Abstract
Many cells latently infected with Epstein-Barr virus (EBV), including certain virus-associated tumors, express latent membrane protein 2A (LMP2A), suggesting an important role for this protein in viral latency and oncogenesis. LMP2A mimics B-cell receptor signaling but can also act as a decoy receptor blocking B-cell receptor (BCR) activation. Studies of peripheral B cells have not resolved this apparent contradiction because LMP2A seems to be dispensable for EBV-induced transformation of these B cells in vitro. We show here that LMP2A is essential for growth transformation of germinal center B cells, which do not express the genuine BCR because of deleterious somatic hypermutations in their immunoglobulin genes. BCR-positive (BCR+) and BCR-negative (BCR−) B cells are readily transformed with a recombinant EBV encoding a conditional, floxed LMP2A allele, but the survival and continued proliferation of both BCR+ and BCR− B cells is strictly dependent on LMP2A. These findings indicate that LMP2A has potent, distinct antiapoptotic and/or transforming characteristics and point to its role as an indispensable BCR mimic in certain B cells from which human B-cell tumors such as Hodgkin lymphoma originate.
Introduction
Epstein-Barr virus (EBV) is a class I carcinogen according to the World Health Organization and is associated with several B-cell lymphomas including Hodgkin lymphoma, Burkitt lymphoma, posttransplantation lymphomas, and certain carcinomas.1 In lymphomas, EBV is found associated with almost all cases of posttransplantation lymphomas and endemic Burkitt lymphoma, whereas approximately 40% and up to 30% of tumor biopsies of Hodgkin lymphoma and sporadic Burkitt lymphoma, respectively, are EBV positive. EBV is also tightly linked to nasopharyngeal carcinoma in Southeast Asia; in contrast, only approximately 20% of all cases of gastric adenocarcinomas are EBV positive.2 These malignancies appear to be relatively rare because more than 95% of the human population is infected with EBV. In these individuals, EBV establishes a benign, latent infection in a small fraction of B cells for a lifetime. In vitro, EBV very efficiently infects all human B lymphocytes in a latent mode, and growth transforms them into lymphoblastoid cell lines (LCLs). Therefore, transformation of primary B cells by EBV is a hallmark of this virus and constitutes a relevant model system for viral transformation.3
In all virus-infected cells, EBV adopts a latent state in which viral gene expression is restricted and de novo viral synthesis is abrogated. In LCLs and cases of posttransplantation lymphomas, 11 so-called latent genes are consistently expressed. These are the EBV nuclear antigens EBNA1, EBNA2, EBNA-LP, EBNA3A, EBNA3B, EBNA3C, and the latent membrane proteins LMP1, LMP2A, and LMP2B, and 2 small, noncoding RNAs.3 Besides these noncoding RNAs, which are expressed in all latently EBV-infected cells, latent viral genes are found in typical combinations in the different EBV-associated tumors. Briefly, in Burkitt lymphoma almost exclusive and invariant expression of EBNA1 is well documented, but recently, LMP2A has also been detected in fresh biopsies of this tumor.2,4 In nasopharyngeal carcinomas and EBV-positive Hodgkin lymphomas, LMP1 is often expressed in conjunction with EBNA1 and LMP2A, and a similar expression profile might also be prevalent in EBV-positive gastric carcinomas.2 Although studies clearly show that EBNA1 and LMP1 definitely contribute to cellular transformation,5,6 the role of LMP2A remains controversial because it has been reported to be dispensable for B-cell transformation in vitro (Speck et al7 and references therein) but has transforming characteristics in epithelial cells.8
LMP2A can constitutively induce several signaling cascades. Its amino-terminal cytoplasmic domain is reminiscent of the signaling domain of the BCR complex because LMP2A as well as the signaling domains of the Igα (CD79A) and Igβ (CD79B) chains of the BCR carry an immunoreceptor tyrosine-based activation motif (ITAM).9 Both LMP2A and BCR signal through Syk, Lyn, Btk, BLNK, AKT, PI3K, and other signaling mediators, and LMP2A can deliver signals similar to those of an activated BCR8 ; however, it can also very efficiently block B-cell activation after BCR triggering,9,10 preventing the induction of EBV's lytic phase11 or abrogating B-cell differentiation in vivo.12 Therefore, it has been proposed that LMP2A might act as a decoy receptor8,13 in that it recruits the protein tyrosine kinases Lyn and Syk causing their proteasomal degradation and subsequent inactivation of the BCR signaling complex.8 Similarly, LMP2A also prevents the BCR from entering lipid rafts after BCR cross-linking blocking both BCR signaling and antigen transport.14 On the other hand, LMP2A expression in the B-cell compartment of BCR-negative (BCR−) mice can replace BCR's function during early B-cell development in the bone marrow12 and does not necessarily interfere with germinal center–like reactions.15 LMP2A seems to act as a surrogate BCR in this model, suggesting that EBV-infected B cells might also be able to bypass BCR-dependent germinal center (GC) differentiation in the human host.12,15 This particular function could be important for proapoptotic GC B cells, which do not receive essential surviving signals because they carry a crippled or low-affinity BCR. Upon EBV infection of these GC B cells, the missing signal(s) could be provided by LMP2A, because its constitutive expression might trigger the same downstream signaling pathways as an activated BCR. Thus LMP2A could enable EBV-infected B cells with nonfunctional immunoglobulin genes to escape negative selection and contribute to the establishment of BCR− germinal center B-cell lymphomas such as Hodgkin lymphoma. A recent survey of data available about crippling mutations in the immunoglobulin genes and the status of EBV in Hodgkin/Reed-Sternberg cells, the tumor cells in classical Hodgkin lymphoma, supports this view. Almost all cases of Hodgkin lymphoma with crippling mutations in the immunoglobulin genes of Hodgkin/Reed-Sternberg cells are EBV positive in contrast to those in which the tumor cells carry an apparently functional BCR.16 In an immunohistochemical analysis, not all EBV-positive Hodgkin lymphomas seem to express LMP2A, but this study did not take the BCR status of the Hodgkin/Reed-Sternberg cells into account and failed to detect LMP2A in “classic” LCLs.17
EBV can rescue from apoptosis GC B cells with crippling mutations in their immunoglobulin genes.18-20 Here, we analyze the role of LMP2A in this process and demonstrate that proapoptotic BCR− B cells are rescued by wild-type EBV, but not by a recombinant EBV lacking the viral LMP2A allele. Surprisingly, survival and continued proliferation of EBV-transformed BCR+ B cells also strictly depend on LMP2A presumably because these cells exhibit low BCR expression in this in vitro model and have been selected to thrive on activating signals that originate from the dominant LMP2A protein. These findings demonstrate that LMP2A can contribute to the survival and therefore transformation of primary B cells. These findings also support the view that LMP2A can provide a survival signal that is used by BCR− B cells during transition from a pretumor cell to a Hodgkin/Reed-Sternberg cell by rescuing EBV-infected GC B cells with disadvantageous mutations in their immunoglobulin genes from apoptosis. Similarly, EBV could sustain Hodgkin lymphoma in those cases in which BCR− Hodgkin/Reed-Sternberg cells continuously express LMP2A.2
Materials and methods
Isolation, separation, and infection of human primary B cells
Human primary B cells were prepared from adenoids as described previously.19 BCR+/− B cells correspond to the unsorted B-cell population. To prepare BCR− B cells, T cells and BCR+ B cells were depleted with magnetic-activated cell sorting (MACS) technology (Miltenyi Biotech, Auburn, CA) and α-CD3 (Miltenyi Biotech), α-λ, and α-κ light chain antibodies (BD Biosciences, San Jose, CA). Cell-cycle analysis was performed as described previously with the BrdU incorporation kit (BD Biosciences).19,21
Construction of viral mutants
Two loxP sites were introduced into the genome of 2089 WT EBV in E coli essentially as described,21 with the promoter and first exon of LMP2A flanked by 2 loxP sites to establish the LMP2A+ 2190 EBV genome. The loxP sites were introduced at nucleotide coordinates nos. 166 170 and 166 938 of the prototype EBV genome B95–8.22 The LMP2A− 2525 mutant EBV is a derivative of 2190 and was established in E coli cells harboring p2190 EBV DNA by transient expression of Cre. Virus stocks were prepared from 293HEK producer cell lines as described previously.21
Deletion of LMP2A in 2190-infected B cells
Established LCLs were transfected by electroporation using the Amaxa Nucleofector (Amaxa, Cologne, Germany). Two different expression plasmids were used. p3233 expresses a bicistronic transcript encoding the truncated version of the NGF receptor (NGF-R) and the site-specific Cre recombinase (Cre) (Figure 2B); as a control, p3232 encodes NGF-R only (not shown). NGF-R expression was monitored 18 hours after transfection by staining with PE-coupled α–NGF-R antibodies (BD Biosciences). Surface-positive, NGF-R–expressing cells coexpress Cre when transfected with p3233. Transfected cells were sorted by FACS into the 5% top fraction of NGF-R–expressing cells with the highest expression level and negative cells using the MoFlo cytometer (Dako Cytomation, Carpinteria, CA). A polymerase chain reaction (PCR) analysis with the primer oligonucleotides P1 (5′-GGAATCGTTTTCCGGGACGCCG) and P2 (5′-GGATCAGTCGCTGGCTGGTGGGC) monitored the transfection efficiency and functionality of the Cre recombinase. A 1220-bp PCR product indicates the floxed LMP2A allele; a 418-bp PCR product is generated after Cre-mediated deletion of the LMP2A promoter and the first exon of LMP2A (Figure 2A). Alternatively, the loxP-flanked LMP2A allele was deleted by transduction of the HTNC protein. This fusion protein consists of an amino-terminal histidine tag, the transduction domain of the retroviral protein TAT, a nuclear localization sequence, and the recombinase Cre.23 The transduction of 1 μM HTNC was performed in the protein-free media CD-CHO (Invitrogen, Carlsbad, CA) for 3 hours. After transient expression or transduction of Cre and deletion of the LMP2A allele, cellular proliferation and the fraction of annexinV-positive cells in each population were assessed. The absolute numbers of cells in the 2 populations were determined with the aid of calibration beads, which were added at a defined concentration as an internal volume standard (BD Biosciences) as described previously.21 Apoptotic cells in each population were stained with the annexinV kit as described by the manufacturer (BioVision, Mountain View, CA). Quantitative PCR analysis (Light Cycler Fast Start DNA Master SYBR Green I; Roche Diagnostics, Mannheim, Germany) assessed the fraction of EBV episomes in which the LMP2A promoter and first exon of LMP2A were successfully deleted after HTNC protein transduction. The primer pair P3 (5′-ATCTGCTTCTGGCTCTTCTGGG) and P4 (5′-CCCGTCATTCCCGTCGTG) was used to quantitatively detect the floxed LMP2A allele by real-time PCR; the primer pair P5 (5′-GATCTCATGCTGGAGTTCTTCGC) and P6 (5′-GATAACATCTCCCGCTAGATGGC) served to determine the absolute number of LMP2A-deleted EBV genomes. This analysis was performed 4 days after HTNC transduction as indicated by an asterisk in Figure 3A. The calculation was based on standard curves using defined amounts of plasmid DNAs for reconstruction.
Detection of BZLF1expression levels via reverse transcription–PCR
Two and 4 days after HTNC protein transduction, RNA was isolated (Qiagen, Hilden, Germany) and transcribed into cDNA (Invitrogen). Primer oligonucleotides complementary to sequences in the first (P7: 5′-GGAAGCCACCCGATTCTTG) and second (P8: 5′-GAGCCTCTGCCACAAGGC) exon of the BZLF1 gene (Figure 3C) were used for PCR amplification. The mRNA-specific cDNA of BZLF1 generates a 400-bp PCR product, whereas the genomic DNA fragment of the corresponding BZLF1 locus is 523 bp in length including the first intron in the BZLF1 gene (Figure 3B,C).
Results and discussion
LMP2A is essential for growth transformation of BCR-negative B cells
To gain insight into the function of LMP2A, we constructed a pair of isogenic viral mutants in E coli as described previously24 ; the constructs differed only with respect to the LMP2A gene. The 2190 mutant EBV is similar to the prototypic 2089 EBV,21 but 2190 carries its LMP2A allele flanked by 2 loxP sites upstream of the LMP2A promoter and downstream of the LMP2A exon, which encodes the amino-terminal signaling domain of LMP2A.8 The 2190 mutant EBV is both genotypically and phenotypically wild type (WT) because in limiting dilution assays21 2089 WT EBV and the 2190 mutant were capable of yielding clonal LCLs at the same rate (data not shown). To generate the LMP2A− mutant 2525 the LMP2A gene in 2190 was deleted by transient expression of Cre in E coli cells harboring the p2190 genomic EBV DNA. Restriction analysis and partial DNA sequencing confirmed the genetic alteration of this EBV mutant that lacks the LMP2A gene but still encodes LMP2B. Virus stocks with LMP2A+ 2190 EBV and LMP2A− 2525 mutant virus were generated in 293HEK cells as described.21
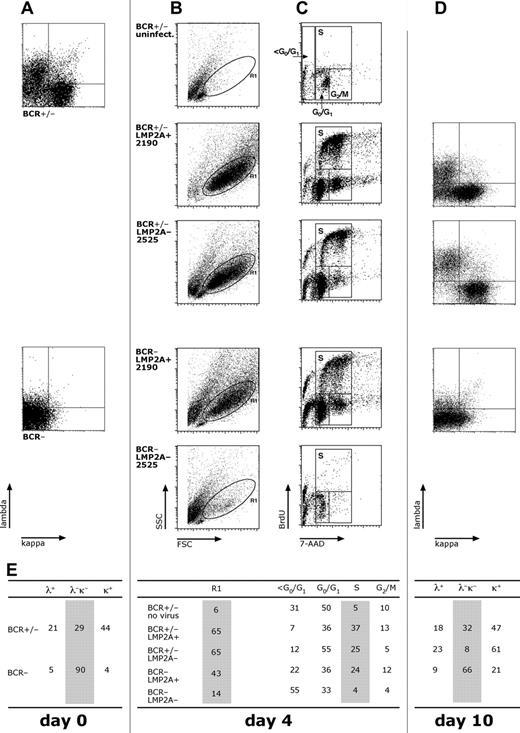
Two different preparations of primary B cells were obtained. A mixed population (BCR+/−) consisting of approximately 30% BCR− and more than 60% BCR+ B cells was directly prepared from adenoids (Figure 1A). Highly purified B cells, which do not express the BCR on their surface (BCR−), were obtained after depletion of T cells and BCR+ B cells with CD3-specific and immunoglobulin light chain–specific antibodies, respectively (Figure 1A). Both BCR+/− and BCR− B-cell populations were independently infected with the 2 different virus stocks, and their cell-cycle status was analyzed 4 days after infection as described.19
BCR+ but not BCR− germinal center B cells infected with LMP2A− 2525 EBV enter the S phase of the cell cycle and continue to proliferate in vitro. (A) Primary lymphocytes were isolated from nasal adenoids. The BCR+/− fraction consisted of approximately 30% immunoglobulin surface-negative cells and more than 60% immunoglobulin light chain–positive B cells. The BCR− fraction consisted of more than 90% CD19+, light chain–negative B cells. (B) Both fractions were infected with virus stocks of LMP2A+ 2190 EBV or LMP2A− 2525 mutant virus and analyzed for forward and sideward scatter characteristics to identify cells in the lymphocyte gate R1 4 days after infection. (C) BCR+/− B cells readily proliferated when infected with either virus (top 2 panels), but cell cycle entry of BCR− B cells was observed with LMP2A+ but not with LMP2A− mutant EBV (bottom 2 panels). Total cells were analyzed for cell-cycle distribution 4 days after infection by dual parameter flow cytometry with an APC-conjugated α-BrdU antibody to detect incorporation of the nucleotide analog BrdU and with 7-AAD to reveal the cellular DNA content as described.19 (D) Cell surface expression of λ and κ light chains of cells as in panel A. Ten days after infection, EBV growth-transformed BCR+/− B cells infected with LMP2A+ 2190 EBV (top panel) maintained the typical initial distribution of BCR+ (>60% of λ or κ light chain staining) and BCR− (approximately 30%) primary B cells as in panel A. In contrast, BCR− B cells were absent 10 days after infection when the same initially heterogeneous BCR+/− fraction was infected with the LMP2A− 2525 virus mutant as indicated by the loss of λ- and κ-positive cells in the lower left quadrant (middle panel). The majority of BCR− cells infected with LMP2+ 2190 EBV maintained their immunoglobulin surface-negative status (bottom panel). No cells survived when BCR− B cells were infected with LMP2A− 2525 mutant EBV. (E) Cell-cycle distribution and immunoglobulin light chain surface expression of cells were analyzed.
BCR+ but not BCR− germinal center B cells infected with LMP2A− 2525 EBV enter the S phase of the cell cycle and continue to proliferate in vitro. (A) Primary lymphocytes were isolated from nasal adenoids. The BCR+/− fraction consisted of approximately 30% immunoglobulin surface-negative cells and more than 60% immunoglobulin light chain–positive B cells. The BCR− fraction consisted of more than 90% CD19+, light chain–negative B cells. (B) Both fractions were infected with virus stocks of LMP2A+ 2190 EBV or LMP2A− 2525 mutant virus and analyzed for forward and sideward scatter characteristics to identify cells in the lymphocyte gate R1 4 days after infection. (C) BCR+/− B cells readily proliferated when infected with either virus (top 2 panels), but cell cycle entry of BCR− B cells was observed with LMP2A+ but not with LMP2A− mutant EBV (bottom 2 panels). Total cells were analyzed for cell-cycle distribution 4 days after infection by dual parameter flow cytometry with an APC-conjugated α-BrdU antibody to detect incorporation of the nucleotide analog BrdU and with 7-AAD to reveal the cellular DNA content as described.19 (D) Cell surface expression of λ and κ light chains of cells as in panel A. Ten days after infection, EBV growth-transformed BCR+/− B cells infected with LMP2A+ 2190 EBV (top panel) maintained the typical initial distribution of BCR+ (>60% of λ or κ light chain staining) and BCR− (approximately 30%) primary B cells as in panel A. In contrast, BCR− B cells were absent 10 days after infection when the same initially heterogeneous BCR+/− fraction was infected with the LMP2A− 2525 virus mutant as indicated by the loss of λ- and κ-positive cells in the lower left quadrant (middle panel). The majority of BCR− cells infected with LMP2+ 2190 EBV maintained their immunoglobulin surface-negative status (bottom panel). No cells survived when BCR− B cells were infected with LMP2A− 2525 mutant EBV. (E) Cell-cycle distribution and immunoglobulin light chain surface expression of cells were analyzed.
Cycling cells could be observed in both BCR+/− and BCR− B-cell populations infected with LMP2A+ 2190 EBV as expected 4 days after infection (Figure 1B,C,E).19 BCR+/− cells were also induced to proliferate when infected with LMP2A− 2525 mutant EBV (Figure 1C middle panel; Figure 1E). When infected with the LMP2A− 2525 mutant EBV, the majority of BCR− B cells had not entered S phase of the cell cycle 4 days after infection (Figure 1C bottom panel) and were not viable as indicated by their sub-G0/G1 DNA content (Figure 1E). Ten days after infection, no BCR− B cells infected with LMP2A− 2525 mutant EBV survived in vitro (data not shown). As expected, BCR− cells (Figure 1A) were readily transformed with 2089 WT EBV as described previously (data not shown)19 or with LMP2A+ 2190 EBV (Figure 1C second panel from bottom), although the majority of proliferating cells did not express immunoglobulins at their surface 10 days after infection (Figure 1D bottom panel). When BCR+/− B-cell populations were infected with LMP2A− 2525 mutant EBV, only surface immunoglobulin-positive B cells of the initially mixed BCR+/− B cells (Figure 1A top panel) survived and proliferated 10 days after infection, as indicated by the disappearance of λ or κ light chain surface-negative cells (ie, the double-negative quadrant in the middle panel of Figure 1D). In marked contrast, both BCR+ as well as BCR− B cells were transformed when infected with LMP2A+ 2190 EBV (Figure 1D top panel). Very consistently, BCR+ cells infected with LMP2A− 2525 mutant EBV also showed higher λ or κ light chain surface expression levels (approximately 2-fold higher median values) than BCR+ B cells infected with LMP2A+ 2190 EBV 10 days after infection (Figure 1D compare top panel with second panel from top).
Together, these results indicated that LMP2A expression is absolutely essential to transform BCR− B cells in vitro. The majority of germinal center B cells has a nonfunctional or low-affinity BCR because of the random process of somatic hypermutation of their immunoglobulin genes and is devoid of BCR signals necessary for their escape from apoptosis (Tze et al25 and references therein). Our results suggest that expression of LMP2A in primary B cells infected with EBV provides a survival signal equivalent to the BCR. This viral signal allows EBV to take advantage not only of BCR+ but also of BCR− and low-affinity B cells for establishment of latency.26
Sustained LMP2A expression prevents apoptotic death of in vitro EBV-transformed B cells
LMP2A is continuously expressed in vivo in many B cells latently infected with EBV, including EBV-associated B-cell tumors and certain carcinomas as described in detail in “Introduction.” Several reports also point to LMP2A's role in establishing or maintaining viral latency because it is expressed in long-lived resting memory B cells,26-31 although a recent report negates these earlier findings in individuals suffering from acute infectious mononucleosis.32 LMP2A has been reported to prevent EBV-infected cells from entering the lytic and productive phase of EBV's life cycle,11 but this issue is controversial.33,34 We therefore asked whether in vitro, growth-transformed LCLs depend on sustained LMP2A signaling. Mixed populations of BCR+/− primary B cells were infected with 2190 EBV, which carries its LMP2A allele flanked by 2 loxP sites (Figure 2A) or with 2089 WT EBV. Two months after infection, 108 cells were transfected with the expression plasmid p3233 encoding the site-specific recombinase Cre and a truncated version of NGF receptor (NGF-R) lacking its signaling domain as a phenotypic marker from a bicistronic transcript (Figure 2B). Cre-mediated deletion of LMP2A could be monitored as early as 2 hours after transfection by PCR. To assess the status of the LMP2A gene, the PCR primers (P1, P2) were used, which gave rise to PCR products of 1220 bp and 418 bp in length with the parental LMP2A+ 2190 EBV and the Cre-deleted LMP2A− variant, respectively (Figure 2C). Approximately 50% of the cells expressed NGF-R at the cell surface (Figure 2D), indicating successful transfection, which allowed FACS sorting of NGF-R–positive and Cre-expressing cells (Cre+/NGF-R+) versus nontransfected Cre−/NGF-R− cells 18 hours after transfection (Figure 2E). As a control, LCLs were used that had been infected with the prototypic 2089 WT EBV lacking loxP sites.
Sustained LMP2A expression prevents apoptotic death of EBV-transformed B cells. (A) Schematic representation of the LMP2A gene locus in 2190 EBV, which carries 2 loxP sites located upstream of the LMP2A promoter (pLMP2A) and downstream of the first exon of LMP2A. This exon is unique to LMP2A and encodes its amino-terminal signaling domain,8 whereas the 12 putative transmembrane domains and a short cytoplasmic terminus are encoded by exons 2 through 8, which are shared by the carboxy coterminal LMP2B protein whose transcript is expressed from the LMP2B promoter (pLMP2B). To monitor the status of the LMP2A gene, the PCR primers (P1, P2) were used, which gave rise to PCR products of 1220 bp and 418 bp in length with the parental LMP2A+ 2190 EBV and the Cre-deleted LMP2A− variant, respectively. (B) The p3233 expression plasmid is schematically shown, which encodes the truncated NGF receptor (NGF-R) as a phenotypic marker35 and the site-specific recombinase Cre from a bicistronic transcript. (C) Cre-mediated deletion of LMP2A could be monitored by PCR as early as 2 hours after transfection of p3233. Lanes indicated as floxed LMP2A, deleted LMP2A, and H2O are PCR controls. The lane indicated as “survivors” refers to PCR analysis of 2190-infected B cells (labeled Cre+ in panel F) at 6 weeks after transfection. (D) Transient transfection of the expression plasmid p3233 and FACS analysis of LMP2A+ 2190 EBV LCLs expressing the truncated NGF-R at their cell surface 18 hours after transfection. LCLs established with the prototypic 2089 WT EBV gave comparable results (data not shown). (E) FACS sorting of successfully transfected LMP2A+ 2190 EBV LCLs with the topmost 5% expression levels of NGF-R (Cre+/NGF-R+) versus NGF-R+–depleted (Cre−/NGF-R−) cells. (F, top panel) The absolute numbers of cells in the different fractions were determined by FACS analysis at indicated days after transfection. BD Biosciences CaliBRITE beads were used as an internal volume standard as described previously.21 Cells in the lymphocyte gate according to forward and sideward scatter criteria were included in the analysis and set to 100% 24 hours after transfection. EBV-infected LCLs infected with LMP2A+ 2190 EBV carrying the floxed LMP2A allele are indicated (2190, black lines) as well as LCLs infected with the prototypic 2089 WT EBV strain (2089, gray lines). Transfected and sorted Cre+/NGF-R+ cell fractions are indicated (Cre+) and compared with cells depleted from NGF-R+ cells by sorting (Cre−). (Bottom panel) Cre-transfected (Cre+/NGF-R+) LCLs infected with either 2089 WT EBV or LMP2A+ 2190 EBV were analyzed for annexinV staining by FACS at the time points indicated.
Sustained LMP2A expression prevents apoptotic death of EBV-transformed B cells. (A) Schematic representation of the LMP2A gene locus in 2190 EBV, which carries 2 loxP sites located upstream of the LMP2A promoter (pLMP2A) and downstream of the first exon of LMP2A. This exon is unique to LMP2A and encodes its amino-terminal signaling domain,8 whereas the 12 putative transmembrane domains and a short cytoplasmic terminus are encoded by exons 2 through 8, which are shared by the carboxy coterminal LMP2B protein whose transcript is expressed from the LMP2B promoter (pLMP2B). To monitor the status of the LMP2A gene, the PCR primers (P1, P2) were used, which gave rise to PCR products of 1220 bp and 418 bp in length with the parental LMP2A+ 2190 EBV and the Cre-deleted LMP2A− variant, respectively. (B) The p3233 expression plasmid is schematically shown, which encodes the truncated NGF receptor (NGF-R) as a phenotypic marker35 and the site-specific recombinase Cre from a bicistronic transcript. (C) Cre-mediated deletion of LMP2A could be monitored by PCR as early as 2 hours after transfection of p3233. Lanes indicated as floxed LMP2A, deleted LMP2A, and H2O are PCR controls. The lane indicated as “survivors” refers to PCR analysis of 2190-infected B cells (labeled Cre+ in panel F) at 6 weeks after transfection. (D) Transient transfection of the expression plasmid p3233 and FACS analysis of LMP2A+ 2190 EBV LCLs expressing the truncated NGF-R at their cell surface 18 hours after transfection. LCLs established with the prototypic 2089 WT EBV gave comparable results (data not shown). (E) FACS sorting of successfully transfected LMP2A+ 2190 EBV LCLs with the topmost 5% expression levels of NGF-R (Cre+/NGF-R+) versus NGF-R+–depleted (Cre−/NGF-R−) cells. (F, top panel) The absolute numbers of cells in the different fractions were determined by FACS analysis at indicated days after transfection. BD Biosciences CaliBRITE beads were used as an internal volume standard as described previously.21 Cells in the lymphocyte gate according to forward and sideward scatter criteria were included in the analysis and set to 100% 24 hours after transfection. EBV-infected LCLs infected with LMP2A+ 2190 EBV carrying the floxed LMP2A allele are indicated (2190, black lines) as well as LCLs infected with the prototypic 2089 WT EBV strain (2089, gray lines). Transfected and sorted Cre+/NGF-R+ cell fractions are indicated (Cre+) and compared with cells depleted from NGF-R+ cells by sorting (Cre−). (Bottom panel) Cre-transfected (Cre+/NGF-R+) LCLs infected with either 2089 WT EBV or LMP2A+ 2190 EBV were analyzed for annexinV staining by FACS at the time points indicated.
After transfection and sorting of Cre+/NGF-R+ versus Cre−/NGF-R− cells, 2 cell populations infected either with 2190 EBV encoding a floxed LMP2A allele or with 2089 WT EBV were analyzed for their phenotypes with respect to cell numbers and annexinV staining to monitor proliferation and apoptosis, respectively. Both Cre−/NGF-R− 2190- and 2089 WT–infected LCLs recovered from transfection after 2 to 3 days and proliferated exponentially (Figure 2F). Cre+/NGF-R+ LCLs infected with 2089 WT EBV showed a certain degree of genotoxicity, presumably resulting from inappropriately high Cre expression levels, as expected,36 and recovered later. Cre+/NGF-R+ LCLs infected with 2190 EBV, which carries a floxed LMP2A allele, did not recover from transfection of Cre for 2 to 3 weeks (Figure 2F top panel). Long-term survivors that grew out 6 to 8 weeks after transfection showed no Cre-mediated deletion of LMP2A as assessed by PCR analysis 6 weeks after transfection (Figure 2C lane labeled “survivors” and data not shown), indicating that they were derived from nontransfected cells contaminating the Cre+/NGF-R+ fraction or from cells that had not undergone Cre-mediated deletion of LMP2A. LCLs transiently transfected with a control expression plasmid encoding NGF-R but not Cre (p3232) and sorted for NGF-R surface expression exhibited a phenotype like that of Cre−/NGF-R− cells in Figure 2F (data not shown), indicating that NGF-R expression had no measurable effect on transfected cells in this experiment. Approximately 30% of cells in the Cre+/NGF-R+ fraction of 2190-infected LCLs became annexinV positive 24 hours after transfection compared with approximately 10% of 2089 WT EBV–infected LCLs (Figure 2F bottom panel). Thus, the dramatic loss of cell numbers in the Cre+/NGF-R+ fraction of 2190-infected LCLs likely resulted from cells that had undergone rapid apoptosis upon Cre-mediated deletion of LMP2A.
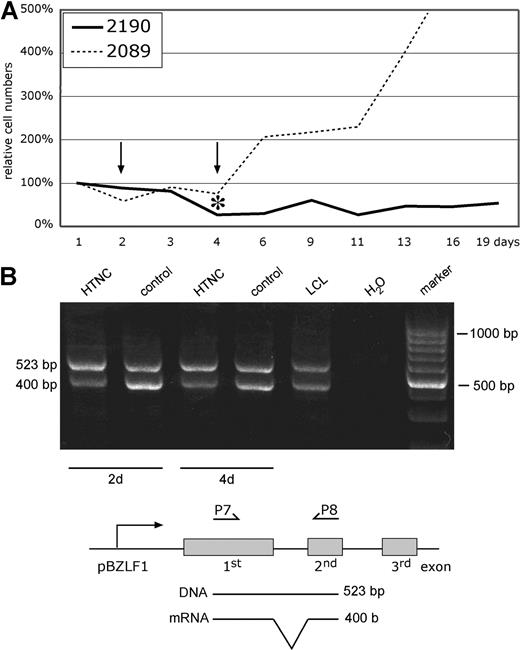
To confirm this finding in an independent experimental approach, we used the recombinant Cre-Tat fusion protein HTNC23 to deliver Cre recombinase activity by direct protein transduction to 2089- and 2190-infected LCLs. Cells infected with the prototypic 2089 WT EBV continued to proliferate upon transduction of HTNC, but 2190-infected LCLs with the loxP-flanked LMP2A allele declined in numbers and did not recover within 3 weeks (Figure 3). As described in “Deletion of LMP2A in 2190-infected B cells,” real-time PCR analyses measuring the total numbers of LMP2A+ and LMP2A− EBV genomes indicated that more than 90% of the Cre-transduced 2190-infected LCLs had their LMP2A gene deleted 4 days after transduction (data not shown).
Continuous proliferation of EBV-transformed cells requires LMP2A. (A) The absolute numbers of B cells infected with 2089 WT EBV or 2190 EBV with its LMP2A gene flanked by 2 loxP sites were determined by FACS analysis as in Figure 2F. The cells were transduced with the E coli–derived Cre protein HTNC23 for 3 hours, and the surviving cells were counted by FACS and set to 100% on day 1. 2089 WT EBV–infected LCLs recovered from this manipulation and started to proliferate, but LCLs infected with LMP2A+ 2190 EBV did not recover within the observation period. The asterisk indicates the time point when cells were removed for quantitative real-time PCR analysis of the LMP2A gene, which revealed that more than 90% of the 2190 EBV–infected cells had lost the LMP2A allele 4 days after transduction (data not shown). (B) Reverse transcription (RT)-PCR analysis of BZLF1 transcripts in HTNC-transduced 2190-infected LCLs 2 and 4 days after Cre transduction. The positions of the primer oligonucleotides P7 and P8 are schematically shown together with the predicted size of the PCR products indicative of mRNA-specific cDNA molecules of the actively transcribed BZLF1 gene and (contaminating) viral DNA. BZLF1 transcription levels did not increase but showed a slight reduction upon deletion of LMP2A, indicating that its loss did not trigger the onset of the lytic phase of EBV's life cycle.
Continuous proliferation of EBV-transformed cells requires LMP2A. (A) The absolute numbers of B cells infected with 2089 WT EBV or 2190 EBV with its LMP2A gene flanked by 2 loxP sites were determined by FACS analysis as in Figure 2F. The cells were transduced with the E coli–derived Cre protein HTNC23 for 3 hours, and the surviving cells were counted by FACS and set to 100% on day 1. 2089 WT EBV–infected LCLs recovered from this manipulation and started to proliferate, but LCLs infected with LMP2A+ 2190 EBV did not recover within the observation period. The asterisk indicates the time point when cells were removed for quantitative real-time PCR analysis of the LMP2A gene, which revealed that more than 90% of the 2190 EBV–infected cells had lost the LMP2A allele 4 days after transduction (data not shown). (B) Reverse transcription (RT)-PCR analysis of BZLF1 transcripts in HTNC-transduced 2190-infected LCLs 2 and 4 days after Cre transduction. The positions of the primer oligonucleotides P7 and P8 are schematically shown together with the predicted size of the PCR products indicative of mRNA-specific cDNA molecules of the actively transcribed BZLF1 gene and (contaminating) viral DNA. BZLF1 transcription levels did not increase but showed a slight reduction upon deletion of LMP2A, indicating that its loss did not trigger the onset of the lytic phase of EBV's life cycle.
LMP2A can block BCR activation by recruiting src family protein tyrosine kinases such as Lyn and Syk.8 This recruitment causes their immediate proteasomal cleavage because LMP2A associates with members of the Nedd4 family ubiquitin protein ligases, which results in rapid degradation of LMP2A and the associated protein tyrosine kinases bound to it.37-39 Depletion of Lyn and Syk from the BCR proximal signaling complex is detrimental for BCR-dependent signaling. BCR activation results in the activation of a number of well-known signaling pathways to support B-cell survival, proliferation, and differentiation. Engagement of the BCR can also lead to transcriptional activation of the viral molecular switch gene BZLF1, inducing EBV's lytic mode, which LMP2A counteracts.11 In our experiments, induced loss of LMP2A signaling by its Cre-mediated deletion could therefore have led to transcriptional activation of BZLF1 and onset of virion synthesis, which is thought to cause cellular death.40 Thus, we assessed transcriptional activation of BZLF1 by reverse-transcription PCR analysis but found no increase in its encoding transcripts upon deletion of LMP2A 2 and 4 days after transduction of Cre (Figure 3B). Similarly, 24 hours after infection primary B cells infected with either LMP2A− 2525 mutant EBV or LMP2A+ 2190 EBV did not differ with respect to BZLF1 or EA-D expression levels (data not shown).
LMP2A appears as a functional analog of the BCR
Immature and mature cells of the B-cell lineage, including pre-B-cells, express immunoglobulins at their surface and form a functional BCR. Even in the absence of its cognate antigen, the expression of a BCR complex mediates what is called “basal” or “tonic” signaling, which is a prerequisite for B-cell survival and maintenance of the developmental state of immature B cells, as has been demonstrated in several in vivo models (Tze et al 25 and references therein). B cells in which BCR expression is abrogated as a result of somatic hypermutation are prone to die by apoptosis during a germinal center reaction because constitutive “tonic” signaling41 of the BCR through protein tyrosine kinases is essential for B-cell survival.25
Our results suggest that LMP2A can replace BCR function(s) because BCR− B cells can be rescued and transformed only with an EBV-expressing LMP2A, not with an LMP2A− mutant (Figure 1). Our results also indicate that LMP2A-derived signals dominate or supersede those that originate from basal BCR signaling. Induced loss of LMP2A in BCR+ B cells leads to immediate induction of apoptosis (Figure 2F). This finding is surprising because several studies have reported that LMP2A is dispensable for growth transformation of peripheral B cells (Speck et al 7 and references therein). LCLs exhibit a low expression of surface immunoglobulins, which are down-regulated by EBV's latent gene EBNA2.42 Our results suggest that LMP2A is only dispensable for transformation when B cells manage to escape from BCR down-regulation and maintain high levels of BCR expression (Figure 1D). Moreover, LMP2A-expressing LCLs might have a proliferative advantage because LMP2A provides a more potent BCR-like signal; this enhanced potency might favor the outgrowth of B cells that express their BCR below levels required for signaling. This scenario might also explain why LMP2A-expressing B cells do not respond to antigen as shown by a lack calcium release.11 In BCR+low B cells, shutoff of LMP2A signaling causes apoptotic death because BCR signaling is not sufficient to maintain survival (Figures 2,3). Therefore LMP2A signaling is fundamental not only in establishing a transformed state in B cells but also in maintaining it.
Taken together, these data show that LMP2A can mimic and replace BCR signaling constitutively supporting an activated, proliferative state in B cells with no or low-affinity BCR characteristics. In vivo, LMP2A is expressed in latently infected germinal center B cells and could rescue those BCR− B cells from apoptosis that had acquired aberrant mutations during somatic hypermutation. Such B cells also constitute a likely reservoir for precursors of EBV-associated tumors such as Hodgkin lymphoma. Because LMP2A is expressed in at least a fraction of EBV-positive Hodgkin lymphoma, this viral gene product might also be a potential therapeutic target. However, the signaling machinery mediating BCR and/or LMP2A signaling is largely down-regulated in Hodgkin/Reed-Sternberg cells,16,43 and the relevance of this approach needs to be assessed.
LMP2A is a remarkable tool for studying BCR signaling. It remains to be demonstrated whether LMP2A- and BCR-derived signals induce the same signaling pathways and address similar cellular target genes because our preliminary data indicate that LMP2A could also provide a proliferative signal akin to an activated BCR.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft Grants SFB455, TRR36, and HA1354/6–1; Deutsche Krebshilfe Grant 107119; and Public Health Service Grant CA70723.
We are grateful to M. Altmann for his valuable input and construction of the mutant EBVs 2190 and 2525. HTNC was kindly provided by F. Edenhofer (University of Bonn, Germany). Cell sorting was supported by J. Ellwart (GSF-Research Center, Munich, Germany).
Authorship
Contribution: C.M. designed and performed research and analyzed data; W.H. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wolfgang Hammerschmidt, GSF-National Research Center for Environment and Health Department of Gene Vectors, Marchioninistr. 25, D-81377 Munich, Germany; e-mail:hammerschmidt@gsf.de.