Abstract
Very low-dose irradiation (2 × 2 Gy) is a new, effective, and safe local treatment for follicular lymphoma. To understand the biologic mechanisms of this extremely effective response, we compared by microarray the gene-expression profile of patients' biopsies taken before and after radiation. In all patients, a major and consistent induction of p53 target genes was seen. p53 targets involved in cell-cycle arrest and apoptosis showed the same mode of regulation, indicating that, in vivo, both are activated simultaneously. p53 up-regulation and p53-mediated proliferation arrest and apoptosis were substantiated using immunohistochemistry, with activation of both the intrinsic and the extrinsic apoptotic pathways. The other induced genes revealed a whole set of biologically meaningful genes related to macrophage activation and TH1 immune response. Immunohistochemical analysis suggested a specific activation or differentiation of resident macrophages by apoptotic cells. These biologic insights are important arguments to advocate the use of low-dose radiotherapy as an effective palliative treatment for follicular lymphoma. Moreover, this study is the first in vivo report of the radiation-induced p53 apoptotic response in patients and suggests that this apoptotic response is not immunologically silent.
Introduction
Follicular lymphoma (FL) represents the second most frequent lymphoma subtype. It is an indolent B-cell lymphoma affecting essentially elderly patients, with a median age of 60 years. The t(14;18) is the cytogenetic hallmark of the disease, present in approximately 85% of patients. It induces Bcl-2 overexpression resulting in the accumulation of B cells that are relatively resistant to the B-cell selection-mediated apoptosis in nonmalignant germinal centers
The disease runs a protracted course in most cases, patients dying of the lymphoma or by the complications of its treatment. Despite numerous efforts and the exploration of different treatment regimens, the prognosis of FL has largely remained unchanged over the last decades, with median survival times of 8 to 10 years.1 All conventional regimens can therefore be considered as palliative. Given the fact that the majority of patients are elderly and have an incurable disease, strategies involving new effective palliative treatment with minimal side effects are very attractive.
In previous studies, we and others demonstrated that very low-dose involved-field radiotherapy (4 Gy) is an effective new therapeutic strategy for symptomatic follicular lymphoma,2,3 with a response rate of 92%, 61% as complete response, with a median time to progression of 14 months and virtually no side effects. This new radiotherapeutic regimen is remarkable because of its low dose, the excellent clinical results, and the rapid onset of the response, which is often complete within one week. This is in sharp contrast to the dose range (up to 40 Gy) and intervals to reach complete regression of several weeks in most hematologic diseases, including Hodgkin lymphoma. Radiotherapy can provide local control and cure in various other malignancies, including carcinoma, but in these solid tumors doses up to 70 Gy in 7 weeks are required. Usually, the local tumor reduction does no start until the very end of the treatment and is complete only at 6 to 8 weeks after radiation. In comparison, FL can therefore be considered as exquisitely radiosensitive. However, the underlying mechanisms are still fully speculative.
Molecular mechanisms of conventional dose radiotherapy have been studied in vitro. First, double-strand breaks, which activate DNA damage checkpoints to initiate signals, ultimately lead to the activation and stabilization of p53. Subsequently, p53 induces the expression of genes involved in cell cycle arrest and DNA repair (eg, CDKN1A, GADD45A). It also induces genes related to apoptosis by both the mitochondria/intrinsic/caspase-9 pathway (eg, BAX, PMAIP1 [or Noxa], BBC3 [or Puma]) and the death receptors/extinsic/caspase-8 pathway (eg, FAS, TNFRSF10 [or Trail-R2]). The level of p53 accumulation results primarily from the extent of DNA damage, and the choice between repair and death is thought to depend on the selectivity in genes activated by p53.4
Low-dose radiotherapy seems to induce apoptosis in FL, since there was a rapid in vivo 99mTc-annexin-V uptake in FL lymph nodes after radiation.5 This contrasts with the notion that Bcl-2 overexpression is able to protect hematopoietic progenitors and lymphoid cells from radiation-induced apoptosis in vitro.6,7 To unravel the molecular mechanisms underlying this hypersensitivity, we compared by microarray global gene expression of FL samples taken before and after radiation.
This is, to our knowledge, the first global expression analysis of the p53 response in vivo in patients. Moreover, results obtained here support the usage of low-dose radiation as clinical and molecular effective palliative treatment for FL and could be of great interest for the study of the immune response in cancer.
Patients, materials, and methods
Patients
Twenty patients were treated with 2 × 2 Gy irradiation (days 1 and 3) on FL-involved lymph node. Excisional or large needle biopsies (3 × 18G) of nodal sites were taken before and 24 hours after the second radiation dose (day 4). Day 4 corresponds to maximal 99mTc-annexin-V uptake by scintigraphy, with still few morphologic signs of apoptosis as assessed in a previous study.8 Part of the biopsy samples was directly snap-frozen in liquid nitrogen, while the remaining material was fixed in buffered formalin. In 15 of the 20 patients high-quality RNA could be isolated from representative biopsy samples from both preradiation and postradiation samples and these were selected for the microarray analysis. Patients characteristics were the following: grades 1 and 2 FL, 9 females and 6 males, median age of 57 years, range 40 to 83 years. The median number of irradiated sites was 2 (range, 1-11 sites), with median lymph node size of 5 cm (range, 3-8 cm). All patients responded well to therapy (12 complete responses [CRs], 3 partial responses [PRs], according to Cheson criteria). Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki, and the NKI-AVL ethical review board approved the study.
Gene-expression profiling
Detailed protocols for RNA isolation, amplification, labeling, and hybridization can be found at http://microarray.nki.nl/download/index.html. In brief, tumor material was snap-frozen in liquid nitrogen within one hour after surgery. Frozen sections were stained with hematoxylin and eosin; only samples that had more than 50% tumor cells were selected for RNA extraction (RNAzolB; Campro Scientific, Veenendaal, the Netherlands). After DNase treatment (RNase-Free DNase Set, Rneasy Mini Kit; QIAGEN, Hilden, Germany) 5 μg total RNA was amplified using the Superscript RNA Amplification System (Invitrogen, Frederick, MD). Amplified RNA (1 μg) was fragmented (Fragmentation reagents; Ambion, Austin, TX), labeled with Cy3 or Cy5 using the ULS-Cy3/5 aRNA Labeling Kit (Kreatech Biotechnology, Amsterdam, the Netherlands), and cohybridized with a fragmented reference RNA labeled with the reverse color. The reference RNA consists of pooled and amplified lymphoid reference RNA isolated from tonsillectomy specimens of patients who underwent routine tonsillectomy for chronic ear/nose/throat infections. A dye swap was done for each sample. Microarray slides were prepared at the central microarray facility (CMF) at the Netherlands Cancer Institute. There were 37 632 70-mer oligonucleotides spotted on amino-silane-coated slides, representing 37 123 gene transcripts. A complete list of genes and controls is available on the CMF website (http://microarrays.nki.nl/index.html). After hybridization, the slides were washed and scanned with a confocal laser scanner (Agilent Technologies, Palo Alto, CA). Fluorescence intensities on scanned images were quantified with the Imagene software (BioDiscovery, El Segundo, CA). Fluorescent intensities were normalized and corrected for a variety of biases that affect the intensity of measurement according to Yang et al.9 Weighted averages and confidence levels were computed according to the Rosetta error model.10
Microarray data analysis
A first step of filtering procedure was applied on the entire gene set. Genes with missing values in more than 3 samples were excluded from the analysis. From the 37 123 transcripts that were present on the microarray, 7087 that were significantly different from the reference in more than 6 samples were selected (significance was based on P < .01 computed with the Rosetta error model11 ). This step removes especially all genes with expression value below the detection level. All analyses were made based on this gene list.
For unsupervised clustering, gene and sample clustering were done independently using agglomerative hierarchic clustering in the software program Genesis (Graz, Austria; http://genome.tugraz.at).12 Complete linkage similarity metrics among genes or samples were calculated on the basis of expression ratio measurement across all samples or significant genes.
To find genes induced or repressed by radiation, supervised analysis was performed on the preradiation and postradiation samples from the 15 patients by using the significance analysis of microarray (SAM).13 SAM uses a modified t test statistic with sample-label permutations to evaluate statistical significance. Calculation was based on the ratio of expression of the 7087 significant genes, expressed in Log2 scale, using a paired SAM analysis. Missing values were imputed with the 10-nearest neighbor. The delta value was set at 1.15 627 to have less than one median false significant gene number for 300 permutations (median = 0.66).
RT-MLPA
Multiplex ligation-dependent amplification procedure (RT-MLPA) for 33 apoptosis regulators was performed as described by Eldering et al.14 This method allows the simultaneous quantification of up to 45 different target sequences with the use of one single primer pair in a final polymerase chain reaction (PCR) amplification. The output, a set of fluorescent fragments, is analyzed via capillary sequencer to quantify the transcript. MLPA was performed on the RNA used for the microarray for the 15 patients. The transcript expression level was expressed in a Log2 scale. A linear normalization was applied for each sample, based on the mean of expression of 2 housekeeping genes (PARN and GUS) included in the reaction.
Immunohistochemistery
Immunohistochemistry was performed according to standard techniques and including citrate-based antigen retrieval on formalin-fixed and paraffin-embedded biopsy samples. Antibodies used are listed in Table 1. T-cell populations and subsets (CD3, CD4, CD8, TIA1, GB7, T-bet) were assessed semiquantitatively and for architectural pattern (intrafollicular, interfollicular, diffuse). MKI67 was assessed semiquantitatively for tumor cells and nontumor cells separately; p53 and p21 were scored per total cell number and for separate cell populations as morphologically recognized (endothelium, macrophages, etc). CD68+ macrophages were counted as absolute cell numbers per follicular high-power field (HPF) at 10 × 60 magnification in 3 representative HPFs. Caspase-3, -8, and -9 were quantified as apoptotic clusters per follicular HPF at 10 × 40 magnification in 3 representative HPFs.
Antibodies used for immunohistochemistry
| Antibody . | Clone . | Source . |
|---|---|---|
| CD20 | L26 | DAKO, Carpinteria, CA |
| CD3 | CD3 | DAKO |
| CD4 | 4B12 | Novacastra, Newcastle upon Tyne, United Kingdom |
| CD8 | C8/144B | DAKO |
| TIA1 | TIA-1 | Coulter, Birmingham, United Kingdom |
| Granzyme-B | GrB-7 | Monosan |
| T-bet | Anti-T-bet | Zymed, South San Francisco, CA |
| CD68 | KP1 | DAKO |
| bcl-2 | 124 | DAKO |
| p53 | DO-7 | DAKO |
| p21 | CP74 | Lab Vision, Fremont, CA |
| Ki-67 | MIB1 | DAKO |
| CD69 | CH11 | Lab Vision |
| Cleaved caspase-3 | 5A1 | Cell Signalling Technology, Beverly, MA |
| Cleaved caspase-8 | 11G10 | Cell Signalling Technology |
| Cleaved caspase-9 | 44-692 | Biosource, Camarillo, CA |
| Antibody . | Clone . | Source . |
|---|---|---|
| CD20 | L26 | DAKO, Carpinteria, CA |
| CD3 | CD3 | DAKO |
| CD4 | 4B12 | Novacastra, Newcastle upon Tyne, United Kingdom |
| CD8 | C8/144B | DAKO |
| TIA1 | TIA-1 | Coulter, Birmingham, United Kingdom |
| Granzyme-B | GrB-7 | Monosan |
| T-bet | Anti-T-bet | Zymed, South San Francisco, CA |
| CD68 | KP1 | DAKO |
| bcl-2 | 124 | DAKO |
| p53 | DO-7 | DAKO |
| p21 | CP74 | Lab Vision, Fremont, CA |
| Ki-67 | MIB1 | DAKO |
| CD69 | CH11 | Lab Vision |
| Cleaved caspase-3 | 5A1 | Cell Signalling Technology, Beverly, MA |
| Cleaved caspase-8 | 11G10 | Cell Signalling Technology |
| Cleaved caspase-9 | 44-692 | Biosource, Camarillo, CA |
Detection of TP53 gene mutations in DLBCL samples
Formalin-fixed, paraffin-embedded biopsy samples from diffuse large B-cell lymphoma patients (DLBCL) patients taken before radiotherapy were analyzed for p53 mutations by direct sequencing. For this, DNA was isolated using the QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer's protocol. Part of the p53 gene, spanning exons 2 to 9, was amplified using the HotStarTaq Master Mix Kit (QIAGEN). First, exon 2, exons 3 to 4, exons 5 to 6, exon 7, and exons 8 to 9 were amplified. Subsequently, the same DNA fragments were reamplified using at least one nested primer. Purified PCR products (QIAquick PCR purification kit; QIAGEN) were directly sequenced using the BigDye Terminator Reaction kit, version 3.1 (Applied Biosystems, Foster City, CA) and an ABI 3700 DNA sequencer. All PCR and sequencing primers used were either newly designed or adapted from previously described studies.15,16 Predicted structural and functional characteristics of the mutations were obtained from the mutation validation tool of the IARC TP53 Mutation Database, version R10, as well as the number of tumors reported with the same TP53 somatic mutation.17–19
Statistical analysis
Gene Ontology analysis was performed using the GOstat (Melbourne, Australia) analysis tool (http://gostat.wehi.edu.au/). GO term overrepresentation is calculated based on a Fisher exact test.
For MLPA, significant change in gene expression was calculated by a paired 2-tailed Student t test for the 15 patients. P < .01 was considered as significant. The comparison of the MLPA and the oligo-arrays was based on the expression of 27 genes measured by both techniques. Paired signal to noise ratio was defined as the mean of the difference of the Log2 expression for each pair, divided by the standard deviation of this difference. Pearson correlation for the SNR obtained by the 2 methods was calculated.
For MKI67, CD68, cleaved caspase-8, and cleaved caspase-9 immunostainings, the number of positive cells per HPF was compared between preradiotherapy and postradiotherapy samples using an unpaired 2-tailed Mann-Whitney test.
For TP53 mutations, the probability of an association of nonfunctional transactivation class predicted by IARC to the response status was calculated by a Fisher exact test.
Results
Low-dose radiation induces 3 biologically meaningful gene-expression profiles
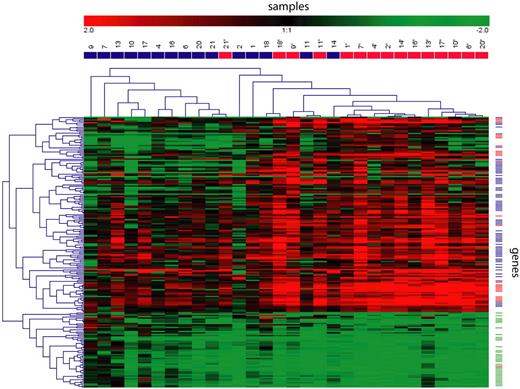
To understand the extreme sensitivity of FL to radiation, we compared gene-expression profiles of FL lymph node biopsies taken before and 24 hours after the second 2-Gy irradiation in 15 patients. An unsupervised hierarchic clustering analysis based on the 7087 transcripts whose expression varied most across samples showed that preradiation and postradiation samples clustered together for 12 of the 15 patients (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), indicating that global gene expression was not essentially modified by radiation. Using a supervised SAM method to detect genes specifically regulated by radiation, we found 180 unique genes represented by 201 transcripts that were significantly differentially expressed with less than 1 false discovered gene (false significant number = 0.66). An unsupervised hierarchal clustering based on these 201 transcripts showed separation of the 2 groups with 80% accuracy (Figure 1; the complete list of genes can be obtained in Table S1). Based on their regulation and function obtained from the literature, the majority of the genes were categorized in 3 biologically meaningful gene profiles: p53 pathway (25 of the 145 induced transcripts, 17%), immune response (69 of the 145 induced transcripts, 48%), and cell cycle (35 of the 56 repressed transcripts, 63%). A Gene Ontology analysis showed a statistically significant overrepresentation of immune response (GO:0006955; P = .001) and cell cycle genes (GO:0007049; P = .001). The majority of remaining genes had no defined function (26 [18%] of the 145 induced transcripts, and 14 [25%] of the 56 repressed transcripts). Induced genes were not significantly different between patients that responded partially (n = 3) or completely (n = 12) to the treatment.
Hierarchic clustering based on transcripts modified by low-dose radiation. We detected 201 transcripts whose expression was significantly modified by low-dose radiation. Hierarchic clustering based on their expression is shown, with samples taken before radiation in blue and samples taken after radiation in red, in the columns. For each gene, the relative expression compared with the reference RNA is represented by a color scale. Based on their regulation and functions, genes (rows) were classified in 3 biologically meaningful profiles: p53 targets in red, immune response in blue, and cell cycle in green.
Hierarchic clustering based on transcripts modified by low-dose radiation. We detected 201 transcripts whose expression was significantly modified by low-dose radiation. Hierarchic clustering based on their expression is shown, with samples taken before radiation in blue and samples taken after radiation in red, in the columns. For each gene, the relative expression compared with the reference RNA is represented by a color scale. Based on their regulation and functions, genes (rows) were classified in 3 biologically meaningful profiles: p53 targets in red, immune response in blue, and cell cycle in green.
Low-dose radiation activates the p53 pathway
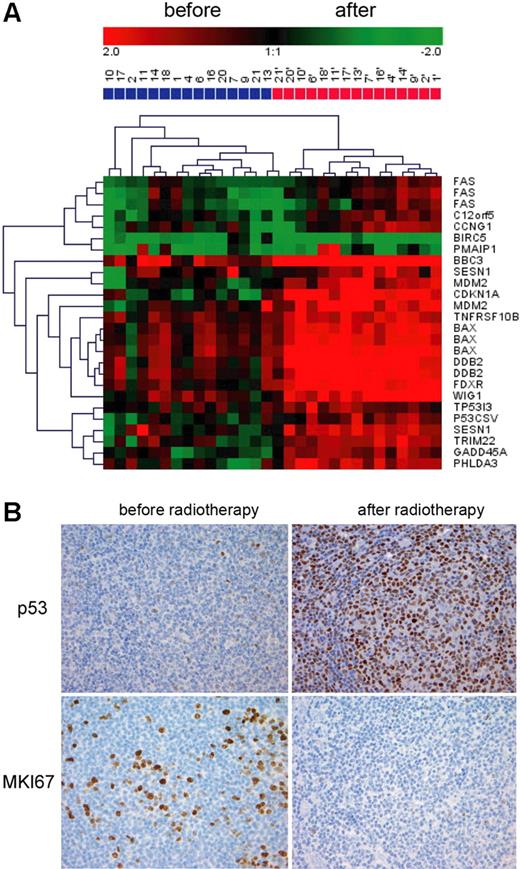
Activation of the p53 pathway was suggested by the strong induction of p53 targets. Indeed, among the top 10 up-regulated transcripts, 7 were known p53 target genes (Table S1). In total, 25 p53 target transcripts were up-regulated by radiation. BIRC5 (or survivin), a classical p53-repressed gene, was down-regulated. p53 activation is illustrated by the hierarchic clustering based only on the expression of these 25 p53 targets, shown on Figure 2A. Unsupervised sample clustering fully separated the preradiation and postradiation samples. Gene clustering showed that typical cell cycle arrest genes (CDKN1A, GADD45A) clustered with apoptosis-related genes (BAX, p53CSV). This shows that, in vivo, genes inducing cell cycle arrest/DNA repair and apoptosis are concomitantly regulated. To confirm p53 activation, we assessed the p53 protein levels using immunohistochemistry on paraffin-embedded preradiation and postradiation samples taken in parallel with the frozen samples for gene-expression profiling. There was a consistent and strong increase of p53 after radiation, confirming p53 pathway activation (from less than 5% positive cells to more than 80% positive cells, n = 5, Figure 2B). Interestingly, this increase was much more pronounced in FL cells compared with T cells and endothelial cells, and was absent in macrophages. Cell cycle arrest, suggested by the coordinate down-regulation of cell cycle related genes, was confirmed by the decrease in MKI67 immunostaining after radiation (Figure 2B, mean of 15.45% of positive cells before radiotherapy, n = 17; and 4.08% after radiotherapy, n = 14, P < .001). CDKN1A was only slightly increased at the protein level, preferentially in FL cells (data not shown).
p53 pathway activation. (A) Hierarchic clustering of the samples taken before (blue) and after (red) radiation based on the 25 known p53 transcripts regulated by low-dose radiation. (B) p53 and MKI67 immunostaining was performed on paraffin sections of FL biopsies take before and after low-dose radiation. Images were acquired with a ColorView (Mitsubishi, Tokyo, Japan) SIS CCD camera (20×, PlanApo, 20×/0.09) and DP-Soft (Muenster, Germany) CVI 3.2 software. One representative field is shown.
p53 pathway activation. (A) Hierarchic clustering of the samples taken before (blue) and after (red) radiation based on the 25 known p53 transcripts regulated by low-dose radiation. (B) p53 and MKI67 immunostaining was performed on paraffin sections of FL biopsies take before and after low-dose radiation. Images were acquired with a ColorView (Mitsubishi, Tokyo, Japan) SIS CCD camera (20×, PlanApo, 20×/0.09) and DP-Soft (Muenster, Germany) CVI 3.2 software. One representative field is shown.
Low-dose radiation activates the intrinsic and the extrinsic apoptotic pathways
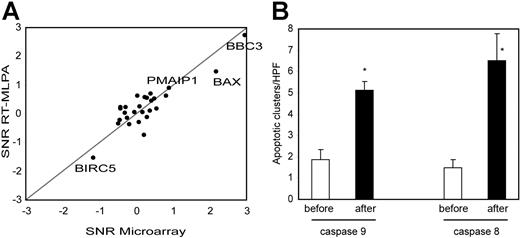
To focus on the apoptotic process, we used RT-MLPA as an independent technique to quantify 33 known apoptosis regulator genes in the samples from the 15 patients (Table S2). This technique confirmed the microarray results, with a good correlation between signal to noise ratio (paired SNR, preradiotherapy compared with postradiotherapy samples) obtained by the 2 methods (Pearson correlation = 0.87, Figure 3A). By MLPA, BBC3 (Puma) was the most up-regulated gene (P < .001). BAX and PMAIP1 (Noxa) were also significantly up-regulated (P < .001, P = .008, respectively). Only BIRC5 (Survivin) was significantly down-regulated (P = .009, 2-tailed paired Student t test). All these genes were p53 targets and no other apoptosis regulators showed significant change in expression.
Apoptotic pathways induced by low-dose radiation. (A) An RT-MLPA quantification of 27 apoptotic regulators was performed on the preradiation and postradiation samples of the 15 patients. SNRs between preradiation and postradiation samples were calculated and compared with the SNRs obtained by microarray. Genes significantly increased based on MLPA data are named. (B) Immunostainings for cleaved caspase-9 (intrinsic apoptotic pathway) and cleaved caspase-8 (extrinsic apoptotic pathway) were performed on preradiation and postradiation samples. Number of apoptotic cell clusters were counted for each sample (mean of 3 representative high-power fields). Mean and SEM are shown for each groups. Differences between preradiation and postradiation samples were highly significant for both stainings (P < .001, unpaired Mann-Whitney test). n for cleaved caspase-8 before = 17, after = 18; n for cleaved caspase-9 before = 17, after = 17.
Apoptotic pathways induced by low-dose radiation. (A) An RT-MLPA quantification of 27 apoptotic regulators was performed on the preradiation and postradiation samples of the 15 patients. SNRs between preradiation and postradiation samples were calculated and compared with the SNRs obtained by microarray. Genes significantly increased based on MLPA data are named. (B) Immunostainings for cleaved caspase-9 (intrinsic apoptotic pathway) and cleaved caspase-8 (extrinsic apoptotic pathway) were performed on preradiation and postradiation samples. Number of apoptotic cell clusters were counted for each sample (mean of 3 representative high-power fields). Mean and SEM are shown for each groups. Differences between preradiation and postradiation samples were highly significant for both stainings (P < .001, unpaired Mann-Whitney test). n for cleaved caspase-8 before = 17, after = 18; n for cleaved caspase-9 before = 17, after = 17.
BBC3, BAX, and PMAIP1 were reported to mediate p53-induced cell death by the intrinsic apoptotic pathway.20–22 To assess the specific induction of the intrinsic apoptotic pathway, we immunostained preradiotherapy and postradiotherapy samples with an antibody specific for cleaved caspase-9. This pathway was significantly activated by low-dose radiotherapy, as shown in Figure 3B.
Death receptors such as TNFRSF10B (TRAIL-R2) and FAS were also among the p53 targets induced by low-dose radiation. To analyze the activation of the death receptor/extrinsic pathway, we performed immunostainings with an antibody recognizing activated caspase-8. A prominent induction of caspase-8–mediated apoptosis was seen, as shown in Figure 3B.
Low-dose radiation influences innate and adaptive immunity
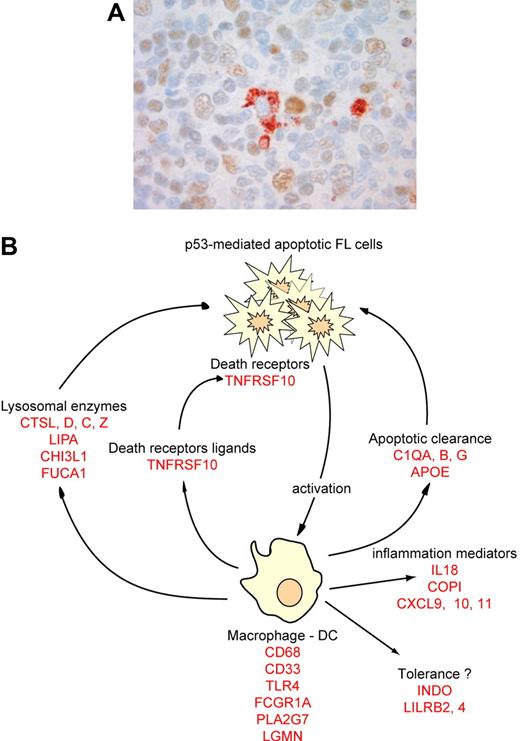
A large proportion of the genes induced by low-dose radiation were related to an immune reaction. Based on the literature, many of these can be associated in categories related to the same cellular subtype or immune function (Table 2). One category of genes was specifically expressed in macrophages and/or dendritic cells (DCs), suggesting that low-dose radiotherapy may alter the number or functional status of mature macrophages and DCs. Immunohistochemistry with anti-CD68 showed no increase in absolute cell numbers (n = 5, mean = 17.8 [range, 7-30] CD68+ cells for preradiotherapy samples, mean = 18.4 [range, 6-30] CD68+ cells for postradiotherapy samples, P = .9), suggesting that resident monocytes are activated or induced to differentiate after radiation. This may be due either to a direct effect of radiation or to an indirect activation by products of neighboring apoptotic cells. Since double-staining for p53 and CD68 showed no CD68+/p53+ macrophages (n = 5, Figure 4), direct activation is not supported and macrophages may rather be activated by specific signals from apoptotic cells to induce apoptotic body clearance. This hypothesis is reinforced by the concordant up-regulation of lysosomal genes necessary for macrophage phagocytic function, and by the up-regulation of genes necessary for macrophage apoptotic clearance such as C1Q or APOE (Table 2). Mice deficient for those genes are defective for apoptotic clearance and have autoimmune diseases.23,24 Interestingly, genes related to tolerance induction by antigen-presenting cells were also induced by radiation (INDO, LILRB4, LILRB2, SIGLEC9, Table 2).
Differentially expressed genes induced by low-dose radiation therapy and related to immunologic processes, listed by function, regulation, or cell-type specificity
| Macrophage . | Lysosomal marker . | Apoptotic clearance . | Death receptor ligand . | Inflammation- IFNγ response . | Tolerance . |
|---|---|---|---|---|---|
| CD68 | CTSL | C1QA | TNFSF10 | IL18 | INDO |
| CD33 | CTSD | C1QB | — | COPI | LILRB2 |
| TLR4 | CTSC | C1QG | — | CXCL9 | LILRB4 |
| PLA2G7 | CTSZ | APOE | — | CXCL10 | SIGLEC9 |
| IGSF6 | LIPA | — | — | CXCL11 | — |
| FCGR1A | CHI3L1 | — | — | GBP1 | — |
| FCER1G | FUCA1 | — | — | GBP5 | — |
| LGMN | — | — | — | ECGF1 | — |
| STAT1 | — | — | — | LST1 | — |
| UBD | — | — | — | INDO | — |
| GDF15 | — | — | — | — | — |
| APOC1 | — | — | — | — | — |
| SAT | — | — | — | — | — |
| TFEC | — | — | — | — | — |
| TCN2 | — | — | — | — | — |
| TYROBP | — | — | — | — | — |
| Macrophage . | Lysosomal marker . | Apoptotic clearance . | Death receptor ligand . | Inflammation- IFNγ response . | Tolerance . |
|---|---|---|---|---|---|
| CD68 | CTSL | C1QA | TNFSF10 | IL18 | INDO |
| CD33 | CTSD | C1QB | — | COPI | LILRB2 |
| TLR4 | CTSC | C1QG | — | CXCL9 | LILRB4 |
| PLA2G7 | CTSZ | APOE | — | CXCL10 | SIGLEC9 |
| IGSF6 | LIPA | — | — | CXCL11 | — |
| FCGR1A | CHI3L1 | — | — | GBP1 | — |
| FCER1G | FUCA1 | — | — | GBP5 | — |
| LGMN | — | — | — | ECGF1 | — |
| STAT1 | — | — | — | LST1 | — |
| UBD | — | — | — | INDO | — |
| GDF15 | — | — | — | — | — |
| APOC1 | — | — | — | — | — |
| SAT | — | — | — | — | — |
| TFEC | — | — | — | — | — |
| TCN2 | — | — | — | — | — |
| TYROBP | — | — | — | — | — |
— indicates not applicable.
Immune response after low-dose radiotherapy in FL. (A) Postradiotherapy biopsies were stained with an anti-p53 antibody, revealed by a brown color, and a macrophage-specific CD68 antibody, revealed by a red color. Image was acquired with a ColorView (Mitsubishi, Tokyo, Japan) SIS CCD camera (400×, PlanApo, 40×/0.95) and DP Soft (Muenster, Germany) CVI 3.2 software. Results are representative of all macrophages found in the 5 patients tested. (B) Based on several genes overexpressed after radiation (in red), we propose a model for the molecular immune-reaction that follows low-dose radiotherapy.
Immune response after low-dose radiotherapy in FL. (A) Postradiotherapy biopsies were stained with an anti-p53 antibody, revealed by a brown color, and a macrophage-specific CD68 antibody, revealed by a red color. Image was acquired with a ColorView (Mitsubishi, Tokyo, Japan) SIS CCD camera (400×, PlanApo, 40×/0.95) and DP Soft (Muenster, Germany) CVI 3.2 software. Results are representative of all macrophages found in the 5 patients tested. (B) Based on several genes overexpressed after radiation (in red), we propose a model for the molecular immune-reaction that follows low-dose radiotherapy.
Beside this macrophage activation, low-dose radiation up-regulates inflammatory and TH-1-related genes. IL18 and its cleaving enzyme, COPI (or procaspase-1), are both up-regulated. Numerous IFNγ-induced genes are also induced, such as chemokines CXCL9, CXCL10, and CXCL11. IFNγ expression was below the level of detection for the microarray sensitivity. This adaptive immune response is not reflected by changes in numeric or architectural alterations in T cells or T-cell subsets as indicated by immunohistochemistry for CD3, CD4, CD8, and Tbet (data not shown).
P53 functional status is associated to the response to LDRT in DLBCL
Since p53 is only very rarely mutated in FL, support for the pivotal role of p53 in the response to LDRT was studied in DLBCL who were treated for various palliative reasons with LDRT. The results are listed in Table 3. Among the 12 DLBCL patients treated with LDRT, 9 showed response (either complete or partial response), and 3 did not respond to the treatment (either stable disease or progression). All p53 exons were sequenced. In the 3 nonresponders, different p53 mutations known to potentially alter the function of p53 were found (IARC TP53 Mutation Database). Only 1 of 9 responder patients had a mutation that most probably alters p53 function. However, because this mutation is heterozygous in the tumor, the normal p53 allele could still be functional. There was a significant association between the predicted p53 functional status and the response to LDRT (P = .018).
Analysis of TP53 mutations in DLBCL samples treated with low-dose radiotherapy
| DLBCL samples . | TP53 mutation analysis from IARC1 . | Conclusion . | ||||
|---|---|---|---|---|---|---|
| Patient . | Response . | Mutation . | No. of tumors with the mutation . | Transactivation class2 . | Structure-based prediction3 . | Predicted TP53 function . |
| 1 | Resp | 46: TCCser -TTCphe | 2 | Functional | NA | Functional |
| 2 | Resp | 55: ACTthr -ATTile | 0 | Functional | NA | Functional |
| 3 | Resp | — | NA | NA | NA | Functional |
| 4 | Resp | 92: CCCpro -CTCleu | 1 | Complex | NA | Functional |
| 5 | Resp | — | NA | NA | NA | Functional |
| 6 | Resp | — | NA | NA | NA | Functional |
| 7 | Resp | 193: CAThis -CGTarg | 61 | Nonfunctional | Nonfunctional | Nonfunctional |
| 8 | Resp | — | NA | NA | NA | Functional |
| 9 | Resp | 142: CCTpro -TCTser | 6 | Complex | Functional | Functional |
| 223: CCTpro -CTTleu | 2 | Complex | Functional | Functional | ||
| 293: GGGgly -GAGglu | 0 | Functional | NA | Functional | ||
| 10 | Non | 135: TGCcys -TACtyr | 63 | Nonfunctional | Nonfunctional | Nonfunctional |
| 11 | Non | 165: CAGglu -TAGstop | 38 | No data | No data | Nonfunctional |
| 12 | Non | 285: GAGglu -AAGlys | 139 | Nonfunctional | Nonfunctional | Nonfunctional |
| DLBCL samples . | TP53 mutation analysis from IARC1 . | Conclusion . | ||||
|---|---|---|---|---|---|---|
| Patient . | Response . | Mutation . | No. of tumors with the mutation . | Transactivation class2 . | Structure-based prediction3 . | Predicted TP53 function . |
| 1 | Resp | 46: TCCser -TTCphe | 2 | Functional | NA | Functional |
| 2 | Resp | 55: ACTthr -ATTile | 0 | Functional | NA | Functional |
| 3 | Resp | — | NA | NA | NA | Functional |
| 4 | Resp | 92: CCCpro -CTCleu | 1 | Complex | NA | Functional |
| 5 | Resp | — | NA | NA | NA | Functional |
| 6 | Resp | — | NA | NA | NA | Functional |
| 7 | Resp | 193: CAThis -CGTarg | 61 | Nonfunctional | Nonfunctional | Nonfunctional |
| 8 | Resp | — | NA | NA | NA | Functional |
| 9 | Resp | 142: CCTpro -TCTser | 6 | Complex | Functional | Functional |
| 223: CCTpro -CTTleu | 2 | Complex | Functional | Functional | ||
| 293: GGGgly -GAGglu | 0 | Functional | NA | Functional | ||
| 10 | Non | 135: TGCcys -TACtyr | 63 | Nonfunctional | Nonfunctional | Nonfunctional |
| 11 | Non | 165: CAGglu -TAGstop | 38 | No data | No data | Nonfunctional |
| 12 | Non | 285: GAGglu -AAGlys | 139 | Nonfunctional | Nonfunctional | Nonfunctional |
Samples from 12 DLBCL patients treated with low-dose radiotherapy (2 × 2 Gy) for various palliative reasons were screened for TP53 gene mutations. Nine patients responded partially or completely to the treatment (Resp). Three patients had a stable or progressive disease (Non). Nonsilent mutations are shown in the table. Predicted structural and functional characteristics of the mutations were obtained from the mutation validation tool of the IARC TP53 Mutation Database.
NA indicates not applicable; —, no mutation.
Discussion
In this study, we show that very low-dose radiation can induce a very efficient p53 response in FL. It is characterized by concomitant induction of several classes of p53 targets genes. p53 is described as the central regulator of the checkpoint activated by DNA damage. Low-dose irradiation is thought to induce low-level DNA damage and a reversible cell-cycle arrest, whereas higher dose irradiation triggers apoptosis. The choice between cell-cycle arrest and apoptosis has been explained by p53 selectively transactivating target genes such as p21 and GADD45A that are cell-cycle regulators and BAX, FAS, or PUMA that are members of the core apoptotic pathway.25 Here, we show that very low radiation dose can, in vivo, induce an efficient apoptotic response, indicating that the intensity of DNA damage is not the only factor determining cell fate. Moreover, in vivo, genes related to cell cycle arrest and apoptosis induction show a similar pattern of induction after radiation. In contrast to the notion based on in vitro systems, this indicates that the choice between cell cycle arrest and DNA repair versus apoptosis is not based on the selective induction of one of the 2 pathways. This choice must rely on mechanisms other than the p53 transcriptional control of target genes.
The second characteristic of the response of FL to low-dose radiotherapy is related to the induction of immune-related genes. A previous microarray study already noticed the induction of immune-related genes in peripheral white blood cells after total body irradiation.26 By contrast, when the gene-expression profile of highly purified chronic lymphocytic leukemic cells was analyzed in vivo after treatment with fludarabine, only the p53 response was observed.27 As far as we can compare those studies, this supports that the immune response seen in FL after LDRT may reflect a response of the microenvironment. This may in a straightforward way be related to tissue damage induced by radiation itself, as in conventional dose radiotherapy. However, several arguments point to alternative mechanisms with a direct role of apoptotic tumor cells in the induction of such a reaction: the radiation dose is too low to be able to induce extensive damage of the normal tissue; p53 was shown not to be directly up-regulated in macrophages; some genes described as important for the clearance of apoptotic cells were regulated by low-dose radiation. Moreover, several lines of evidence support the notion that FL represents immunologically functional B cells in which nonmalignant tumor-infiltrating immune cells, in concert with tumor cell features determined by genomic alterations, regulate growth properties and clinical behavior.28
A model on the role of the immune response in low-dose radiotherapy in FL is illustrated in Figure 4. At 24 hours after radiotherapy, the apoptotic pathway has come to the stage of the exposure of phosphatidylserine at the surface of dying cells, with few morphologic signs of apoptosis.29 Triggered by phosphatidylserine receptor on macrophages, “apoptotic clearance” activation, reflected by the up-regulation of C1q, ApoE, and lysosomal enzymes, is induced. The TH1 and inflammatory reaction that takes place may be either part of a normal apoptotic clearance process or could result from the amount of dying cells. Gene-expression data also suggest induction of immunologic tolerance as reflected by INDO, LILRB2, and SIGLEC9. This suggests a homeostatic regulatory response to avoid autoimmune disease caused by the presence of abundant numbers of apoptotic cells. These insights provide several pathways that may be modulated to improve the results of combinations of radiotherapy and immunotherapy. Especially modulation of the TH1 response may be an attractive approach to implement in radioimmunotherapy in FL.
The exceptional feature of FL as an immunologically active disease may also be part of the explanation of the exquisite sensitivity of FL to radiotherapy. In contrast to many malignancies, p53 is only very rarely mutated in FL. A functional p53 is a prerequisite for radiosensitivity, since the p53 pathway is responsible for the apoptotic process, as confirmed by the dependency on a functional p53 gene and protein in the response of DLBCL to LDRT. Aberrantly overexpressed bcl-2 in FL would somehow protect the tumor cells from radiation-induced apoptosis via the intrinsic pathway by stabilizing the mitochondrial membranes. The fact that the functional composition of the immune response is modified after radiotherapy in this tumor that depends on its immunologic environment for growth support and that the extrinsic apoptotic pathway is prominently activated after radiation may explain the rapid apoptotic collapse after radiotherapy. The simultaneous induction of TRAIL-R2, a p53 target, and TRAIL, a ligand frequently expressed on immune cells, illustrates this hypothesis. In vitro studies mimicking this situation could confirm this hypothesis, but are hampered by the absence of relevant murine models and the virtual impossibility to propagate FL cells in vitro for more than a few days.
This study shows that, beside its clinical efficiency, low-dose radiation is able to induce a very effective p53 response leading to the rapid death and clearance of FL cells. This provides a novel, biology-based argument to advocate this type of treatment in symptomatic and recurrent FL. Obviously, this type of low-dose radiotherapy with a palliative intent should not replace standard dose-involved field radiotherapy (30 Gy in 3 weeks) as first-line treatment of localized FL with a curative intent. However, for the vast majority of patients current therapeutic modalities have not yet improved outcome despite improvement in progression-free and relapse-free survival. Therefore, for the large proportion of advanced stage patients there is still an important need for palliative regimens that lack side effects, which are effective and inexpensive to control their repeatedly recurring disease.
It is also not surprising that FL is the first disease where radiolabeled antibodies have shown their efficiency.30 Future clinical studies should focus on the specific advantage of using labeled antibodies, rather than an effect related to the extreme sensitivity of the disease. The results of studies performed on less radiosensitive tumors will certainly be interesting in that respect.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Dutch National Genomics Initiative, Cancer genomic center. L.K. was supported by a grant from the Belgian Federation against Cancer, nonprofit organization.
We thank Arno Velds, Mike Heimerikx, and Ron M. Kerkhoven from the Central Microarray Facility at the Netherlands Cancer Institute (NKI) for technical assistance and fruitful discussions; Rene Bernards for very helpful comments; Dr Philippe Poortmans, Verbeeten Institute for Radiotherapy, Tilburg, the Netherlands, for providing clinical information on the series of diffuse large B-cell lymphoma; the pathologists from the Pathology Laboratory Kennemerland, Haarlem, Pathology Laboratory Central Brabant, Tilburg, Departments of Pathology of the Amphia Hospital, Breda and Jeroen Bosch Hospital, Den Bosch, the Netherlands, for providing tissue samples for this study.
Authorship
Contribution: L.K., R.H., and D.J. designed the project, collected, analyzed, and interpreted the data, and wrote the paper; S.K., D.M., and C.O. collected and analyzed the data; A.B., E.E., J.P.B., M.V., A.V., and L.V. analyzed and interpreted the data. L.K. and R.H. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daphne de Jong, Department of Pathology, The Netherlands Cancer Institute, Amsterdam, the Netherlands; e-mail: d.d.jong@nki.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal