Abstract
Gene expression profiling of acute myeloid leukemia (AML) allows the discovery of previously unrecognized molecular entities. Here, we identified a specific subgroup of AML, defined by an expression profile resembling that of AMLs with mutations in the myeloid transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα), while lacking such mutations. We found that in these leukemias, the CEBPA gene was silenced, which was associated with frequent promoter hypermethylation. The leukemias phenotypically showed aberrant expression of T-cell genes, of which CD7 was most consistent. We identified 2 mechanisms that may contribute to this phenotype. First, absence of Cebpa led to up-regulation of specific T-cell transcripts (ie, Cd7 and Lck) in hematopoietic stem cells isolated from conditional Cebpa knockout mice. Second, the enhanced expression of TRIB2, which we identify here as a direct target of the T-cell commitment factor NOTCH1, suggested aberrantly activated Notch signaling. Putatively activating NOTCH1 mutations were found in several specimens of the newly identified subgroup, while a large set of control AMLs was mutation negative. A gene expression prediction signature allowed the detection of similar cases of leukemia in independent series of AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease with respect to its underlying genetic abnormalities and clinical biology.1 It has therefore been postulated that AML, although always characterized by a malignant accumulation of myeloid progenitor cells, in fact represents not a single disease, but a group of neoplasms. Cytogenetic and molecular analyses, studying specific chromosomal translocations and mutations, are used to identify subgroups of AML with distinct biologic and clinical behavior. Recent developments in microarray research have resulted in further improvements in the characterization of the molecular heterogeneity of AML,2 and gene expression profiling (GEP) studies have demonstrated that specific chromosomal aberrations, such as the common translocations t(8;21), t(15;17), and inv(16), correlate with unique expression signatures.3-5
In a GEP study of 285 cases of de novo AML, we found that specific expression profiles are associated with other recurrent genetic aberrations such as mutations in NPM1 and alterations involving MLL, and with overexpression of the proto-oncogene EVI1 as well.4,6 Some AMLs lacking these particular abnormalities were found to express the same characteristic profiles, suggesting that deregulation of the same pathways had occurred through yet-unknown mechanisms. This study also showed that 2 distinct expression clusters (no. 4 and no. 15) consisted primarily of AML cases with mutations in CEBPA, the gene encoding the basic leucine zipper transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα). C/EBPα is a critical regulator of hematopoietic stem cell (HSC) homeostasis and myeloid differentiation,7-10 and multiple studies have shown that C/EBPα function is frequently abrogated in AML. Besides mutations in the CEBPA gene itself, C/EBPα inhibition may result from functional inactivation due to dysregulation by oncogenes, such as AML1-ETO, BCR-ABL, or FLT3-ITD.11-14 While both CEBPA clusters appeared sharply defined, a subset of AMLs in cluster no. 4 did not carry CEBPA mutations, nor were any other common genetic aberrations found, which was suggestive of C/EBPα interference by other mechanisms.
Recently, we found that Tribbles homolog 2 (TRIB2) was highly expressed in several of the cluster no. 4 leukemias lacking CEBPA mutations.15 Trib2 causes fatal transplantable AML when introduced in murine hematopoietic stem cells in vivo.15 Significantly, the leukemogenic potential of Trib2 was associated with its ability to promote degradation of C/EBPα, thus providing further evidence suggesting that disruption of normal C/EBPα function was an important characteristic of this leukemia subgroup. TRIB2 had been initially identified in an unbiased search for downstream effectors of the NOTCH1 pathway.15 NOTCH1 encodes a membrane receptor and transcriptional regulator that plays a critical role in T-cell development.16 Activating NOTCH1 mutations are observed in approximately 50% of individuals with T-cell acute lymphoblastic leukemia (T-ALL), ranking them among the most frequent abnormalities observed in this type of malignancy.17,18 In contrast, NOTCH1 mutations are rare in AML and other malignancies.19-21 The selectively high mRNA levels of the downstream NOTCH1 target TRIB2 pointed to a possible role for activated Notch signaling in these AMLs.
Here, we present the results of experiments that indicate that the leukemias in cluster no. 4 lacking CEBPA mutations represent a previously unidentified subset of AML, of which CEBPA silencing is a key hallmark. Furthermore, we found in these leukemias a strong association with putatively activating mutations in NOTCH1. We established a link between these genetic characteristics and the partially T-lymphoid phenotype of the AMLs, and describe an approach to identify similar leukemia cases in a second cohort of AMLs using gene expression data.
Patients, materials, and methods
This research has been approved by the Institutional Review Board of Erasmus University Medical Center. Informed consent was obtained in accordance with the Declaration of Helsinki.
AML patients, gene expression profiling, and data analysis
We made use of the leukemic cell specimens of 2 independent and representative series of AML patients. The first cohort of de novo AML patients has been described previously.4 The second series is comparable with the first cohort with respect to mean age, white blood cell and bone marrow blast counts, French-American-British (FAB) classification (with the exception of FAB-M3, which is underrepresented in this second cohort), distribution of the main cytogenetic abnormalities (with the exception of t(15;17)), and mutations in FLT3, CEBPA, NPM1, and NRAS (P.J.M.V., B.L., and R.D., manuscript in preparation). Gene expression profiling of 285 AML specimens from the first cohort using Affymetrix (Santa Clara, CA) HGU133A GeneChips has been described previously.4 Further details on data analysis are available in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Nucleotide sequencing of NOTCH1 and CEBPA
RNA isolation and synthesis of complementary DNA (cDNA) were performed as described previously.4 The strategy for determining CEBPA mutations in AML has been described.22 Regions of NOTCH1 cDNA in the heterodimerization domain (HD, exons 26 and 27) and proline-glutamate-serine-threonine–rich domain (PEST, exon 34) found mutated in T-ALL previously17 were analyzed for the presence of mutations (Document S1).
Immunophenotyping and T-cell receptor gene rearrangement analysis
Immunophenotyping was performed by multicolor flow cytometry using an LSR II flow cytometer (BD Biosciences, San Jose, CA). Details on labeling combinations are given in Document S1. Data analysis was performed using FACSDiva software, version 4.1.2 (BD Biosciences). Polymerase chain reaction (PCR) for T-cell receptor (TCR) gene rearrangement analysis was performed in an automated thermocycler (model ABI 9600/2700; Applied Biosystems, Foster City, CA) using primers and conditions as defined in the BIOMED-2 multiplex PCR protocol.23 All BIOMED-2 multiplex PCR kits were obtained from InVivoScribe Technologies (http://www.invivoscribe.com; San Diego, CA). After PCR amplification of TCRG and TCRD gene rearrangements, products were subjected to either heteroduplex analysis (TCRD) or GeneScan analysis (TCRG) for identification of monoclonal gene rearrangements.23
CebpaF/F mice and poly I:C treatment, and staining and sorting of murine bone marrow HSCs
Adult CebpaF/FMx1-Cre conditional knockout mice and Cebpawt/wtMx1-Cre control mice were treated with polyinosinic-polycytidylic acid (poly I:C; Sigma, Steinheim, Germany). Poly I:C (450 μg) was administrated by intraperitoneal injection every other day for a total of 7 injections. Cebpa excision was determined as previously described.7 Hematopoietic stem cells were isolated from murine bone marrow as previously described.24 Briefly, bone marrow cells were stained with rat antibodies specific for lineage markers (ie, CD11b, CD3, CD4, CD8, Gr1, B220 and CD19; Caltag Laboratories, Carlsbad, CA). Lineage-positive cells were partially removed with sheep anti–rat IgG-conjugated immunomagnetic beads (Dynabeads M-450; Dynal, Oslo, Norway). The remaining bone marrow cells were stained with TC-conjugated lineage markers (Caltag Laboratories), APC-conjugated c-Kit and PE-conjugated Sca-1 (BD Biosciences Pharmingen, San Diego, CA). Lin−c-Kit+Sca1+ (KSL) cells were sorted in PBS and resorted in RTL buffer (Qiagen, Valencia, CA) for RNA isolation or culture medium for retroviral transduction.
Cebpa expression construct and retroviral transduction
Murine Cebpa was cloned into the Murine stem cell virus (MSCV)-IRES-GFP retroviral vector. Bosc23 cells were cotransfected using lipofectamin with either empty vector or MSCV-Cebpa-IRES-GFP, a Gag-Pol construct and an ecotropic Env construct. Virus containing supernatants was collected at 48 and 72 hours after transfection, filtered though 0.45-μm filter, and centrifuged at 87275g for 2 hours at 4°C in a Beckman Coulter XL-90 ultracentrifuge (Beckman Coulter, Fullerton, CA) using an SW28 rotor. Pellets were resuspended in PBS containing 0.1% BSA and stored at −80°C. For retroviral transduction, sorted KSL cells were prestimulated in CellGenix SCGM medium (CellGenix, Freiburg, Germany) supplemented with murine Flt3 (100 ng/mL), murine IL3 (20 ng/mL), human IL6 (20 ng/mL), murine Scf (100 ng/mL), and murine Tpo (100 ng/mL) (PeproTech, Rocky Hill, NJ) for 24 hours. Transductions were performed in culture dishes (Falcon 1008; Becton Dickinson, Lincoln Park, NJ) coated with 12 μg/mL retronectin during 2 consecutive days. Transduced cells were sorted twice for GFP expression.
RNA isolation from murine HSCs and real-time quantitative (RQ)–PCR
RNA was extracted with an RNeasy Micro kit (Qiagen) and reverse transcribed with SuperScriptII RNase H-Reverse Transcriptase (Invitrogen, Carlsbad, CA). Complete absence of Cebpa expression was confirmed by RQ-PCR using TaqMan technology on an ABI Prism 7700 Sequence Detector (Applied Biosystems) with primers, probe, and conditions as described previously.25 For Cd7 and Lck, RQ-PCR was performed with SYBR Green (Applied Biosystems) with the following parameters: 50°C for 2 minutes; 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Expression levels were normalized to Gapdh. In the PCRs involving Cd7, the normalized expression levels for samples from same groups (eg, F/F cre+, or wt cre+) were then directly combined for subsequent analysis. In the Lck PCRs, additional normalization to a calibrator sample was carried out according to the ΔΔCt method22 to correct for PCR-dependent variability. Student t test (2-tailed) was performed using Microsoft Excel 2002 (Microsoft, Redmond, WA) to compare expression levels.
NOTCH1 expression construct and retroviral transduction
The intracellular domain of human NOTCH1 (ICN1, amino acids 1760–2555) was subcloned into an MSCV–based retroviral vector that coexpresses truncated human nerve growth factor receptor (tNGFR) as a surrogate marker. Scid.adh cells were grown in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and 1 mM MEM nonessential amino acids. For gamma secretase inhibitor (GSI) rescue experiments, 5 × 105 Scid.adh cells were transduced with NGFR-normalized retroviral supernatants obtained following transfection of 293T cells with either NGFR or NGFR-ICN1 and sorted 48 hours later. Cells were subsequently treated with 1 μM GSI (compound E, synthesized in the laboratory of M. S. Wolfe, Harvard Medical School and Brigham and Women's Hospital, Boston, MA26 ) or DMSO vehicle control for 15 hours. RNA, isolated using the RNEasy kit (Qiagen), was digested with DNaseI and used for reverse transcription according to the manufacturer's instructions (Superscript IITM kit; Invitrogen). Standard semiquantitative polymerase chain reaction (PCR) techniques were used, and murine Hprt was used as an internal control.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed using ChIP assay kits (Upstate Biotechnology, Lake Placid, NY). Scid.adh cells (2.5 × 107) were fixed with 1% paraformaldehyde at room temperature for 10 minutes, washed, and lysed with SDS lysis buffer (50 mM Tris-HCL, 1% SDS, 10 mM EDTA, protease inhibitor cocktail; Sigma, St Louis, MO). The lysates were sonicated for 30 seconds × 5 at 50% output. The soluble fraction was diluted and precleared with salmon sperm DNA/protein A-agarose. The precleared lysate was split and incubated with either Notch1, acetylated histone 4, or Myc antibodies. The immune complexes were then precipitated with protein A-agarose, washed, and eluted with elution buffer. The eluate was reverse cross-linked and treated with proteinase K (20 μg/mL). DNA was purified using a PCR purification kit (Qiagen) and eluted in water (5 × 106 cell equivalents/50 μL). (Semi)quantitative PCR was performed (see Document S1 for primers). For RQ-PCR, the precleared lysate was split and incubated with either Notch1 or control normal rabbit IgG. DNA sequences were quantified using SYBR green. Nonimmunoprecipitated input DNA was serially diluted and used as a standard curve to quantify levels of DNA recovered after immunoprecipitation (IP).
Results
CEBPA silencing and promoter hypermethylation are associated with AMLs sharing a CEBPA mutant gene expression signature
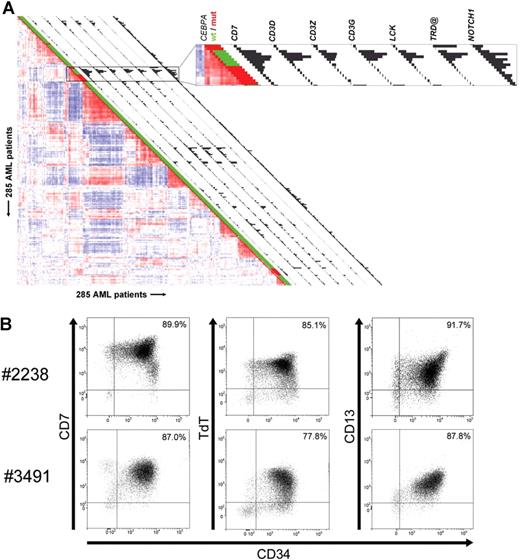
Unsupervised cluster analysis previously revealed 16 subclasses of AML with distinctive gene expression signatures.4 AMLs with mutations in the CEBPA gene were predominantly found in 2 clusters (Figure 1A; Table S1).4 Among cases in cluster no. 4, a subset of 6 AMLs without CEBPA mutations was apparent (Figure 1A). We applied significance analysis of microarrays (SAM)27 to determine gene array probe sets distinguishing these 6 cases from the specimens with CEBPA mutations in the same cluster. One of the strongest discriminating genes was CEBPA itself (Table S2). Whereas CEBPA levels were very high in CEBPA mutant cases, expression was minimal or nonexistent in the nonmutant specimens (Figure 1A; Table S3A,B). CEBPA expression levels were confirmed by real-time quantitative polymerase chain reaction (RQ-PCR) (data not shown). Bisulfite genomic sequencing revealed that in 4 of the 6 patient samples, a proximal region in the CEBPA promoter was densely CpG hypermethylated (Figure 1B). In contrast, no methylation of the same region was observed in 38 control AMLs from variable clusters (Table S3B).
CEBPA silencing and promoter hypermethylation are associated with AMLs sharing a CEBPA mutant gene expression signature in GEP cluster no. 4. (A) Pairwise correlations between gene expression profiles of 285 AML samples calculated on the basis of 2856 probe sets are displayed as described.4 Colors of boxes visualize Pearson correlation coefficient: deeper red indicates higher positive correlation; deeper blue indicates higher negative correlation. Sixteen distinct clusters were previously distinguished, which can be recognized by the red blocks showing high correlation along the diagonal.4 Cluster no. 4 and cluster no. 15, associated with CEBPA mutations, are enlarged. The bar and histogram next to each patient represent CEBPA mutation status and CEBPA expression level, respectively. CEBPA mutation status: presence (“mut,” red) or absence (“wt,” green) of mutations in bZIP region and/or N-terminus. These data indicate that in leukemias in cluster no. 4 lacking mutations, CEBPA expression is low or absent. In cluster no. 4, the order of samples, from top to bottom, is as follows: no. 3327, no. 2242 (both CEBPA mutant); no. 2668, no. 2238, no. 3314, no. 2686, no. 3483, no. 3491 (all 6 without CEBPA mutation); no. 2218, no. 1316, no. 2273, no. 2545, no. 2169, no. 2753, and no. 2192 (all 7 CEBPA mutant). (B, upper part) Schematic representation of the chromosomal region surrounding the transcriptional start of the CEBPA gene. Numbers indicate position relative to CEBPA transcriptional start. Vertical lines represent CpG dinucleotides; “transcription” indicates transcriptional start; “ATG,” translational start site. (Lower part) Level of cytosine methylation in the region surrounding the CEBPA transcriptional start site of the 6 AML cases in cluster no. 4 with low CEBPA expression, with patient numbers on the left. Every cytosine in a CpG dinucleotide is depicted as a circle. For each of these cytosines, the fraction of methylated residues was determined, which is visualized by the color of the circle: methylated (> 75% of all cytosines methylated, black), partly methylated (25%-75% methylated, gray), unmethylated (< 25% methylated, white).
CEBPA silencing and promoter hypermethylation are associated with AMLs sharing a CEBPA mutant gene expression signature in GEP cluster no. 4. (A) Pairwise correlations between gene expression profiles of 285 AML samples calculated on the basis of 2856 probe sets are displayed as described.4 Colors of boxes visualize Pearson correlation coefficient: deeper red indicates higher positive correlation; deeper blue indicates higher negative correlation. Sixteen distinct clusters were previously distinguished, which can be recognized by the red blocks showing high correlation along the diagonal.4 Cluster no. 4 and cluster no. 15, associated with CEBPA mutations, are enlarged. The bar and histogram next to each patient represent CEBPA mutation status and CEBPA expression level, respectively. CEBPA mutation status: presence (“mut,” red) or absence (“wt,” green) of mutations in bZIP region and/or N-terminus. These data indicate that in leukemias in cluster no. 4 lacking mutations, CEBPA expression is low or absent. In cluster no. 4, the order of samples, from top to bottom, is as follows: no. 3327, no. 2242 (both CEBPA mutant); no. 2668, no. 2238, no. 3314, no. 2686, no. 3483, no. 3491 (all 6 without CEBPA mutation); no. 2218, no. 1316, no. 2273, no. 2545, no. 2169, no. 2753, and no. 2192 (all 7 CEBPA mutant). (B, upper part) Schematic representation of the chromosomal region surrounding the transcriptional start of the CEBPA gene. Numbers indicate position relative to CEBPA transcriptional start. Vertical lines represent CpG dinucleotides; “transcription” indicates transcriptional start; “ATG,” translational start site. (Lower part) Level of cytosine methylation in the region surrounding the CEBPA transcriptional start site of the 6 AML cases in cluster no. 4 with low CEBPA expression, with patient numbers on the left. Every cytosine in a CpG dinucleotide is depicted as a circle. For each of these cytosines, the fraction of methylated residues was determined, which is visualized by the color of the circle: methylated (> 75% of all cytosines methylated, black), partly methylated (25%-75% methylated, gray), unmethylated (< 25% methylated, white).
AML cases with silenced CEBPA in GEP cluster no. 4 express myeloid as well as T-lymphoid genes
Further comparison of CEBPA silenced leukemias to CEBPA mutant cases in cluster no. 4 revealed that a considerable number of significantly overexpressed genes in the first group was related to T-cells and T-lymphoid development (Tables S2,S4,S5; Figure S1). In Figure 2A, the expression of a selection of those significantly elevated T-lymphoid genes is depicted as a snapshot of the GEP correlation plot of 285 AML cases,4 and illustrates expression of CD7, CD3D, CD3Z, CD3G, the T-cell–specific Src-family kinase LCK, TRD@, encoding the T-cell receptor (TCR) delta locus, and NOTCH1. Multicolor flow cytometric analysis demonstrated that blast cells of the 6 cases coexpressed myeloid and T-lymphoid lineage markers (Table 1; Figure 2B). CD7 was consistently highly expressed in all 6 specimens, while in 4 cases the expression of one or more additional T-cell marker(s) was seen. CD3 protein surface expression was observed only weakly in one specimen, which contrasted with high mRNA expression in multiple cases. Rearrangements in T-cell receptor genes (ie, TCRD and/or TCRG) were detected in 4 of 6 cases (Table 1). Importantly, all 6 specimens with silenced CEBPA showed expression of several myeloid lineage markers, including CD13 and CD33. The immature stage of the myeloid/T-lymphoid leukemias was confirmed by membrane presence of CD34 protein.
Immunophenotypes and T-cell receptor rearrangements of AMLs with silenced CEBPA in GEP cluster no. 4
| Immunophenotypes and T-cell receptor rearrangements . | Patient no. . | |||||
|---|---|---|---|---|---|---|
| 2668 . | 2238 . | 3314 . | 2686 . | 3483 . | 3491 . | |
| T-lymphoid antigens | ||||||
| CD7 | + | + | + | + | + | + |
| SmCD3 | – | – | – | +/– | – | – |
| CyCD3 | +/– | + | – | +/– | – | – |
| CD2 | +/– | – | – | +/– | – | – |
| TdT | – | + | – | – | – | + |
| Myeloid antigens | ||||||
| CD13 | + | + partly | +/– | +/– | + | + |
| CD33 | +/– | +/– | + | +/– | +/– | + |
| CD14 | +/– | – | – | +/– | – | – |
| CD64 | – | + | – | – | – | + |
| CD11b | + | – | + | + | – | – |
| MPO | – | – | + | + | + | + |
| Stem-cell antigens | ||||||
| CD34 | + | + | + | +/– | + | + |
| CD117 | + | – | +/– | +/– | + | + |
| CD133 | – | – | – | – | +/– | +/– |
| Other antigens | ||||||
| CD45 | +/–, +/− | +/– | +/– | +/– | +/– | +/– |
| CD56 | partly | ND | + | +/– | + | ND |
| DNAs* | ||||||
| TCRD rearrangement | + | + | + | – | + | – |
| TCRG rearrangement | + | + | – | – | + | – |
| Immunophenotypes and T-cell receptor rearrangements . | Patient no. . | |||||
|---|---|---|---|---|---|---|
| 2668 . | 2238 . | 3314 . | 2686 . | 3483 . | 3491 . | |
| T-lymphoid antigens | ||||||
| CD7 | + | + | + | + | + | + |
| SmCD3 | – | – | – | +/– | – | – |
| CyCD3 | +/– | + | – | +/– | – | – |
| CD2 | +/– | – | – | +/– | – | – |
| TdT | – | + | – | – | – | + |
| Myeloid antigens | ||||||
| CD13 | + | + partly | +/– | +/– | + | + |
| CD33 | +/– | +/– | + | +/– | +/– | + |
| CD14 | +/– | – | – | +/– | – | – |
| CD64 | – | + | – | – | – | + |
| CD11b | + | – | + | + | – | – |
| MPO | – | – | + | + | + | + |
| Stem-cell antigens | ||||||
| CD34 | + | + | + | +/– | + | + |
| CD117 | + | – | +/– | +/– | + | + |
| CD133 | – | – | – | – | +/– | +/– |
| Other antigens | ||||||
| CD45 | +/–, +/− | +/– | +/– | +/– | +/– | +/– |
| CD56 | partly | ND | + | +/– | + | ND |
| DNAs* | ||||||
| TCRD rearrangement | + | + | + | – | + | – |
| TCRG rearrangement | + | + | – | – | + | – |
The following lineage markers were also analyzed, and found negative on all 6 samples: TCRα/β, TCRγ/δ, CD4, CD8, CD10, CD1a, CD65, and CD15. CD5, CD19, and CD22 were tested in 4 samples (all except for no. 2238 and no. 3491) and were negative in all these 4 cases.
+ indicates positive;–, negative; +/–, weakly positive; partly, part of population positive; and ND, not determined.
As a control, 6 GEP cluster no. 4 cases with CEBPA mutations were analyzed for T-cell receptor rearrangements as well. Patients no. 1316, no. 2242, no. 2273, no. 2545, and no. 3327 were negative for TCRD and TCRG rearrangements. Patient no. 2218 was positive for TCRG rearrangement and negative for TCRD rearrangement.
AML cases with silenced CEBPA in cluster no. 4 simultaneously express myeloid- and T-lymphoid–specific genes and lineage markers. (A) Pairwise correlations between samples are displayed as explained in the legend to Figure 1, and GEP cluster no. 4 is enlarged in the box on the right. Histograms next to each patient display expression of selected genes with significantly elevated expression in cluster no. 4 cases with silenced CEBPA. Expression levels for probe sets of the following genes are visualized: CD7, CD3D, CD3Z, CD3G, LCK, TRD@, and NOTCH1. Corresponding expression levels and Affymetrix probe set identifiers are depicted in Table S4. (B) Representative dot plot images from flow cytometric analysis of samples obtained from 2 individual patients (ie, patients no. 2238 and no. 3491) demonstrating that the majority of cells from these patients simultaneously express CD34, CD13, CD7, and terminal deoxynucleotidyltransferase (TdT). The tumor population was identified by its weak expression of CD45, depicted in black, whereas CD45high cells, which are considered to be mature lymphocytes, are colored in gray.
AML cases with silenced CEBPA in cluster no. 4 simultaneously express myeloid- and T-lymphoid–specific genes and lineage markers. (A) Pairwise correlations between samples are displayed as explained in the legend to Figure 1, and GEP cluster no. 4 is enlarged in the box on the right. Histograms next to each patient display expression of selected genes with significantly elevated expression in cluster no. 4 cases with silenced CEBPA. Expression levels for probe sets of the following genes are visualized: CD7, CD3D, CD3Z, CD3G, LCK, TRD@, and NOTCH1. Corresponding expression levels and Affymetrix probe set identifiers are depicted in Table S4. (B) Representative dot plot images from flow cytometric analysis of samples obtained from 2 individual patients (ie, patients no. 2238 and no. 3491) demonstrating that the majority of cells from these patients simultaneously express CD34, CD13, CD7, and terminal deoxynucleotidyltransferase (TdT). The tumor population was identified by its weak expression of CD45, depicted in black, whereas CD45high cells, which are considered to be mature lymphocytes, are colored in gray.
Excision of Cebpa in hematopoietic stem cells induces expression of Cd7 and Lck
To investigate whether lack of the myeloid master regulator Cebpa could be sufficient to induce T-lymphocyte–associated transcripts, we made use of a conditional Cebpa knockout mouse model.7 CebpaF/F (CebpaFlox/Flox) Mx1-Cre conditional knockout mice and Cebpawt/wt (Cebpawild type/wild type) Mx1-Cre control mice were treated with polyinosinic-polycytidylic acid (poly I:C), and Lin−c-Kit+Sca1+ hematopoietic stem cells (HSCs) were isolated from bone marrow 2 weeks after the last injection. We investigated expression levels of 4 particular T-cell–associated genes by RQ-PCR. Excision of Cebpa resulted in up-regulation of Cd7 and Lck compared with control animals (P < .01 and P < .05, respectively) (Figure 3A). Moreover, reintroduction of Cebpa in HSCs isolated from poly I:C–treated CebpaF/FMx1-Cre animals strongly reduced mRNA levels of both genes (Figure 3B), further demonstrating that Cebpa plays a role in repression of these T-lymphocyte–associated transcripts. However, transcript levels of Cd3d and Cd3g did not change in Cebpa knockout HSCs (data not shown), suggesting that additional T-cell regulatory pathways had been dysregulated in AML cases with silenced CEBPA in GEP cluster no. 4.
Absence of Cebpa in murine HSCs up-regulates Cd7 and Lck. (A) Lin−c-Kit+Sca1+ hematopoietic stem cells (HSCs) were isolated and purified from poly I:C–treated Cebpawt/wtMx1-Cre mice (wt cre+) and CebpaF/FMx1-Cre knockout mice (F/F cre+). mRNA expression levels of Cd7 and Lck were determined by RQ-PCR. Data are presented relative to expression of wt cre+ animals as mean plus standard deviation of at least 3 individual mice in both groups. (B) Purified HSCs from poly I:C–treated CebpaF/FMx1-Cre mice were transduced with either empty vector (vector) or Cebpa expression construct (Cebpa). Cd7 and Lck mRNA expression levels were determined in GFP+ infected cells 1 day after transduction. Data are presented relative to expression of empty vector–transduced cells as mean plus standard deviation of 4 reactions.
Absence of Cebpa in murine HSCs up-regulates Cd7 and Lck. (A) Lin−c-Kit+Sca1+ hematopoietic stem cells (HSCs) were isolated and purified from poly I:C–treated Cebpawt/wtMx1-Cre mice (wt cre+) and CebpaF/FMx1-Cre knockout mice (F/F cre+). mRNA expression levels of Cd7 and Lck were determined by RQ-PCR. Data are presented relative to expression of wt cre+ animals as mean plus standard deviation of at least 3 individual mice in both groups. (B) Purified HSCs from poly I:C–treated CebpaF/FMx1-Cre mice were transduced with either empty vector (vector) or Cebpa expression construct (Cebpa). Cd7 and Lck mRNA expression levels were determined in GFP+ infected cells 1 day after transduction. Data are presented relative to expression of empty vector–transduced cells as mean plus standard deviation of 4 reactions.
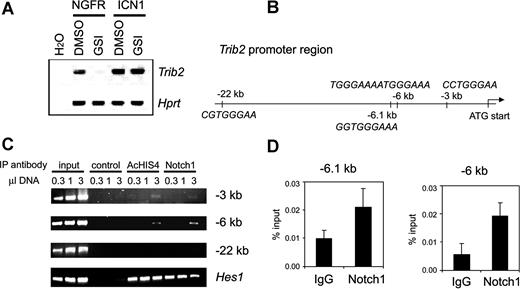
TRIB2 overexpression points to aberrant Notch activation in AMLs with silenced CEBPA, and associates with NOTCH1 mutations
The leukemias with silenced CEBPA in cluster no. 4 frequently showed high expression of the myeloid oncogene TRIB2 and NOTCH1 (Figure 2A), suggesting potential activation of Notch signaling. Treatment of Notch1-dependent murine cell lines T6E28 and Scid.adh15,29 with gamma secretase inhibitors (GSIs) led to Trib2 down-regulation (Figure 4A and not shown). Moreover, Trib2 expression was restored when these cells were transduced with a retroviral vector expressing intracellular NOTCH1 (ICN1), which is GSI resistant (Figure 4A). Likewise, introduction of ICN1 into the myeloid leukemia cell line U937 led to up-regulation of TRIB2 levels (Figure S2). To gain further insight, we investigated whether TRIB2 could be directly regulated by NOTCH1. ICN1 binds the transcriptional repressor protein CSL, converting it into an activator. Several putative CSL-binding sites were identified, at − 22 kb, − 6.1 kb, − 6 kb, and − 3 kb from the Trib2 translational start site, respectively (Figure 4B). Chromatin immunoprecipitation (ChIP) showed binding of the ICN1/CSL complex to both the − 6.1-kb/− 6-kb regions, containing a single conserved as well as a tandem CSL-binding site, and to a weaker extent to the single − 3-kb site (Figure 4C,D). Both regions were also immunoprecipitated by antibodies against acetylated histone 4 (AcHIS4), consistent with transcriptional activity. Together, these results indicate that Trib2 is a direct transcriptional target of Notch1, and further suggest that AML cases with selectively increased levels of TRIB2 may be subject to aberrant induction of Notch signaling. Activating mutations in NOTCH1, as found in T-ALL, target 2 regions of the gene: the heterodimerization (HD) and the proline-glutamate-serine-threonine–rich (PEST) domain. Nucleotide sequencing showed 4 similar, putatively activating mutations in cDNA and genomic DNA in 3 of the 6 AMLs with high TRIB2 expression (Table 2). In one case, an HD and a PEST mutation were found in cis. In contrast, we did not identify NOTCH1 mutations in 100 control AML samples, including several AMLs from each GEP cluster, and the CEBPA mutant AML cases from clusters no. 4 and no. 15 (Table S3B,C).
NOTCH1 mutations identified in patients from 2 independent AML cohorts
| Patient no. . | AML cohort . | NOTCH1 mutation* . | Predicted amino acid change . |
|---|---|---|---|
| 2238 | First | 4741-4743del | In-frame deletion of M1581 |
| 3314 | First | 5036T>A, 7376ins37 bp | L1679Q, L2458fsX2463 |
| 2686 | First | 7552ins13 bp | P2518fsX2523 |
| 6947 | Second | 4790T>C | L1597P |
| 7053 | Second | 4820del22/ins13 bp | Mutation at aa 1607 FKRDAHGQ into SGRRP |
| Patient no. . | AML cohort . | NOTCH1 mutation* . | Predicted amino acid change . |
|---|---|---|---|
| 2238 | First | 4741-4743del | In-frame deletion of M1581 |
| 3314 | First | 5036T>A, 7376ins37 bp | L1679Q, L2458fsX2463 |
| 2686 | First | 7552ins13 bp | P2518fsX2523 |
| 6947 | Second | 4790T>C | L1597P |
| 7053 | Second | 4820del22/ins13 bp | Mutation at aa 1607 FKRDAHGQ into SGRRP |
The first AML cohort represents the initial series of 285 cases. The second cohort represents the independent series of 268 AMLs, which was interrogated using a gene expression prediction signature.
Nucleotide numbering according to Entrez nucleotide accession number NM_017617 (version NM_017617.2). The mutations in patients no. 2238, no. 6947, and no. 7053 are located in the NOTCH1 HD domain (exons 26 and 27). The mutation in patient no. 2686 is located in the NOTCH1 PEST domain (exon 34). Patient no. 3314 harbors in cis mutations both in the HD domain as well as the PEST domain. All mutations identified are heterozygous.
Trib2 is a direct target gene of Notch1. (A) Reverse transcriptase (RT)–PCR analysis of Trib2 expression in Scid.adh cells treated with GSI (+) or DMSO as vehicle control (−). Cells were transduced with an empty vector expressing only the NGFR selection marker (NGFR) or a vector expressing ICN1 (ICN1). Trib2 mRNA expression was assessed. Hprt is an internal loading control. Results are representative of triplicate experiments. (B) Schematic representation of the Trib2 promoter region. Four putative CSL-binding sites were identified in the region shown, including a tandem CSL site at − 6 kb and 3 single binding sites at − 22 kb, − 6.1 kb, and − 3 kb relative to the translational start site. (C) Notch1 binds to CSL-binding sites in the Trib2 promoter. Chromatin immunoprecipitates were performed on cross-linked fragmented DNAs prepared from Scid.adh cells. Immunoprecipitations were carried out with antibodies against Myc as a negative control (control), acetylated histone 4 (AcHIS4), and Notch1. PCR was performed using primers directed against indicated CSL-binding site regions at − 22, − 6, and − 3 kb from the ATG translational start site of Trib2. PCR for the Hes1 promoter region is shown as a positive control. The figure shown is representative of duplicate experiments. For the − 6-kb region, which shows the strongest enrichment, results are representative of duplicate samples and triplicate experiments. (D) After ChIP as described in panel C, using either Notch1 or normal rabbit control IgG antibody, RQ-PCR was performed using primer sets flanking putative CSL-binding sites in the Trib2 promoter. RQ-PCR was performed using the primers at − 6 kb to quantify enrichment in this region, and also using primers flanking a conserved CSL-binding site at − 6.1 kb. Graphs represent mean of the ratios of the amount of IP DNA/input from values of duplicate wells plus or minus standard deviation. Data are representative of 3 independent experiments.
Trib2 is a direct target gene of Notch1. (A) Reverse transcriptase (RT)–PCR analysis of Trib2 expression in Scid.adh cells treated with GSI (+) or DMSO as vehicle control (−). Cells were transduced with an empty vector expressing only the NGFR selection marker (NGFR) or a vector expressing ICN1 (ICN1). Trib2 mRNA expression was assessed. Hprt is an internal loading control. Results are representative of triplicate experiments. (B) Schematic representation of the Trib2 promoter region. Four putative CSL-binding sites were identified in the region shown, including a tandem CSL site at − 6 kb and 3 single binding sites at − 22 kb, − 6.1 kb, and − 3 kb relative to the translational start site. (C) Notch1 binds to CSL-binding sites in the Trib2 promoter. Chromatin immunoprecipitates were performed on cross-linked fragmented DNAs prepared from Scid.adh cells. Immunoprecipitations were carried out with antibodies against Myc as a negative control (control), acetylated histone 4 (AcHIS4), and Notch1. PCR was performed using primers directed against indicated CSL-binding site regions at − 22, − 6, and − 3 kb from the ATG translational start site of Trib2. PCR for the Hes1 promoter region is shown as a positive control. The figure shown is representative of duplicate experiments. For the − 6-kb region, which shows the strongest enrichment, results are representative of duplicate samples and triplicate experiments. (D) After ChIP as described in panel C, using either Notch1 or normal rabbit control IgG antibody, RQ-PCR was performed using primer sets flanking putative CSL-binding sites in the Trib2 promoter. RQ-PCR was performed using the primers at − 6 kb to quantify enrichment in this region, and also using primers flanking a conserved CSL-binding site at − 6.1 kb. Graphs represent mean of the ratios of the amount of IP DNA/input from values of duplicate wells plus or minus standard deviation. Data are representative of 3 independent experiments.
Gene expression prediction signature allows identification of similar leukemias in an independent cohort of AMLs
To validate our findings, we determined a gene expression prediction signature to identify additional leukemias with the same characteristics. We made use of Affymetrix HGU133 Plus2.0 GeneChips, and based prediction analysis on data obtained from 262 of 285 specimens, including all cases in cluster no. 4. A signature of 9 probe sets was defined by the prediction analysis of microarrays (PAM) class prediction algorithm.30 This signature accurately detected all 6 CEBPA silenced cases in cross-validation experiments, while the average number of false positives was minimal using the same detection criteria (2/256, 0.8%; Figure 5A). In a second cohort of 268 AML cases, this predictor detected 6 new cases sharing the same profile (Figure 5B). Four of these 6 new leukemias showed no or low amounts of CEBPA as well as elevated levels of TRIB2 and NOTCH1 mRNA, consistent with our prior findings—in spite of the fact that the prediction signature itself did not contain probe sets for these genes (Figure 5). Analysis of available diagnostic flow cytometric data for these 4 leukemias revealed myeloid lineage marker expression in combination with CD7 in all, while 2 of 4 leukemias expressed additional T-lymphoid markers (Table S6). NOTCH1 mutations were found in 2 of these leukemias, and hypermethylation of the CEBPA promoter was apparent in 2 cases (Table 2; Figure 5B). In contrast, elevated levels of CEBPA mRNA were observed in the remaining 2 of 6 leukemias predicted in the second series of AML (Figure 5B). Both cases showed mutations in the CEBPA coding region, explaining the similar expression profile.
A gene expression prediction signature identifies new leukemias with the silenced CEBPA phenotype in an independent cohort of AML. (A) For 262 samples analyzed on Affymetrix HGU133Plus2.0 GeneChips, log-transformed (base 2) and mean-centered expression levels for 13 probe sets are depicted (left panel) for an arbitrary range from − 4 to + 4 (corresponding to 16-fold lower to 16-fold higher expression relative to the mean, respectively). The ordering of patients in the figure is arbitrary. In the right panel, these data are enlarged for 31 of these 262 leukemias, representing 3 groups: (I) the 6 cases with silenced CEBPA previously identified, with from top to bottom cases no. 2668, no. 2238, no. 3314, no. 2686, no. 3483, and no. 3491; (II) a variable selection of 10 AMLs from distinct GEP clusters for which also NOTCH1 mutational analysis and CEBPA promoter bisulfite sequencing were performed; and (III) 15 AMLs with CEBPA mutations, originating from either GEP cluster no. 4 (upper 9 samples) or cluster no. 15 (lower 6 samples). The 9 probe sets on the left side constitute the most predictive gene expression signature for group I, as determined by PAM: 216191_s_at (TRA@/TRD@ 1), 217143_s_at (TRA@/TRD@ 2), 213830_at (TRD@), 225681_at (CTHRC1), 1565809_x_at (no annotation), 1560018_at (ARPP-21), 210844_x_at (CTNNA1 1), 200764_s_at (CTNNA1 2), and 200765_x_at (CTNNA1 3). To the right, 4 additional probe sets are indicated, that is, 204039_at (CEBPA), 218902_at (NOTCH1), 202478_at (TRIB2), and 214551_s_at (CD7). Mutational data for NOTCH1 and CEBPA, and methylation status of the CEBPA promoter are depicted next to the normalized hybridization intensities of the probe sets. (B) Two hundred sixty-eight samples obtained from a second cohort of AML were hybridized to HGU133Plus2.0 GeneChips. The 9 probe set signature was used to identify leukemias with a profile similar to group I (A), resulting in the detection of group IV (from top to bottom cases no. 6376, no. 6735, no. 6947, no. 7053, no. 7076, and no. 7120).
A gene expression prediction signature identifies new leukemias with the silenced CEBPA phenotype in an independent cohort of AML. (A) For 262 samples analyzed on Affymetrix HGU133Plus2.0 GeneChips, log-transformed (base 2) and mean-centered expression levels for 13 probe sets are depicted (left panel) for an arbitrary range from − 4 to + 4 (corresponding to 16-fold lower to 16-fold higher expression relative to the mean, respectively). The ordering of patients in the figure is arbitrary. In the right panel, these data are enlarged for 31 of these 262 leukemias, representing 3 groups: (I) the 6 cases with silenced CEBPA previously identified, with from top to bottom cases no. 2668, no. 2238, no. 3314, no. 2686, no. 3483, and no. 3491; (II) a variable selection of 10 AMLs from distinct GEP clusters for which also NOTCH1 mutational analysis and CEBPA promoter bisulfite sequencing were performed; and (III) 15 AMLs with CEBPA mutations, originating from either GEP cluster no. 4 (upper 9 samples) or cluster no. 15 (lower 6 samples). The 9 probe sets on the left side constitute the most predictive gene expression signature for group I, as determined by PAM: 216191_s_at (TRA@/TRD@ 1), 217143_s_at (TRA@/TRD@ 2), 213830_at (TRD@), 225681_at (CTHRC1), 1565809_x_at (no annotation), 1560018_at (ARPP-21), 210844_x_at (CTNNA1 1), 200764_s_at (CTNNA1 2), and 200765_x_at (CTNNA1 3). To the right, 4 additional probe sets are indicated, that is, 204039_at (CEBPA), 218902_at (NOTCH1), 202478_at (TRIB2), and 214551_s_at (CD7). Mutational data for NOTCH1 and CEBPA, and methylation status of the CEBPA promoter are depicted next to the normalized hybridization intensities of the probe sets. (B) Two hundred sixty-eight samples obtained from a second cohort of AML were hybridized to HGU133Plus2.0 GeneChips. The 9 probe set signature was used to identify leukemias with a profile similar to group I (A), resulting in the detection of group IV (from top to bottom cases no. 6376, no. 6735, no. 6947, no. 7053, no. 7076, and no. 7120).
Discussion
In the present study, we define a specific subgroup of AML with a discriminative gene expression profile and expression of T-cell genes. These specific AMLs associate with silencing of CEBPA and aberrant activation of NOTCH1 by mutations. Our data provide evidence for the involvement of C/EBPα in the active repression of several of the T-cell–associated genes, supporting a cooperating role for CEBPA silencing in development of the leukemic phenotype. Using gene expression data, we determined a predictor of 9 probe sets by which we have been able to identify similar leukemia cases in a second cohort of AMLs.
We initially studied these specific AMLs because their gene expression profiles were similar to leukemias with mutations in the transcription factor CEBPA. This similarity suggested another mechanism of CEBPA deregulation. Indeed, all 6 cases revealed complete or near-complete absence of CEBPA mRNA. Importantly, CEBPA mutant cases in this cluster carried either homozygous mutations or mutations in both CEBPA N-terminus and basic leucine zipper region, which are typically biallelic (Table S1).10,22 Therefore, none of these mutant cases was predicted to express wild-type CEBPA, and lack of wild-type CEBPA appeared to be the common hallmark of all specimens in GEP cluster no. 4. Down-regulation or functional inactivation of CEBPA has been implicated in development of myeloid malignancies through several mechanisms previously.10-12 Aberrant CpG methylation of the CEBPA promoter is involved in carcinogenesis of solid tissue such as lung,31 and has recently been described sporadically in AML as well.32,33 Because of lack of other recurrent molecular lesions, we addressed this particular mechanism of gene silencing. Indeed, extensive CEBPA promoter hypermethylation associated with 4 of 6 of these cases and with 2 of 4 cases of the second cohort. Similar experiments in a set of control AMLs by us and experiments by other groups suggest that CEBPA promoter hypermethylation is generally not very common in AML,32,33 and no other clusters appeared to be characterized by unified CEBPA silencing (Figure 1A). Although we cannot rule out that CEBPA promoter hypermethylation may occur in other sporadic cases, these observations suggest that this mechanism is significantly associated with this specific subset. Interestingly, in 4 of the 10 cases identified in the 2 cohorts, silencing was not associated with hypermethylation, suggesting a possible other, yet-unknown mechanism of CEBPA mRNA repression in those.
The leukemias with silenced CEBPA simultaneously expressed myeloid and T-lymphoid genes. Mixed myeloid/T-lymphoid leukemias have been described previously and are thought to represent a variety of diseases with a large genetic heterogeneity, ranging from myeloid leukemias with aberrant expression of only one or few T-cell markers, to “true” biphenotypic leukemias.34-37 The T-cell gene most consistently associated with the patient group described here was CD7, which is known to be expressed by a considerable proportion of immature AMLs,38-40 suggesting that the precursor cell in which transformation occurred already had undergone some initial myeloid differentiation. Interestingly, CD7 expression is frequently associated with AMLs carrying CEBPA mutations.40,41 Expression of additional T-cell lineage markers was apparent in 4 of 6 cases of the subgroup with silenced CEBPA. A previously proposed scoring system based on immunophenotype would classify the majority of the cases identified by us as myeloid malignancies with aberrant T-cell expression, rather than as “true” biphenotypic leukemias.42 In line with previous observations, most leukemias in the subgroup described here showed several cytogenetic abnormalities, however, none of these appeared common to all (Tables S1,S6). Our observations classify this specific heterogeneous population as a homogeneous subset based on their gene expression profiles.
We found that in a Cebpa conditional knockout model, transcript levels in HSCs of Lck and Cd7 appeared to be dependent on the absence of C/EBPα, suggesting that C/EBPα is directly involved in negative regulation of these genes. These observations mirror those from previous studies that showed that CEBPA has lineage switch potential when expressed in lymphoid cells.43-47 Although the role of C/EBPα in lymphoid cell fate decision may not be fully understood yet, as highlighted by recent observations indicating that overexpression of C/EBPα due to chromosomal translocations may be involved in B-cell ALLs,48,49 our studies show that decreased expression of C/EBPα in hematopoietic stem cells can be sufficient to induce T-cell transcripts.
In addition to loss of C/EBPα function, the experiments performed in murine HSCs also suggested dysregulation of additional pathways leading to aberrant T-cell commitment in the clinical leukemias, as Cebpa excision failed to explain the expression of all T-cell transcripts. As an additional explanation for the phenotype observed, we found evidence for aberrant Notch activation in several of these leukemias. In the present study, we identified TRIB2 as a direct target of NOTCH1, which led us to investigate whether activating NOTCH1 mutations had occurred in the AMLs. Several cases exhibited mutations in NOTCH1 PEST and/or HD domains, which were all predicted to result in enhanced NOTCH1 signaling (Table 2).50,51 In contrast, we did not detect similar mutations in a large cohort of AML control samples, concordant with recent literature.19,20 Our data therefore suggest that in AML, NOTCH1 mutations are largely restricted to this particular subset of myeloid/T-lymphoid leukemias. At present, it is not known what the precise mechanism of transformation in these leukemias is or why NOTCH1 mutations specifically associate with this type of AML. Expression of activated Notch1 in murine bone marrow cells in vivo leads exclusively to T-cell neoplasms,28 whereas the tumors described in the current study had myeloid features, possibly suggesting that the leukemia-initiating cell was a committed myeloid progenitor. Interestingly, overexpression of Trib2 in murine HSCs induced AML,15 suggesting an alternative scenario in which TRIB2 plays an important role in leukemic transformation. Trib2 inactivates C/EBPα p42, implying that high TRIB2 expression may be an alternative mechanism to interfere with C/EBPα function in AMLs lacking hypermethylation of the CEBPA promoter. TRIB proteins may also enhance ERK phosphorylation, which might be suggestive of an additional mechanism involved in transformation15,52 Several leukemias of this subgroup did not reveal NOTCH1 HD and PEST mutations. It is possible that in these cases aberrant Notch activation occurred by other mechanisms, explaining the elevated expression of TRIB2 in those tumors. This could involve other Notch receptors, ligands, or other mutations in NOTCH1. Cytogenetic analysis did not reveal translocations involving the NOTCH1 locus on chromosome 9q34.3 (Table S1),18 nor did we observe overexpression of NOTCH1 ligand coding genes (data not shown).
We evaluated our observations in an independent representative series of AMLs. We successfully defined a gene expression predictor signature for the leukemias with silenced CEBPA, and identified 4 new cases with highly similar molecular and phenotypic characteristics. The 6 classifying genes, represented by 9 probe sets, have previously been associated with immature thymocytes (T-cell receptor genes TRA@ and TRD@ and the cyclic AMP-regulated phosphoprotein ARPP-2153,54 ) or with malignant transformation (alpha-E-catenin [CTNNA1]55-57 and collagen triple helix repeat containing 1 [CTHRC1]58 ), while another probe set (1565809_x_at) maps to an uncharacterized gene. We propose that a simple algorithm to diagnose this subclass of leukemias could make use of a combination of this prediction signature with subsequent CEBPA mutational analysis. Our data do not rule out the possibility that some leukemias with silenced CEBPA and/or NOTCH1 mutations were not detected by the predictor. Although the low prevalence of NOTCH1 mutations and CEBPA promoter hypermethylation in AML argues against many overlooked cases, additional studies are warranted to test these findings in a more diagnostic setting. Additional studies will also need to address whether our preliminary observations concerning clinical outcome, which suggest a poor response to treatment (Table S7), will hold in larger series.
In conclusion, our data delineate a specific subset of AMLs within the population of myeloid malignancies with aberrant expression of T-cell genes. This subtype of leukemia is defined primarily by its gene expression profile, which is likely to be determined by silencing of the CEBPA gene, and is associated with recurring mutations in NOTCH1. Importantly, we have demonstrated that these leukemias can be predicted in independent series of AML using expression data from a limited set of genes. Together, these observations emphasize the applicability and power of in-depth exploration of GEP data for characterization of previously unrecognized subgroups of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Dutch Cancer Society “Koningin Wilhelmina Fonds” (EMCR 2006–3522) to R.D., B.L., and P.J.M.V.; a Fellow Award from the Leukemia and Lymphoma Society to K.K.; grants from the National Institutes of Health (NIH) to D.G.T. and R.D. (CA118316) and to W.S.P. (PO1 CA119070); a SCOR Award from the Leukemia and Lymphoma Society to W.S.P., and an EHA research fellowship from The European Hematology Association to M.A.J.
We thank Kirsten van Lom (Erasmus University Medical Center, Rotterdam, The Netherlands) and Gerrit-Jan Schuurhuis (Free University Medical Center, Amsterdam, the Netherlands) for help in providing clinical data; Carlos Rodriguez for help with the Trib2 experiments; our colleagues from the bone marrow transplantation group in the Department of Hematology at Erasmus University Medical Center for storage of samples; and Marieke von Lindern for critical reading of the paper.
National Institutes of Health
Authorship
Contribution: B.J.W., B.L., and R.D. designed the study; B.J.W., M.A.J., K.K., I.L., D.T., C.J.H., C.A.J.E.-V., Y.H., and Y.Y.-O. performed research; B.J.W., M.A.J., K.K., D.T., A.W.L., and R.G.W.V. analyzed data; P.Z., and D.G.T. contributed an essential mouse model; B.L., W.S.P., P.J.M.V., M.A.J., D.G.T., and K.K. revised the paper; B.J.W. and R.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Erasmus University Medical Center, Department of Hematology, Room Ee1342, PO Box 2040, 3000 CA Rotterdam, the Netherlands; e-mail:h.delwel@erasmusmc.nl.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal