM antibody. FA protein signals were quantified and normalized against tubulin-β signals. Data represent means ± SE from 3 independent experiments (bottom graphs). (C) Dose-dependent down-regulation of FANCA by 17-AAG. HeLa and MCF7 cells were treated with various concentrations of 17-AAG for 2 hours. Cell lysates were immunoblotted with the indicated antibodies (upper panels). FANCA signals were quantified and normalized against tubulin-β signals. Data represent means ± SD from 3 independent experiments (bottom graphs). (D) After treatment with vehicle (−) or 17-AAG (+), lysates from K562 cells (500 nM, 4 hours), Jurkat cells (250 nM, 2 hours), RPMI8226 cells (1 μM, 2 hours), and NIH3T3 cells (1 μM, 4 hours) were immunoblotted with appropriate antibodies (lanes 1-8). HeLa cells were treated with 250 nM 17-AAG, 2 μM radicicol, or 2 mM novobiocin for 2 hours (lanes 9-12). Cell lysates were immunoblotted with anti-FANCA and anti–tubulin-β antibodies. Numbers at the bottom of sample lanes 9-12 represent relative FANCA protein levels normalized against control.
17-AAG induces a rapid down-regulation of FANCA. HeLa/FH-FANCA (A) and parental HeLa (B) cells were treated with 250 nM 17-AAG for the indicated times. Lysates were immunoblotted with the indicated antibodies. To assess total cellular levels of FANCM, whole-cell lysates were immunoprecipitated (IP) and immunoblotted with anti-FANCM antibody. FA protein signals were quantified and normalized against tubulin-β signals. Data represent means ± SE from 3 independent experiments (bottom graphs). (C) Dose-dependent down-regulation of FANCA by 17-AAG. HeLa and MCF7 cells were treated with various concentrations of 17-AAG for 2 hours. Cell lysates were immunoblotted with the indicated antibodies (upper panels). FANCA signals were quantified and normalized against tubulin-β signals. Data represent means ± SD from 3 independent experiments (bottom graphs). (D) After treatment with vehicle (−) or 17-AAG (+), lysates from K562 cells (500 nM, 4 hours), Jurkat cells (250 nM, 2 hours), RPMI8226 cells (1 μM, 2 hours), and NIH3T3 cells (1 μM, 4 hours) were immunoblotted with appropriate antibodies (lanes 1-8). HeLa cells were treated with 250 nM 17-AAG, 2 μM radicicol, or 2 mM novobiocin for 2 hours (lanes 9-12). Cell lysates were immunoblotted with anti-FANCA and anti–tubulin-β antibodies. Numbers at the bottom of sample lanes 9-12 represent relative FANCA protein levels normalized against control.
Because 17-AAG is known to accelerate degradation of Hsp90-interacting clients, we assessed the effect of 17-AAG on stability of FANCA by monitoring FANCA protein levels after blocking protein synthesis (Figure 3A-B). The half-life of FANCA was about 8 hours in HeLa/FH-FANCA cells, whereas it was reduced to about 1 hour in 17-AAG–treated cells, suggesting that the drug accelerated degradation of FANCA (Figure 3A). Similarly, the half-life of endogenous FANCA was markedly reduced from about 8 hours to about 2 hours after 17-AAG treatment of parental HeLa cells (Figure 3B). We next examined effects of proteasome inhibitors. As shown in Figure 3C, MG132 and lactacystin inhibited 17-AAG–induced reduction of FANCA. Polyubiquitination precedes proteasomal degradation of a wide variety of proteins. We therefore examined the effect of 17-AAG on ubiquitination of FANCA. For this purpose, cell lysates of HeLa/FH-FANCA were immunoprecipitated using an anti-FLAG antibody, and immunoblotted with an antiubiquitin antibody. Drug treatment increased polyubiquitinated forms of FANCA, detected as a smear with slower mobility, particularly in the presence of MG132 (Figure 3D left panels). The presence of ubiquitinated FANCA in cells treated with MG132 alone suggested that FH-FANCA was ubiquitinated at basal states. To study ubiquitination of endogenous FANCA, we transiently transfected HeLa cells with HA-ubiquitin and treated the cells with 17-AAG or MG132 or both. FANCA was immunoprecipitated and immunoblotted with an anti-HA antibody. Consistent with the results obtained in HeLa/FH-FANCA, treatment with 17-AAG with or without MG132 promoted polyubiquitination of FANCA (Figure 3D right panels). Taken together, these results suggest that Hsp90 inhibition induces FANCA degradation through the ubiquitin-proteasome pathway.
17-AAG induces degradation of FANCA via the ubiquitin-proteasome pathway. (A-B) FANCA degradation induced by 17-AAG. HeLa/FH-FANCA (A) and parental HeLa (B) cells were treated with 100 μg/mL CHX alone (CHX) or with 250 nM 17-AAG (CHX + 17-AAG) for the indicated times. Cell lysates were immunoblotted with the indicated antibodies (upper blots). FANCA signals were quantified and normalized against tubulin-β signals. Data represent means ± SE from 3 independent experiments (bottom graphs). (C) Proteasome inhibitors block 17-AAG–induced FANCA down-regulation. HeLa/FH-FANCA cells were treated with 17-AAG or MG132, at appropriate concentrations for 4 hours (lanes 1-6). Cell lysates prepared using SDS-sample buffer were immunoblotted with anti-FLAG and anti–tubulin-β antibodies. HeLa cells were treated with 17-AAG and proteasome inhibitors, MG132 or lactacystin, at appropriate concentrations for 4 hours (lanes 7-15). Cell lysates prepared using SDS-sample buffer were immunoblotted with anti-FANCA and anti–tubulin-β antibodies. (D) Enhancement of polyubiquitination of FANCA by 17-AAG. HeLa/FH-FANCA cells were treated with vehicle (−) or 250 nM 17-AAG (+), in the absence (−) or presence (+) of 10 μM MG132 for 1 hour (lanes 1-4). Cell lysates prepared using ubiquitin lysis buffer were immunoprecipitated with anti-FLAG antibody and immunoblotted with antiubiquitin and anti-HA antibodies to detect polyubiquitinated FANCA (Ubn-FANCA). The arrow indicates nonubiquitinated FANCA. HeLa cells were transfected with empty vector (−; lane 5) or a plasmid encoding HA-ubiquitin (HA-Ub; +; lanes 6-10). After 24 hours of transfection, cells were treated with 17-AAG and MG132, as described. Cell lysates were immunoprecipitated with either anti-FANCA antibody (lanes 5-9) or control rabbit IgG (lane 10) and immunoblotted with anti-HA and anti-FANCA antibodies. (E) Association of CHIP with FANCA. HeLa/FH-FANCA cells were treated with 17-AAG and MG132 alone or in combination (lanes 1-4), as described in Figure 3D. Cell lysates were immunoprecipitated using anti-FLAG M2 agarose and immunoblotted with the indicated antibodies (upper panels). The same lysates were immunoblotted with the indicated antibodies (lower panels).
17-AAG induces degradation of FANCA via the ubiquitin-proteasome pathway. (A-B) FANCA degradation induced by 17-AAG. HeLa/FH-FANCA (A) and parental HeLa (B) cells were treated with 100 μg/mL CHX alone (CHX) or with 250 nM 17-AAG (CHX + 17-AAG) for the indicated times. Cell lysates were immunoblotted with the indicated antibodies (upper blots). FANCA signals were quantified and normalized against tubulin-β signals. Data represent means ± SE from 3 independent experiments (bottom graphs). (C) Proteasome inhibitors block 17-AAG–induced FANCA down-regulation. HeLa/FH-FANCA cells were treated with 17-AAG or MG132, at appropriate concentrations for 4 hours (lanes 1-6). Cell lysates prepared using SDS-sample buffer were immunoblotted with anti-FLAG and anti–tubulin-β antibodies. HeLa cells were treated with 17-AAG and proteasome inhibitors, MG132 or lactacystin, at appropriate concentrations for 4 hours (lanes 7-15). Cell lysates prepared using SDS-sample buffer were immunoblotted with anti-FANCA and anti–tubulin-β antibodies. (D) Enhancement of polyubiquitination of FANCA by 17-AAG. HeLa/FH-FANCA cells were treated with vehicle (−) or 250 nM 17-AAG (+), in the absence (−) or presence (+) of 10 μM MG132 for 1 hour (lanes 1-4). Cell lysates prepared using ubiquitin lysis buffer were immunoprecipitated with anti-FLAG antibody and immunoblotted with antiubiquitin and anti-HA antibodies to detect polyubiquitinated FANCA (Ubn-FANCA). The arrow indicates nonubiquitinated FANCA. HeLa cells were transfected with empty vector (−; lane 5) or a plasmid encoding HA-ubiquitin (HA-Ub; +; lanes 6-10). After 24 hours of transfection, cells were treated with 17-AAG and MG132, as described. Cell lysates were immunoprecipitated with either anti-FANCA antibody (lanes 5-9) or control rabbit IgG (lane 10) and immunoblotted with anti-HA and anti-FANCA antibodies. (E) Association of CHIP with FANCA. HeLa/FH-FANCA cells were treated with 17-AAG and MG132 alone or in combination (lanes 1-4), as described in Figure 3D. Cell lysates were immunoprecipitated using anti-FLAG M2 agarose and immunoblotted with the indicated antibodies (upper panels). The same lysates were immunoblotted with the indicated antibodies (lower panels).
Because Hsp90 inhibition promotes Hsp70-mediated association of several clients with the chaperone-associated ubiquitin ligase CHIP leading to their proteasomal degradation,6,7,51 we studied its interaction with FANCA. Our results showed that interactions of CHIP with FANCA and Hsp70 were markedly enhanced after 17-AAG treatment in HeLa/FH-FANCA cells (Figure 3E). The proteasome inhibitor MG132 stabilized the CHIP-Hsp70-FANCA complex, in the absence and presence of 17-AAG. These results suggest that CHIP may be involved in the 17-AAG-induced proteasomal degradation of FANCA.
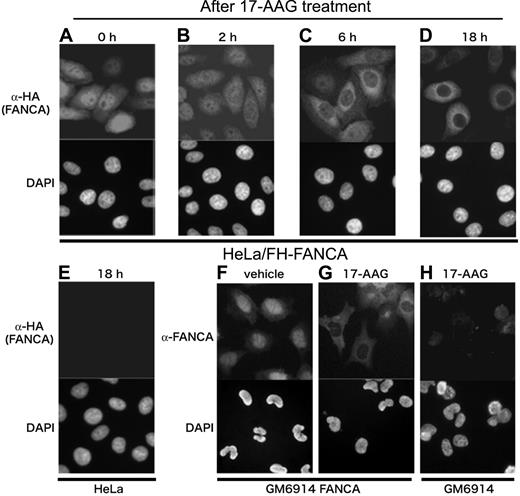
Nuclear localization of FANCA is critical for its function and appears to be regulated by its tertiary conformation of the C-terminus18,27,39 ; in addition, Hsp90 regulates transmembrane trafficking of some client proteins.2 We therefore studied the effect of 17-AAG on subcellular localization of FANCA. Anti-HA immunostaining of HeLa/FH-FANCA cells (Figure 4A-D) showed that although transduced FANCA was predominant in the nucleus at basal states, as previously described,18,20,27,39 it relocalized to the cytoplasm, particularly to the perinuclear cytoplasm after cellular exposure to 17-AAG (Figure 4C-D). Because FH-FANCA was reduced during drug treatment, exposure time for image acquisition was adjusted for optimal visualization of each specimen. Few signals were observed in the nuclei and the cytoplasm of control HeLa cells (Figure 4E), supporting the specificity of fluorescence signals in this system. To further confirm these results, GM6914 fibroblasts stably transduced with wild-type FANCA were immunostained with an anti-FANCA antibody, and again the drug-induced cytoplasmic relocalization of FANCA was observed (Figure 4F-G). Most of control GM6914 cells gave only a few background signals (Figure 4H).
17-AAG induces cytoplasmic relocalization of FANCA. HeLa/FH-FANCA cells (A-D) and control HeLa cells (E) were treated with 250 nM 17-AAG for the indicated times, and then stained with anti-HA antibody. GM6914 cells stably expressing wild-type FANCA (GM6914/FANCA) were cultured with vehicle (F) or 250 nM 17-AAG (G) for 18 hours, and then stained with anti-FANCA antibody. As a control, GM6914 cells treated with 250 nM 17-AAG for 18 hours were stained with anti-FANCA antibody (H). For optimal visualization of fluorescence signals, exposure time was adjusted. Cell nuclei were visualized with DAPI staining. Images were obtained on an Olympus AX70 microscope equipped with UPlan Apo 20×/0.70 NA and WH10×/22 lenses (Olympus, Tokyo, Japan) using a PXL charged-coupled device camera (model CH1; Photometrics, Osnabruck, Germany).
17-AAG induces cytoplasmic relocalization of FANCA. HeLa/FH-FANCA cells (A-D) and control HeLa cells (E) were treated with 250 nM 17-AAG for the indicated times, and then stained with anti-HA antibody. GM6914 cells stably expressing wild-type FANCA (GM6914/FANCA) were cultured with vehicle (F) or 250 nM 17-AAG (G) for 18 hours, and then stained with anti-FANCA antibody. As a control, GM6914 cells treated with 250 nM 17-AAG for 18 hours were stained with anti-FANCA antibody (H). For optimal visualization of fluorescence signals, exposure time was adjusted. Cell nuclei were visualized with DAPI staining. Images were obtained on an Olympus AX70 microscope equipped with UPlan Apo 20×/0.70 NA and WH10×/22 lenses (Olympus, Tokyo, Japan) using a PXL charged-coupled device camera (model CH1; Photometrics, Osnabruck, Germany).
Hsp90 stabilizes cytoplasmic FANCA independently of FANCG
To further characterize the interaction between Hsp90 and FANCA, we determined the subcellular localization of the FANCA/Hsp90 complex. Cell fractionation studies using HeLa/FH-FANCA cells (Figure 5A) showed that the FANCA/Hsp90 complex was mainly detectable in the cytoplasm, whereas the FANCA/C/F complex was predominant in the nucleus, as previously described.20,23 Consistently, the association between Hsp90 and endogenous FANCA was mostly attributed to the cytoplasmic Hsp90/FANCA complex in parental HeLa cells (Figure 5B). These results suggest that the major target of Hsp90 is a cytoplasmic fraction of FANCA.
Hsp90 stabilizes cytoplasmic FANCA in a FANCG-independent manner. (A) Cell fractionation study of HeLa/FH-FANCA. Cytoplasmic extracts (Cyto) and nuclear extracts (Nuc) prepared from control HeLa and HeLa/FH-FANCA cells were immunoprecipitated (IP) with anti-FLAG antibody followed by immunoblotting with the indicated antibodies (upper panels). The same extracts were directly immunoblotted with the indicated antibodies (lower panels). Topoisomerase II (Top II) and tubulin-β are nuclear and cytoplasmic markers, respectively. (B) Cell fractionation study of parental HeLa cells. Whole-cell lysates (Whole) and cytoplasmic extracts (Cyto) and nuclear extracts (Nuc), prepared from HeLa cells (1 ×107 cells) were immunoprecipitated with either control rabbit IgG or anti-FANCA antibody, and then immunoblotted with anti-FANCA and anti-Hsp90 antibodies (upper panels). The same extracts were directly immunoblotted with the indicated antibodies (lower panels). Topoisomerase II (Top II) and tubulin-β are nuclear and cytoplasmic markers, respectively. (C) In vitro interaction of FANCA with Hsp90. In vitro transcription/translation reactions were programmed with empty vector (control), pcDNA3 Myc-FANCA (Myc-FANCA), or pcDNA3 Myc-FANCG (Myc-FANCG) in the absence (−) or presence (+) of 10 μM 17-AAG (lanes 1-4). Reaction mixtures were immunoprecipitated with anti-Myc antibody and immunoblotted with the indicated antibodies (upper panels), or immunoprecipitated with anti-Hsp90 antibody and immunoblotted with the indicated antibodies (middle panels). The arrow indicates Myc-FANCA. A portion (10%) of the input material was directly immunoblotted with anti-Myc antibody to detect synthesized FA proteins (lower panels). FANCG was synthesized alone or with Myc-FANCA, in the absence (−) or presence (+) of 10 μM 17-AAG, as described (lanes 5-7). Reaction mixtures were immunoprecipitated with anti-Myc antibody and immunoblotted with the indicated antibodies. (D) Recognition of different regions of FANCA by Hsp90 and FANCG. FANCA full-length protein and deletion mutants with a FLAG-tag at their N-termini were synthesized in vitro. Structures of the deletion mutants are schematically shown at the bottom of the figure. Reaction mixtures were immunoprecipitated with anti-FLAG antibody, followed by immunoblotting with anti-FLAG and anti-Hsp90 antibodies. Arrows indicate synthesized FANCA polypeptides. The same FANCA polypeptides were cosynthesized with Myc-FANCG in vitro, and reaction mixtures were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG (data not shown) and anti-Myc antibodies. Results of binding studies are summarized on the right for each mutant (bottom, ranging from negative, −, to strongly positive, ++). (E) Interaction of Hsp90 with FANCA mutants. Lysates from GM6914 cells stably expressing FLAG-tagged wild-type (WT) or mutant FANCA proteins were immunoprecipitated with anti-FLAG antibody (upper panels) or anti-Hsp90 antibody (lower panels) and immunoblotted with the indicated antibodies. (F) Sensitivity of ΔNLS FANCA mutant to 17-AAG. GM6914 cells expressing wild-type FANCA (WT) or ΔNLS mutant were treated with 100 μg/mL CHX alone, (CHX) or with 250 nM 17-AAG (CHX + 17-AAG) for the indicated times. Cell lysates were immunoblotted with anti-FANCA and anti–tubulin-β antibodies. FANCA signals were quantified and normalized against tubulin-β signals. Data represent means ± SE from 3 independent experiments (bottom graph).
Hsp90 stabilizes cytoplasmic FANCA in a FANCG-independent manner. (A) Cell fractionation study of HeLa/FH-FANCA. Cytoplasmic extracts (Cyto) and nuclear extracts (Nuc) prepared from control HeLa and HeLa/FH-FANCA cells were immunoprecipitated (IP) with anti-FLAG antibody followed by immunoblotting with the indicated antibodies (upper panels). The same extracts were directly immunoblotted with the indicated antibodies (lower panels). Topoisomerase II (Top II) and tubulin-β are nuclear and cytoplasmic markers, respectively. (B) Cell fractionation study of parental HeLa cells. Whole-cell lysates (Whole) and cytoplasmic extracts (Cyto) and nuclear extracts (Nuc), prepared from HeLa cells (1 ×107 cells) were immunoprecipitated with either control rabbit IgG or anti-FANCA antibody, and then immunoblotted with anti-FANCA and anti-Hsp90 antibodies (upper panels). The same extracts were directly immunoblotted with the indicated antibodies (lower panels). Topoisomerase II (Top II) and tubulin-β are nuclear and cytoplasmic markers, respectively. (C) In vitro interaction of FANCA with Hsp90. In vitro transcription/translation reactions were programmed with empty vector (control), pcDNA3 Myc-FANCA (Myc-FANCA), or pcDNA3 Myc-FANCG (Myc-FANCG) in the absence (−) or presence (+) of 10 μM 17-AAG (lanes 1-4). Reaction mixtures were immunoprecipitated with anti-Myc antibody and immunoblotted with the indicated antibodies (upper panels), or immunoprecipitated with anti-Hsp90 antibody and immunoblotted with the indicated antibodies (middle panels). The arrow indicates Myc-FANCA. A portion (10%) of the input material was directly immunoblotted with anti-Myc antibody to detect synthesized FA proteins (lower panels). FANCG was synthesized alone or with Myc-FANCA, in the absence (−) or presence (+) of 10 μM 17-AAG, as described (lanes 5-7). Reaction mixtures were immunoprecipitated with anti-Myc antibody and immunoblotted with the indicated antibodies. (D) Recognition of different regions of FANCA by Hsp90 and FANCG. FANCA full-length protein and deletion mutants with a FLAG-tag at their N-termini were synthesized in vitro. Structures of the deletion mutants are schematically shown at the bottom of the figure. Reaction mixtures were immunoprecipitated with anti-FLAG antibody, followed by immunoblotting with anti-FLAG and anti-Hsp90 antibodies. Arrows indicate synthesized FANCA polypeptides. The same FANCA polypeptides were cosynthesized with Myc-FANCG in vitro, and reaction mixtures were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG (data not shown) and anti-Myc antibodies. Results of binding studies are summarized on the right for each mutant (bottom, ranging from negative, −, to strongly positive, ++). (E) Interaction of Hsp90 with FANCA mutants. Lysates from GM6914 cells stably expressing FLAG-tagged wild-type (WT) or mutant FANCA proteins were immunoprecipitated with anti-FLAG antibody (upper panels) or anti-Hsp90 antibody (lower panels) and immunoblotted with the indicated antibodies. (F) Sensitivity of ΔNLS FANCA mutant to 17-AAG. GM6914 cells expressing wild-type FANCA (WT) or ΔNLS mutant were treated with 100 μg/mL CHX alone, (CHX) or with 250 nM 17-AAG (CHX + 17-AAG) for the indicated times. Cell lysates were immunoblotted with anti-FANCA and anti–tubulin-β antibodies. FANCA signals were quantified and normalized against tubulin-β signals. Data represent means ± SE from 3 independent experiments (bottom graph).
The FANCA/G complex is detectable in the cytoplasm probably as an early intermediate subcomplex during assembly of the FA core complex.20,22,27,32 In addition, FANCG has a tetratricopeptide repeat domain,52 which serves as an Hsp90-interacting consensus motif in various cochaperones.1 Thus, it is hypothesized that Hsp90 directly binds to FANCG and stabilizes FANCA through promoting the FANCA/G interaction. To address this question, we expressed FANCA or FANCG by in vitro translation in RRL. RRL is a rich source of Hsp90 and its cofactors such as Hsp70 and p23, which form the Hsp90-based multichaperone complex. In these reactions, FANCA associated with Hsp90 in a 17-AAG–sensitive manner, whereas FANCG did not (Figure 5C upper panels). When FANCA and FANCG were cotranslated in RRL, the 2 proteins formed a complex, which was not affected in the presence of 17-AAG (Figure 5C lower panels). Thus, contrary to this hypothesis, the Hsp90 chaperone complex directly bound FANCA but not FANCG in vitro and had little effect on the FANCA/G interaction.
To determine the region of FANCA that was responsible for the interaction with Hsp90, we expressed a series of FANCA deletion mutants and examined their interactions with Hsp90 in the same system. As shown in Figure 5D, Hsp90 could interact with full-length, 1-1200, and 1-900 fragments of FANCA, whereas the binding of 1-600 FANCA to Hsp90 was drastically diminished, and no interaction was observed between 1-300 FANCA and Hsp90. The N-terminal deletion mutants 301-1455 and 601-1455 FANCA bound to Hsp90. The Hsp90-binding of 901-1455 and 1201-1455 FANCA was reduced and not observed, respectively. Further analysis showed that both of 601-900 and 901-1200 FANCA moderately bound to Hsp90, and 601-1200 FANCA intensely bound to Hsp90. Thus, both amino acid residues 601-900 and 901-1200 of FANCA are required for full binding to Hsp90. On the other hand, 1-300 of FANCA was responsible for binding to FANCG, consistent with previous reports.20,21 These results suggest that Hsp90 and FANCG independently recognize different regions of FANCA.
In a further attempt to identify amino acid residues responsible for interaction with Hsp90, we examined association of Hsp90 with several patient-derived mutant proteins (H492R, H1110P, F1263del, and W1302R), in which mutations were located on and surrounding the Hsp90-binding fragments of FANCA. These mutants, expressed in GM6914 FANCA-null cells, fail to complement the cells and enter the nucleus.27 However, these mutations markedly enhanced interaction between Hsp90 and FANCA (Figure 5E). Although these results may appear to be paradoxical to the notion that Hsp90 promotes FANCA, enhanced binding to client proteins with aberrant conformation is a physiologic behavior of the chaperone. Further work is required to elucidate the structural basis of the interaction between Hsp90 and FANCA. The previous observation that these mutant proteins interact with FANCG to a similar extent with the wild-type protein27 further supports the notion that FANCG and Hsp90 independently interact with FANCA.
Taken together, these results suggest that Hsp90 stabilizes cytoplasmic FANCA in a FANCG-independent manner. To confirm this notion, we stably expressed ΔNLS FANCA mutant lacking an N-terminal region (amino acid residues 1-35) in GM6914 cells, and studied its turnover. The mutant protein fails to bind FANCG and fails to enter the nucleus.18,20,39 The results showed that the ΔNLS mutant was degraded at a similar rate to wild-type FANCA (Figure 5E). Unexpectedly, the basal half-life of the ΔNLS mutant protein was also similar to the wild-type protein. One explanation is that FANCG-binding plays a minor role in the stability of the cytoplasmic pool, although it is required for maintaining total cellular levels of FANCA through stabilization of the FA core complex in the nucleus. Thus, Hsp90 and FANCG play distinct roles in cellular homeostasis of FANCA.
17-AAG suppresses activation of FANCD2
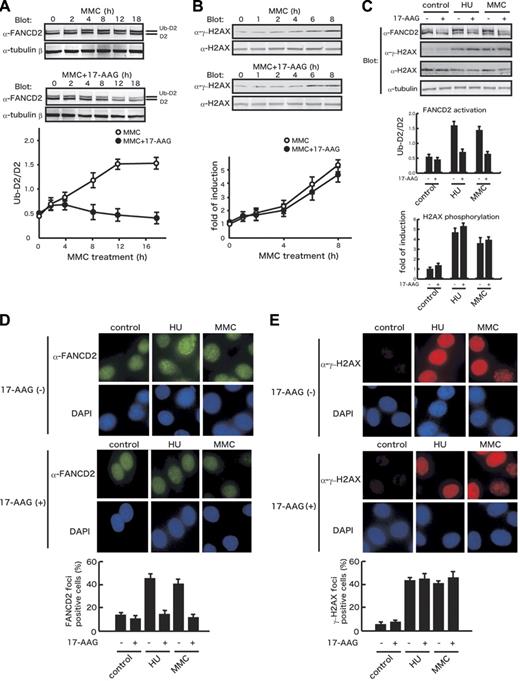
The 17-AAG–induced reduction and cytoplasmic relocalization of FANCA are expected to impair downstream activation of FANCD2 through depletion of the FA core complex in the nucleus. Therefore, we examined the effect of 17-AAG on FANCD2 monoubiquitination following cellular treatment with MMC. In HeLa cells, FANCD2 monoubiquitination began to increase 2 hours after MMC treatment, reaching a maximal plateau at 12 to 16 hours (Figure 6A). Addition of 17-AAG suppressed MMC-stimulated FANCD2 monoubiquitination, which was evident from 8 hours. It should be noted that 17-AAG treatment induced slow decrease in total FANCD2 levels, which may contribute to suppression of FANCD2 monoubiquitination in the late phase. A dose-response study showed that 17-AAG suppressed FANCD2 monoubiquitination at a similar range of concentrations (50-500 nM) to those where FANCA was down-regulated (data not shown). Moreover, the drug suppressed FANCD2 monoubiquitination in HU-treated cells (Figure 6C). FANCD2 monoubiquitination seems to be triggered by the formation of double-strand breaks (DSBs) during DNA replication after cellular exposure to DNA cross-linkers and replication inhibitors.53 Thus, it could be possible that the 17-AAG–mediated suppression of FANCD2 activation was secondary to suppressed DSB formation due to the drug-induced growth arrest. To exclude this possibility, we examined the effect of 17-AAG on phosphorylation of a histone H2A variant H2AX, which is a sensitive marker of DSBs.54 Immunoblot studies showed that 17-AAG treatment had little effects on MMC- or HU-induced increases of phosphorylated H2AX (γ-H2AX; Figure 6B-C), suggesting that the drug impedes a process from DSB formation to FANCD2 activation. We next studied the effect of 17-AAG on formation of FANCD2 nuclear foci representing recruitment of monoubiquitinated FANCD2 to the chromatin,25,35 in comparison with formation of γ-H2AX nuclear foci. Consistent with results of the immunoblot studies, 17-AAG treatment markedly suppressed MMC- or HU-induced FANCD2 nuclear focus formation but had little effect on γ-H2AX nuclear focus formation (Figure 6D-E).
17-AAG inhibits FANCD2 activation but not H2AX phosphorylation. (A) Effects of 17-AAG on MMC-induced FANCD2 monoubiquitination. HeLa cells were treated with 40 ng/mL MMC alone (MMC) or with 250 nM 17-AAG (MMC + 17-AAG) for the indicated times. Cell lysates were immunoblotted with anti-FANCD2 and anti–tubulin-β antibodies. Protein bands corresponding to FANCD2 (D2) and a monoubiquitinated form of FANCD2 (Ub-D2) were quantified, and ratios of the 2 isoforms (Ub-D2/D2) were determined for each sample. Data represent means ± SE from 3 independent experiments (bottom graph). (B) Effects of 17-AAG on MMC-induced H2AX phosphorylation. HeLa cells were treated with 40 ng/mL MMC alone (MMC) or with 250 nM 17-AAG (MMC + 17-AAG) for the indicated times. Cell lysates were immunoblotted with anti-γ-H2AX and anti-H2AX. Protein bands were quantified and fold inductions of γ-H2AX normalized against H2AX were determined for each sample. Data represent means ± SE from 3 independent experiments (bottom graph). (C) Effects of 17-AAG on FANCD2 monoubiquitination and H2AX phosphorylation in control and HU- and MMC-treated HeLa cells. HeLa cells were treated with 1 mM HU or 40 ng/mL MMC in the absence (−) or presence (+) of 250 nM 17-AAG for 8 hours. Cell lysates were immunoblotted with the indicated antibodies. Ratios of FANCD2 isoforms (Ub-D2/D2) and fold inductions of γ-H2AX normalized against H2AX were determined for each sample as described. Data represent means ± SD from 3 independent experiments. (D-E) Effects of 17-AAG on formation of FANCD2 and γ-H2AX nuclear foci. HeLa cells were treated with 1 mM HU or 40 ng/mL MMC in the absence (−) or presence (+) of 250 nM 17-AAG for 8 hours. Cells were stained with anti-FANCD2 (D) or anti-γ-H2AX (E) antibodies. Cell nuclei were visualized with DAPI staining. In each sample, at least 200 nuclei were examined at original magnification ×200. Nuclei containing more than 10 bright foci were scored as FANCD2- and γ-H2AX foci-positive cells. Data represent means ± SD from 3 independent experiments (bottom graphs). Images were obtained on an Olympus AX70 microscope equipped with UPlanApo 20×/0.70 NA and WH 10×/22 objectives (Olympus, Tokyo, Japan) using a PXL charge-coupled device camera (model CH1; Photometrics, Osnabrück, Germany).
17-AAG inhibits FANCD2 activation but not H2AX phosphorylation. (A) Effects of 17-AAG on MMC-induced FANCD2 monoubiquitination. HeLa cells were treated with 40 ng/mL MMC alone (MMC) or with 250 nM 17-AAG (MMC + 17-AAG) for the indicated times. Cell lysates were immunoblotted with anti-FANCD2 and anti–tubulin-β antibodies. Protein bands corresponding to FANCD2 (D2) and a monoubiquitinated form of FANCD2 (Ub-D2) were quantified, and ratios of the 2 isoforms (Ub-D2/D2) were determined for each sample. Data represent means ± SE from 3 independent experiments (bottom graph). (B) Effects of 17-AAG on MMC-induced H2AX phosphorylation. HeLa cells were treated with 40 ng/mL MMC alone (MMC) or with 250 nM 17-AAG (MMC + 17-AAG) for the indicated times. Cell lysates were immunoblotted with anti-γ-H2AX and anti-H2AX. Protein bands were quantified and fold inductions of γ-H2AX normalized against H2AX were determined for each sample. Data represent means ± SE from 3 independent experiments (bottom graph). (C) Effects of 17-AAG on FANCD2 monoubiquitination and H2AX phosphorylation in control and HU- and MMC-treated HeLa cells. HeLa cells were treated with 1 mM HU or 40 ng/mL MMC in the absence (−) or presence (+) of 250 nM 17-AAG for 8 hours. Cell lysates were immunoblotted with the indicated antibodies. Ratios of FANCD2 isoforms (Ub-D2/D2) and fold inductions of γ-H2AX normalized against H2AX were determined for each sample as described. Data represent means ± SD from 3 independent experiments. (D-E) Effects of 17-AAG on formation of FANCD2 and γ-H2AX nuclear foci. HeLa cells were treated with 1 mM HU or 40 ng/mL MMC in the absence (−) or presence (+) of 250 nM 17-AAG for 8 hours. Cells were stained with anti-FANCD2 (D) or anti-γ-H2AX (E) antibodies. Cell nuclei were visualized with DAPI staining. In each sample, at least 200 nuclei were examined at original magnification ×200. Nuclei containing more than 10 bright foci were scored as FANCD2- and γ-H2AX foci-positive cells. Data represent means ± SD from 3 independent experiments (bottom graphs). Images were obtained on an Olympus AX70 microscope equipped with UPlanApo 20×/0.70 NA and WH 10×/22 objectives (Olympus, Tokyo, Japan) using a PXL charge-coupled device camera (model CH1; Photometrics, Osnabrück, Germany).
The DNA damage-sensitive kinase, ataxia telangiectasia and RAD3-related protein (ATR), is required for DNA damage-induced FANCD2 activation.55 However, 17-AAG treatment had little effect on ATR activation, as assessed by phosphorylation of Chk1, a substrate of ATR (data not shown), in agreement with previously published data.11 Collectively, our results suggest that the 17-AAG–induced suppression of FANCD2 activation can be explained by the drug-induced severe reduction and cytoplasmic retention of FANCA.
17-AAG enhances DNA cross-linker–induced cytotoxicity and chromosomal abnormalities
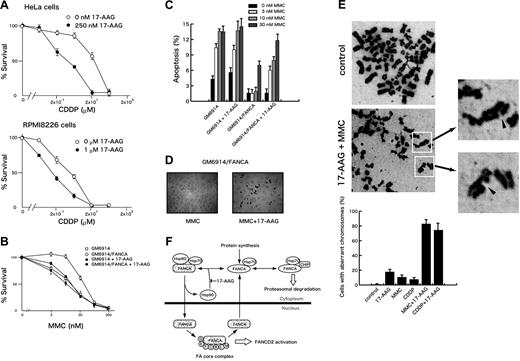
To test whether 17-AAG-induced inactivation of the FA pathway affected cellular sensitivity to DNA cross-linkers, we studied effects of 17-AAG on cisplatin-induced cytotoxicity in HeLa cervical cancer and RPMI8226 myeloma cells. As shown in Figure 7A, 17-AAG sensitized both cells to cisplatin. 17-AAG showed similar effects on MMC-induced cytotoxicity in these cells (data not shown). We next examined whether the effect of 17-AAG on DNA cross-linker–induced cytotoxicity was mediated through suppression of the FA pathway. For this purpose, we studied 17-AAG–induced sensitization to MMC in FANCA-deficient fibroblasts (GM6914) and those complemented by wild-type FANCA (GM6914/FANCA). As shown in Figure 7B, 17-AAG sensitized GM6914/FANCA cells to MMC-induced cytotoxicity, but this effect was much less pronounced in GM6914 cells. To confirm these results, we determined percentages of apoptotic cells by TUNEL assay (Figure 7C-D). 17-AAG significantly increased MMC-induced apoptotic cells in GM6914/FANCA cells, but this effect was not seen in GM6914 cells (Figure 7C). Together, these results suggest that the effect of 17-AAG depends on the presence of an intact FA pathway.
17-AAG enhances DNA cross-linker–induced cytotoxicity and chromosome abnormalities. (A) HeLa cells were treated with various concentrations of cisplatin (CDDP) alone or with 250 nM 17-AAG for 14 hours. RPMI8226 cells were treated with various concentrations of cisplatin (CDDP) alone or with 1 μM 17-AAG for 20 hours. Cells were washed and then incubated in drug-free culture medium. After 72 hours from the time of initial drug application, cell survival was colorimetrically determined. Data represent means ± SD from triplicate measurements. (B) FANCA-deficient (GM6914) and complemented (GM6914/FANCA) cells were treated with various concentrations of MMC alone or with 250 nM 17-AAG for 14 hours. Cells were washed and incubated in drug-free culture medium. After 72 hours from the time of initial drug application, cell survival was colorimetrically measured. (C-D) FANCA-deficient (GM6914) and complemented (GM6914/FANCA) cells were treated with MMC (0, 3, 10, 30 nM) and 250 nM 17-AAG for 14 hours. Cells were washed and then incubated in drug-free culture medium to complete a total of 40 hours from the time of initial drug application. Apoptotic cells were detected by TUNEL staining. In each sample, at least 400 cells were examined at original magnification ×100, and percentages of apoptotic cells were determined. Data represent means ± SD from triplicate measurements. TUNEL stainings of GM6914/FANCA cells treated with 10 nM MMC alone (MMC) or with 250 nM 17-AAG (MMC + 17-AAG) are shown in panel D. Images were obtained on an Olympus AX70 microscope equipped with UPlanApo 10×/0.40 NA and WH 10×/22 lenses (Olympus) using a PXL charge-coupled device camera (model CH1; Photometrics). (E) HeLa cells were treated with 100 nM MMC or 2 μM cisplatin (CDDP) with or without 250 nM 17-AAG for 24 hours. Cells in metaphase were assessed as aberrant if they presented chromatid breaks. Data represent means ± SD from 3 independent experiments. Arrowheads indicate chromatid breaks. Images were obtained on a Leica DM3000 microscope equipped with HI PLAN 100x/1.25 NA and HC PLAN 10x/22 lenses (Leica, Bensheim, Germany) using a DFC280 digital camera (Leica). (F) A model illustrating how Hsp90 regulates intracellular stability and trafficking of FANCA. FANCA shuttles between the cytoplasm and the nucleus. Cytoplasmic FANCA, newly synthesized and exported from the nucleus, is folded into proper conformation required for nuclear entry by interacting with the Hsp90-based multichaperone complex.1,2 Hsp90 is probably recycled to form a complex with FANCA in the cytoplasm. The 17-AAG–mediated inhibition of the chaperone cycle promotes proteasomal degradation of FANCA, at least in part, through Hsp70-mediated association with CHIP. In the nucleus, FANCA, -B, -C, -E, -F, -G, -L, and -M are assembled into a multisubunit complex (FA core complex) that is required for FANCD2 activation.
17-AAG enhances DNA cross-linker–induced cytotoxicity and chromosome abnormalities. (A) HeLa cells were treated with various concentrations of cisplatin (CDDP) alone or with 250 nM 17-AAG for 14 hours. RPMI8226 cells were treated with various concentrations of cisplatin (CDDP) alone or with 1 μM 17-AAG for 20 hours. Cells were washed and then incubated in drug-free culture medium. After 72 hours from the time of initial drug application, cell survival was colorimetrically determined. Data represent means ± SD from triplicate measurements. (B) FANCA-deficient (GM6914) and complemented (GM6914/FANCA) cells were treated with various concentrations of MMC alone or with 250 nM 17-AAG for 14 hours. Cells were washed and incubated in drug-free culture medium. After 72 hours from the time of initial drug application, cell survival was colorimetrically measured. (C-D) FANCA-deficient (GM6914) and complemented (GM6914/FANCA) cells were treated with MMC (0, 3, 10, 30 nM) and 250 nM 17-AAG for 14 hours. Cells were washed and then incubated in drug-free culture medium to complete a total of 40 hours from the time of initial drug application. Apoptotic cells were detected by TUNEL staining. In each sample, at least 400 cells were examined at original magnification ×100, and percentages of apoptotic cells were determined. Data represent means ± SD from triplicate measurements. TUNEL stainings of GM6914/FANCA cells treated with 10 nM MMC alone (MMC) or with 250 nM 17-AAG (MMC + 17-AAG) are shown in panel D. Images were obtained on an Olympus AX70 microscope equipped with UPlanApo 10×/0.40 NA and WH 10×/22 lenses (Olympus) using a PXL charge-coupled device camera (model CH1; Photometrics). (E) HeLa cells were treated with 100 nM MMC or 2 μM cisplatin (CDDP) with or without 250 nM 17-AAG for 24 hours. Cells in metaphase were assessed as aberrant if they presented chromatid breaks. Data represent means ± SD from 3 independent experiments. Arrowheads indicate chromatid breaks. Images were obtained on a Leica DM3000 microscope equipped with HI PLAN 100x/1.25 NA and HC PLAN 10x/22 lenses (Leica, Bensheim, Germany) using a DFC280 digital camera (Leica). (F) A model illustrating how Hsp90 regulates intracellular stability and trafficking of FANCA. FANCA shuttles between the cytoplasm and the nucleus. Cytoplasmic FANCA, newly synthesized and exported from the nucleus, is folded into proper conformation required for nuclear entry by interacting with the Hsp90-based multichaperone complex.1,2 Hsp90 is probably recycled to form a complex with FANCA in the cytoplasm. The 17-AAG–mediated inhibition of the chaperone cycle promotes proteasomal degradation of FANCA, at least in part, through Hsp70-mediated association with CHIP. In the nucleus, FANCA, -B, -C, -E, -F, -G, -L, and -M are assembled into a multisubunit complex (FA core complex) that is required for FANCD2 activation.
FA pathway-defective cells characteristically show a high frequency of chromosomal aberrations after exposure to DNA cross-linkers.14–16 Our results show that chromosome aberrations were detected in a small fraction of HeLa cells after exposure to DNA cross-linkers or 17-AAG, whereas percentages of cells containing chromatid breaks markedly increased after combined treatment with DNA cross-linkers and 17-AAG (Figure 7E). However, radial chromosomes, typical aberrations observed in FA cells, were not detected in these cells. One likely explanation is that Hsp90 inhibition suppresses other DNA repair pathways through which DNA cross-linker–induced DSBs are misrepaired to generate radial chromosomes.14 To support this notion, activation of DNA-dependent protein kinase, a critical component for nonhomologous end joining, is suppressed by Hsp90 inhibitors.13
Discussion
In the present work, we have demonstrated that the Hsp90 chaperone machinery regulates the FA pathway. Our results showed that FANCA associated with Hsp90, in vivo and in vitro, in a 17-AAG–sensitive manner. Furthermore, 17-AAG–mediated disruption of the association between Hsp90 and FANCA induced rapid proteasomal degradation and cytoplasmic retention of FANCA. Together, these results indicate that FANCA is a client of Hsp90. Because nuclear levels of FANCA have profound effect on FANCD2 activation,25,27,29,30 the 17-AAG–induced dramatic effects on intracellular turnover and localization of FANCA could be expected to compromise FANCD2 activation. Indeed, 17-AAG markedly suppressed DNA damage-induced monoubiquitination and nuclear focus formation of FANCD2. Furthermore, the drug enhanced cytotoxicity of DNA cross-linkers, apparently in an FA pathway-dependent manner. Even more remarkably, 17-AAG enhanced DNA cross-linker–induced chromosome aberrations. Recent studies suggest that Hsp90 inhibitors sensitize cells to genotoxic agents through depletion of Chk1 and consequent abrogation of cell cycle checkpoint activation. However, since 17-AAG induced similar decreases of Chk1 in FA-defective (GM6914) and complemented (GM6914/FANCA) cells (data not shown), 17-AAG–induced Chk1 depletion is unlikely to explain selective sensitization of the complemented cells. Taken together, our results suggest that Hsp90 is required for full activation of the FA pathway through maintaining intracellular homeostasis of FANCA (Figure 7F), although we cannot exclude the possibility that 17-AAG–induced down-regulation of other FA proteins contribute to suppression of the FA pathway.
As illustrated in our model (Figure 7F), the present work provides several new insights into the regulation of turnover and trafficking of FANCA. First, the disruption of the interaction between Hsp90 and a cytoplasmic fraction of FANCA rapidly induces a large reduction of total cellular levels of FANCA, despite its predominant localization in the nucleus. One explanation is that a nuclear pool of FANCA is rapidly exchangeable with a cytoplasmic pool that is in continuous dynamic cycles of assembly/disassembly with Hsp90 to be protected from proteolysis. To support this notion, nucleocytoplasmic shuttling of FANCA was recently reported.40 Second, Hsp90 binds and stabilizes cytoplasmic FANCA in a FANCG-independent manner. Although FANCG forms an intermediate subcomplex with FANCA in the cytoplasm,20,22,27,32 this interaction does not seem critical for stability of cytoplasmic FANCA. FANCG-mediated stabilization of FANCA is probably through promoting the FA core complex formation in the nucleus. Third, our results suggested that Hsp90 inhibition markedly accelerated FANCA degradation through the ubiquitin-proteasome pathway. Our preliminary data suggest that Hsp90 inhibition enhances Hsp70-mediated association between FANCA and CHIP,6,7,51 which may contribute to proteasomal targeting of FANCA. Finally, Hsp90 is required for nuclear localization of FANCA, although the underlying mechanism remains unknown. Previous studies suggested that N-terminal NLS as well as C-terminus were required for its nuclear localization.18,27,39 Hence, one likely explanation is that Hsp90-assisted folding of cytoplasmic FANCA is required for the function of NLS or proper conformation of the C-terminal region. An alternative, but not mutually exclusive, explanation is that Hsp90 and its cochaperones link FANCA to the microtubule-based nuclear import machinery, as documented for glucocorticoid receptors and p53.56,57
The present findings have important implications for cancer chemotherapy. DNA cross-linkers are a major class of antitumor agents used for standard therapeutics.58 Increasing evidence suggests that the FA pathway plays an important role in tumor sensitivity to DNA cross-linkers. For example, epigenetic inactivation of FANCF by promoter hypermethylation is associated with cisplatin hypersensitivity in primary ovarian cancers, whereas reactivation of the FANCF gene causes cross-linker resistance.59 In another case, enhancement of the FA pathway is involved in melphalan resistance in myeloma cells.60 Accordingly, pharmacologic inhibition of the FA pathway is expected to sensitize tumor cells to DNA cross-linkers. Recently, Chirnomas et al reported that curcumin inhibits the FA pathway and can be used as a chemosensitizer of cisplatin, although a molecular target of curcumin remains unknown.61 Geldanamycin analogues are now in clinical trials, and new analogues are being developed.3,4 In addition, another group of antitumor agents, histone deacetylase inhibitors, induce hyperacetylation of Hsp90, thereby disrupting its chaperone activity.62–65 These Hsp90 inhibitors are promising for the combined use with DNA cross-linkers. On the other hand, our findings suggest that Hsp90 inhibitors suppress the FA pathway, causing genomic instability in tumor cells, which may lead to malignant progression in vivo. This important question should be addressed in future studies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports and Culture of Japan, and by grants from the Ministry of Health, Labor and Welfare of Japan.
We thank Dr Johan de Winter, Dr Weidong Wang, Dr Maureen Hoatlin, Dr Hiroshi Handa, and Dr Hirokazu Murakami for kindly providing us reagents. We thank Dr Tatsutoshi Nakahata for generous support, Dr Yuko Sato and Dr Miharu Yabe for helpful discussion, and Keiko Nakazato for technical assistance.
Authorship
Contribution: T.O. performed research and wrote the paper; T.H. performed research and analyzed data; H.M. performed research; N.T. analyzed data; and T.Y. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Takayuki Yamashita, Laboratory of Molecular Genetics, Institute for Molecular and Cellular Regulation, Gunma University, 3-39-15 Showa-machi, Maebashi, Gunma 371-8512, Japan; e-mail: y-taka@showa.gunma-u.ac.jp.