Abstract
Mutation in the target oncoprotein is a common mechanism of resistance to tyrosine kinase inhibitors, as exemplified by the many BCR/ABL mutations that thwart imatinib activity in patients with chronic myelogenous leukemia. It remains unclear whether normal cellular protein targets of chemotherapeutics will evolve drug resistance via mutation to a similar extent. We conducted an in vitro screen for resistance to lonafarnib, a farnesyl protein transferase inhibitor that blocks prenylation of a number of proteins important in cell proliferation, and identified 9 mutations clustering around the lonafarnib binding site. In patients treated with a combination of imatinib and lonafarnib, we identified farnesyl protein transferase mutations in residues identified in our screen. Substitutions at Y361 were found in patients prior to treatment initiation, suggesting that these mutants might confer a proliferative advantage to leukemia cells, which we were able to confirm in cell culture. In vitro mutagenesis of normal cellular enzymes can be exploited to identify mutations that confer chemotherapy resistance to novel agents.
Introduction
Rational drug development against tyrosine kinases such as Her2/Neu, BCR/ABL, and EGFR has produced a number of promising new chemotherapeutic compounds. The most dramatic clinical success is the treatment of chronic myelogenous leukemia (CML) patients with the BCR/ABL inhibitor imatinib, which induces disease remission in more than 95% of chronic-phase patients.1 Imatinib response is durable in CML patients treated during the chronic phase of the disease, but is invariably transient in patients treated in the advanced stages. Drug resistance is due mainly to the development of mutations in the BCR/ABL target protein. A number of second-generation compounds are currently under development to target mutant forms of BCR/ABL known to cause imatinib resistance. Clinical trials using these new agents have shown great promise.2 Although development of target-specific cancer therapy has focused on tyrosine kinase inhibitors, enzymes in other signal transduction pathways represent important targets, including the farnesyl protein transferase (FPTase).
FPTase is responsible for a posttranslational modification that is required for the function of a number of proteins acting in signal transduction pathways. FPTase attaches an isoprenyl moiety to the C-terminus of its substrate proteins. This prenylation has been reported to be necessary for the activity of proteins such as Ras, Rheb, and CENP-E, among others.3–5 Although the original rationale for the clinical development of FTIs was the inhibition of Ras protein function, it has since been demonstrated that K-Ras maintains its activity in the presence of FTIs due to alternative prenylation by the geranylgeranyl protein transferase.6,7 The antiproliferative effect of FTIs on various cell lines (including ones with K-Ras mutations) and in patients is therefore attributable to inhibition of other cellular targets such as CENP-E, CENP-F, Rheb, and others (reviewed in Basso et al8 ).
The broad antitumor activity of FTIs has led to clinical trials against both solid tumors and hematopoietic malignancies. Several agents are under development, including lonafarnib, tipifarnib, and BMS214662, and moderate activity has been reported in phase 1 and 2 trials using FTIs as monotherapy.9–12 Recently, the focus of clinical trials has shifted to the use of combination therapy, based on successful preclinical models that show synergy with other agents (eg, David et al,13 Jorgensen et al,14 Adjei et al,15 and Hoover et al16 ). Promising results have been published for using FTIs in combination with imatinib for the treatment of CML, and in combination with taxanes for the treatment of breast cancer (reviewed in Basso et al8 and Jabbour et al17 ). In addition to the activity of FTIs against cancer, preclinical results have demonstrated the sensitivity of a number of eukaryotic pathogens to FPTase inhibition (eg, Plasmodium falciparum and Trypanosoma brucei; reviewed in Buckner et al18 ). Interestingly, recent studies of 2 separate Hutchinson-Gilford progeria mouse models (associated with accumulation of farnesylated prelamin A) have shown that administration of FTIs reversed disease symptoms and improved over all survival.19,20
Resistance to farnesyltransferase inhibitors has previously been described in cell culture by mechanisms independent of FPTase mutation.21,22 The overexpression of the ATP11A transporter protein was reported to cause resistance both to FTIs and to a geranylgeranyl protein transferase inhibitor,23 and Bruzek et al report FTI resistance associated with mTOR phosphorylation that was reverted by the use of the mTOR inhibitor rapamycin.24 Mutations in residues homologous to the human Y361 FPTase beta subunit were reported to cause FTI resistance both in a yeast model and in P. falciparum.25,26 In addition, a G612A mutation (homologous to the human G250 residue) was recently reported to cause resistance to the BMS339941 FTI in P. falciparum as well.27
The development of drug resistance due to mutations in the target oncoprotein has been described as the main mechanism of drug resistance for a number of tyrosine kinase inhibitors.28–30 We reasoned that this phenomenon might also apply to other cellular targets of drugs. For this purpose, we developed an in vitro mutagenesis method to search for FPTase mutations causing drug resistance in cells treated with the FTI lonafarnib. We identified a number of mutations that confer varying degrees of FTI resistance on cells in vitro, and subsequently found some of the same mutations in lonafarnib-treated patients. Prompted by finding FTI-resistance mutations in some patients, even before the initiation of lonafarnib treatment, we tested whether these FPTase mutations confer growth advantage in the absence of lonafarnib, and indeed have demonstrated that the most drug-resistant amino acid substitution also appears to confer a growth advantage on cells. These data will be useful for monitoring the clinical resistance that will inevitably develop in patients treated with FTIs.
Patients, materials, and methods
Plasmids
The beta subunit of FPTase was cloned into the EcoR1 sites of the pEYK3.1 retroviral vector31 generating pEYK-FTB, which was used for the random mutagenesis, and into the EcoR1 sites of the pBabe-puro retroviral vector32 generating pB-FTB, which was used for verification of resistance by de novo mutation generation. K-Ras61L (a constitutively active form of K-Ras containing a substitution of glutamine to leucine at position 61) was cloned into the EcoR1 sites of the MSCV-IRES-GFP retroviral vector.33
Cell lines
BaF3 cells are a murine IL3-dependent pro-B-cell line that can be made IL3 independent by the expression of oncogenes such as BCR/ABL and KRas-61L. We found that the BaF3-KRas-61L cells grown in the absence of IL3 had increased sensitivity to lonafarnib compared with BaF3 cells grown in the presence of IL3, and thus these conditions were used for testing drug sensitivity.
Random mutagenesis
pEYK-FTB plasmid was transformed into XL-1 Red E Coli according to manufacturer's directions (Stratagene, La Jolla, CA). Cells were plated on zeocin agar and incubated at 37°C for 24 to 30 hours. Bacterial colonies were then collected by scraping of the plates and the mutated plasmid library was isolated as previously described.34 Then 1 μg of the plasmid library was used along with 1 μg of the pCL/Eco viral packaging construct35 to transfect 106 293t cells for virus production. Media were changed at 24 hours and viral supernatant was collected at 48 hours after transfection and used to infect 106 BaF3-KRas-61L cells with a multiplicity of infection (MOI) of 0.3. The MOI was kept low in order to minimize multiple retroviral integrations and confounding effects of insertional mutagenesis on the screen. Cells were plated 24 hours after infection in 6-well plates at a density of 5 × 104 cells per well in 4 mL soft agar media (54% RPMI, 16% DME, 10% inactivated FCS, and 20% agar solution: 1.2% agar in PBS) in the presence of varying drug concentrations (diluent [DMSO], 1 μM, 5 μM, and 10 μM lonafarnib). Plated cells were incubated at 37°C for 14 days. Individual drug-resistant colonies were then picked and expanded in liquid media. Genomic DNA was then isolated followed by a polymerase chain reaction (PCR) amplification of vector DNA using vector-specific oligonucleotides (1785F 5′-CACCCCCACCGCCCTCAAAGTAG-3′ and Zeo52R 5′-TAGTTCCTCACCTTGTCGTATTAT-3′). This PCR product was then sequenced for the identification of mutations.
Verification by site-directed mutagenesis
Mutations identified in the initial screen were recreated in the pB-FTB vector by site-directed mutagenesis according to manufacturer's instructions (Quick change kit; Stratagene). 293t cells were transfected with mutant pB-FTB and pCL/Eco. BaF3 cells were infected with viral supernatant with an MOI of 0.3 to 0.4 (as described in “Random mutagenesis”). FPTase β-expressing cells were then selected in puromycin. The ability of mutant FPTase β to confer drug resistance was assessed by 2 assays: (1) A colony-counting assay was used in which mutant and wild-type control pB-FTB-expressing cells were plated in soft agar (44% RPMI, 16% DME, 10% WeHi-3B conditioned media [a source of IL3], 10% inactivated FBS, 20% agar solution [1.2% agar in PBS]) in the presence of varying drug concentration. Cell colonies were counted after 14 days. (2) An immunoblot assay was used in which 106 cells were plated in RPMI, 10% IFS, 10% WeHi-3B conditioned media, at varying drug concentrations (diluent, 0.1 μM, 0.5 μM, 1 μM, 2.5 μM, 5 μM, 7 μM, and 10 μM). Cells were collected after 48 hours and lysed (using 150 mM NaCl, 20 mM Tris [pH 7.4], 10 mM NaF, 1 mM EDTA, 1 mM ZnCl, 1 mM MgCl, 1% NP-40 [Sigma, St Louis, MO], 10% glycerol). Then, 70 μg total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a PVDF membrane. Membranes were then incubated with HDJ2 primary antibody (NeoMarkers, Fremont, CA). Prenylated HDJ2 proportion was assessed by the ImageJ software.36,37 All independent experiments mentioned refer to independent infection by viral supernatant.
Patients
Patients were participants in a pilot study of SCH66336 and imatinib mesylate in chronic myelogenous leukemia conducted at the M. D. Anderson Hospital (Houston, TX). The study protocol was approved by institutional review boards of M. D. Anderson, the Brigham and Women's Hospital, and Children's Hospital Boston and was conducted in accordance with the Declaration of Helsinki. All patients signed an informed consent permitting the use of peripheral blood and bone marrow samples in this research. Peripheral blood (10 mL) and bone marrow (2 mL) samples were collected from 10 patients and used to isolate RNA and genomic DNA (using the RNeasy and DNeasy kits, respectively; Qiagen, Hilden, Germany). cDNA was made from 1 μg RNA using the SuperScript III kit (Invitrogen, Carlsbad, CA).
Patient FPTase β sequencing
Patient FPTase β was PCR amplified from either RNA or genomic DNA, cloned into the TOPO cloning vector according to manufacturer's instructions (Invitrogen), and used to transform E coli. Individual E coli colonies, each harboring a single PCR amplicon, were then isolated and sequenced.
Y361 and C95R cell proliferation assays
Mutant cells (106; infected with either Y361H, Y361L, or Y361S, at MOI of 0.3) were combined with wild-type cells (106; infected with wild-type FPTase β, at MOI of 0.3). Twenty-four thousand of the combined cells were then plated in 12 wells at a density of 2000 cells per well in RPMI media, containing 1% and 3% inactivated FBS, and 10% WeHi-3B conditioned media, and cultured for 14 to 30 days (37°C). The remainder of the cells was harvested directly after they were combined and used as time 0 controls. Genomic DNA was extracted from cultured cells and controls (DNeasy kit; Qiagen) and used as template for a PCR reaction using the following primers: FTBF 5′-TACTATTGCCCTCCATCTTCCTCC-3′, FTBR 5′CTCTGCCGATGTCTCATCCTTAAG-3′ and sequenced (by direct sequencing). Sequence chromatograms were analyzed for the relative contribution of wild-type versus mutant cells. Results of individual wells were scored by comparing the relative size (M) of the mutant chromatogram peak of cultured cells (ie, M = mutant/[mutant + wild type] chromatogram peak area) to the relative size (M0) of the mutant chromatogram peak at time 0. If M was greater than M0, the well was scored as predominately mutant. The result of each experiment is the relative number of predominantly mutant wells (ie, if 9/12 wells scored as predominately mutant the result for the experiment would be 0.75). Consistent results were obtained in 6 independent experiments that entailed independent retroviral infections, thereby minimizing the potential that artifacts from retroviral insertion account for the proliferative phenotype. The results of the 6 independent experiments were averaged and graphed in Figure 2 for each of the mutant/wild-type mixtures.
Immunoblot assays were carried out to verify equal FPTase expression levels in the pure cell populations (before equal mixing of wild-type and mutants), using the FTβ (X-28) antibody (Santa Cruz Biotechnology, Santa Cruz, CA; see above for immunoblot methodology).
Results
In vitro mutagenesis screen for the identification of FPTase β mutations causing lonafarnib resistance
In order to identify mutations in FPTase that cause lonafarnib resistance, we applied a methodology previously developed to identify BCR/ABL mutations that cause imatinib resistance34,38 (for strategy outline, see Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Briefly, we cloned the β subunit of FPTase into the pEYK3.1 retroviral vector, generating pEYK-FTB. We next transformed XL1-Red E coli cells that are deficient in DNA repair functions, thus generating a library of randomly mutated pEYK-FTB. This library was introduced by retroviral infection into BaF3 cells expressing K-Ras 61L. The introduction of K-Ras-61L, a constitutively active form of K-Ras, allows BaF3 cells to survive in the absence of IL3. Treatment of K-Ras-61L BaF3 cells with lonafarnib inhibits cell proliferation and induces apoptosis. BaF3/K-Ras-61L cells infected with the pEYK-FTB mutagenized library were grown in soft agar in the presence of varying concentrations of lonafarnib (1 μM and 5 μM). Resistant cell colonies were picked and expanded in liquid culture. Genomic DNA was isolated from the cell expansions, and FPTase β was sequenced, yielding 17 mutations: C95R, W106R, P152S, A155S, V195D, G196R, G224S, G241E, V242I, E265K, M282V, E285K, A305T, F360S, Y361H, Y361L, Y361S. Fourteen of these mutations are located on the surface of the active site of FPTase (C95R, W106R, P152S, A155S, G241E, V242I, E265K, M282V, E285K, A305T, F360S, Y361H, Y361L, Y361S).
Verification of random mutagenesis screen results
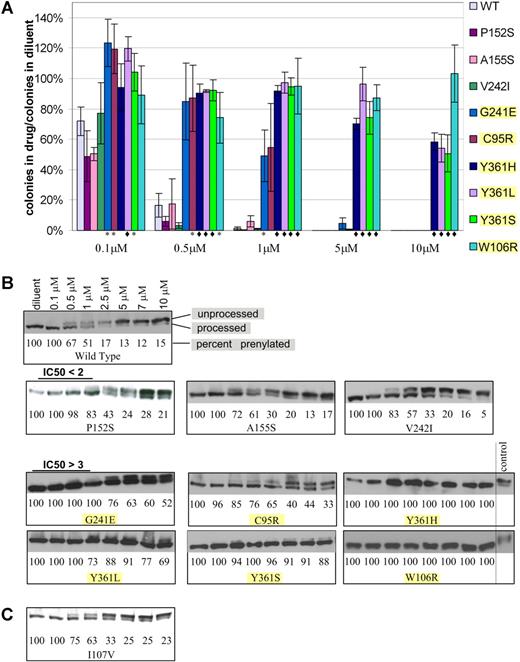
To verify the ability of the mutations identified in the random mutagenesis screen to confer lonafarnib resistance, wild-type FPTase β was cloned into the pBabe-puro retroviral vector, generating pB-FTB. Each of the mutations identified in the screen was recreated de novo in pB-FTB by site-directed mutagenesis. BaF3 cells (without K-Ras 61L expression) were infected with mutant pB-FTB or wild-type control, selected in puromycin, and grown in the presence of lonafarnib. Lonafarnib resistance of each mutant was verified by 2 assays. The first is a soft agar plating assay where cells were plated in the presence of varying lonafarnib concentrations and allowed to proliferate for 14 days. Drug resistance was measured as a ratio between the number of colonies formed in drug to the number of colonies formed in diluent alone (Figure 1A). Control cells were infected with wild-type pB-FTB and thus serve as controls for protein overexpression as well as positional proviral integration effect. The second assay is an immunoblot analysis of mutation-harboring cells grown in varying drug concentrations. Farnesylated proteins undergo an obligate postfarnesylation truncation of 3 C-terminal amino acids. Thus, both the processed and unprocessed proteins can be visualized by immunoblot. Proteins that have completed farnesylation will have a faster migration, while unprocessed proteins will migrate slower, and their ratio will reflect FPTase inhibition. In this assay, we assessed the effect of varying drug concentrations on the proportion of processed HDJ2 protein, a farnesylated chaperone protein used here as a convenient biomarker (Figure 1B). Of the 17 mutations originally identified, 6 (C95R, W106R, G241E, Y361L, Y361H, Y361S) showed drug resistance by soft agar proliferation assay (t test of significance P < .05) and by immunoblot. In addition, 3 mutations (P152S, A155S, and V242I) showed very weak resistance seen only by the immunoblot assay (Table 1). This mild resistance was observed in 2 independent experiments (which included independent vector infection and immunoblot analysis). All 9 verified mutations were located at the active site of FPTase, and 5 of them were found to be in direct contact of lonafarnib (Table 1). The remaining 8 mutations did not confer FTI resistance when reproduced in isolation and their mechanism of action is unexplained.
Resistance of BaF3-FPTase β mutants to lonafarnib. (A) Soft agar colony formation in the presence of varying lonafarnib concentrations. Colony formation is shown as a ratio of colonies formed in the presence of lonafarnib to colonies formed in diluent alone. t test: *P < .05; ♦P < .01. Error bars are standard errors. (B) Immunoblot of the HDJ2 biomarker. Prenylated (processed) versus unprenylated (unprocessed) HDJ2 can be discerned at varying lonafarnib concentrations. Mutations with P < .05 in the soft agar colony assay are highlighted. (C) Immunoblot of the I107V mutation identified in a patient sample, and not in the random mutagenesis.
Resistance of BaF3-FPTase β mutants to lonafarnib. (A) Soft agar colony formation in the presence of varying lonafarnib concentrations. Colony formation is shown as a ratio of colonies formed in the presence of lonafarnib to colonies formed in diluent alone. t test: *P < .05; ♦P < .01. Error bars are standard errors. (B) Immunoblot of the HDJ2 biomarker. Prenylated (processed) versus unprenylated (unprocessed) HDJ2 can be discerned at varying lonafarnib concentrations. Mutations with P < .05 in the soft agar colony assay are highlighted. (C) Immunoblot of the I107V mutation identified in a patient sample, and not in the random mutagenesis.
FPTase β mutation description
| Mutation . | Cellular IC50, μM . | Immunoblot IC50, μM . | Smallest distance from lonafarnib, Å . |
|---|---|---|---|
| Wild-type | 0.15 | 0.8 | — |
| P152S | 0.05 | 1.7 | 7.3 |
| I107V* | 0.13 | 1 | 10.1 |
| A155S | 0.13 | 1.1 | 7.6 |
| V242I | 0.36 | 1.3 | 13.5 |
| G241E† | 1.1 | 5 | 11.8 |
| C95R† | 1.1 | 4 | 3.1‡ |
| Y361H† | > 10 | > 10 | 3.1‡ |
| Y361L† | 10.3 | > 10 | 3.1‡ |
| Y361S† | > 10 | > 10 | 3.1‡ |
| W106R† | > 10 | > 10 | 3.4‡ |
| Mutation . | Cellular IC50, μM . | Immunoblot IC50, μM . | Smallest distance from lonafarnib, Å . |
|---|---|---|---|
| Wild-type | 0.15 | 0.8 | — |
| P152S | 0.05 | 1.7 | 7.3 |
| I107V* | 0.13 | 1 | 10.1 |
| A155S | 0.13 | 1.1 | 7.6 |
| V242I | 0.36 | 1.3 | 13.5 |
| G241E† | 1.1 | 5 | 11.8 |
| C95R† | 1.1 | 4 | 3.1‡ |
| Y361H† | > 10 | > 10 | 3.1‡ |
| Y361L† | 10.3 | > 10 | 3.1‡ |
| Y361S† | > 10 | > 10 | 3.1‡ |
| W106R† | > 10 | > 10 | 3.4‡ |
— indicates not applicable.
Identified in a patient—not by FPTase mutagenesis.
Significant resistance in soft agar colony-forming assay (P< .05).
Distance less than 4 Å from drug to residue.
Mutation modeling
FPTase β mutations were modeled onto the FPTase-lonafarnib cocrystal structure using the PyMol (De Lano Scientific, Palo Alto, CA) and DS ViewerPro (Accelyrs, San Diego, CA) software.
Patient studies
Based on the ability of FPTase β mutations to confer lonafarnib resistance in cell culture, we hypothesized that we might find the same mutations emerging in lonafarnib-treated patients. For that reason, we collected blood and bone marrow samples from patients enrolled in a clinical study of a combination treatment of lonafarnib and imatinib (Table 2). All patients entering the study were imatinib refractory, with the rationale that the addition of lonafarnib might resensitize imatinib response.16 Samples were collected at baseline, and every 3 months thereafter. We searched for FPTase β mutations in patient samples by PCR amplification of FPTase β from cDNA and from exons 4 and 11 of genomic DNA (where the majority of the mutations defined by our screen could be found). We sequenced between 20 and 50 separate PCR amplicons for each patient (as described in “Patients, materials, and methods”), and have identified a number of mutations previously seen in our in vitro screen, both in baseline samples and in samples taken after initiation of treatment. Patient 1 had a I107V mutation found twice independently in a sample taken 3 months after treatment initiation. This mutation was not identified in our screen; however, mutation in the adjacent residue W106R was found to confer strong resistance. We have recreated I107V de novo, and did not find it to show resistance in a soft agar assay. The immunoblot analysis showed a very mild increase in the IC50 (Figure 1C). We found 2 mutations, Y361L and C95R, in patient 2 at a sample taken at baseline and 3 months after treatment initiation. A Y361H mutation was also found in a single baseline sample from an additional patient (patient 10). No further samples were collected due to the patient's decision to withdraw from the study.
Patient description
| Patient no. . | Disease stage . | Response to therapy . | Response duration, mo . | BCR/ABL positivity, % . | BCR/ABL mutations . | FTase mutations . |
|---|---|---|---|---|---|---|
| 1 | AP | Partial HR | 10 | 100 | E292V | I107V |
| 2 | AP | None | 0 | 100 | G250E, M351T | Y361L C95R |
| 3 | CP | Partial CR | <15 | 88.33 | None | None |
| 4 | CP | Partial HR | 12 | 30.24 | None | None |
| 5 | AP | None | 0 | NA | None | None |
| 6 | CP | None | 0 | 100 | None | None |
| 7 | CP | None | 0 | 100 | G250E | None |
| 8 | AP | CHR | 2 | NA | F359V | None |
| 9 | AP | SD | 3 | 9.148 | E279L | None |
| 10* | NA | NA | NA | NA | NA | Y361H |
| Patient no. . | Disease stage . | Response to therapy . | Response duration, mo . | BCR/ABL positivity, % . | BCR/ABL mutations . | FTase mutations . |
|---|---|---|---|---|---|---|
| 1 | AP | Partial HR | 10 | 100 | E292V | I107V |
| 2 | AP | None | 0 | 100 | G250E, M351T | Y361L C95R |
| 3 | CP | Partial CR | <15 | 88.33 | None | None |
| 4 | CP | Partial HR | 12 | 30.24 | None | None |
| 5 | AP | None | 0 | NA | None | None |
| 6 | CP | None | 0 | 100 | None | None |
| 7 | CP | None | 0 | 100 | G250E | None |
| 8 | AP | CHR | 2 | NA | F359V | None |
| 9 | AP | SD | 3 | 9.148 | E279L | None |
| 10* | NA | NA | NA | NA | NA | Y361H |
AP indicates accelerated phase; CP, chronic phase; CHR, complete hematologic response; SD, stable disease; and NA, not available.
Patient 10 decided not to enter the study. Only a baseline sample was collected.
Y361 mutations confer growth advantage in the absence of lonafarnib
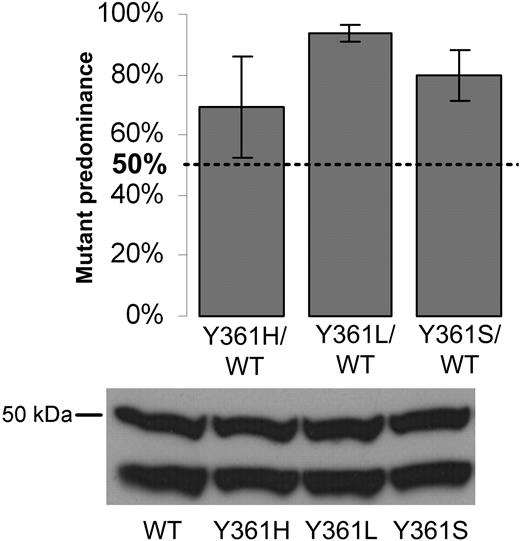
The presence of an FPTase mutation in a patient sample taken before the initiation of lonafarnib treatment suggested that the mutation might confer a growth advantage on cells, even in the absence of drug exposure. This prompted us to assess the effect of amino acid substitutions of the Y361 residue on cell proliferation. For that purpose, BaF3 cells expressing a mutant FPTase β allele (Y361H, Y361L, Y361S) were mixed in equal proportion with cells expressing the wild-type FPTase β (wild-type pB-FTB), plated in low serum conditions (1% and 3%), and allowed to proliferate to confluency over 14 to 30 days. Cells were then collected and FPTase β was sequenced. Similar results were observed for cells cultured in 1% and 3% serum. Cells expressing all Y361 mutants had a clear predominance over the wild-type cells in 6 independent experiments, with the effect especially pronounced for the Y361L and Y361S mutations (Figure 2).
Competitive proliferation assay. Wild-type and Y361 mutant BaF3-FPTase β cells were mixed at equal ratio, plated in 12 separate wells, and allowed to proliferate in culture to confluency. Individual wells were harvested and FPTase β was sequenced and compared with time 0 controls. Results are a compilation of the relative number of predominantly mutant wells in 6 independent experiments (“Patients, materials, and methods”). A mutant contribution of 50% would indicate no proliferative advantage. Error bars are standard errors. Bottom: immunoblot of FPTase β in the pure wild-type and mutant cells. FPTase β is a 46-kDa protein. The FPTase β antibody appears as doublet with an additional 50 kDa band.
Competitive proliferation assay. Wild-type and Y361 mutant BaF3-FPTase β cells were mixed at equal ratio, plated in 12 separate wells, and allowed to proliferate in culture to confluency. Individual wells were harvested and FPTase β was sequenced and compared with time 0 controls. Results are a compilation of the relative number of predominantly mutant wells in 6 independent experiments (“Patients, materials, and methods”). A mutant contribution of 50% would indicate no proliferative advantage. Error bars are standard errors. Bottom: immunoblot of FPTase β in the pure wild-type and mutant cells. FPTase β is a 46-kDa protein. The FPTase β antibody appears as doublet with an additional 50 kDa band.
Discussion
Lonafarnib is a highly specific small molecule inhibitor of FPTase that is currently being evaluated in clinical trials for a number of solid tumors and malignant bone marrow disorders, both as monotherapy and in combination with other agents. We have reasoned that the highly specific nature of this inhibitor is likely to render it susceptible to resistance mutations in the FPTase target that will prevent drug binding. Mutations causing drug resistance have been well documented for the BCR/ABL inhibitor imatinib, and recently also for inhibitors of other protein kinases such as gefitinib, erlotinib, and PKC412.28–30 Here we have demonstrated that mutations in a normal cellular protein target can cause drug resistance; thus, this phenomenon is not restricted to oncogenic protein kinases. The drug-resistance mutations identified in our in vitro screen were also found in patients, confirming the utility of the in vitro strategy for discovering mutations that are likely to be observed clinically.
Seventeen lonafarnib-resistant mutations were isolated in our initial random mutagenesis screen. Each of these mutations was then generated de novo by site-directed mutagenesis and tested for lonafarnib resistance by 2 assays: (1) colony formation in soft agar, which measures the ability of cells to proliferate in the presence of drug and enables calculation of a cellular IC50; and (2) immunoblot analysis of HDJ2 farnesylation, which reflects FPTase activity (Table 1; Figure 1). The immunoblot assay was included here as a cellular indicator of FPTase activity. It, therefore, cannot be regarded as equivalent to an in vitro enzyme activity test. HDJ2 farnesylation is commonly used as a biomarker of FPTase activity in clinical trials,9,39–41 although it has not been shown to correlate with clinical outcome. We were unable to recapitulate resistance for 8 mutations, suggesting that mechanisms other than FPTase β protein mutations can account for resistance to FTIs in our screen. While 6 of the validated mutants show robust lonafarnib resistance both by soft agar assay and by immunoblot (G241E, C95R, Y361L, Y361S, Y361H, and W106R), 3 additional mutations (P152S, A155S, and V242I) appeared only weakly resistant by the immunoblot assay. While we cannot conclude that this appearance of mild resistance is biologically relevant, the calculated IC50 values were similar in 2 independent experiments entailing independent vector infection and immunoblot analysis.
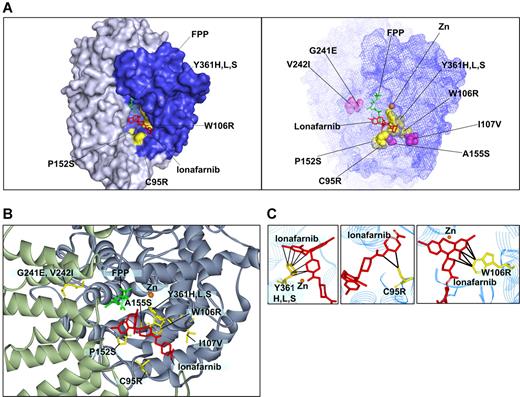
We modeled the validated mutations onto the FPTase-lonafarnib cocrystal structure42 (Figure 3). Of greatest interest are the W106R and Y361 mutations that confer the highest drug resistance of all mutants found. The growth of cells harboring the W106R mutation in all drug concentrations was comparable with that of control cells grown in diluent alone. In addition, the immunoblot analysis of HDJ2 of the W106R and Y361H mutants shows complete protein farnesylation in all drug concentrations. Modeling of the W106R mutation onto the cocrystal structure of lonafarnib and FPTase reveals a close contact between the tryptophan residue and lonafarnib. A substitution of this amino acid with an arginine is, thus, predicted to disrupt the van der Waals interactions that appear critical for anchoring lonafarnib to the FPTase β subunit (Figure 3C). Similarly, the 3 Y361 substitutions (to leucine, serine, and histidine) come into close contact with lonafarnib, explaining the critical role they play in lonafarnib binding (Figure 3C). Mutations in this residue have previously been reported to confer resistance to tricyclic FPTase inhibitors similar to lonafarnib. In addition, Y361 is located in proximity to the zinc ion required for enzymatic activity. The zinc ion is coordinated by the 3 beta subunit amino acids: D297, C299, and H362, and the Y361L mutation was shown to increase FPTase sensitivity to zinc chelators.25,43 It should also be mentioned that FPTase activity was shown to be enhanced by several hundred fold in the presence of magnesium ion bound to the D352 residue of the beta subunit.44 However, none of the mutations identified in our screen is located in proximity to this magnesium binding site. C95 comes into contact with lonafarnib only at the tip of the molecule. Hence, drug resistance caused by the substitution to arginine can be completely overcome by increasing the drug concentration to 5 μM (Figure 3C). The other 4 mutations at residues P152, A155, G241, and V242 do not come into direct contact with the drug. Therefore, their effect on drug resistance may be a result of conformational changes to the active site. We find that these substitutions cause only a mild drug resistance that may still play a clinical role with trough plasma concentrations reported to reach only 1.5 μM.12 Interestingly, mutations in the yeast homologues of amino acids 152 and 361 were previously reported to alter FPTase substrate specificity (in yeast, amino acid 159 and 362, respectively). Such mutants had increased ability to farnesylate substrates terminating with a leucine, which are typically prenylated by another prenyltransferase—geranylgeranyl transferase I (GGTase I).45 Therefore, these mutations may result in increased farnesylation efficiency of some substrates.
Modeling of FTase β mutations onto the lonafarnib/FPTase cocrystal structure. (A, left) Four of the mutated residues found are on the molecular surface of the active site (C95R, W106R, P152S, and Y361H,L,S; surfaces of the corresponding amino acid side chains are denoted in yellow). (A, right) Mapping of all the residues for which mutations were identified. Purple indicates residues that are not directly on the surface of the active site. (B) Close-up view of the residues identified. (C) A close-up view of residues in close proximity with lonafarnib (W106, Y361, and C95). Black lines mark distances of less than 4 Å.
Modeling of FTase β mutations onto the lonafarnib/FPTase cocrystal structure. (A, left) Four of the mutated residues found are on the molecular surface of the active site (C95R, W106R, P152S, and Y361H,L,S; surfaces of the corresponding amino acid side chains are denoted in yellow). (A, right) Mapping of all the residues for which mutations were identified. Purple indicates residues that are not directly on the surface of the active site. (B) Close-up view of the residues identified. (C) A close-up view of residues in close proximity with lonafarnib (W106, Y361, and C95). Black lines mark distances of less than 4 Å.
To assess the clinical implications that mutations in FPTase may have for patients treated with lonafarnib, we analyzed blood samples from CML patients participating in a clinical trial of imatinib-refractory patients who were switched to a combination treatment with lonafarnib and imatinib. We speculated that treatment with lonafarnib might provide a growth advantage to leukemic cells harboring FPTase β mutations. For this purpose, we sequenced FPTase β both from RNA and genomic DNA isolated from patient blood samples and found mutations of interest in 3 patients.
Patient 1 had no mutations that were previously identified in our screen; however, this patient did have a I107V mutation that we identified in 2 independent samples. Because this mutation is adjacent to the W106 residue that we discovered in our screen to confer high-grade lonafarnib resistance, we generated I107V de novo, and found it to confer only a very weak lonafarnib resistance similar to the one seen for mutations P152S, A155S, and V242I (Figure 1C). As with these mutations, we cannot conclude that this weak resistance is biologically relevant. Identification of this mutant in patients but not in our screen suggests that our in vitro screen is not fully comprehensive and that other mutants remain to be identified.
Patient 2 had 2 FPTase β mutations previously identified in the in vitro screen: C95R and Y361L. Both of these mutations were detected at baseline and in samples acquired 3 months past treatment initiation. This patient was taken off the study after 6 months due to lack of clinical response, but neither of the FPTase β mutations was detected by cloning and sequencing of samples taken at 6 months after treatment initiation, suggesting that other mechanisms accounted for the loss of treatment response. This patient entered the study with 2 BCR/ABL mutations that have both been associated with imatinib resistance (G250E and M351T). We tested the combination of lonafarnib and imatinib on BaF3 cells harboring G250E and M351T, and found that M351T was partially resistant and G250E fully resistant to combination treatment (Figure S2). Indeed, lonafarnib will not synergize with imatinib in cells transformed by the strongest mutants (eg, T315I16 ). Therefore, the presence of the strong BCR/ABL mutation G250E may account for the lack of treatment response in patient 2.
In none of our patients did we identify dominant clones of leukemic cells with FPTase β mutations, thereby suggesting that FTI resistance was not the primary cause of treatment failure and that other mechanisms were contributory. Indeed, the patient population was selected to be imatinib refractory, and lonafarnib was added in an attempt at salvage. It is likely that mutations in BCR/ABL accounted for much of the treatment resistance in this small pilot study, and thus it is not surprising that FTI-resistant mutations did not dominate.
A mutation affecting residue Y361 was detected in patient 2 and patient 10 prior to initiation of lonafarnib treatment, suggesting that the presence of mutations in this residue may confer growth advantage to cells even in the absence of lonafarnib. To investigate this possibility, we preformed a competitive proliferation assay whereby cells carrying specific mutants were mixed with cells expressing a native form of FPTase β. Cultures containing cell mixtures of either one of the Y361 mutations showed predominance of the mutant form after a short period in culture. This predominance was strongest in cultures containing the Y361L mutation, slightly less pronounced in cultures containing the Y361S mutation, and only mild for cultures of the Y361H mutant (Figure 2). Interestingly, the Y361L FPTase mutant was shown to have lower enzymatic activity than the wild-type molecule. We speculate that its growth advantage over the wild-type form may be the result of the change in substrate specificity previously reported45 (as discussed above). We were unable to show any proliferative advantage of cells expressing the C95R mutation (results not shown).
The ongoing development of target-specific cancer therapy requires an evaluation of mechanisms that can contribute to the emergence of drug resistance. The identification of mutations in the target protein was shown to be a major mechanism of drug resistance to a number of tyrosine kinase inhibitors. Here we show that this phenomenon is not limited to tyrosine kinases and is relevant to the FPTase inhibitors as well, and pertains to cellular targets of drugs. In addition, the identification of FPTase mutations in patients prior to treatment initiation suggests that FPTase mutations are likely to prove clinically relevant. Modeling of FPTase β mutations onto the FPTase-lonafarnib cocrystal structure reveals molecular mechanisms for the development of resistance, and can assist in the design of additional compounds that might be active against resistant targets. By this iterative process of mutagenesis and identification of drug resistance, followed by identification of compounds that are active against resistant target proteins, one can endeavor to identify a series of drugs directed against a particular target that can be used in combination chemotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the NIH and the NIH Director's Pioneer Award of the NIH Roadmap for Medical Research and in part by a grant from the Schering Plough Research Institute. G.Q.D. is a recipient of the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research. T.R. is a recipient of an NIH NRSA grant CA101505.
We wish to acknowledge Russ Hoover for the K-Ras61L MSCV vector and Piotr Sliz for discussion on the effect of FPT mutations on protein structure.
National Institutes of Health
Authorship
Contribution: T.R. designed the experiments, performed the research, and wrote the paper; V.N. performed the research and contributed to experimental design; M.A. contributed to development of methodology design; J.C. contributed the patient samples and clinical information; G.Q.D. conceptualized the study, contributed to methodology design, and wrote the paper.
Conflict-of-interest disclosure: T.R., M.A., and G.Q.D. hold a patent related to the present work. All other authors declare no competing financial interests.
Correspondence: George Q. Daley, Karp family building 7214, Children's Hospital, 300 Longwood Ave, Boston, MA 02115; e-mail: george.daley@childrens.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal