Mantle cell lymphoma (MCL) has a chromosomal translocation resulting in the expression of the cyclin D1 gene driven by the powerful enhancer of the immunoglobulin heavy chain gene, leading to uncontrolled, overexpressed cyclin D1 protein. We showed that suberoylanilide hydroxamic acid (SAHA; vorinostat), one of the histone deacetylase inhibitors derived from hydroxamic acid, caused a dramatic decrease (90%) in protein levels of cyclin D1 after 8-hour exposure to SAHA (5 μM) in MCL lines (SP49, SP53, Jeko1). mRNA levels and protein stability of cyclin D1 were minimally affected by SAHA over 8 hours. In contrast, metabolic labeling assays showed that SAHA decreased incorporation of [35S]methionine into cyclin D1 protein. The drug also decreased levels of phosphorylated Akt, mammalian target of Rapamycin (mTOR), and eukaryotic translation initiation factor 4E binding protein (eIF4E-BP) and lowered the cap site binding activity of eIF4E in the MCL cells. In vitro phosphatidyl inositol (PI) kinase assay demonstrated that SAHA directly inhibited kinase activity of PI 3′ kinase. Taken together, SAHA caused a rapid decrease of cyclin D1 in MCL by blocking the translation of cyclin D1 by inhibiting the phosphatidylinositol 3-kinase (PI3K)/Akt/mTOR/eIF4E-BP pathway, probably by PI3K inhibition.
Introduction
Mantle cell lymphoma (MCL) is a distinct subtype of B-cell non-Hodgkin lymphoma that shows specific pathologic features and has the t(11;14)(q13;q32) chromosomal abnormality.1,2 This chromosomal translocation juxtaposes the immunoglobulin heavy chain gene (IgH) on 14q32 to the cyclin D1 gene on 11q13, leading to overexpression of cyclin D1 mRNA.1,2
Transgenic mice overexpressing cyclin D1 in their lymphoid tissues develop lymphoid hyperplasia, not lymphomas, which suggests that overexpression of cyclin D1 is not sufficient for development of MCL, and additional genetic events may be necessary for lymphomagenesis of MCL.3,4 New techniques, including gene expression profiling, genetic comparative hybridization, and protein expression profiling, have elucidated novel genetic abnormalities in MCL.5,,,,–10 Many apoptosis-associated genes, including ATM, p53, p16INK4/p14ARF, Bim1, and MDM2, are dysregulated in this disease.2
MCL is usually resistant to standard-dose chemotherapeutic reagents; patients with this disease are often treated with high-dose chemotherapy either with or without stem cell transplantation.11 We previously found that histone deacetylase (HDAC) inhibitors, including suberoylanilide hydroxamic acid (SAHA; vorinostat), had profound antiproliferative activity against MCL cell lines.12 We have also found that cyclin D1 protein levels in MCL cells decrease rapidly after their treatment with SAHA.12 Another group of investigators also has shown that SAHA can lower levels of cyclin D1.13 In this study, we explore how SAHA decreases levels of cyclin D1.
Materials and methods
Cell lines and reagents
Jeko1 cells (MCL cell line) were a gift from Dr Sven deVos (Department of Hematology/Oncology, UCLA); SP49 and SP53 cells (MCL cell lines) were generously provided by Dr M. Daibata (Kochi University, Kochi, Japan). K562 (chronic myelogenous leukemia in blastic crisis cell line) and PC3 (prostate cancer cell line) were purchased from American Type Culture Collection (Manassas, VA). All cell lines except PC3 were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) with 20% fetal bovine serum (FBS). PC3 was cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% FBS. PS341 (also known as bortezomib [Velcade]) was kindly provided by Millennium Pharmaceuticals (Cambridge, MA). SAHA (Vorinostat) was generously provided by Dr Victoria Richon (Merck Research Laboratories, Boston, MA). Sodium butyrate (NaBu), valproic acid sodium (VPA), 5-fluorouracil (5FU), trichostatin A (TSA), and cycloheximide were purchased from Sigma-Aldrich (St Louis, MO). The phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 was obtained from Cell Signaling Technology (Danvers, MA). The mammalian target of Rapamycin (mTOR) inhibitor RAD001 was kindly provided by the Novartis Institutes for BioMedical Research Basel, Oncology (Basel, Switzerland).
Immunoprecipitation and Western blot analysis
Western blot analysis was performed as previously reported.12 Cells were treated with cell lysis buffer and incubated on ice for 15 minutes. Cell lysates were fractionated in SDS-PA gels (Bio-Rad Laboratories, Hercules, CA) and transferred to nitrocellulose membrane (Sigma). Membranes were incubated with primary antibodies (anti-cyclin D1, anti-PI3Kp110, anti-PI3Kp85 [Santa Cruz Bioscience, Santa Cruz, CA], anti-beta-actin [Sigma], anti-eIF4E, anti-eIF4BP, anti-phospho-eIF4BP, anti-phospho-mTOR, anti-phospho-Akt (Ser473), and anti-acetyl-histone H3 [Cell Signaling Technology]).
Secondary antibodies included horseradish peroxidase (HRP) conjugated anti-rabbit and anti-mouse immunoglobulin antibodies (GE Healthcare, Chalfont St. Giles, United Kingdom), HRP-conjugated anti-goat immunoglobulin antibody (Santa Cruz Biotechnology). The membranes were reacted using SuperSignal Western blotting kits (Pierce Biotechnology, Rockford, IL) and exposed to X-ray film according to the manufacturer's protocol.
Cap site binding activity assay
Assay for cap site binding activity was performed as described previously.14 In brief, Jeko1 cells treated either with or without SAHA (5 μM) were lysed and suspended in cap binding buffer.14 The mixture was precleared for 1 hour at 4°C with Protein A Sepharose. The supernatant was incubated with m7GTP-sepharose 4B resin (GE Healthcare), then bound proteins were fractionated in a sodium dodecyl sulfate-polyacrylamide (SDS-PA) gel and transferred to nitrocellulose membrane. eIF4E protein bound to m7GTP was detected with an anti-eIF4E antibody.
Northern blot analysis
Total RNAs were extracted using TRIzol reagents (Invitrogen) according to the manufacturer's protocol. RNAs were fractionated in formamide agarose gel and transferred to nylon membrane, Hybond N+ (Amersham Bioscience) according to the standard protocol.
Cyclin D1 radioactive probes were generated using RadPrime DNA Labeling System (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Prehybridization, hybridization, and washing were performed according to the protocol. Signals were exposed to X-ray film (BioMax; Eastman Kodak, Rochester, NY) for 24 hours. Intensity of bands was quantified using National Institutes of Health (NIH) Image software, and cyclin D1 transcript levels relative to S18 rRNA are shown.
Pulse-chase analysis and 35S protein labeling
Jeko1 cells were preincubated in methionine-free medium including 10% dialyzed fetal calf serum for 1 day, then labeled with [35S]methionine (0.2 mCi/mL; PerkinElmer Life and Analytical Sciences, Waltham, MA) as described previously.15 For pulse-chase analysis, pulse labeling was for 4 hours, followed by 3 washings with standard medium and then culturing in standard medium for 15 and 45 (chase). Labeled cells were lysed and subjected to immunoprecipitation with anti-cyclin D1 antibody. The immunoprecipitated proteins were fractionated in SDS-PA gels and exposed to X-ray film. Intensity of cyclin D1 protein bands was quantified by NIH Image software. For metabolic labeling of Jeko1, 1 × 106 cells were labeled with [35S]methionine as described previously for 2 and 4 and lysed in 30 μL of cell lysis buffer.
Radioactivity of 5 μl of whole lysates was counted by the LS 3801 liquid scintillation counter (Beckman Coulter, Fullerton, CA) and plotted. Lysates (10 μL) were fractionated in SDS-PA gel and transferred to nitrocellulose membrane. The transferred proteins were stained with Ponceau S and photographed. The membranes were exposed to X-ray film.
Plasmids and transfection
FOXO1a expression plasmid was provided by Dr C. Daly (Regeneron Pharmaceuticals. Tarrytown, NY)16 ; a constitutively active PI3K expression plasmid (Myr-delta 72-c-P3k) was from Dr P. Vogt (the Scripps Research Institute, La Jolla, CA)17 ; a constitutively active Akt expression plasmid (pSG5 HA-PH-Akt/PKB) was from Dr IM Kamer (Imperial College, London, United Kingdom).18 293T cells were transfected using Effectene Transfection Reagent (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Forty-eight hours after the transfection, the cells were treated either with or without SAHA (5 μM) for 6 hours followed by Western blot analysis.
Luciferase assay
Cyclin D1-internal ribosome entry site (IRES) and empty reporter plasmids19 were kindly provided by Dr J. Gera (UCLA). These plasmids are bicistronic expression vectors containing Renilla reniformis and firefly luciferase genes in one vector (Figure 5, top). The gene expression was driven by SV40 promoter. R reniformis luciferase proteins are normally translated, but when IRES sequence is inserted in front of the firefly luciferase gene, firefly luciferase is also translated under the control of the IRES sequence.
Transfection of SP49 cells was performed using the Electro Square Porator T820 (BTX, Holliston, MA) according to the manufacturer's protocol. In brief, 1 × 107 cells were suspended in 300 μL culture medium and mixed with 10 μg of each plasmid, then incubated on ice for 10 minutes. Electroporation conditions are as follows: 300 V, 20 ms, 1 pulse; after pulsation, the cells were incubated on ice for 10 minutes, then mixed with 3 mL fresh medium containing 20% fetal calf serum. After electroporation, cells were incubated in fresh medium for 39 hours, then treated either with or without SAHA (5 μM) for 9 hours. The cells were lysed with 100 μL of passive lysis buffer (Promega, Madison, WI) according to the manufacturer's protocol. Luciferase activity was measured using Dual-luciferase reporter system (Promega) on Luminometer, AutoLumat (Berthold, Oak Ridge, TN).
In vitro PI3K assay
In vitro PI3K assay was performed as described previously.20,21 In brief, Jeko1 cells were lysed, and PI3K was immunoprecipitated with anti-p85 PI3K antibody (Cell Signaling Technology). Precipitated proteins were mixed with phosphatidyl inositol (PI; Sigma) and [γ-32P]ATP (final concentration, 0.08 mCi/mL; PerkinElmer Life and Analytical Sciences). The PI3K reaction was carried out at room temperature for 20 minutes, and the labeled phosphatidylinositol monophosphate (PIP) was fractionated on a silica gel TLC plate (EM Science, Gibbstown, NJ) and exposed to X-ray film.
Results
SAHA rapidly decreased cyclin D1 protein levels in MCL lines but not in the K562 acute myeloid leukemia cell line
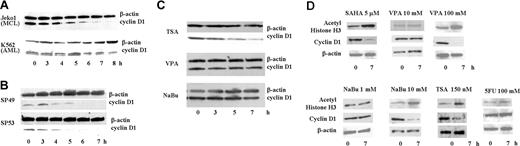
The MCL cell line, Jeko1, and a leukemia cell line, K562, were treated with SAHA (5 μM) for 8 hours. In Jeko1 cells, cyclin D1 levels rapidly decreased, with a 50% decrease by 4 hours (Figure 1A). In contrast, cyclin D1 protein levels in K562 cells were less affected by SAHA (Figure 1A). 2 additional MCL lines, SP49 and SP53, showed the same rapid decline of cyclin D1 levels when cultured with SAHA (5 μM) (Figure 1B). To determine whether other HDAC inhibitors were also able to decrease cyclin D1 protein expression, Jeko1 cells were exposed to 3 other HDAC inhibitors: TSA (150 nM), NaBu (1 mM), and VPA (5 mM) for 7 hours. TSA rapidly decreased cyclin D1 protein level comparable with SAHA (Figure 1C). In contrast, cyclin D1 protein levels were not affected by either NaBu or VPA (Figure 1C) even though these concentrations produced the same antiproliferative activity as SAHA against these cells12 (data not shown).
SAHA rapidly decreased cyclin D1 protein in MCL cells. (A) Jeko1 cells (MCL line) and K562 cells (AML line) and (B) 2 other MCL cell lines (SP49, SP53) were cultured with SAHA (5 μM) for the indicated times, and cyclin D1 and β-actin protein levels were measured by Western blot analysis. (C) Jeko1 cells were treated with either TSA (150 nM), VPA (5 mM), or NaBu (1 mM) for the indicated times and analyzed for expression of cyclin D1 and β-actin by Western blot. (D) Jeko 1 cells were treated with HDAC inhibitors (SAHA, VPA, NaBu, TSA) or 5FU at indicated concentrations for 7 hours. Levels of cyclin D1, acetyl-histone H3 (a known target of SAHA), and β-actin (loading control) were examined by Western blot analysis.
SAHA rapidly decreased cyclin D1 protein in MCL cells. (A) Jeko1 cells (MCL line) and K562 cells (AML line) and (B) 2 other MCL cell lines (SP49, SP53) were cultured with SAHA (5 μM) for the indicated times, and cyclin D1 and β-actin protein levels were measured by Western blot analysis. (C) Jeko1 cells were treated with either TSA (150 nM), VPA (5 mM), or NaBu (1 mM) for the indicated times and analyzed for expression of cyclin D1 and β-actin by Western blot. (D) Jeko 1 cells were treated with HDAC inhibitors (SAHA, VPA, NaBu, TSA) or 5FU at indicated concentrations for 7 hours. Levels of cyclin D1, acetyl-histone H3 (a known target of SAHA), and β-actin (loading control) were examined by Western blot analysis.
SAHA, an HDAC inhibitor, causes acetylation of histones. As shown on Figure 1A, SAHA (5 μM) rapidly decreased expression of cyclin D1 in MCL cells and, as expected, increased levels of acetyl-histone H3 (Figure 1D). VPA and NaBu at a low concentration (5 and 1 mM, respectively) neither decreased levels of cyclin D1 nor increased levels of acetyl-histone H3. However, at higher concentrations (VPA or NaBu [100 and 10 mM, respectively]), these drugs lowered levels of cyclin D1 and increased levels of acetyl-histone H3 (Figure 1D). MCL cells were also treated with high concentrations (100 mM) of 5FU. 5FU changed neither levels of cyclin D1 nor acetyl-histone H3, suggesting that the changes in cyclin D1 levels mediated by HDAC inhibitors was not caused by a cytotoxic effect (Figure 1D). SAHA and TSA showed a similar effect on cyclin D1 levels in MCL cells; these 2 reagents are HDAC inhibitors derived from hydroxamic acid. Our further experiments focused on SAHA.22
Levels of cyclin D1 mRNA as well as stability of cyclin D1 protein in MCL cells were minimally affected by SAHA
To examine the mechanism of rapid decrease of cyclin D1 after treatment with SAHA, we examined levels of cyclin D1 transcripts before and after exposure to the drug. Northern blotting analysis of Jeko1 MCL cells showed that SAHA (5 μM) minimally affected mRNA levels of cyclin D1 at 8 hours (Figure 2A). In addition, using the protein synthesis inhibitor cycloheximide, stability of cyclin D1 in Jeko1 cells was slightly enhanced by SAHA (5 and 50 μM) (Figure 2B). Furthermore, pulse-chase analysis also showed that stability of cyclin D1 protein in Jeko1 cells was not decreased by SAHA (5 μM) (Figure 2C).
SAHA minimally affected cyclin D1 mRNA levels and stability of cyclin D1 protein. (A) Northern blot analysis of cyclin D1 in Jeko1 cells treated with SAHA (5 μM, 8 hours). Levels of 18S ribosomal RNA (18S) are shown as an internal control. Intensity of bands was quantified, and levels of cyclin D1 relative to 18S rRNA are shown. Cyclin D1 mRNA/S18 in nontreated cells is regarded as 1.00. (B) Jeko1 cells were pretreated with SAHA (0, 5, or 50 μM; 4 hours); then, the protein synthesis inhibitor cycloheximide was added to each well (CHX, 100 μM). Cells were collected at indicated time points, and cyclin D1 and β-actin proteins were detected by Western blot analysis. Levels of cyclin D1 protein decreased, whereas β-actin did not change. In the presence of SAHA (5 or 50 μM), stability of cyclin D1 slightly increased (0 μM SAHA, 34% protein levels at 30 minutes; 5 μM SAHA, 47% protein at 30 minutes; 50 μM SAHA, 69% protein at 30 minutes.) Protein levels at 30 minutes are expressed as a percentage of levels at time 0, and the data are shown graphically. (C) Result of pulse-chase assay. Both SAHA and [35S]methionine were added to culture media containing Jeko 1 cells at the beginning of the incubation. Jeko1 cells plus or minus SAHA (5 μM) were labeled with [35S]methionine for 4 hours (Pulse), washed 3 times with culture medium, and released in normal culture medium plus or minus SAHA (5 μM) for indicated times (Chase) followed by immunoprecipitation of the cyclin D1 protein with anti-cyclin D1 antibody and fractionated in SDS-PA gel. Signal of cyclin D1 proteins was captured on a X-ray film, and intensity of the bands were quantified and plotted graphically. Intensity at time 0 is regarded as 1.0. (D) Jeko1 cells were treated with SAHA (5 μM) and/or the proteosome inhibitor, PS341 (also known as bortezomib [Velcade]) (10 μM) for 6 hours. Cellular lysate was Western-blotted and probed with antibodies to cyclin D1 and β-actin. No tx, cellular lysate of cells treated with diluent.
SAHA minimally affected cyclin D1 mRNA levels and stability of cyclin D1 protein. (A) Northern blot analysis of cyclin D1 in Jeko1 cells treated with SAHA (5 μM, 8 hours). Levels of 18S ribosomal RNA (18S) are shown as an internal control. Intensity of bands was quantified, and levels of cyclin D1 relative to 18S rRNA are shown. Cyclin D1 mRNA/S18 in nontreated cells is regarded as 1.00. (B) Jeko1 cells were pretreated with SAHA (0, 5, or 50 μM; 4 hours); then, the protein synthesis inhibitor cycloheximide was added to each well (CHX, 100 μM). Cells were collected at indicated time points, and cyclin D1 and β-actin proteins were detected by Western blot analysis. Levels of cyclin D1 protein decreased, whereas β-actin did not change. In the presence of SAHA (5 or 50 μM), stability of cyclin D1 slightly increased (0 μM SAHA, 34% protein levels at 30 minutes; 5 μM SAHA, 47% protein at 30 minutes; 50 μM SAHA, 69% protein at 30 minutes.) Protein levels at 30 minutes are expressed as a percentage of levels at time 0, and the data are shown graphically. (C) Result of pulse-chase assay. Both SAHA and [35S]methionine were added to culture media containing Jeko 1 cells at the beginning of the incubation. Jeko1 cells plus or minus SAHA (5 μM) were labeled with [35S]methionine for 4 hours (Pulse), washed 3 times with culture medium, and released in normal culture medium plus or minus SAHA (5 μM) for indicated times (Chase) followed by immunoprecipitation of the cyclin D1 protein with anti-cyclin D1 antibody and fractionated in SDS-PA gel. Signal of cyclin D1 proteins was captured on a X-ray film, and intensity of the bands were quantified and plotted graphically. Intensity at time 0 is regarded as 1.0. (D) Jeko1 cells were treated with SAHA (5 μM) and/or the proteosome inhibitor, PS341 (also known as bortezomib [Velcade]) (10 μM) for 6 hours. Cellular lysate was Western-blotted and probed with antibodies to cyclin D1 and β-actin. No tx, cellular lysate of cells treated with diluent.
Cyclin D1 protein is degraded by the proteosome complex under physiologic conditions.23,24 We treated MCL cells with SAHA and/or the proteosome inhibitor, PS341 (bortezomib [Velcade], 10 μM) for 6 hours.25 The proteosome inhibitor alone decreased cyclin D1 protein levels in MCL cells. This compound plus SAHA markedly decreased protein levels of cyclin D1 (Figure 2D). It is unclear why the proteosome inhibitor PS341 decreased levels of cyclin D1 in MCL. The mechanism causing degradation of cyclin D1 might be altered in MCL cells and lead to this paradoxical phenomenon.
Because mRNA levels and protein stability of cyclin D1 were minimally affected by SAHA over 8 hours, we speculated that protein translation of cyclin D1 was diminished by SAHA. Therefore, we performed metabolic labeling assays using [35S]methionine. Jeko1 cells were metabolically labeled either in the presence or absence of SAHA, lysed, and cyclin D1 protein was immunoprecipitated (Figure 3A). SAHA treatment (3 hours) decreased incorporation of [35S]methionine into cyclin D1 proteins (Figure 3A left panel). Intensity of bands of incorporated radioisotope in cyclin D1 was measured and plotted on a graph (Figure 3A right panel). In contrast, mRNA and whole protein levels of cyclin D1 were not affected after 3 hours of SAHA exposure (Figure 3B). These data suggested that cyclin D1 translation is decreased by SAHA treatment.
Incorporation of [35S]methionine into cyclin D1 was decreased by exposure to SAHA. (A) Jeko 1 cells were metabolically labeled in the absence of SAHA for 2 hours, followed by an additional 3 hours of metabolic labeling of the cells either with or without SAHA. At each time point, cells were collected and lysed, and cyclin D1 protein was immunoprecipitated and fractionated in a SDS-PA gel. Signal of cyclin D1 protein was detected on X-ray film (left); intensity of the bands were quantified and graphically plotted. (right). (B) To measure the immunoprecipitated cyclin D1 protein and the mRNA levels, the same experiment was performed in the absence of radioisotope; the proteins and mRNA were extracted after a 3-hour culture either with or without SAHA (5 μM) and analyzed by Western and Northern blot analysis.
Incorporation of [35S]methionine into cyclin D1 was decreased by exposure to SAHA. (A) Jeko 1 cells were metabolically labeled in the absence of SAHA for 2 hours, followed by an additional 3 hours of metabolic labeling of the cells either with or without SAHA. At each time point, cells were collected and lysed, and cyclin D1 protein was immunoprecipitated and fractionated in a SDS-PA gel. Signal of cyclin D1 protein was detected on X-ray film (left); intensity of the bands were quantified and graphically plotted. (right). (B) To measure the immunoprecipitated cyclin D1 protein and the mRNA levels, the same experiment was performed in the absence of radioisotope; the proteins and mRNA were extracted after a 3-hour culture either with or without SAHA (5 μM) and analyzed by Western and Northern blot analysis.
Some chemicals and/or culture conditions can diminish translation of proteins in whole cells.26,27 We analyzed the global rate of translation of proteins in MCL cells after their culture with SAHA using a radioisotope protein labeling assay (Figure 4). Amount of proteins applied onto the lanes was quantified by staining the membrane with Ponceau S (Figure 4B). Incorporated radioisotope activity was measured and plotted on a graph (Figure 4A) and fractionated in a gel (Figure 4C). Global protein translation in MCL cells was not affected by SAHA as examined by both techniques.
SAHA did not affect translation of cellular proteins in Jeko1 cells. (A) Jeko1 cells were metabolically labeled ([35S]methionine) in either the presence or the absence of SAHA (5 μM). Labeled cells were lysed, and activity of incorporated radioisotope into proteins was measured by scintillation counts. CPM, counts per minute. (B,C) Jeko1 cells were metabolically labeled either in the presence or absence of SAHA (5 μM) for the indicated times. Labeled cells were lysed, and proteins were extracted, fractionated on SDS-PA gel, transferred to membranes, and stained with Ponceau S to determine the amounts of proteins loaded in the lanes (B). Amount of proteins loaded in each lane was well-balanced. (C) The membranes were exposed on X-ray film to determine the levels of incorporated radioisotope in the proteins. Levels of incorporated radioisotopes into proteins increased depending on the duration of labeling, but those levels were not affected by the presence of SAHA.
SAHA did not affect translation of cellular proteins in Jeko1 cells. (A) Jeko1 cells were metabolically labeled ([35S]methionine) in either the presence or the absence of SAHA (5 μM). Labeled cells were lysed, and activity of incorporated radioisotope into proteins was measured by scintillation counts. CPM, counts per minute. (B,C) Jeko1 cells were metabolically labeled either in the presence or absence of SAHA (5 μM) for the indicated times. Labeled cells were lysed, and proteins were extracted, fractionated on SDS-PA gel, transferred to membranes, and stained with Ponceau S to determine the amounts of proteins loaded in the lanes (B). Amount of proteins loaded in each lane was well-balanced. (C) The membranes were exposed on X-ray film to determine the levels of incorporated radioisotope in the proteins. Levels of incorporated radioisotopes into proteins increased depending on the duration of labeling, but those levels were not affected by the presence of SAHA.
SAHA did not affect translation from IRES in cyclin D1 mRNA
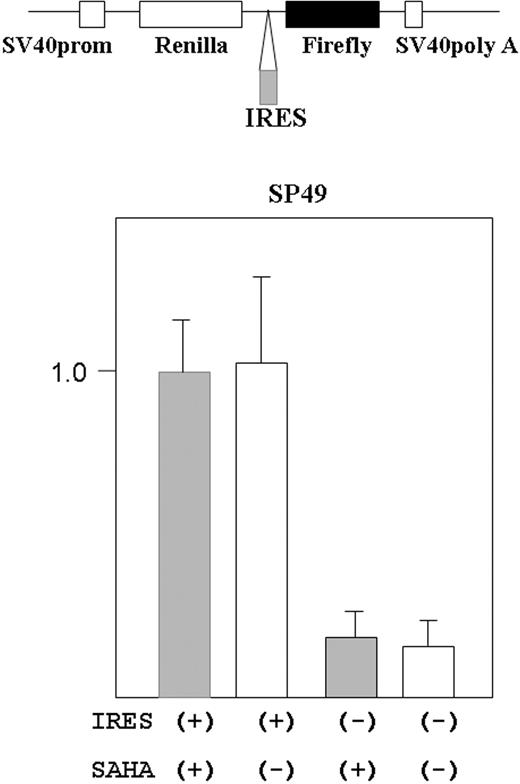
Cyclin D1 translation is regulated by cap-site-dependent and -independent (IRES-mediated initiation) manner.19 We examined if SAHA affected IRES-mediated initiation in cyclin D1. A reporter plasmid containing cyclin D1-IRES sequences was transfected into MCL cells (SP49), and the cells were treated either with or without SAHA (5 μM) for 9 hours (Figure 5). SAHA did not affect the translation dependent on IRES-mediated initiation.
SAHA did not affect translation from cyclin D1 IRES site in MCL cells. Structure of the bicistronic reporter gene is shown (top panel). Plasmid contains both the R reniformis (Renilla) and firefly luciferase genes. Gene expression was driven by the simian virus 40 promoter (prom). Without the IRES sequence between the R reniformis and firefly luciferase, only R reniformis luciferase protein was translated. When an IRES sequence was inserted in front of the firefly luciferase gene, the firefly gene was also translated dependently on the IRES sequence. The reporter construct that contained cyclin D1 IRES region in front of the firefly luciferase gene was transfected into SP49 MCL cells. Transfected cells were treated either with or without SAHA (5 μM, 9 hours); cells were lysed and luciferase activity was measured. Firefly luciferease activity was normalized by R reniformis luciferase activity. Constructs without the IRES sequence were used as a negative control. Results represent mean value and SD of 3 experiments done independently. Results are normalized to the mean value of experiments using the IRES (+) construct and SAHA.
SAHA did not affect translation from cyclin D1 IRES site in MCL cells. Structure of the bicistronic reporter gene is shown (top panel). Plasmid contains both the R reniformis (Renilla) and firefly luciferase genes. Gene expression was driven by the simian virus 40 promoter (prom). Without the IRES sequence between the R reniformis and firefly luciferase, only R reniformis luciferase protein was translated. When an IRES sequence was inserted in front of the firefly luciferase gene, the firefly gene was also translated dependently on the IRES sequence. The reporter construct that contained cyclin D1 IRES region in front of the firefly luciferase gene was transfected into SP49 MCL cells. Transfected cells were treated either with or without SAHA (5 μM, 9 hours); cells were lysed and luciferase activity was measured. Firefly luciferease activity was normalized by R reniformis luciferase activity. Constructs without the IRES sequence were used as a negative control. Results represent mean value and SD of 3 experiments done independently. Results are normalized to the mean value of experiments using the IRES (+) construct and SAHA.
SAHA blocked the signal transduction pathway of Akt/mTOR/eIF4E-BP/eIF4E
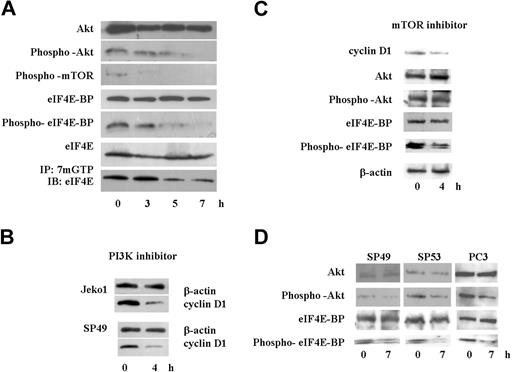
Because translation of cyclin D1 is regulated by the activity of eIF4E,28 we analyzed the cap binding activity of eIF4E using m7GTP-Sepharose beads. SAHA rapidly diminished cap binding activity of eIF4E in Jeko1 cells (Figure 6A bottom lane). Cap binding activity of eIF4E is regulated by eIF4E-BP activity, which is regulated by phosphorylation of eIF4E-BP.29,–31 Protein levels and phosphorylation status of eIF4E-BP was examined by immunoblotting analysis (Figure 6A). Although protein levels of eIF4E-BP were not affected by SAHA at 8 hours, phosphorylation of eIF4E-BP was rapidly decreased by exposure of Jeko1 cells to SAHA (Figure 6A). Phosphorylation of eIF4E-BP is regulated by mTOR.29,–31 SAHA rapidly decreased the phosphorylation of this protein in Jeko1 cells (Figure 6A). Phosphorylation of mTOR is modulated by Akt1.29,–31 SAHA quickly decreased phosphorylation of this protein in Jeko1 cells, even though whole protein levels of Akt1 were not affected by SAHA at 8 hours (Figure 6A). These data suggested that SAHA rapidly blocked the Akt/mTOR/eIF4E-BP/eIF4E signal transduction pathway and might diminish translation of cyclin D1, leading to a rapid decrease of protein levels of cyclin D1 in MCL cells after their treatment with SAHA.
SAHA diminished Akt/mTOR/eIF4E-BP/eIF4E signal pathway, and a PI3K inhibitor, LY294002, decreased cyclin D1 protein levels in MCL cells. (A) Jeko1 cells were cultured with SAHA (5 μM) for the indicated times. Levels of phospho-Akt, Akt, phospho-mTOR, phospho-eIF4E-BP, eIF4E-BP, eIF4E, and active eIF4E (bottom; see “Materials and methods, Immunoprecipitation and Western blot analysis”) were detected by Western blot analysis. (B) 2 MCL lines (Jeko1 and SP49) were treated with the PI3K inhibitor, LY294002 (50 μM) for 4 hours. Levels of cyclin D1 were determined by Western blot analysis. β-Actin was used as an internal control. (C) Jeko 1 cells were treated with the mTOR inhibitor RAD001 (200 nM) for 4 hours. Levels of cyclin D1, Akt, phospho-Akt, eIF4E-BP, and phospho-eIF4E-BP were detected by Western blot analysis. (D) 2 additional MCL lines (SP49, SP53) and a prostate cancer cell line PC3, in which Akt/mTOR/eIF4BP pathway is active because the PI3K inhibitory protein (PTEN) is deleted, were treated with SAHA (5 μM, 7 hours). Phosphorylation status of Akt and eIF4E-BP as well as whole Akt and eIF4E-BP levels were examined by Western blot. SAHA treatment decreased phosphorylated Akt and eIF4E-BP, whereas total levels of these proteins were not affected.
SAHA diminished Akt/mTOR/eIF4E-BP/eIF4E signal pathway, and a PI3K inhibitor, LY294002, decreased cyclin D1 protein levels in MCL cells. (A) Jeko1 cells were cultured with SAHA (5 μM) for the indicated times. Levels of phospho-Akt, Akt, phospho-mTOR, phospho-eIF4E-BP, eIF4E-BP, eIF4E, and active eIF4E (bottom; see “Materials and methods, Immunoprecipitation and Western blot analysis”) were detected by Western blot analysis. (B) 2 MCL lines (Jeko1 and SP49) were treated with the PI3K inhibitor, LY294002 (50 μM) for 4 hours. Levels of cyclin D1 were determined by Western blot analysis. β-Actin was used as an internal control. (C) Jeko 1 cells were treated with the mTOR inhibitor RAD001 (200 nM) for 4 hours. Levels of cyclin D1, Akt, phospho-Akt, eIF4E-BP, and phospho-eIF4E-BP were detected by Western blot analysis. (D) 2 additional MCL lines (SP49, SP53) and a prostate cancer cell line PC3, in which Akt/mTOR/eIF4BP pathway is active because the PI3K inhibitory protein (PTEN) is deleted, were treated with SAHA (5 μM, 7 hours). Phosphorylation status of Akt and eIF4E-BP as well as whole Akt and eIF4E-BP levels were examined by Western blot. SAHA treatment decreased phosphorylated Akt and eIF4E-BP, whereas total levels of these proteins were not affected.
We speculated that the PI3K/Akt/mTOR/eIF4E-BP/eIF4E pathway was active, and translation of cyclin D1 was dependent on the activity of this pathway in MCL cells. MCL cells (Jeko1, SP94) were cultured with a PI3K inhibitor, LY294002 (50 μM, 4 hours), which resulted in a rapid decreased cyclin D1 protein level in MCL cells (Figure 6B). Furthermore, we treated Jeko1 cells with the mTOR inhibitor RAD001 (200 nM). RAD001 did not change phosphorylation status of Akt, but decreased phosphorylation of eIF4E-BP and cyclin D1 level (Figure 6C). Another 2 MCL lines (SP49, SP53) were treated with SAHA, and phosphorylated Akt/eIF4E-BP levels were examined (Figure 6D). SAHA decreased phosphorylation of these proteins in both cell lines (Figure 6D).
To examine whether SAHA blocked activated PI3K/Akt/mTOR/eIF4E-BP/eIF4E pathway in other cells, we treated the prostate cancer cell line PC3. These cells lack PTEN, a negative regulator of PI3K; therefore, this signal pathway is constitutively active.32,33 Treatment of PC3 cells with SAHA (5 μM, 7 hours) rapidly decreased phosphorylated Akt levels leading to a rapid decrease of phosphorylated eIF4E-BP (Figure 6D).
SAHA blocked activity of PI3K
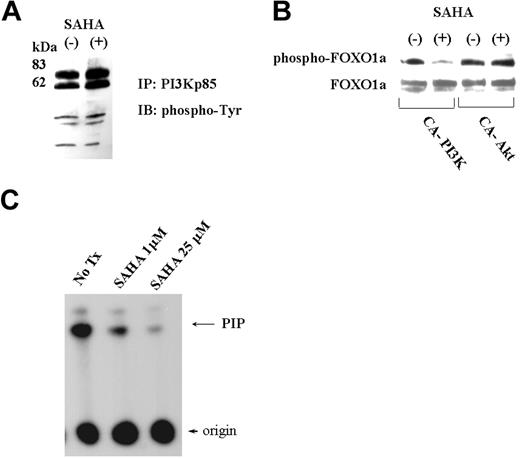
We analyzed the effect of SAHA on binding of phosphotyrosine proteins to PI3K. MCL cells were cultured either with or without SAHA; proteins binding to PI3K were immunoprecipitated using anti-PI3K p85 antibody, run on a gel, blotted, and probed with an antibody that recognizes phosphotyrosine-containing proteins. SAHA did not change the phosphotyrosine proteins binding to PI3K (Figure 7A).
SAHA did not change phospho-tyrosine proteins bound to PI3K but did directly inhibit PI3K activity. (A) Phospho-tyrosine proteins bound to PI3K. Cell lysates of Jeko1 cells cultured either with or without SAHA (5 μM, 6 hours), were immunoprecipitated with anti-PI3K p85 antibody. Precipitated proteins were fractionated on a SDS-PA gel and transferred to a membrane that was probed with an anti-phosphotyrosine antibody. (B) FOXO1a expression vector plus a vector expressing constitutive active (CA)-Akt or -PI3K were transfected into 293T. Cells were cultured in either the presence or the absence of SAHA (5 μM, 6 hours). Cellular lysate was Western blotted, and phosphorylated FOXO1a and whole FOXO1a were detected with either phospho-FOXO1a or FOXO1a antibodies. (C) PI3K in vitro assay. PI3K was precipitated using anti-p85 PI3K antibody from Jeko1 cells. The precipitated PI3K was subjected to in vitro PI3K assay using PI as substrate. Kinase assay was performed either in the presence or absence of SAHA (1 μM, 25 μM). The phosphorylated PI (PIP) was separated by chromatography. No tx, control Jeko1 cells with no treatment.
SAHA did not change phospho-tyrosine proteins bound to PI3K but did directly inhibit PI3K activity. (A) Phospho-tyrosine proteins bound to PI3K. Cell lysates of Jeko1 cells cultured either with or without SAHA (5 μM, 6 hours), were immunoprecipitated with anti-PI3K p85 antibody. Precipitated proteins were fractionated on a SDS-PA gel and transferred to a membrane that was probed with an anti-phosphotyrosine antibody. (B) FOXO1a expression vector plus a vector expressing constitutive active (CA)-Akt or -PI3K were transfected into 293T. Cells were cultured in either the presence or the absence of SAHA (5 μM, 6 hours). Cellular lysate was Western blotted, and phosphorylated FOXO1a and whole FOXO1a were detected with either phospho-FOXO1a or FOXO1a antibodies. (C) PI3K in vitro assay. PI3K was precipitated using anti-p85 PI3K antibody from Jeko1 cells. The precipitated PI3K was subjected to in vitro PI3K assay using PI as substrate. Kinase assay was performed either in the presence or absence of SAHA (1 μM, 25 μM). The phosphorylated PI (PIP) was separated by chromatography. No tx, control Jeko1 cells with no treatment.
To determine whether SAHA blocked either upstream or downstream of the Akt pathway, an expression vector coding for FOXO1a, a substrate of Akt, and either a constitutive active (CA)-Akt or CA-PI3K were transfected into 293T cells. These transfected cells were treated with either SAHA or vehicle for 4 hours. Protein lysates were blotted and the phosphorylation status of FOXO1a was analyzed by probing with a phospho-FOXO1a specific antibody (Figure 7B). SAHA did not diminish phosphorylation of FOXO1a after cotransfection of CA-Akt (Figure 7B), which suggested that SAHA blocked at or upstream of Akt in the signal transduction pathway of PI3K/Akt. Consistent with this notion, SAHA clearly diminished phosphorylation of FOXO1a after cotransfection with CA-PI3K, which suggested that SAHA blocked either downstream of the signal pathway of PI3K activity or PI3K itself (Figure 7B). To determine whether SAHA directly inhibited kinase activity of PI3K, in vitro PI3K kinase assay was performed. SAHA profoundly diminished activity of PI3K in vitro (Figure 7C).
Discussion
Our findings showed that SAHA dramatically decreased cyclin D1 protein levels in MCL cells. This initially surprised us, because the gene is translocated and under the control of the powerful immunoglobulin heavy chain gene in MCL. Further studies showed that SAHA affected neither the level of cyclin D1 mRNA nor the stability of cyclin D1 protein but was associated with an inhibition of translation of cyclin D1 protein. Because the half-life of this protein is short (≈30 minutes) in MCL cells, blockade of translation by SAHA resulted in a rapid decrease of the cyclin D1 protein level. Additional experiments demonstrated that the block of translation of cyclin D1 occurred by inhibiting the PI3K/Akt/mTOR/eIF4E-BP pathway.
PI3K phosphorylates the third position of the inositol ring.29,–31 The pleckstrin homology domain of Akt binds to PI3K lipid products, leading to translocation of Akt to the cellular membrane, where it becomes active and phosphorylates Ser2448 of mTOR protein.29,–31 This phosphorylation activates mTOR kinase, which then phosphorylates Thr37/Thr46 of eIF4E-BP.29,–31 This is followed by phosphorylation of Ser65/Thr70 of eIF4E-BP.29,–31 The phosphorylated eIF4E-BP releases eIF4E, leading to initiation of translation.29,–31 Our data suggest that this pathway is active and associated with translation of cyclin D1 protein in MCL cells.
The blockade of this pathway by either PI3K or mTOR inhibitors also decreased cyclin D1 protein levels in MCL cells. Nevertheless, over the 8-hour exposure to either SAHA, PI3K, or mTOR inhibitors, MCL cells were morphologically unchanged (data not shown). Our studies showed decreased activation (phosphorylation) of the proteins involved in this pathway. Using dominant active expression vectors for Akt and PI3K, as well as, direct measurement of PI3K activity, we showed that SAHA probably decreased levels of cyclin D1 in MCL by inhibiting the PI3K pathway.
Several other interesting findings occurred in this study. We showed that the PI3K/Akt/mTOR/eIF4E-BP/eIF4E pathway is activated in MCL. Recently, Rizzatti et al34 detected the activation of the Akt/mTOR pathway in MCL cells using the gene expression profiling technique. Overexpression of cyclin D1 alone is not sufficient for development of MCL and other genetic event(s) are necessary.3,4 An active eIF4E has been reported to be important for lymphomagenesis, including the ability of this protein to cooperate with c-Myc in development of lymphomas in mice.35,36 Perhaps an active eIF4E induced by an up-regulated PI3K/Akt/mTOR pathway can cooperate with the overexpressed cyclin D1 protein in the development of MCL.
Several other investigators have also reported that HDAC inhibitors can inhibit the PI3K/Akt pathway in cancer cells.37,38 We do not know how SAHA affects PI3K activity. We examined changes in acetylation of this protein complex but did not find any change (data not shown), even though we easily detected acetylation of histone H3 (Figure 1D). Genes modulated by SAHA may be able to indirectly affect the molecules involved in this pathway.
This study found that several phosphotyrosine proteins were immunoprecipitated with PI3K, including 70- and 55-kDa phosphoproteins (Figure 7). These proteins may be ZAP70 and lck because expression of both has been detected in MCL cells.39 Receptor- and/or non-receptor-type tyrosine kinases may be involved in the activation of PI3K in MCL cells. For example, activation of CD40 and its associated protein has been reported in MCL cells.40,41 In contrast to MCL cells, we found that SAHA did not affect levels of cyclin D1 in HEK293T cells that had been transiently transfected with cyclin D1 cDNA (data not shown). Therefore, the ability of SAHA to inhibit levels of cyclin D1 depends on the cell type.
Gera et al42 reported that translation of cyclin D1 was dependent on the Akt/mTOR pathway when this pathway was active; in contrast, translation of cyclin D1 was independent of this pathway when Akt/mTOR pathway was inactive. In addition, Shi et al19 reported that translation of cyclin D1 was regulated by 2 mechanisms, cap-dependent and cap-independent (IRES-dependent) translation. We found that SAHA did not affect the cap-independent (IRES-mediated) translation pathway in MCL cells. Perhaps, the translational control of cyclin D1 is unique in MCL cells, and other cancer cells overexpressing cyclin D1.
One observation made in this study deserves further exploration. In normal cells, cyclin D1 protein levels are not abundant and fluctuate during the cell cycle.23,24 Proteosomal degradation plays a prominent role in this process.23,24 Paradoxically, the proteosome inhibitor PS341 (Bortezomib [Velcade]) decreased levels of cyclin D1 in MCL cells, and the combination of the proteosome inhibitor and SAHA dramatically decreased cyclin D1 to almost undetectable levels. This was an unexpected and unexplained finding suggesting that degradation of cyclin D1 in MCL is markedly different from that in normal cells. Furthermore, this combination of SAHA and PS341 could represent a novel therapeutic approach.
In summary, our data showed that SAHA inhibited translation of cyclin D1 by affecting activity of PI3K and its downstream proteins important for efficient translation of proteins. In addition, we found that SAHA and a proteosome inhibitor markedly decreased cyclin D1 levels in MCL cells. The data provide a strong rationale for a clinical trial of SAHA alone or with bortezomib in persons with MCL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the generous support of NIH as well as the Lymphoma Foundation of America, Sheryl and David Weissberg Lymphoma Research Fund, Inger Fund, and C. and H. Koeffler Foundation. N.K. is supported by a fellowship from the Tower Cancer Research Foundation. H.P.K. holds the Mark Goodson Endowed Chair of Oncology Research at Cedars-Sinai Medical Center and is a member of the Jonsson Cancer Center and the Molecular Biology Institute of UCLA.
National Institutes of Health
Authorship
Contribution: N.K. designed the research, analyzed data, and wrote the manuscript. J.C. analyzed data. H.P.K. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Norihiko Kawamata, Cedars-Sinai Medical Center/UCLA School of Medicine, 8700 Beverly Blvd, Los Angeles, CA 90048; e-mail:kawamatan@cshs.org.

![Figure 2. SAHA minimally affected cyclin D1 mRNA levels and stability of cyclin D1 protein. (A) Northern blot analysis of cyclin D1 in Jeko1 cells treated with SAHA (5 μM, 8 hours). Levels of 18S ribosomal RNA (18S) are shown as an internal control. Intensity of bands was quantified, and levels of cyclin D1 relative to 18S rRNA are shown. Cyclin D1 mRNA/S18 in nontreated cells is regarded as 1.00. (B) Jeko1 cells were pretreated with SAHA (0, 5, or 50 μM; 4 hours); then, the protein synthesis inhibitor cycloheximide was added to each well (CHX, 100 μM). Cells were collected at indicated time points, and cyclin D1 and β-actin proteins were detected by Western blot analysis. Levels of cyclin D1 protein decreased, whereas β-actin did not change. In the presence of SAHA (5 or 50 μM), stability of cyclin D1 slightly increased (0 μM SAHA, 34% protein levels at 30 minutes; 5 μM SAHA, 47% protein at 30 minutes; 50 μM SAHA, 69% protein at 30 minutes.) Protein levels at 30 minutes are expressed as a percentage of levels at time 0, and the data are shown graphically. (C) Result of pulse-chase assay. Both SAHA and [35S]methionine were added to culture media containing Jeko 1 cells at the beginning of the incubation. Jeko1 cells plus or minus SAHA (5 μM) were labeled with [35S]methionine for 4 hours (Pulse), washed 3 times with culture medium, and released in normal culture medium plus or minus SAHA (5 μM) for indicated times (Chase) followed by immunoprecipitation of the cyclin D1 protein with anti-cyclin D1 antibody and fractionated in SDS-PA gel. Signal of cyclin D1 proteins was captured on a X-ray film, and intensity of the bands were quantified and plotted graphically. Intensity at time 0 is regarded as 1.0. (D) Jeko1 cells were treated with SAHA (5 μM) and/or the proteosome inhibitor, PS341 (also known as bortezomib [Velcade]) (10 μM) for 6 hours. Cellular lysate was Western-blotted and probed with antibodies to cyclin D1 and β-actin. No tx, cellular lysate of cells treated with diluent.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/7/10.1182_blood-2005-11-026344/2/m_zh80210708410002.jpeg?Expires=1767730540&Signature=WEdgwuSUEDdIxxrNS2UTHFy5nl~aoh1ZIdJh~F9tBEUteP-Zbj6Dyc44auYGXihaPDTmG9z6QLHRwxj4ctGrhe1dhiTHSZHviuodk~Bd5GGesccO-rmfCnQfIE0vVQ5GMJXerNgrbNGXcEF6UV9gKPfZmWYnxMDA0OZJQ7UfM5HXBOZWiHVWe5pgkcAKB07GWK~eguBvUmeOjowdEe4dAIJetGqhvSn9P4KK~J4wkChVAX4W0MqAKhEdaih1EVO37nkCh8BtPLXq~cnVbpbWQInhXSCrkbHQ9q7D-~bEDIFDMiT9RTCx4F9UadA4NCwgO4~iSIa-vQ4OgVCZzqRDEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Incorporation of [35S]methionine into cyclin D1 was decreased by exposure to SAHA. (A) Jeko 1 cells were metabolically labeled in the absence of SAHA for 2 hours, followed by an additional 3 hours of metabolic labeling of the cells either with or without SAHA. At each time point, cells were collected and lysed, and cyclin D1 protein was immunoprecipitated and fractionated in a SDS-PA gel. Signal of cyclin D1 protein was detected on X-ray film (left); intensity of the bands were quantified and graphically plotted. (right). (B) To measure the immunoprecipitated cyclin D1 protein and the mRNA levels, the same experiment was performed in the absence of radioisotope; the proteins and mRNA were extracted after a 3-hour culture either with or without SAHA (5 μM) and analyzed by Western and Northern blot analysis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/7/10.1182_blood-2005-11-026344/2/m_zh80210708410003.jpeg?Expires=1767730540&Signature=QG70eQ89hUZdd17kbTNqFqjJdBsGejyxGl07RvxH833ajLGv~eEkgVwIoC88Bhub-6ozjGs~RYPuNtGZM0matcNDCVasQ2W6ERlSGNA1dW8VhdYemR~-A3Dxd3KemTFo-LOFDI1BConLsKNn1KrnffEG0f0nC32RQGLUj-Rw1fApGuKTYUOHC4vzfLnhKH8oqddZMUZKLWLVPeMt~ab0ZdwMf8~49kqBVn7hDQC8xW3l0vlS8hKP56eaHthmP43q68tG0WG7jB-Zurvou4XzX3r9yEvbu3oaiQL0UgGR9eBOko3JJVOtxTvMq43nEvrg874GSI49zJKEgqQTI0vJ5Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. SAHA did not affect translation of cellular proteins in Jeko1 cells. (A) Jeko1 cells were metabolically labeled ([35S]methionine) in either the presence or the absence of SAHA (5 μM). Labeled cells were lysed, and activity of incorporated radioisotope into proteins was measured by scintillation counts. CPM, counts per minute. (B,C) Jeko1 cells were metabolically labeled either in the presence or absence of SAHA (5 μM) for the indicated times. Labeled cells were lysed, and proteins were extracted, fractionated on SDS-PA gel, transferred to membranes, and stained with Ponceau S to determine the amounts of proteins loaded in the lanes (B). Amount of proteins loaded in each lane was well-balanced. (C) The membranes were exposed on X-ray film to determine the levels of incorporated radioisotope in the proteins. Levels of incorporated radioisotopes into proteins increased depending on the duration of labeling, but those levels were not affected by the presence of SAHA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/7/10.1182_blood-2005-11-026344/2/m_zh80210708410004.jpeg?Expires=1767730540&Signature=YsA3y7Whxgq4-Cpe3vX0sct~4DG1EHlLuFpeL-DcT0byJHdXlmk6~51bUFW38fVDp5AgoPV1z0DAoYUmYNeNA6XotPXT6ItVtM-m35mCvvjXcQZu3Fo35sbTuq8kY-~K2l~HH-JuNuMOiGzH6EQOs86mfZvAvnueueFlob2bcP-DO9XVUEyhObqHFbIbq8WTZ50UK~oPYzXlt0i8gxSE4VFkTc-c423pMngJP~MWfyIif17XtGF25rH5EszrtJGqTzJIXmGDUEAVra-e90wazJWpJ9SElObXgcv~O3xI~INuAYgaClGXRyNlnq7u5K48bOB488p8ZxPY58Jc7SlIbw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal