Abstract
How receptor acquisition correlates with the functional maturation of natural killer (NK) cells is poorly understood. We used quantitative real-time polymerase chain reaction (PCR) assays to compare NKG2 and killer immunoglobulin-like receptor (KIR) gene expression in NK cells from allogeneic transplant recipients and their donors. Marked differences were observed in the NK subsets of recipients who had 8-fold more CD56bright cells, diminished KIR expression (except 2DL4), and increased NKG2A. In normal blood not all CD56dim cells express KIR, and a novel subpopulation of cells committed to the NK-cell lineage was defined. These cells, which comprise 19.4% ± 2.8% of the CD56dim NK population in healthy donors, express the activating NKG2D and NKG2E receptors but no KIR or NKG2A. Although the CD56dim NKG2A− KIR− NK cells lack “at least one” inhibitory receptor for autologous MHC class I, they are not fully responsive, but rather functionally immature cells with poor cytotoxicity and IFN-γ production. Upon culture with IL-15 and a stromal cell line, CD56dim and CD56bright KIR− NK cells proliferate, express KIR, and develop cytotoxicity and cytokine-producing potential. These findings have implications for the function of NK cells reconstituting after transplantation and support a model for in vivo development in which CD56bright cells precede CD56dim cells.
Introduction
Natural killer (NK) cells use several families of NK-cell receptors (NKRs) to detect MHC class I. NK-cell recognition of targets that lose MHC class I expression, “missing self,” is mediated by inhibitory MHC class I receptors and renders them susceptible to NK-cell lysis. In humans, such inhibitory receptors include the conserved CD94/NKG2A receptor, which recognizes leader peptides derived from classical HLA-A, -B, or -C presented by nonclassical class I molecule HLA-E, and the variable killer immunoglobulin-like receptors (KIRs) that recognize polymorphic determinants of HLA-A, -B, and -C. Analysis of functional specificity and its correlation with NKR expression showed that NK-cell clones are prevented from killing healthy autologous cells by inhibitory receptors that engage autologous MHC class I molecules.1 Such observations led to the proposition that mature peripheral blood NK cells must express at least one inhibitory NKR for self-MHC class I to prevent autoreactivity.2,3 Recent studies of mouse4 and human5 NK cells that lack inhibitory receptors for self-MHC class I raise questions about this model
NKRs are expressed at late stages in NK development: first CD94/NKG2A and then KIR. Although human NK cells arise from marrow-derived progenitors,6 their subsequent maturation, which may occur in the bone marrow, lymph nodes, or other peripheral sites,7,8 is poorly understood. CD56bright NK cells typically express CD94/NKG2 at high frequency and KIR at low frequency; they are weakly cytotoxic, produce high levels of cytokines and may be precursors of CD56dim NK cells.9 Development of CD56dim NK cells includes the variegated expression of KIR. This process, by which NK cells express different combinations of KIR, involves the transcriptional regulation of KIR genes by competing promoters under epigenetic control.10,11 Interactions between inhibitory KIR and cognate HLA class I ligands are central to this stage of NK-cell development and affect both the frequencies of NKR expression and their functional potential.5,12 In the absence of a defined mechanism, this effect of NKR–MHC class I interactions on NK- cell education alternatively has been described as “calibration,”13 “licensing,”14 “disarming,”15 or “education.”5
In patients recovering from hematopoietic cell transplantation (HCT), NK cells have higher CD94/NKG2A expression and decreased KIR expression compared with healthy adults.16,17 Such differences correlate with clinical outcome.16,17 Impaired NKR expression after HCT may be due to dysregulation of receptor expression or to incomplete NK-cell development, but in either case could limit the potential of a patient's NK cells to control infection and malignancy. And in general the use of NK cells in clinical applications18-21 is predicated on the assumption that therapeutic benefit will be mediated by functionally mature NK cells.
To explore the significance of these differences in patients' NK cells after HCT we have studied the function and phenotype of mature NK cells after normal development. Because flow cytometric investigation of KIR expression is limited by the lack of monoclonal antibodies (mAbs) specific for individual KIR, we developed quantitative real-time reverse-transcription–polymerase chain reaction (Q-RT-PCR) assays that improved the assessment of KIR and NKG2 expression by NK cells. Combining this novel Q-RT-PCR approach with functional assays we evaluated NKR expression by different subsets of cells in the NK-cell lineage.
Materials and methods
Cell isolation, function, and cell culture
Blood was obtained from 58 healthy volunteers and 32 donor/recipient pairs from the National Marrow Donor Program (NMDP) Research Repository after informed consent, using guidelines approved by the Committee on the Use of Human Subjects in Research at the University of Minnesota and the NMDP in accordance with the Declaration of Helsinki. NK cells were isolated from peripheral blood mononuclear cells (PBMCs) by magnetic bead selection to enrich for CD56+ cells (Miltenyi Biotech, Auburn, CA) and then sorted into subpopulations according to KIR and NKG2A expression on a FACS Diva (BD Biosciences, San Jose, CA). Cells were cultured with cytokines in the presence or absence of the murine embryonic liver cell line EL08–1D2.22 Proliferation and cytotoxicity were tested as described previously.6,23
Flow cytometry
Immunophenotypic analysis of cells was performed using 4-color analysis on a FACSCalibur (Becton Dickinson, San Jose, CA) with CELLQuest Pro software (Becton Dickinson). Cells were stained with the following monoclonal antibodies (mAbs): fluorescein isothiocyanate (FITC)– or phycoerythrin (PE)–conjugated DX9 (anti-CD158e), EB6 (anti-CD158a/h), GL183 (anti-CD158b/j); peridinin chlorophyll A protein (PerCP)–conjugated SK7 (anti-CD3); allophycocyanin (APC)–conjugated NCAM16.2 (anti-CD56); and FITC-conjugated anti-CD16, CD161, CD2, CD94, CD44, CD62L, CD7, CD244, CD18, and CD34 (Becton Dickinson). FITC-conjugated anti–hIL-12Rβ1 and PE-conjugated CD117 were obtained from (R&D Systems, Minneapolis, MN). PE-conjugated Z199 (anti-NKG2A) (Immunotech, Brea, CA) was used as indicated.
NK IFN-γ production and degranulation detected by intracellular flow cytometry
Freshly isolated or cultured NK cells (106) were incubated overnight with IL-12 (10 ng/mL) and IL-18 (100 ng/mL; both from R&D Systems) to stimulate IFN-γ production24 and for the last 5 hours brefeldin A (0.4 mg/mL) was added. Intracellular IFN-γ was stained with allophycocyanin (APC)–conjugated antihuman IFN-γ (clone 25723.11; Becton Dickinson). In degranulation assays, NK cells were incubated overnight at 37°C in 5% CO2 with or without 1000 U/mL recombinant interleukin-2 (rIL2; Chiron, Emeryville, CA). The following day the NK cells were resuspended at 1 × 106 cells/mL with or without K562 or 721.221 class I–deficient target cells added at a 1:1 ratio. Antihuman CD107a-FITC (BD Biosciences Pharmingen, San Jose, CA) was added to the tubes at 20 μL/mL. After 1 hour of incubation, monensin (GolgiStop; BD Biosciences) was added to a final concentration of 6 μL/mL, after which the incubation was continued for an additional 5 hours. In both assays, NKR mAbs were added as described in “Flow cytometry.”
KIR genotyping and real-time quantitative PCR
Preparation of genomic DNA from PBMCs and KIR genotyping by PCR—sequence-specific primer (SSP) were as described.25 Total RNA was extracted with the RNeasy Mini Kit (QIAgene, Valencia, CA) and digested with RNase-free DNase (Invitrogen, Carlsbad, CA). First-Strand cDNA synthesis used SuperScript II; reverse transcription was at 42°C for 50 minutes followed by heat inactivation at 70°C for 15 minutes; RNA was degraded by the addition of 1 μL (2 units) of Escherichia coli RNase H (Invitrogen) and incubation at 37°C for 20 minutes. Synthetic oligonucleotides (http://www.idtdna.com; Integrated DNA Technologies, Coralville, IA) were designed from the publicly available KIR sequences (GenBank [http://www.ncbi.nlm.nih.gov/] and the IPD-KIR Sequence Database [http://www.ebi.ac.uk/ipd/kir/]). PCR products were sequenced (BioMedical Genomics Center, St Paul, MN) to confirm the specificity of each reaction. We used SYBR green to label Q-RT-PCR products and found that it performed well with homogeneous cell populations: IL-2 activated NK cells or NK92 cells. With PBMCs, in which NK cells are diluted by other cell types, and the amounts of KIR mRNA are lower, primer-dimers were detected on melting point dissociation curves. Consequently, the nonspecific binding of SYBR green to double-stranded DNA, labeling both amplicons and primer-dimers, rendered many reactions unreliable (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). Instead we designed TaqMan Probes such that their annealing temperatures were 8°C to 10°C higher than their respective amplification primers (Primer Express v2.0 software; Applied Biosystems, Foster City, CA). The human 18s rRNA used to control for RNA quantity and NCAM (CD56) was measured using standard primer and probe sets (Applied Biosystems). Reactions were performed using the TaqMan Universal PCR Master Mix and a 7500 Q-PCR System (Applied Biosystems).
Statistical analysis
A classification and regression tree (CART) model was used to define the optimal Q-RT-PCR cycle threshold (Ct) at which the presence of mRNA accurately predicts KIR genotypes.26 A receiver-operating characteristic (ROC) curve was used to plot the true positive rate (sensitivity) and false-positive rate (1 − specificity) of Ct values and to calculate the positive predictive value (PPV) and Youden index.27 The Youden index (sensitivity + specificity − 1) represents the distance between points on the ROC curve and the best possible value. To analyze the KIR expression data for sorted cell populations, a Wilcoxon test was performed to derive P values comparing KIR mRNA expression levels between 2 populations and a Student t test was used for other subset comparisons. To compare the abundance of KIR mRNA with that of surface protein we used a simple linear regression model.
Results
Q-RT-PCR assays of NKR expression are sensitive and specific
To study NKR expression by peripheral blood NK cells we developed Q-RT-PCR assays specific for individual KIR genes. In designing the primers the goal was to combine gene specificity with coverage of all its alleles: a total of 203 variants being currently listed for all human KIR genes.28 Because we found SYBR green, a nonspecific stain for double-stranded DNA, to be unreliable, specific TaqMan probes were designed for each KIR gene (Table 1) that increased the specificity and reliability of the assay. To provide further validation of our reactions, amplification standard curves were run with limiting dilutions of RNA for each KIR to verify the relative linear efficiency of the PCR reaction (Figure S2).
Oligonucleotide primers and probes of KIR amplification
| Gene . | Forward primer, 5′-3′ . | Reverse primer, 5′-3′ . | Amplicon size, bp . | Taqman MGB probes, 6FAM 5′-3′ MGBNFQ . | Primer conc, nM . | Probe conc, nM . |
|---|---|---|---|---|---|---|
| 2DL1* | GCAGCACCATGTCGCTCT | GTCACTGGGAGCTGACAC | 355 | ATGCTGACGAACAAGAG | 200 | 50 |
| 2DL2 | GGAGGGGGAGGCCCATGAAT | GTCGGGGGTTACCGGTTTTA | 241 | CCAAGGTCAACGGAACA | 100 | 100 |
| 2DL3 | CCACTGAACCAAGCTCCG | CAGGAGACAACTTTGGATCA | 351 | CTGGTGCTGCAACAA | 150 | 100 |
| 2DL4 | AAACCTTCGCTTACAGCCCG | CACTGAGTACCTAATCACAG | 374 | TGCCCAGCATCAAT | 150 | 100 |
| 2DL5 | CTCTACAACAAAATATTCTGGAAGAGCA | CCGGCTGGGCTGAGAGT | 180 | TCACAGGTCTATTTGGGAAA | 200 | 100 |
| 2DS1* | TCTCCATCA GTCGCATGR | AGGGCCCAGAGGAAAGTT | 313 | AGGTCTATATGAGAAACCT | 300 | 50 |
| 2DS2 | TGCACAGAGAGGGGAAGTA | CACGCTCTCTCCTGCCAA | 256 | GTCATCACAGGTCTATATGA | 100 | 100 |
| 2DS3* | TCACTCCCCCTATCAGTTT | GCATCTGTAGGTTCCTCCT | 279 | GCCCCACGGTTCT | 300 | 200 |
| 2DS4* | CTGGCCCTCCCAGGTCA | GGAATGTTCCGTTGATGC | 450 | AAGTTTAACAACACTTTGCACC | 500 | 50 |
| 2DS5 | GCTCATGGTCATCAGCATGG | CGACCGATGGAGAAGTTGC | 260 | CACATGAGGGATTCC | 150 | 100 |
| 3DL1 (1278) | GCCTGCAGGGAACAGAACAGCC | AGAGAGGCACCAGATTTGTGGC | 365 | CTGAAGGCGTGAGTCTT | 150 | 100 |
| 3DL1 (ALL) | AGAACAGCCAACAGCGAGG | ATCTGGGCTTAGCATTTGGAAG | 171 | CCTACAGATACCATCTTGTACA | 100 | 100 |
| 3DL2* | CGGTCCCTTGATGCCTGT | GACCACACGCAGGGCAG | 368 | TGATCACAGGTCTATATGAG | 150 | 50 |
| 3DL3 | AGGGACCTACAGATGTTGCA | CTCTCTGTGCAGAAGGAAGC | 204 | CCAGCAACCCTGTGGTGATCAT | 300 | 150 |
| 3DS1* | GCACCCAGCAACCCCA | TAGGTCCCTGCAAGGGCAC | 256 | AATTTCTCCATCGGTTCCATGA | 300 | 100 |
| NKG2A | CTCCAGAGAAGCTCATTGTTGG | CACCAATCCATGAGGATGGTG | 329 | CGATAGTTGTTATTCCCTCTACA | 250 | 100 |
| NKG2D | ACAGCTGGGAGATGAGTGAATTTC | TGACTTCACCAGTTTAAGTAAATCCTG | 421 | TAGAGAAAATGCATCTCCA | 550 | 100 |
| NKG2E | GCCTGTGCTTCAAAGAACTCTTCT | CACACTGGTCTGATATAAGTCCACG | 231 | CAAACATGAGATAAAAGAC | 250 | 50 |
| Gene . | Forward primer, 5′-3′ . | Reverse primer, 5′-3′ . | Amplicon size, bp . | Taqman MGB probes, 6FAM 5′-3′ MGBNFQ . | Primer conc, nM . | Probe conc, nM . |
|---|---|---|---|---|---|---|
| 2DL1* | GCAGCACCATGTCGCTCT | GTCACTGGGAGCTGACAC | 355 | ATGCTGACGAACAAGAG | 200 | 50 |
| 2DL2 | GGAGGGGGAGGCCCATGAAT | GTCGGGGGTTACCGGTTTTA | 241 | CCAAGGTCAACGGAACA | 100 | 100 |
| 2DL3 | CCACTGAACCAAGCTCCG | CAGGAGACAACTTTGGATCA | 351 | CTGGTGCTGCAACAA | 150 | 100 |
| 2DL4 | AAACCTTCGCTTACAGCCCG | CACTGAGTACCTAATCACAG | 374 | TGCCCAGCATCAAT | 150 | 100 |
| 2DL5 | CTCTACAACAAAATATTCTGGAAGAGCA | CCGGCTGGGCTGAGAGT | 180 | TCACAGGTCTATTTGGGAAA | 200 | 100 |
| 2DS1* | TCTCCATCA GTCGCATGR | AGGGCCCAGAGGAAAGTT | 313 | AGGTCTATATGAGAAACCT | 300 | 50 |
| 2DS2 | TGCACAGAGAGGGGAAGTA | CACGCTCTCTCCTGCCAA | 256 | GTCATCACAGGTCTATATGA | 100 | 100 |
| 2DS3* | TCACTCCCCCTATCAGTTT | GCATCTGTAGGTTCCTCCT | 279 | GCCCCACGGTTCT | 300 | 200 |
| 2DS4* | CTGGCCCTCCCAGGTCA | GGAATGTTCCGTTGATGC | 450 | AAGTTTAACAACACTTTGCACC | 500 | 50 |
| 2DS5 | GCTCATGGTCATCAGCATGG | CGACCGATGGAGAAGTTGC | 260 | CACATGAGGGATTCC | 150 | 100 |
| 3DL1 (1278) | GCCTGCAGGGAACAGAACAGCC | AGAGAGGCACCAGATTTGTGGC | 365 | CTGAAGGCGTGAGTCTT | 150 | 100 |
| 3DL1 (ALL) | AGAACAGCCAACAGCGAGG | ATCTGGGCTTAGCATTTGGAAG | 171 | CCTACAGATACCATCTTGTACA | 100 | 100 |
| 3DL2* | CGGTCCCTTGATGCCTGT | GACCACACGCAGGGCAG | 368 | TGATCACAGGTCTATATGAG | 150 | 50 |
| 3DL3 | AGGGACCTACAGATGTTGCA | CTCTCTGTGCAGAAGGAAGC | 204 | CCAGCAACCCTGTGGTGATCAT | 300 | 150 |
| 3DS1* | GCACCCAGCAACCCCA | TAGGTCCCTGCAAGGGCAC | 256 | AATTTCTCCATCGGTTCCATGA | 300 | 100 |
| NKG2A | CTCCAGAGAAGCTCATTGTTGG | CACCAATCCATGAGGATGGTG | 329 | CGATAGTTGTTATTCCCTCTACA | 250 | 100 |
| NKG2D | ACAGCTGGGAGATGAGTGAATTTC | TGACTTCACCAGTTTAAGTAAATCCTG | 421 | TAGAGAAAATGCATCTCCA | 550 | 100 |
| NKG2E | GCCTGTGCTTCAAAGAACTCTTCT | CACACTGGTCTGATATAAGTCCACG | 231 | CAAACATGAGATAAAAGAC | 250 | 50 |
Primers from Uhrberg et al.47
conc indicates concentration.
Performance of the Q-RT-PCR assay was tested by analyzing the expression of 14 functional KIR genes by PBMCs from 36 healthy donors of known KIR genotype (2DL5A and B were tested together). For 12 of the 14 genes, expression was reliably detected by Q-RT-PCR whenever the gene was present. For 2 genes, 3DL3 and 2DS4, correlations between gene presence and gene expression were less strong. For 3DL3, a component of all KIR haplotypes, expression was undetectable in approximately 80% of the donors. This is consistent with others' observations showing the 3DL3 promoter is normally turned off by methylation, and that surface protein expression is detected only after in vitro demethylation.11,29 For 2DS4, we also obtained negative Q-RT-PCR results from most humans having the gene (74%). This pattern was not fully explained by the presence of 2DS4*003, a common impaired allele giving very low levels of mRNA.30,31 We therefore excluded 2DS4 and 3DL3 from further analysis, which focused on the Q-RT-PCR reactions for 2DL1–5, 2DS1–3, 2DS5, 3DL1/S1, and 3DL2 (Table 1).
Using the KIR genotype as the standard, a classification and regression tree (CART) analysis was used to determine the optimal Q-RT-PCR criteria for assignment of KIR gene presence and expression. Using a cutoff of less than 38 for the cycle threshold (Ct) at which the transcript expression was first detected, we correctly predicted 412 (95.3%) of 432 genotypes with specificity (0.96), sensitivity (0.94), PPV (0.978), and Youden index (0.9). In a previous Q-RT-PCR study using SYBR green, only 75% correlation between genotype and mRNA expression was obtained for the 4 KIR genes analyzed.32 We also compared Q-RT-PCR assessment of mRNA to the abundance of KIR protein at the cell surface determined by flow cytometry. We tested the 3DL1-specific mAb DX9 (anti-CD158e) against a Q-RT-PCR reaction that detects all 3DL1. Although a significant correlation was obtained between the abundance of 3DL1 mRNA expression and cell surface protein (R = 0.42, P = .014), we repeated the comparison using an alternative 3DL1 reaction that detects most of the commonly expressed alleles but not 3DL1*004,33 for which protein is made but does not reach the cell surface. With this approach, the correlation improved (r = .61, P = .001). Using similar methods, Q-RT-PCR reactions were developed for NKG2A, NKG2D, and NKG2E (Table 1).
NK-cell development is altered after hematopoietic cell transplantation
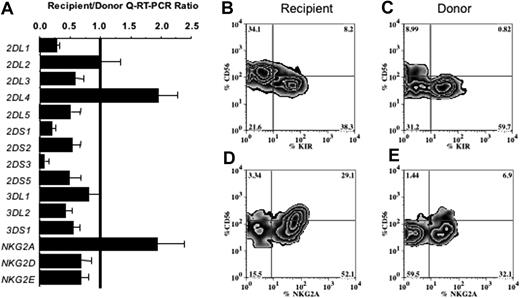
We previously reported decreased surface KIR expression by NK cells after HCT.17 Here the same samples were analyzed by Q-RT-PCR to determine whether individual KIRs were affected equally. NKR expression was compared in normal donor and donor-derived recipient blood samples collected before and approximately 100 days after unmanipulated, T-cell–replete marrow transplantation facilitated through the NMDP. Relative mRNA expression data were calculated for each transplant as a recipient-donor ratio, allowing each recipient their own paired control. As the Q-RT-PCR assays use unsorted mononuclear cells, it was necessary to interpret results based on the proportion of NK cells in each sample. To do so the amounts of NKR mRNA were normalized to the amount of CD56 (NCAM) mRNA in each sample, as also assayed by Q-RT-PCR. CD56 expression was considerably increased (11.6 ± 4.2-fold) in recipient versus donor samples, consistent with the higher proportion of NK cells seen after HCT.16,17 After transplantation all KIR had an average mRNA expression ratio of less than or equal to 1, with the exception of 2DL4, which was expressed more in recipient than donor samples (Figure 1A). NKG2A was increased significantly after transplantation but NKG2D and NKG2E were not.
NK cells reconstituted 100 days after allogeneic transplantation are more frequently CD56bright and display a distinctive repertoire of NK-cell receptors compared with the donor's NK cells. (A) Comparison of NKR expression in blood mononuclear cells of recipients 100 days after transplantation and their donors, as assessed by Q-RT-PCR of 15 individual NKR genes. For each gene the mean recipient/donor ratio obtained from analysis of 32 transplant pairs is shown. Not all KIR were analyzed for all transplant pairs, because one or more KIR genes were absent from the donor KIR genotype. Variation in the number of NK cells in the mononuclear cell fractions was corrected by normalization of the NKR transcript numbers to the NCAM (CD56) transcript number. Error bars indicate standard error of the mean (SEM). (B-E) Flow cytometric analysis comparing the surface expression of KIR (B-C) and NKG2A (D-E) on CD56+CD3− gated NK cells from one representative transplant recipient (panels B,D) and the donor (panels C,E). The proportion of cells in each of the 4 quadrants is given as a percentage. The KIR analysis used a cocktail of the EB6, GL183, and DX9 mAbs.
NK cells reconstituted 100 days after allogeneic transplantation are more frequently CD56bright and display a distinctive repertoire of NK-cell receptors compared with the donor's NK cells. (A) Comparison of NKR expression in blood mononuclear cells of recipients 100 days after transplantation and their donors, as assessed by Q-RT-PCR of 15 individual NKR genes. For each gene the mean recipient/donor ratio obtained from analysis of 32 transplant pairs is shown. Not all KIR were analyzed for all transplant pairs, because one or more KIR genes were absent from the donor KIR genotype. Variation in the number of NK cells in the mononuclear cell fractions was corrected by normalization of the NKR transcript numbers to the NCAM (CD56) transcript number. Error bars indicate standard error of the mean (SEM). (B-E) Flow cytometric analysis comparing the surface expression of KIR (B-C) and NKG2A (D-E) on CD56+CD3− gated NK cells from one representative transplant recipient (panels B,D) and the donor (panels C,E). The proportion of cells in each of the 4 quadrants is given as a percentage. The KIR analysis used a cocktail of the EB6, GL183, and DX9 mAbs.
To determine whether the decreased KIR expression was limited to a particular NK-cell subset, flow cytometry was used to evaluate NK receptor expression on CD56bright and CD56dim populations. KIR expression was measured using a cocktail of 3 mAbs (GL183, EB6, DX9) that recognizes 6 of the 15 KIRs (2DL1, 2DS1, 2DL2, 2DL3, 2DS2, and 3DL1). There was an average 8.2 ± 2.8-fold increase in the proportion of CD56bright NK cells in recipients (38% ± 5.3% of NK cells) compared with donors (9% ± 1.6%, P < .0001). Furthermore, KIR expression on reconstituting NK cells was significantly decreased after transplantation (25% ± 3.5% of total NK cells) compared with donor cells (42% ± 4%, P = .003, Figure 1B-C). A reciprocal pattern was seen for NKG2A, with 92% ± 1.5% of recipient NK cells expressing this receptor compared with 54% ± 4.4% of donor NK cells (Figure 1D-E). After transplantation the predominant NK-cell phenotype was NKG2A+KIR− (which includes both CD56bright and CD56dim cells). As is seen in normal NK cells, the recipient CD56bright cells expressed less KIR than the CD56dim NK cells. Because of the increased NKG2A seen after transplantation, only 8.4% ± 1.2% of the CD56dim cells lacked both NKG2A and KIR after HCT, a significantly smaller number than the 27% ± 3.1% of such cells measured in the donors. To understand the significance of these findings, we performed experiments to test the function and developmental potential of NK-cell subsets from the peripheral blood of healthy donors.
A subpopulation of blood CD56dim NK cells lacks NKG2A and KIR
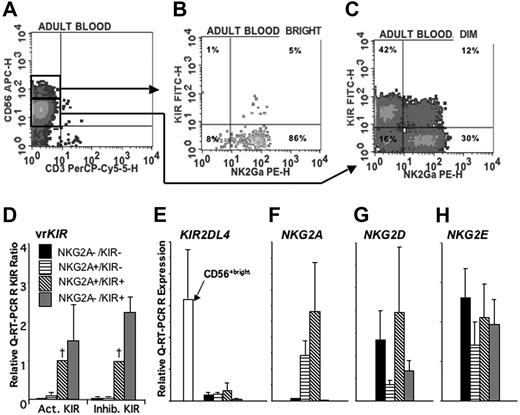
For this analysis KIR+ peripheral blood NK cells were defined by flow cytometry using the same KIR mAb cocktail. Study of 8 healthy donors showed that 55% ± 2.2% of CD56dim NK cells expressed KIR, compared with 7.8% ± 1.1% CD56bright NK cells (P < .001, Figure 2A-C). We first sorted the CD56dim NK cells into KIR+ and KIR− subpopulations, which were then compared for KIR gene expression by Q-RT-PCR assays. As expected, the abundance of 2DL1, 2DS1, 2DL2, 2DL3, 2DS2, and 3DL1 mRNA was much higher in the KIR+ subpopulation. The median ratio of mRNA levels for those KIR was 221.3 (n = 43, P < .001) in KIR+ relative to KIR− cells. Expression of genes (2DS4, 2DL4, 3DL2, 2DL5, 2DS3, 3DS1 and 2DS5) encoding KIR not recognized by the antibody cocktail was also higher in CD56dim KIR+ cells than CD56dim KIR− cells, with a median ratio of 5.05 (P < .001). That CD56dim NK cells expressing one KIR gene are more likely to express the others is consistent with the variegated KIR expression by NK-cell clones.3 For all KIR genes except 2DL4, the abundance of mRNA was less in CD56bright than CD56dim KIR+ cells, with a median expression ratio of 0.04 (P = .001; n = 29). 2DL4 mRNA levels were 6.6-fold higher in CD56bright KIR− cells than in CD56dim KIR+ cells (P = .03; n = 6). These results are consistent with knowledge of the common regulatory elements controlling KIR gene expression during NK-cell development and the apparent need for NK cells to transcribe 2DL4 before any other KIR genes.11,34
NKG2A and KIR expression distinguishes populations of CD56dim NK cells. PBMCs were enriched for NK cells using a negative immunomagnetic bead depletion strategy. A representative example of a 4-color analysis shows gating of (A) CD56bright and CD56dim cells and analysis of the expression of NKG2A and KIR (using a cocktail of EB6, GL183, and DX9 mAbs) on (B) CD56bright and (C) CD56dim cells. Each of the 4 subpopulations shown in panel C was sorted and then analyzed by Q-RT-PCR for the expression of (D) KIR exhibiting variegated expression (vr) (n = 33 reactions), (E) KIR2DL4 (n = 6), (F) NKG2A (n = 7), (G) NKG2D (n = 7), and (H) NKG2E (n = 7). The numbers of NKG2 and KIR2DL4 transcripts in each subpopulation are compared with the positive control of IL-2 activated NK cells to give a relative expression. For comparison of vrKIR expression in the 4 subpopulations, the data for each KIR were normalized to those of the NKG2A+KIR+ (designated by a †). These values were used to calculate a mean relative expression for the activating (KIR2DS1–3, 2DS5, 3DS1) and inhibitory (KIR2DL1–3, 2DL5, 3DL1, 3DL2). When a KIR was not expressed in the NKG2A+KIR+ population, indicating absence of the gene, it was excluded from the calculation of mean relative expression. Error bars indicate SEM.
NKG2A and KIR expression distinguishes populations of CD56dim NK cells. PBMCs were enriched for NK cells using a negative immunomagnetic bead depletion strategy. A representative example of a 4-color analysis shows gating of (A) CD56bright and CD56dim cells and analysis of the expression of NKG2A and KIR (using a cocktail of EB6, GL183, and DX9 mAbs) on (B) CD56bright and (C) CD56dim cells. Each of the 4 subpopulations shown in panel C was sorted and then analyzed by Q-RT-PCR for the expression of (D) KIR exhibiting variegated expression (vr) (n = 33 reactions), (E) KIR2DL4 (n = 6), (F) NKG2A (n = 7), (G) NKG2D (n = 7), and (H) NKG2E (n = 7). The numbers of NKG2 and KIR2DL4 transcripts in each subpopulation are compared with the positive control of IL-2 activated NK cells to give a relative expression. For comparison of vrKIR expression in the 4 subpopulations, the data for each KIR were normalized to those of the NKG2A+KIR+ (designated by a †). These values were used to calculate a mean relative expression for the activating (KIR2DS1–3, 2DS5, 3DS1) and inhibitory (KIR2DL1–3, 2DL5, 3DL1, 3DL2). When a KIR was not expressed in the NKG2A+KIR+ population, indicating absence of the gene, it was excluded from the calculation of mean relative expression. Error bars indicate SEM.
NKG2A expression by CD56dim KIR+ and CD56dim KIR− subpopulations of NK cells was examined next. Flow cytometry showed a higher frequency of NKG2A expression by CD56dim KIR− cells compared with CD56dim KIR+ cells (75.3% ± 2.5% vs 32.0% ± 6.6%; P = .038; n = 3). Such inverse correlationbetween KIR and NKG2A expression is typical,35 and reflects the analogous self-recognition functions of CD94:NKG2A and the inhibitory HLA class I–specific KIR. To distinguish the subsets, a further sorting analysis was performed that included an anti-NKG2A mAb. As in the posttransplantation samples, a distinct subpopulation of CD56dim NK cells that had neither NKG2A nor KIR at the cell surface was identified. These CD56dim NKG2A−KIR− NK cells were present in the blood of all donors (Figure 2A-C). Q-RT-PCR confirmed their lack of KIR expression: mRNA levels for 6 inhibitory and 5 activating KIR genes were 25- to 50-fold lower in the KIR− NKG2A+ or NKG2A− populations compared with either KIR+ population (Figure 2D). Separate measurement of 2DL4 mRNA showed it was present at uniformly low level in all of the CD56dim populations, but at significantly higher level in CD56bright cells (Figure 2E). Although NKG2A gene transcription was essentially absent from the CD56dim NKG2A−KIR− NK cells, Q-RT-PCR analysis revealed consistent expression of NKG2D and NKG2E, which encode activating receptors (Figure 2F-H). In summary, the circulating pool of normal human CD56dim NK cells divides into 4 subpopulations according to KIR and NKG2A expression: NKG2A−KIR− (19% ± 2.8% of CD56dim NK cells, n = 26); NKG2A+KIR− (29% ± 2.6%); NKG2A+KIR+ (15% ± 1.8%); NKG2A−KIR+ (36% ± 3.2%) (Figure 2C).
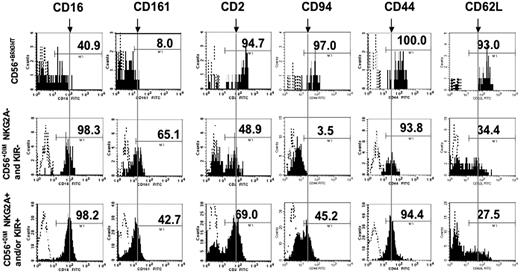
To evaluate the NKG2A−KIR− NK-cell subset further, we performed flow cytometry with a panel of antibodies recognizing surface antigens of NK cells or their precursors. Shown in Figure 3 are the expression profiles of 3 subsets: CD56bright, CD56dimNKG2A−KIR−, and all other CD56dim cells expressing NKG2A and/or KIR. All subsets uniformly expressed CD7, 2B4, CD18, and IL12Rβ1 and lacked CD117 and CD34 (not shown). Apart from CD94, which heterodimerizes with NKG2, the expression profile of the CD56dimNKG2A−KIR− subset was similar to the mature KIR+ NK-cell populations. CD56bright NK cells differed significantly from CD56dim cells by expression of more CD2, CD94, CD44, and CD62L and less CD16 and CD161. The surface antigens present on the CD56dimNKG2A−KIR− subset are consistent with their assignment to the NK-cell lineage rather than cells of a nonlymphoid lineage that have acquired aberrant expression of CD56.
NK-cell subsets differentially express surface antigens. PBMCs were stained with anti-CD56, anti-CD3, a cocktail of NKG2A and KIR antibodies, and FITC-conjugated antibodies against CD2, 16, 44, 62L, 94, and 161. NK-cell subsets were gated on the expression of CD56, and on the presence or absence of NKG2A and KIR. Shown are representative histograms (n = 5-11) for the surface antigens indicated. To facilitate comparison of antigen density, a vertical line (designated by a downward arrow above the CD56bright plots) is drawn through the approximate mean channel fluorescence of the subpopulation of CD56dim cells expressing NKG2A and/or KIR.
NK-cell subsets differentially express surface antigens. PBMCs were stained with anti-CD56, anti-CD3, a cocktail of NKG2A and KIR antibodies, and FITC-conjugated antibodies against CD2, 16, 44, 62L, 94, and 161. NK-cell subsets were gated on the expression of CD56, and on the presence or absence of NKG2A and KIR. Shown are representative histograms (n = 5-11) for the surface antigens indicated. To facilitate comparison of antigen density, a vertical line (designated by a downward arrow above the CD56bright plots) is drawn through the approximate mean channel fluorescence of the subpopulation of CD56dim cells expressing NKG2A and/or KIR.
CD56dim NKG2A−KIR− NK cells have diminished effector functions
The CD56bright and CD56dim NK cells were compared to determine whether the CD56dim NKG2A−KIR− subpopulation that lacks self-inhibitory receptors has normal NK-cell functions. First we measured the proliferation of KIR− (CD56bright and CD56dim) and CD56dim KIR+ cells. Because cytokine-driven proliferation of immature NK cells is greatly enhanced by contact with accessory cells,6 these experiments also tested whether a novel murine embryonic liver cell line (EL08–1D2)22 could costimulate NK-cell growth in the presence of IL-15. Whereas minimal expansion of CD56dim KIR− or CD56dim KIR+ cells was observed after 14 days culture with IL-15 alone (not shown), inclusion of EL08–1D2 cells facilitated proliferation of all subpopulations (Figure 4). This proliferation was significantly less when the NK cells were separated from EL08–1D2 cells by a microporous membrane, showing its dependence on cell-cell contact. As expected, the CD56bright cells proliferated significantly better than the CD56dim populations. We found also that the CD56dim KIR− cells proliferated better than the CD56dim KIR+ cells. To examine the former population further, CD56dim KIR− cells were sorted by their NKG2A expression and tested in a 6-day thymidine incorporation assay with IL-15 alone (Figure 4 inset). The CD56dim NKG2A−KIR− population proliferated about 4-fold less the NKG2A+KIR− population, showing that it was hyporesponsive to IL-15 (P < .002).
CD56dim NK cells proliferate less than CD56bright NK cells. Enriched NK cells were sorted into CD56bright KIR−, CD56dim KIR−, and CD56dim KIR+ NK cells and assayed for proliferation after 14 days of culture with the mouse stromal cell EL08–1D2 and exogenous IL-15 (n = 4). NK cells were either in direct contact with the stromal cells (black bars: EL Contact) or separated from them by a Transwell membrane (hatched bars: EL TW). These data are shown in the lower part of the figure: CD56bright KIR− NK cells proliferated more than either CD56dim subset (*P < .01) and the CD56dim KIR− population proliferated more than CD56dim KIR+ cells (P = .001). In the experiment shown in the boxed, upper part of the figure the CD56dim KIR− subset was further divided into CD56dim NKG2A−KIR− and NKG2A+KIR− subsets (n = 4) and tested in a 6-day thymidine incorporation assay where the NKG2A−KIR− subset showed less short-term responsiveness to IL-15 (10 ng/mL) (P = .002). Error bars indicate SEM.
CD56dim NK cells proliferate less than CD56bright NK cells. Enriched NK cells were sorted into CD56bright KIR−, CD56dim KIR−, and CD56dim KIR+ NK cells and assayed for proliferation after 14 days of culture with the mouse stromal cell EL08–1D2 and exogenous IL-15 (n = 4). NK cells were either in direct contact with the stromal cells (black bars: EL Contact) or separated from them by a Transwell membrane (hatched bars: EL TW). These data are shown in the lower part of the figure: CD56bright KIR− NK cells proliferated more than either CD56dim subset (*P < .01) and the CD56dim KIR− population proliferated more than CD56dim KIR+ cells (P = .001). In the experiment shown in the boxed, upper part of the figure the CD56dim KIR− subset was further divided into CD56dim NKG2A−KIR− and NKG2A+KIR− subsets (n = 4) and tested in a 6-day thymidine incorporation assay where the NKG2A−KIR− subset showed less short-term responsiveness to IL-15 (10 ng/mL) (P = .002). Error bars indicate SEM.
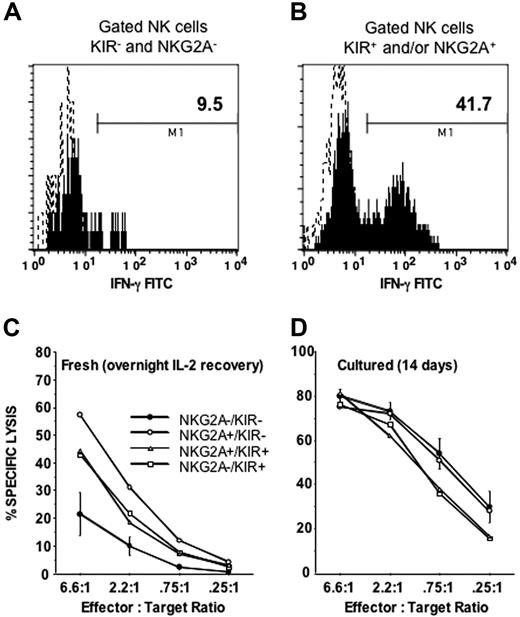
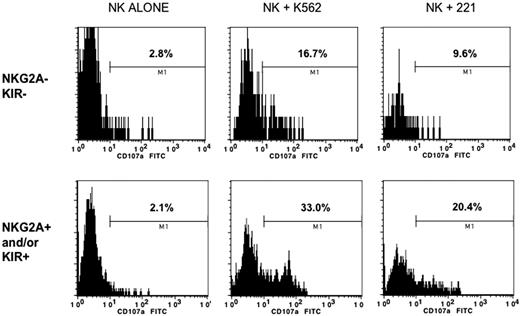
The 4 CD56dim subpopulations were next compared for cytotoxicity and IFN-γ production. Compared with the NKG2A+ and/or KIR+ subsets, NKG2A−KIR− cells expressed IFN-γ poorly after overnight stimulation with IL-12 and IL-18 (Figure 5A-B). Furthermore, the NKG2A−KIR− subset was the only subpopulation with diminished killing of K562 cells (P < .05 compared with all other populations; Figure 5C). Consistent with these results the CD56dim NKG2A−KIR− population had the least degranulation, as measured by CD107a expression, when stimulated with class I–deficient target cells (K562 and 721.22) (Figure 6).
CD56 dim NK cells lacking NKG2A and KIR are functionally immature. PBMCs were incubated overnight with IL-12 and IL-18 and then stained with anti-CD56, anti-CD3, a cocktail of NKG2A and KIR antibodies, and IFN-γ for (A) CD56dim NKG2A−KIR− cells or (B) CD56dim NKG2A+ and/or KIR+ cells. Shown is a representative example of 12 experiments that all gave similar results. Sorted NKG2A−KIR−, NKG2A+KIR−, NKG2A−KIR+, and NKG2A+KIR+ populations were tested for cytolysis of K562 target cells in a 4-hour chromium release assay after (C) a 16-hour incubation with IL-2 (n = 6), to allow recovery of cytolytic machinery, or (D) after 14 days of maturation on EL08–1D2 and IL-15 (n = 4). After short-term culture with IL-2 the NKG2A−KIR− population exhibited a marked lack of cytolytic activity compared with the other populations (all P < .05). After maturation in culture for 14 days this difference was no longer apparent.
CD56 dim NK cells lacking NKG2A and KIR are functionally immature. PBMCs were incubated overnight with IL-12 and IL-18 and then stained with anti-CD56, anti-CD3, a cocktail of NKG2A and KIR antibodies, and IFN-γ for (A) CD56dim NKG2A−KIR− cells or (B) CD56dim NKG2A+ and/or KIR+ cells. Shown is a representative example of 12 experiments that all gave similar results. Sorted NKG2A−KIR−, NKG2A+KIR−, NKG2A−KIR+, and NKG2A+KIR+ populations were tested for cytolysis of K562 target cells in a 4-hour chromium release assay after (C) a 16-hour incubation with IL-2 (n = 6), to allow recovery of cytolytic machinery, or (D) after 14 days of maturation on EL08–1D2 and IL-15 (n = 4). After short-term culture with IL-2 the NKG2A−KIR− population exhibited a marked lack of cytolytic activity compared with the other populations (all P < .05). After maturation in culture for 14 days this difference was no longer apparent.
CD56dim NKG2A− KIR− cells are hyporesponsive to HLA class I–deficient target cells. Enriched NK cells were incubated alone, or after 4 hours of exposure to class I–deficient K562 or 221 cells. Cells were then stained for CD56, CD3, NKG2A, and KIR, and the degranulation antigen CD107a. Shown is a representative example gated on receptor-negative or -positive cells based on 6 experiments showing a similar pattern. The percents listed (M1 gate) represent positive events based on an isotype-matched control antibody.
CD56dim NKG2A− KIR− cells are hyporesponsive to HLA class I–deficient target cells. Enriched NK cells were incubated alone, or after 4 hours of exposure to class I–deficient K562 or 221 cells. Cells were then stained for CD56, CD3, NKG2A, and KIR, and the degranulation antigen CD107a. Shown is a representative example gated on receptor-negative or -positive cells based on 6 experiments showing a similar pattern. The percents listed (M1 gate) represent positive events based on an isotype-matched control antibody.
CD56dim NKG2A−KIR− cells are precursors of mature NKG2A+ and KIR+ NK cells
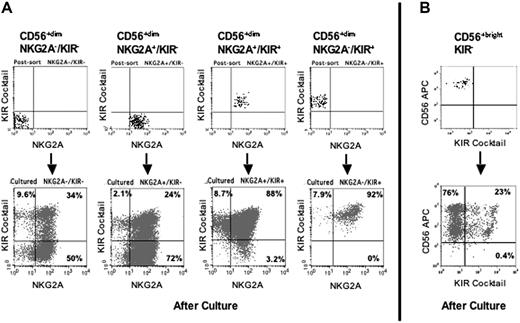
Because the NKG2A−KIR− cells comprise approximately 20% of the circulating CD56dim NK population in healthy humans, we hypothesized that these poorly functional cells are precursors to NKG2A and KIR-bearing functional NK cells. To test the hypothesis we sorted fresh CD56dim NKG2A−KIR−, NKG2A+KIR−, NKG2A+KIR+, and NKG2A−KIR+ NK cells and cultured them with IL-15 and EL08–1D2 cells for 14 days (n = 12). The culture of cells that already expressed KIR or NKG2A produced progeny that predominantly retained expression of their respective receptors (Figure 7A). A small population lost expression, demonstrating that NKR expression can be down-regulated. For cells that initially lacked NKG2A, the culture produced cells that expressed NKG2A on a majority of cells. Likewise, the culture of cells that lacked KIR produced cells with a polyclonal distribution of KIR as detected by individual anti-KIR antibodies (data not shown). Culture of the NKG2A−KIR− subpopulation resulted in 29% ± 3.9% of the cells expressing any KIR and 85% ± 3.8% of the cells expressing NKG2A. Similarly, culture of the NKG2A+KIR− subpopulation resulted in 22% ± 3.7% of the cells expressing KIR. In addition to the change in surface phenotype, culturing the CD56dim NKG2A−KIR− cells with IL-15 and EL08–1D2 cells gave rise to cells with cytolytic and cytokine-producing functions (Figure 5D and data not shown). These results demonstrate that despite a surface phenotype demonstrating commitment to the NK-cell lineage, the human peripheral blood CD56dim NKG2A−KIR− NK cell is functionally immature, and can be induced to proliferate and differentiate in vitro, gaining KIR and NKG2A receptors and effector functions.
CD56dim and CD56bright KIR− cells are precursors to receptor-expressing NK cells. CD56dim and CD56bright NK cells were sorted according to NKG2A and KIR expression, analyzed for NKR expression, cultured for 14 days on EL08–1D2 cells with IL-15, and then reanalyzed for NKR expression. (A) Shown is a representative analysis of the 4 subpopulations of CD56dim cells. The postsort analysis of each starting population (top row) is compared with similar analysis after culture to induce further maturation (bottom row). (B) Shown is an analysis of the CD56bright population in which essentially all cells were NKG2A+ (not shown). The postsort analysis (top) is compared with that after culture with EL08–1D2 cells and IL-15 (bottom).
CD56dim and CD56bright KIR− cells are precursors to receptor-expressing NK cells. CD56dim and CD56bright NK cells were sorted according to NKG2A and KIR expression, analyzed for NKR expression, cultured for 14 days on EL08–1D2 cells with IL-15, and then reanalyzed for NKR expression. (A) Shown is a representative analysis of the 4 subpopulations of CD56dim cells. The postsort analysis of each starting population (top row) is compared with similar analysis after culture to induce further maturation (bottom row). (B) Shown is an analysis of the CD56bright population in which essentially all cells were NKG2A+ (not shown). The postsort analysis (top) is compared with that after culture with EL08–1D2 cells and IL-15 (bottom).
CD56bright NK cells, all expressing high-density NKG2A, were then sorted and tested for their capacity to acquire KIR in vitro. Although almost all CD56bright cells are KIR−, a cocktail of anti-KIR was included in the antibodies used for sorting to minimize the possibility of contamination with KIR+ cells. After culture of CD56bright KIR− cells with IL-15 and EL08–1D2 cells for 14 days 23% ± 7% of their progeny acquired KIR, significantly different from the starting population, which, as seen in the postsort analysis (Figure 7B), was more than 99% KIR− (n = 4, P = .027). Another important change was in the expression of CD56, which was no longer homogeneously high; after culture the cells consisted of 2 subpopulations having high (CD56bright) and low (CD56dim) expression of CD56. Taken together, these results directly support a model in which CD56bright NK cells are more immature than CD56dim NK cells, and can give rise to CD56dim KIR-expressing cells in an in vitro culture with stromal cells.
Discussion
We identified a unique subset of human peripheral blood CD56dim NKG2A−KIR− NK cells that is hyporesponsive to cells lacking MHC class I, but has the developmental potential to express NKR, acquire cytolytic and cytokine-secreting effector functions, and become responsive to cells lacking MHC class I. We conclude that lineage-committed peripheral blood NK cells do not require inhibitory self-tolerance mechanisms until they reach a late stage of differentiation and acquire either NKG2A or KIR. Our results agree with those presented in a recent report on CD56dim NKG2A−KIR− peripheral blood cells in humans.5 In addition, we demonstrate that peripheral blood CD56bright NK cells can develop into CD56dim cells, with or without KIR expression, a finding that advances knowledge of NK-cell development.7,8,36 These data support a model in which the perturbations to NKR expression seen after hematopoietic cell transplantation, including the higher frequencies of CD94/NKG2A+ and CD56bright NK cells, reflect disruptions to normal NK-cell development, both in lymphoid tissue and peripheral blood.
In healthy human donors the CD56dim NKG2A−KIR− subpopulation comprises about 20% of peripheral blood NK cells. Although committed to the NK-cell lineage and having a mature surface phenotype, these cells lack expression of “at least one” inhibitory receptor that engages MHC class I to provide self-tolerance of the type identified by studies of NK-cell cytolysis and alloreactivity.37 Because several of the inhibitory KIRs cannot be detected with the available mAb, we also measured KIR expression with robust Q-RT-PCR methods that confirmed the relative absence of inhibitory KIR transcripts in the CD56dim NKG2A−KIR− subset defined by surface phenotype. Mouse NK cells that lack inhibitory receptors for MHC class I and have a mature phenotype (CD11b+/DX5+) were shown to be hyporesponsive to cells of the same MHC phenotype, but maintained their capacity to produce cytokines after cross-linking with NKG2D and anti-Ly49D or stimulation with PMA/ionomycin.4 In contrast to the selective hyporesponsiveness of those mouse cells, the human CD56dim NKG2A−KIR− cells described here exhibit a general and nonselective hyporesponsiveness, suggesting they are developmentally immature. That such hyporesponsive NKG2A−KIR− cells were not described in a previous analysis of human NK-cell clones3 is not surprising. In that study, only NK-cell clones exhibiting a strong cytotoxic response to class I–deficient 221 cells were included in the analysis.
The developmental immaturity of NKG2A−KIR− CD56dim NK cells was demonstrated by our finding that culture with a stromal cell line and IL-15 drove these cells to acquire both effector functions and the expression of KIR and/or NKG2A. To our knowledge this is the first report that a subpopulation of lineage-committed NK cells in the peripheral blood can be induced to proceed to another developmental stage. A recent model of human NK-cell development in lymphoid tissue proposes 5 phenotypically different stages.7,8,36 In this model, stage III NK cells are CD16− CD56−/+dim, lack NKG2A and KIR, and are functionally immature; stage IV cells are CD56bright with high NKG2A and minimal KIR expression; and the proposed stage 5 cells are CD56dim CD16+ KIR+ and/or NKG2A+ and are fully functional. Overall, our results fit well with this model, but there are some differences. In lymphoid tissue CD117 is required for CD56 expression and all CD56+ cells express either CD117 and/or CD94, whereas in peripheral blood we found essentially all CD56dim NKG2A−KIR− NK cells are CD117−. Maturation in the different tissues surely involves distinct cell-cell interactions and altered exposures to cytokines, which may cause subtle differences in the expression of CD56 and KIR during NK- cell differentiation and their traffic through different tissues. Regardless, there is agreement that functional maturity is preceded by the expression of NKG2A (or CD94), which is a prerequisite for IFN-γ production.
To expand on the model of Freud and Caligiuri,7,8,36 we propose that acquisition of KIR marks an additional, key step in NK-cell differentiation. CD56bright NK cells highly express KIR2DL4, produce the most IFN-γ, and can give rise to KIR+ and CD56dim populations on in vitro culture. We suggest that CD56dim cells be divided according to their expression of CD56 and KIR, with stage 5 corresponding to CD56dim/KIR− cells and stage 6 corresponding to CD56dim/KIR+ cells. NKG2A is a less definitive marker of developmental stage, because although its expression precedes that of KIR, it subsequently varies. Reduction of NKG2A and KIR2DL4 expression is associated with expression of the other KIRs and a reduced level of CD56. For NKG2A, the reduced expression can be mediated by the transcription factor GATA-3.38 The decrease in IFN-γ production may be linked to the loss of KIR2DL4 expression.39 Not yet explained by this model is the loss of IFN-γ production between stages 4 and 5, and its return between stages 5 and 6, when KIR is acquired. Finding small subpopulations of cells (such as the KIR+ CD56bright cells seen here and also described by Caligiuri's group [Freud et al8 ]) that overlap these developmental stages may reflect their activation status in response to cytokines, such as IL-15, which is responsible for homeostatic proliferation.
Various developmental models for the acquisition of self-tolerance have been proposed. Signaling via stimulatory receptors that is unopposed by an inhibitory signal via a self-MHC receptor may “disarm” potentially alloreactive NK cells via a mechanism similar to the development of anergy in T cells.40,41 Alternatively, ligation of an inhibitory receptor by self-MHC may precede a terminal differentiation step whereby cells acquire function in a process described as “licensing.”14,42 Or the expression of NKR and the acquisition of function may be developmentally synchronized, with the genetic program disallowing the maturation of functional cells until they express adequate self-inhibitory molecules.43 The data presented here show that the effector function in mature peripheral blood NK cells correlates with NKR expression (NKG2A with or without KIR) and that in our culture system, the acquisition of NKR and mature function are temporally linked. These experiments on NK subsets neither support nor refute a particular mechanism for the regulation of self-tolerance, but suggest that NK cells do not require inhibitory MHC class I self-tolerance mechanisms until they reach a late stage of differentiation. One possibility is that self-tolerance is maintained by other mechanisms before the acquisition of KIR and NKG2A. Several MHC-independent inhibitory receptors have been described,44 including 2B4, which mediates NK-cell self-recognition in mice.45 Our in vitro developmental model should be useful to address further these mechanistic questions.
In summary, we have shown that KIR expression is impaired after HCT and propose that in this setting NK-cell development is transiently delayed at the CD56bright stage, which has low KIR expression and poor cytotoxic function. This observation has important implications for the use of KIR-ligand mismatched NK cells in clinical practice. The identification of circulating NK cells that lack inhibitory self-MHC receptors after HCT does not necessarily define an alloreactive population, as they may be developmentally immature. The differences between the NK-cell populations in healthy donors and in recipients at 100 days after allogeneic transplantation may be explained by a synchronization of developing cells by factors that drive homeostatic expansion after lymphodepleting therapy such as IL-15.46 Ongoing prospective studies evaluating NK-cell subsets at additional time points after transplantation will address this issue more definitively. Future studies should address the regulation of KIR and NKG2A, and how the interaction between expressed receptors and self-MHC affects the acquisition of effector function and self-tolerance in normal development and after HCT.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health Grants P01-CA-65493 (J.S.M.), P01-CA-111412–01 (J.S.M., P.P.), R01-AI-22039 (P.P.), R01-HL-55417 (J.S.M.), and K12 RR023247-01 (S.C.). Additional support was through the NMDP, the Health Resources and Services Administration no. 240–97-0036 and Office of Naval Research N00014–93-1–0658 to the NMDP.
The views expressed in this article do not reflect the official policy or position of the Department of the Navy, the Department of Defense, or the U.S. Government.
We would like to thank Greg Veltri from the University of Minnesota Flow Cytometry Core for his assistance in cell sorting.
National Institutes of Health
Authorship
Contribution: S.C. constructed the study plan, performed experiments, data analysis, and interpretation, and wrote the paper; F.X. performed experiments, designed PCR reactions, and performed data analysis and interpretation; M.P. performed Q-PCR and data analysis; M.G. performed experiments with NK-cell subsets and data analysis; V.M. performed laboratory analysis and flow cytometry; T.L.B. was responsible for all statistical analysis; K.L.M. performed KIR genotyping, data analysis, interpretation; L.A.G. performed data analysis, interpretation, and paper preparation; P.P. performed data analysis, interpretation, and paper preparation; J.S.M. constructed the study plan, performed data analysis and paper preparation, and was responsible for all aspects of the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey S. Miller, Professor of Medicine, University of Minnesota Cancer Center, MMC 806, Division of Hematology, Oncology, and Transplantation, Harvard Street at East River Road, Minneapolis, MN 55455; e-mail: mille011@umn.edu.