Abstract
Cell-based immunotherapy in settings of allogeneic stem cell transplantation or donor leukocyte infusion has curative potential, especially in hematologic malignancies. However, this approach is severely restricted due to graft-versus-host disease (GvHD). This limitation may be overcome if target antigens are molecularly defined and effector cells are specifically selected. We chose formin-related protein in leukocytes 1 (FMNL1) as a target antigen after intensive investigation of its expression profile at the mRNA and protein levels. Here, we confirm restricted expression in peripheral blood mononuclear cells (PBMCs) from healthy donors but also observe overexpression in different leukemias and aberrant expression in transformed cell lines derived from solid tumors. We isolated allorestricted T-cell clones expressing a single defined TCR recognizing a particular HLA-A2–presented peptide derived from FMNL1. This T-cell clone showed potent antitumor activity against lymphoma and renal cell carcinoma cell lines, Epstein-Barr virus (EBV)–transformed B cells, and primary tumor samples derived from patients with chronic lymphocytic leukemia (CLL), whereas nontransformed cells with the exception of activated B cells were only marginally recognized. Allorestricted TCRs with specificity for naturally presented FMNL1-derived epitopes may represent promising reagents for the development of adoptive therapies in lymphoma and other malignant diseases.
Introduction
Although it is controversially discussed whether adaptive immune responses in tumor-bearing hosts play a role in controlling growth and recurrence of human tumors,1,2 T cells can be converted to highly efficient killers of tumors. Donor leukocyte infusions (DLIs) are responsible for a graft-versus-leukemia effect (GvL) after allogeneic stem-cell transplantation (SCT), representing a therapeutic option with curative potential in different diseases at advanced stages.3 Although GvL responses have also been shown for low-grade lymphomas, improvement of long-term survival after allogeneic transplantation has not been demonstrated in these patients.4-8 This can be attributed primarily to the high treatment-associated mortality due to transfer of T cells recognizing allogeneic minor histocompatibility (MHC) antigens, thereby causing potentially life-threatening graft-versus-host disease (GvHD). For broader applicability of this therapeutic option, it is therefore essential to reduce the risk of detrimental GvHD.
One approach to gain a tumor-specific effect while significantly reducing the risk of GvHD is to generate allorestricted T cells with specificity for epitopes derived from tumor-associated antigens (TAAs).9 Such allorestricted peptide-specific T cells may display high avidity toward MHC-presented TAAs, because they have not been negatively selected in the thymus. Moreover, they can be isolated by tetramers and cloned by limiting dilution to identify the specific TCR responsible for tumor-selective killing.10 The isolation of a TCR with defined specificity for TAAs facilitates genetic TCR transfer into T-cell lines,11-14 allowing major expansion of tumor-specific T cells.
The choice of an appropriate target antigen is of fundamental importance for the success of an antitumor immunotherapeutic approach. So far, there is no universal antigen that serves as a target antigen in a broad range of malignancies. Many tumor-specific antigens are derived from aberrant expression of cell type–specific proteins and are expressed only in a restricted number of tumor types, limiting the tumors that can be treated. However, even the presence of a suitable antigen does not guarantee effective antigen recognition, because tumor cells evade immune recognition by down-regulation of the specific target or by inhibition of MHC class I–restricted antigen presentation. Thus, the level of epitope presentation in the context of classical or nonclassical MHC molecules is probably the most important predictor for tumor-specific recognition.15,16 Selecting specificities for a number of defined epitopes of different target antigens may therefore be of fundamental importance for the development of effective therapies to treat a broad variety of tumors and to reduce the risk of evasion from immune-mediated destruction.
We previously identified 14 novel lymphoma-associated antigens using the SEREX approach in chronic lymphocytic leukemia (CLL).17 One of them, KW13, is homologous to the formin-related protein in leukocytes 1 (FMNL1). FMNL1 belongs to a conserved family of formin-related proteins, which are present in all eukaryotes and regulate actin dynamics.18 Human FMNL1 mRNA has been shown to be selectively expressed in peripheral blood mononuclear cells (PBMCs) and overexpressed in CLL samples as well as in malignant cell lines.17,19 Recently, Gomez et al identified FMNL1 to play a role in centrosome polarity and cytotoxicity of T cells.20
Here, we demonstrate expression of FMNL1 in different subtypes of blood-derived cells and aberrant expression in tumor cells, making FMNL1 an attractive target antigen for allorestricted immune responses. In addition, we identified a candidate peptide epitope derived from FMNL1 that is specifically recognized by allorestricted cytotoxic T-cell clones that all display an identical TCR. This T-cell clone, originating from an HLA-A2− donor, specifically recognized HLA-A2+ tumor cell lines derived from lymphomas and other malignant cells as well as primary tumor material specimens from HLA-A2+ CLL patients. We observed high peptide specificity and restriction to HLA-A2. Several HLA-A2 subtypes have been recognized, whereas cross-reactivity against HLA-A2− allotypes has been observed in only 2 of 16 virus-transformed lymphoblastoid cell lines (LCLs). Altogether, the selected T-cell receptor may be a suitable tool for the development of allorestricted target-specific therapies in lymphomas and other malignant diseases.
Materials and methods
Cells and cell lines
PBMCs from healthy donors as well as patients with CLL, T- and B-cell acute lymphoblastic leukemia (T-ALL and B-ALL, respectively), and acute myeloid leukemia (AML) were collected with donors' and patients' informed consent in accordance with the Declaration of Helsinki and after approval by the institutional review board of Ludwig-Maximilians-Universität, Munich, Germany. Patients had diagnosis of CLL, ALL, and AML by morphology, flow cytometric analysis, and cytogenetics. PBMCs were obtained by density gradient centrifugation on Ficoll/Hypaque (Biochrom, Berlin, Germany). T cells and other PBMC subpopulations from healthy donors were isolated by negative or positive magnetic bead depletion (Invitrogen, Karlsruhe, Germany), and high purity was confirmed by flow cytometric analysis.
Peptide-pulsed T2 cells (ATCC CRL-1992; American Type Culture Collection, Manassas, VA) were used for priming and restimulation of HLA-A2− T cells. The following cell lines were used for analysis of FMNL1 expression and as targets for FMNL1-specific T cells: HLA-A2+; Epstein-Barr virus–negative (EBV−) lymphoma cell lines BJ18 and DG75 (kindly provided by J. Mautner); HLA-A2− lymphoma cell lines Daudi (ATCC CCL-213) and Raji (ATCC CCL86); chronic myelogenous leukemia cell line K562 (ATCC CCL-243); HLA-A2+ renal cell carcinomas RCC26 and RCC5321 ; HLA-A2− renal cell carcinoma KT187 (DKFZ, Heidelberg, Germany); HLA-A2+ 143 TK− lung fibroblasts (kindly provided by R. Mocikat); 293 HEK embryonal kidney cells (ATCC CRL-1573); HLA-A2+ and HLA-A2− EBV-transformed B-cell lines; and different homozygous EBV-transformed B-cell lines.22 The human B-cell lines C1R untransfected and transfected with HLA-A*0201, HLA-A*3303, and HLA-A*6601 (kindly provided by S. Stevanovic)23 were used for analysis of cross-reactivity.
Unspecific stimulation of PBMCs was induced by IL-2 (Chiron Vaccines International, Marburg, Germany) and OKT3 (ATCC) for 3 days. CLL and normal B cells were stimulated by CD40 ligand–transfected NIH3T3 fibroblasts.24
Real-time polymerase chain reaction for FMNL1 expression
To evaluate quantitative mRNA expression of FMNL1, total RNA was extracted from primary tumor cell material, normal PBMCs, and cell lines,25 and cDNA was synthesized by Superscript II reverse transcriptase (Invitrogen) following the manufacturer's instructions. Detection of FMNL1 mRNA was conducted using the Light Cycler PCR Master Mix (Roche, Grenzach-Wyhlen, Germany). Exon-overlapping primers for FMNL1 detection were used (5′-CAAGAACCCCAGAACCAAGGCTCTGG and 3′-CTGCAGGTCGTACGCCTCAATGGC) resulting in a 299-bp amplicon. The amplicon was detected by fluorescence (530 nm) using a double-stranded (ds) DNA binding dye, SYBR Green I with the Light Cycler Instrument (Roche). 18S rRNA was processed as a control for relative quantification. RNA from normal tissue pooled from different donors (Clontech, Palo Alto, CA) was used for determination of FMNL1 expression in nontransformed tissues. Relative quantitative expression was calculated using the delta-delta Ct (cycle threshold) method.26 Skeletal muscle was used as reference tissue because it showed the highest Ct for detection of FMNL1.
FMNL1 cloning, protein expression, monoclonal antibody production, and Western blot
FMNL1 cDNA was amplified by polymerase chain reaction (PCR) and cloned into a modified pTris-His–tagged vector (Invitrogen) resulting in an additional N-terminal His-Tag. XLI (Stratagene, La Jolla, CA) and Top10 E coli bacteria (Invitrogen) were used for transformation and protein expression. Purification of the His-tagged protein from supernatants of lysed bacteria was performed by incubation with nickel-NTA-agarose (Qiagen, Hilden, Germany) overnight, followed by imidazole elution and dialysis against PBS over 24 hours. The identity of the purified FMNL1 protein was verified by mass spectrometry (matrix-assisted laser desorption/ionization time-of-flight [MALDI-TOF]) (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article).
For eukaryotic expression, FMNL1 cDNA was amplified by PCR and cloned into an eukaryotic pCMV-His–tagged expression vector (Invitrogen).
A peptide comprising the following amino acids “KKEAAAQEAGADT” of the C-terminal end of the FH2 domain of FMNL1 was synthesized and coupled to KLH or OVA (PSL, Heidelberg, Germany). After immunization of Lou/C rats, myeloma cell line P3X63-Ag8.653 (ATCC) was fused to rat immune spleen cells.27 Bound monoclonal antibodies were detected with biotinylated mouse anti–rat IgG subclass-specific antibodies (TIB 170, TIB 173, TIB 174 [ATCC]; R2c, GSF) in a peroxidase reaction (horseradish peroxidase avidin D; Vector, Burlingame, CA). An irrelevant peptide coupled to OVA was used as a negative control. Hybridoma supernatants (dilution, 1:10) were screened by Western blot analysis for reactivity against the recombinant FMNL1 protein using a secondary peroxidase-conjugated goat antirat antibody at 1:2000 (Jackson ImmunoResearch, West Grove, PA). Proteins were detected by chemiluminescence. Anti-Xpress antibody (Invitrogen) and anti-His antibody (Invitrogen) were used for size comparison of bands detected by supernatants containing anti-FMNL1–specific antibodies. Several specific antibodies were identified, and the antibody 6F2 (rat IgG2b subclass) was used further in this study (Figure S1B). Actin served as loading control, using a rabbit antihuman actin antibody (Sigma, St Louis, MO) and a secondary antirabbit antibody (Pharmacia, North Peapack, NJ). Prepared cell lysates of cell lines and primary tumor samples were used for FMNL1 protein detection.
Peptides
FMNL1-derived peptides were selected using different prediction algorithms for immunogenic epitopes including binding to HLA-A2 (genotype A*0201), TAP transportation, and cleavage by the immune proteasome (Table S1).28-32 We chose the following FMNL1-derived peptides for pulsing of antigen-presenting cells: FMNL1-PP1 (VLLEYLAFA), FMNL1-PP2 (RLPERMTTL), FMNL1-PP6 (CVNEIALSL), and FMNL1-PP8 (TLLHYLVKV). The peptide HDAC6 (residues 862-871: RLAERMTTR) derived from histone deacetylase 6 (HDAC6) with high homology to FMNL1-PP2, the HLA-A2–restricted influenza-associated peptide Flu (MP58: GILGFVFTL), the HLA-A2–binding Her2-derived peptide 369 (residues 369-378: KIFGSLAFL), the HLA-A2–binding tyrosinase-derived peptide 369 (residues 369-378: YMNGTMSQV), the predicted BCR-derived peptide BCR (residues 205-214: GLQGTYQDV), and the HLA-B35–binding cytomegalovirus-derived peptide IPS (IPSINVHHY)33 were used as control antigens. Peptides were synthesized by standard fluorenylmethoxycarbonyl (Fmoc) synthesis (Metabion, Martinsried, Germany; Biosyntan, Berlin, Germany). Purity was more than 90% as determined by reverse-phase high-performance liquid chromatography (RP-HPLC) and verified by mass spectrometry. Lyophilized peptides were dissolved in DMSO (Sigma) for 2-mM stock solutions.
Peptide binding to HLA-A2
A peptide competition assay was used to assess binding of synthetic peptides derived from FMNL1.34 T2 cells were prepulsed with the specific FMNL1-derived peptide (100 μM) followed by pulsing with the tyrosinase-derived peptide 369 (1 μM). The tyrosinase 369-specific T-cell clone IVSB (kindly provided by T. Wölfel)35 was assayed at various E/T ratios for lytic activity in response to peptide- and nonpeptide-prepulsed T2 targets in a 4-hour 51Cr-release assay as previously described (Figure S2).17 The HLA-A2–binding Flu peptide served as a positive control and the HLA-B35–binding cytomegalovirus-derived IPS peptide, as negative control for binding competition.
Tetramers and antibodies
Tetramers were synthesized as previously reported.36 Specific tetramers were used for the following peptides: A2-FMNL1-PP1, A2-FMNL1-PP2, A2-FMNL1-PP6, and A2-FMNL1-PP8 as well as the control tetramers detecting T cells specific for cytomegalovirus (CMV) pp65, A2-pp65, and B7-pp65.37 Tetramer-binding assays were performed essentially as described38 and analyzed by flow cytometry. The following antibodies were used to characterize PBMC-derived cells, primary tumor cells, and malignant cell lines: anti–CD3-FITC (UCHT1; BD, Heidelberg, Germany), anti–CD4-FITC (RPA-T4; BD), anti–CD8-FITC (V 5T-HIT8a; BD), anti–CD8-PE and -APC (RPA-T8; BD), anti–CD5-FITC (BL1a; Beckman Coulter, Krefeld, Germany), anti–CD19-FITC and -PE (HIB19; BD), anti–CD14-PE (M5E2; BD), anti–CD56-PE (B159; BD), anti–HLA-A2-FITC (BB7.2; ATCC), anti–HLA-A2 unlabeled (BB7.2; ATCC), and anti–αβ-TCR-FITC (T10B9.1A-31; BD). The anti–HLA-A2 antibody HB54 (ATCC) was used for blocking experiments. A Vβ14-specific antibody (Dako, Hamburg, Germany) was used for TCR staining of the isolated T-cell clone.
CTLs
Cytotoxic T lymphocyte (CTL) lines were generated using peptide-pulsed T2 cells for specific stimulation. T2 cells were pulsed with specific peptides (10 μM) and used for CTL priming and restimulation at a stimulator–effector cell ratio = 1:10. Cytokines were added as follows: IL-2 (50 U/mL) (Chiron Vaccines International), IL-7 (10 ng/mL; Peprotech, London, United Kingdom), and IL-15 (10 ng/m; Peprotech). Peptide-specific T cells were detected by flow cytometry using PE-conjugated peptide-presenting HLA-A2+ tetramers and sorted by a high-performance cell sorter (MoFlo; Dako). Sorted cells were nonspecifically restimulated using pooled allogeneic irradiated PBMCs together with OKT3, IL-2, IL-7, and IL-15. Sorted cells were cultured after limiting dilution in 96-well plates. T cells were investigated for their cytotoxic potential by 4-hour 51Cr-release assays. For cold target assays, T cells were exposed concurrently to 51Cr-labeled and unlabeled targets in different ratios.
BioPlex analysis and ELISA
For cytokine detection, effector and target cells were incubated at a ratio of 1:5 or 1:2 for 24 hours. Supernatants were collected and stored at −20°C until analysis. The presence of cytokines was analyzed using standard enzyme-linked immunosorbent assay (ELISA) for IFN-γ (BD) following the recommendations of the manufacturer and the BioPlex system (as described by de Jager et al39 ; Bio-Rad, Munich, Germany). For BioPlex analysis, the following cytokines were investigated: IL-2, IL-4, IL-6, IL-8, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, and TNF-α.
TCR analysis
PCR analysis of expressed TCR chains was performed as previously described.40 Total RNA from T-cell clones and lines was extracted according to the manufacturer's recommendation (Trizol reagent; Invitrogen). cDNA was synthesized using superscript II reverse transcriptase (Invitrogen) and oligo dT primers. Subfamily-specific TCR-PCR was performed using 34 Vα and 37 Vβ primers followed by gel isolation (NucleoSpin; Macherey-Nagel, Düren, Germany) and direct DNA sequencing of the amplified products. The T-cell receptor nomenclature was used according to the WHO-IUIS nomenclature subcommittee on TCR designation.41 In addition to the single V gene segment-specific primer approach, 2 degenerate primers covering 98% of the 55 Vβ region gene segments were used for analysis of TCR β-chains.42
Results
Expression of FMNL1 in normal and malignant tissue
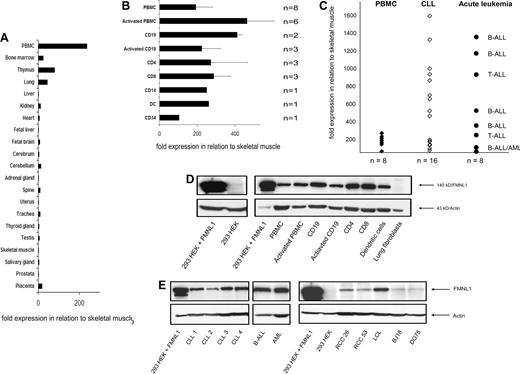
FMNL1 is a formin-related protein in leukocytes that has been shown to be selectively expressed at the mRNA level in PBMCs, thymus, and spleen of healthy donors and overexpressed in CLL samples and malignant cell lines.17 We confirmed and extended these data by quantitative RT-PCR and Western blot using a monoclonal antibody directed against a FMNL1-derived C-terminal peptide. Here, we show detailed data of FMNL1 expression in normal and malignant tissues (Figure 1A-E). In normal tissue, FMNL1 mRNA is almost exclusively expressed in PBMCs and homing tissues of lymphocytes and myeloid cells, such as bone marrow and thymus (Figure 1A). We then investigated the expression of FMNL1 mRNA in PBMCs more in detail. We tested the expression in PBMCs stimulated with IL-2 and OKT3; PBMC subpopulations including CD19+ cells and CD19+ cells activated with CD40 ligand; and CD4+, CD8+, CD14+, and mature DCs as well as CD34+ cells (Figure 1B), demonstrating the highest expression of FMNL1 mRNA in stimulated PBMCs and CD19 cells, whereas other populations showed slightly lower expression. In comparison with normal PBMCs, FMNL1 mRNA is overexpressed in malignant cells in more than 60% of CLL samples tested and also in acute B- and T-lymphoblastic leukemia cells (Figure 1C). Furthermore, FMNL1 protein expression correlates largely to the mRNA expression data in healthy and malignant tissues as determined by Western blot analysis (Figure 1D,E). Unstimulated and stimulated PBMCs as well as PBMC subpopulations showed distinct expression of FMNL1 protein. There was no expression of FMNL1 protein in lung fibroblasts and nontransfected 293 HEK cells (Figure 1D) as well as 293 HEK cells transfected with the control vector (Figure S1B). High protein expression was observed in native malignant cells from patients with lymphatic as well as myeloid leukemias and EBV-transformed B cells. Moreover, FMNL1 protein expression has been also detected in lymphoma-derived cell lines (BJ18, DG75), and aberrant protein expression was found in renal cell carcinoma lines (RCC26, RCC53) compared with nontransfected embryonal kidney cells (293 HEK; Figure 1E).
Expression of FMNL1 in normal and malignant tissue. (A) Relative quantitative mRNA expression of FMNL1 in different tissues pooled from different healthy donors was measured using quantitative real-time PCR. The relative quantitative expression compared with skeletal muscle was calculated using the delta-delta Ct method. (B) Relative quantitative mRNA expression of FMNL1 in stimulated PBMCs and PBMC-derived subpopulations is shown. Subpopulations were isolated by negative (CD4, CD8, CD14, CD19) and positive (CD34) magnetic bead depletion. Monocyte-derived dendritic cells (DCs) were isolated by adhesion, cultured in presence of IL-4 and GM-CSF, and matured with IL1β, IL-6, TNF-α, and prostaglandin E2. For activation of PBMCs, cells were incubated for 3 days using IL-2 and OKT3; B-cell activation was induced by incubation with CD40L-transfected NIH3T3 cells for at least 3 weeks. Error bars indicate the standard deviation of different samples tested. (C) Relative quantitative mRNA expression of FMNL1 in CLL samples and tumor cells derived from patients with acute B- and T-ALL as well as AML is shown in comparison with normal PBMCs. (D) FMNL1 protein expression in PBMC-derived cells, lung fibroblasts, and nontransfected 293 HEK cells by Western blot analysis. Total protein (50 μg) was used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). 293 HEK cells transfected with FMNL1 were used as positive control. Probing with antiactin antibody served as loading control. (E) FMNL1 protein expression in native tumor cells and transformed cell lines by Western blot analysis. Total protein of native leukemic cells (50 μg) and total protein of lysed cell lines (100 μg) was used for SDS-PAGE. 293 HEK cells transfected with FMNL1 were used as positive control. Probing with antiactin antibody served as loading control.
Expression of FMNL1 in normal and malignant tissue. (A) Relative quantitative mRNA expression of FMNL1 in different tissues pooled from different healthy donors was measured using quantitative real-time PCR. The relative quantitative expression compared with skeletal muscle was calculated using the delta-delta Ct method. (B) Relative quantitative mRNA expression of FMNL1 in stimulated PBMCs and PBMC-derived subpopulations is shown. Subpopulations were isolated by negative (CD4, CD8, CD14, CD19) and positive (CD34) magnetic bead depletion. Monocyte-derived dendritic cells (DCs) were isolated by adhesion, cultured in presence of IL-4 and GM-CSF, and matured with IL1β, IL-6, TNF-α, and prostaglandin E2. For activation of PBMCs, cells were incubated for 3 days using IL-2 and OKT3; B-cell activation was induced by incubation with CD40L-transfected NIH3T3 cells for at least 3 weeks. Error bars indicate the standard deviation of different samples tested. (C) Relative quantitative mRNA expression of FMNL1 in CLL samples and tumor cells derived from patients with acute B- and T-ALL as well as AML is shown in comparison with normal PBMCs. (D) FMNL1 protein expression in PBMC-derived cells, lung fibroblasts, and nontransfected 293 HEK cells by Western blot analysis. Total protein (50 μg) was used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). 293 HEK cells transfected with FMNL1 were used as positive control. Probing with antiactin antibody served as loading control. (E) FMNL1 protein expression in native tumor cells and transformed cell lines by Western blot analysis. Total protein of native leukemic cells (50 μg) and total protein of lysed cell lines (100 μg) was used for SDS-PAGE. 293 HEK cells transfected with FMNL1 were used as positive control. Probing with antiactin antibody served as loading control.
Isolation of FMNL1-specific T cells
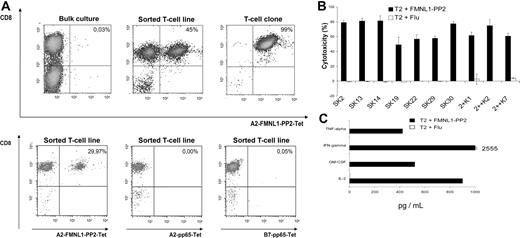
Several prediction algorithms, including binding to HLA-A2, TAP transportation, and proteasomal cleavage, were used for selection of FMNL1-derived peptides (Table S1). Binding to HLA-A2 was investigated by a peptide-competition assay (Figure S2), and 4 peptides were selected for stimulation assays (FMNL1-PP1, FMNL1-PP2, FMNL1-PP6, and FMNL1-PP8). Peptide-pulsed T2 cells were then used for priming and restimulation of T cells. After 2 stimulations, specific T cells were detected by tetramers only at very low numbers (0.01%-0.1%; Figure 2A) and could not be enriched in bulk cultures by additional stimulations (data not shown). However, T cells with specificity toward one peptide, FMNL1-PP2, could be enriched up to 45% by tetramer sorting (Figure 2A) and were subsequently cloned by limiting dilution (Figure 2A). T cells positive for the tetramer A2-FMNL1-PP2 did not react with A2-pp65– or B7-pp65–specific tetramers (Figure 2A). To investigate functional specificity of T-cell lines and clones, we performed a 51Cr-release assay to detect peptide-specific killing. Peptide-specific killing was observed in 10 T-cell clones that have been cloned from the primary sorted cell line showing cytotoxicity against peptide FMNL1-PP2 (Figure 2B) but not against the control peptide Flu loaded on T2 cells. We also detected cytokine secretion of IL-2, GM-CSF, IFN-γ, and TNF-α in culture supernatants of clone SK22 stimulated with T2 cells pulsed with the specific peptide but not T2 cells pulsed with the control peptide Flu (Figure 2C), representative of 9 other clones.
Isolation of allorestricted FMNL1-specific T cells. (A) FMNL1-PP2–specific CD8+ T cells in bulk cultures after 2 stimulations with peptide-pulsed T2 cells, in sorted T-cell lines and in T cells cloned by limiting dilution, were detected by flow cytometry using anti-CD8 monoclonal antibodies and FMNL1-PP2–specific HLA-A2 tetramers. The numbers in the fluorescence-activated cell sorting (FACS) plot represent percentage of cells in that quadrant. As control, staining with HLA-A2 and HLA-B7 pp65-specific tetramers was performed. (B) Peptide recognition of cloned T cells was investigated using T2 cells pulsed with FMNL1-PP2 (■) and T2 cells pulsed with Flu (□) by 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate standard deviation of tested duplicates. (C) Cytokine secretion of the FMNL1-PP2–specific T-cell clone SK22 was investigated in response to T2 cells pulsed with the peptide FMNL1-PP2 (■) and pulsed with the Flu peptide (□) at an effector-target ratio of 1:5. Culture supernatants were harvested after 24 hours and assessed by BioPlex multicytokine analysis.
Isolation of allorestricted FMNL1-specific T cells. (A) FMNL1-PP2–specific CD8+ T cells in bulk cultures after 2 stimulations with peptide-pulsed T2 cells, in sorted T-cell lines and in T cells cloned by limiting dilution, were detected by flow cytometry using anti-CD8 monoclonal antibodies and FMNL1-PP2–specific HLA-A2 tetramers. The numbers in the fluorescence-activated cell sorting (FACS) plot represent percentage of cells in that quadrant. As control, staining with HLA-A2 and HLA-B7 pp65-specific tetramers was performed. (B) Peptide recognition of cloned T cells was investigated using T2 cells pulsed with FMNL1-PP2 (■) and T2 cells pulsed with Flu (□) by 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate standard deviation of tested duplicates. (C) Cytokine secretion of the FMNL1-PP2–specific T-cell clone SK22 was investigated in response to T2 cells pulsed with the peptide FMNL1-PP2 (■) and pulsed with the Flu peptide (□) at an effector-target ratio of 1:5. Culture supernatants were harvested after 24 hours and assessed by BioPlex multicytokine analysis.
TCR analysis of FMNL1-PP2–specific T-cell clones
To test the clonal origin of the different T-cell clones, we performed a TCR repertoire of all peptide-specific clones. Thirty-four single V alpha gene segment–specific primers were used for the α-repertoire revealing only one single in-frame Vα14 chain (Figure S3A) for all clones tested (n = 10). Sequencing of gel-purified bands revealed one sequence identical in all clones tested. For the TCR β-chain repertoire, 2 clones were tested using 37 single V-beta segment–specific primers (Figure S3B); for the other clones, degenerative primers were applied (data not shown). Sequencing of all gel-purified bands revealed only one single in-frame sequence coding for a specific Vβ14 chain, identical in all clones tested (n = 10). We additionally stained T cells with a Vβ14-specific antibody revealing a single Vβ14+ population (99%) (Figure S3C). These data demonstrate that all clones derived from the same clonal T cell.
Functional activity and avidity of the isolated FMNL1-PP2–specific T-cell clone
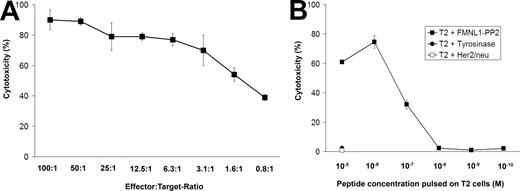
We next investigated the functional activity of the isolated clone and detected activity of the T-cell clone down to an effector-target ratio of 0.8:1 (Figure 3A). We examined the functional avidity of the T-cell clone by titrating the peptide concentration on target cells necessary for peptide-specific killing. Half-maximal lysis of peptide-pulsed T2 cells was observed with a peptide concentration of 0.1 μM (Figure 3B) representative of all clones tested (n = 8) and corresponding to an intermediate functional avidity.
The FMNL1-PP2–specific T-cell clone shows high functional activity and an intermediate functional avidity. (A) T2 cells pulsed with FMNL1-PP2 (10 μM) were used as targets for the FMNL1-PP2–specific T-cell clone at different effector-target ratios in a 51Cr-release assay. Error bars indicate the standard deviation of tested duplicates. (B) T2 cells were pulsed with FMNL1-PP2 at different concentrations and used as targets for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Tyrosinase- and Her2/neu-derived peptides were used as negative controls. Error bars indicate the standard deviation of tested duplicates.
The FMNL1-PP2–specific T-cell clone shows high functional activity and an intermediate functional avidity. (A) T2 cells pulsed with FMNL1-PP2 (10 μM) were used as targets for the FMNL1-PP2–specific T-cell clone at different effector-target ratios in a 51Cr-release assay. Error bars indicate the standard deviation of tested duplicates. (B) T2 cells were pulsed with FMNL1-PP2 at different concentrations and used as targets for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Tyrosinase- and Her2/neu-derived peptides were used as negative controls. Error bars indicate the standard deviation of tested duplicates.
Detection of natural target cells by the FMNL1-PP2-specific T-cell clone
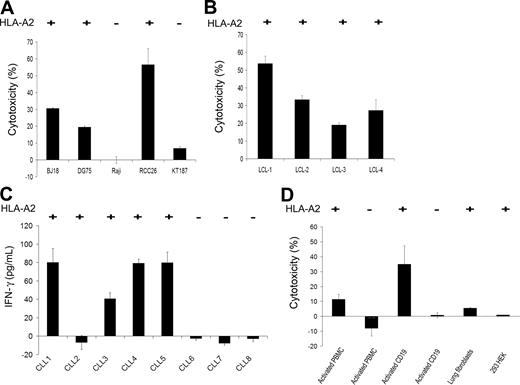
To investigate whether the FMNL1-PP2–specific T-cell clone also detects natural targets, different tumor targets, including lymphoma cell lines, solid tumor cell lines, as well as native leukemia cells, EBV-transformed B cells, and nontransformed cells were tested. The FMNL1-PP2–specific T-cell clone specifically lysed HLA-A*0201+ targets such as EBV-negative lymphoma cell lines, BJ18 and DG75, and HLA-A*0201+ renal cell carcinoma cell line RCC26 (Figure 4A). In contrast, the HLA-A2− lymphoma cell line Raji and the HLA-A2− renal cell carcinoma cell line KT187 were not recognized (Figure 4A). In addition, HLA-A*0201+ EBV-transformed cell lines were lysed (Figure 4B). Moreover, 4 of 5 CD40L-activated HLA-A2+ CLL cell samples were recognized, as seen by IFN-γ release, whereas there was no IFN-γ release in response to 3 of 3 CD40L-activated HLA-A2− CLL cell samples (Figure 4C).
Specific recognition of natural targets by the FMNL1-PP2–specific T-cell clone. The FMNL1-PP2–specific T-cell clone was tested against the HLA-A2+ lymphoma cell lines BJ18 and DG75; the HLA-A2− lymphoma cell line Raji; the HLA-A2+ renal cell carcinoma cell line RCC26; the HLA-A2− renal cell carcinoma cell line KT187 (A); and HLA-A2+ EBV-transformed cell lines (B), in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of at least 3 experiments. (C) IFN-γ release was investigated by ELISA to test the FMNL1-PP2–specific T-cell clone against CD40L-activated CLL cells (HLA-A2+ and HLA-A2−) at an effector-target ratio of 1:2. Error bars indicate the standard deviation of tested triplicates. (D) PBMCs (HLA-A2+ and HLA-A2−) activated with IL-2 and OKT3, CD19+ B cells (HLA-A2+ and HLA-A2−) stimulated with CD40L, as well as HLA-A2+ lung fibroblasts and HLA-A2+ embryonal kidney cells (293 HEK) were used as target cells for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 5 experiments.
Specific recognition of natural targets by the FMNL1-PP2–specific T-cell clone. The FMNL1-PP2–specific T-cell clone was tested against the HLA-A2+ lymphoma cell lines BJ18 and DG75; the HLA-A2− lymphoma cell line Raji; the HLA-A2+ renal cell carcinoma cell line RCC26; the HLA-A2− renal cell carcinoma cell line KT187 (A); and HLA-A2+ EBV-transformed cell lines (B), in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of at least 3 experiments. (C) IFN-γ release was investigated by ELISA to test the FMNL1-PP2–specific T-cell clone against CD40L-activated CLL cells (HLA-A2+ and HLA-A2−) at an effector-target ratio of 1:2. Error bars indicate the standard deviation of tested triplicates. (D) PBMCs (HLA-A2+ and HLA-A2−) activated with IL-2 and OKT3, CD19+ B cells (HLA-A2+ and HLA-A2−) stimulated with CD40L, as well as HLA-A2+ lung fibroblasts and HLA-A2+ embryonal kidney cells (293 HEK) were used as target cells for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 5 experiments.
In healthy tissue, we observed only minor killing of activated HLA-A2+–activated PBMCs, whereas CD40L-activated HLA-A2+ CD19+ cells were well recognized (Figure 4D). HLA-A2− PBMCs and HLA-A2− CD19+ cells were not recognized. Similar results were obtained using PBMC-derived cells from different HLA-A2+ and HLA-A2− donors. HLA-A2+ cultured lung fibroblasts and HLA-A2+ embryonal kidney cells (293 HEK) were not targeted (Figure 4D).
Specificity, TCR dependency, and cross-reactivity of the FMNL1-PP2–specific T-cell clone
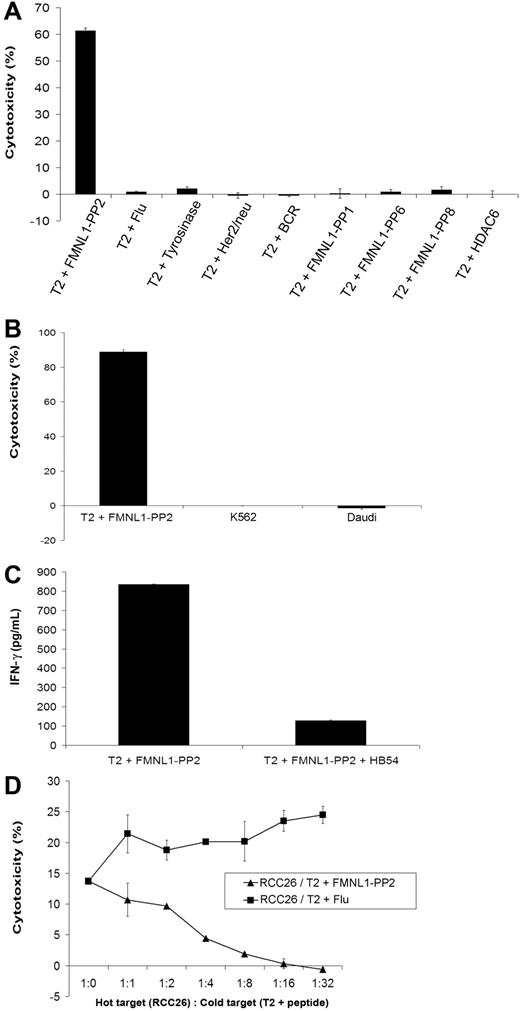
To evaluate the specificity of the FMNL1-PP2–specific T-cell clone more in detail, we used T2 cells as target cells pulsed with a number of different peptides. Cytotoxicity of peptide-pulsed T2 cells was detected only against FMNL1-PP2–pulsed T2 cells. In contrast, T2 cells pulsed with a set of irrelevant peptides including the highly homologous peptide RLAERMTTR derived from histone-deacetylase 6 (HDAC6) were not recognized (Figure 5A). To further exclude NK- or non-MHC–restricted killing, K562 and Daudi cells were used as targets in a 51Cr-cytotoxicity assay, neither of which was recognized by FMNL1-PP2–specific T-cell clone (Figure 5B). Specific killing of peptide-pulsed T2 cells could be inhibited by adding a monoclonal blocking antibody against HLA-A2 (Figure 5C). In addition, recognition of natural targets could be also inhibited by addition of T2 cells pulsed with FMNL1-PP2 as cold targets but not pulsed with Flu (Figure 5D).
The FMNL1-PP2–specific T-cell clone shows peptide specificity and TCR dependency. (A) T2 cells were pulsed with a set of different peptides at 10 μM and investigated as targets for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 2 experiments. (B) K562 and Daudi cells were used as target cells for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 2 experiments. (C) Recognition of T2 cells pulsed with FMNL1-PP2 was inhibited by the monoclonal anti–HLA-A2 antibody (HB54) investigated by ELISA (IFN-γ release). Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 2 experiments. (D) The cytotoxicity of the isolated T-cell clone against RCC26 could be inhibited by addition of cold T2 cells pulsed with FMNL1-PP2 but not pulsed with Flu. Error bars indicate the standard deviation of tested duplicates.
The FMNL1-PP2–specific T-cell clone shows peptide specificity and TCR dependency. (A) T2 cells were pulsed with a set of different peptides at 10 μM and investigated as targets for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 2 experiments. (B) K562 and Daudi cells were used as target cells for the FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 2 experiments. (C) Recognition of T2 cells pulsed with FMNL1-PP2 was inhibited by the monoclonal anti–HLA-A2 antibody (HB54) investigated by ELISA (IFN-γ release). Error bars indicate the standard deviation of tested duplicates. Results shown are representative of 2 experiments. (D) The cytotoxicity of the isolated T-cell clone against RCC26 could be inhibited by addition of cold T2 cells pulsed with FMNL1-PP2 but not pulsed with Flu. Error bars indicate the standard deviation of tested duplicates.
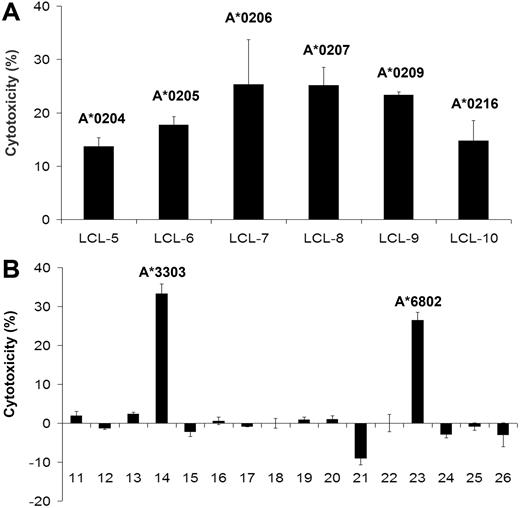
As alloreactive T cells may recognize different epitopes,43 we tested 26 different EBV-transformed cell lines with different HLA allotypes. Several HLA-A2 subtypes were recognized (Figure 6A), whereas HLA-A2–negative samples (Table S2) were only recognized in 2 of 16 samples (Figure 6B). One of them (LCL-23) had the allotype HLA-A*6802 also belonging to the HLA-A2 supertype family.44 The other restriction element was HLA-A*3303 (LCL-14), and restriction to HLA-A*3303 was confirmed using C1R cells transfected with HLA-A*3303 (Figure S4).
Cross-reactivity of the isolated FMNL1-PP2–specific T-cell clone. LCLs with different HLA-A2 subtypes (A) and HLA-A2− LCLs (B) were used as target cells for FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Corresponding results were obtained by IFN-γ ELISA.
Cross-reactivity of the isolated FMNL1-PP2–specific T-cell clone. LCLs with different HLA-A2 subtypes (A) and HLA-A2− LCLs (B) were used as target cells for FMNL1-PP2–specific T-cell clone in a 51Cr-release assay at an effector-target ratio of 7.5:1. Error bars indicate the standard deviation of tested duplicates. Corresponding results were obtained by IFN-γ ELISA.
Discussion
Substantial numbers of TAAs recognized by T cells have been identified and described.45 However, most of these antigens are expressed in selected tissues, and universal applicability of any antigen has not been proved so far. Moreover, it is likely that a variety of antigens needs to be targeted to reduce the risk of tumor escape. Thus, the identification of novel target antigens is of fundamental importance for the development of effective immunotherapies for various types of malignancies.
FMNL1 is a formin-related protein in leukocytes and belongs to the family of diaphanous-related formins (DRFs).46 Most recently, FMNL1 has been shown to play a role in polarization of T cells.20 We demonstrate expression of FMNL1 also in other PBMC-derived cells such as healthy B cells, monocytes, and dendritic cells, as well as overexpression in leukemic cells of different origin and aberrant expression in tumor cell lines. Whereas the function of FMNL1 needs to be further elucidated, formins have been suggested to represent a family of attractive drug targets for the treatment of actin-dependent processes such as inflammation, metastasis, and invasive diseases.46
As most tumor-associated antigens are self-antigens and therefore are not recognized by autologous T cells, we used the allorestricted approach to generate T cells with specificity for epitopes derived from FMNL1.9 Such allorestricted peptide-specific T cells might display high avidity toward MHC-presented TAAs, because they have not been negatively selected in the thymus.47 In this study, we isolated a peptide-specific allorestricted T-cell clone recognizing the peptide FMNL1-PP2 derived from FMNL1. The isolated T-cell clone showed peptide-specific functional activity at very low effector-target ratios. Functional avidity for peptide-pulsed T2 cells was intermediate (half-maximal lysis of 100 nM) which is in common with the functional avidity observed for many foreign antigens.43 However, the reduced capacity of FMNL1-PP2 to bind to HLA-A2 that we repeatedly observed in the peptide-competition assay in comparison with other tested peptides may have an impact on the functional avidity of this clone. Nevertheless, the isolated T-cell clone showed potent cytotoxic activity against cell lines derived from several malignant tissues as well as primary tumor material, suggesting that the selected peptide is not only a naturally presented epitope but also an attractive tumor-associated target antigen. T cells specifically killed malignant cell lines derived from lymphomas and renal cell carcinoma aberrantly expressing FMNL1. The T-cell clone also specifically recognized FMNL1-expressing EBV-transformed B cells and tumor cells from CLL patients after stimulation with CD40L. In healthy donors, activated B cells were best recognized, whereas activated PBMCs were killed to a lesser extent. No cytotoxic activity was detected against lung fibroblasts and embryonal kidney cells not expressing FMNL1. The isolated T-cell clone showed high peptide specificity as it did not recognize a number of peptides including the highly homologous peptide RLAERMTTR derived from HDAC6. HLA-A2 restriction was confirmed using a blocking antibody against HLA-A2 on peptide-pulsed T2 cells. Investigating cross-reactivity, we observed a broader recognition of LCLs with different subtypes of HLA-A2. This is in common with previous reports demonstrating that a majority of peptides binding to the subtype of HLA-A*0201 may cross-react with other alleles belonging to the A2-like supertype and that HLA-A*0201–restricted CTLs are able to recognize TAAs in the context of different HLA-A2 subtypes.44,48-50 However, FMNL1-PP2–specific T cells also cross-reacted with 2 of 16 HLA-A2− LCLs. One of them is likely to be HLA-A*6802 as one other HLA-A*6802–positive sample has been recognized (data not shown) and this HLA-A allele also belongs to the A2 supertype family.44 The other recognized HLA-A2− LCL was positive for HLA-A*3303. HLA-A*3303 could be confirmed as a highly specific restriction element because the T cells specifically lysed HLA-A*3303–transfected C1R cells but showed no reactivity against other HLA-A alleles belonging to the A3-like supertype such as HLA-A03, HLA-A31, or the genotype HLA-A*3301 expressed on other tested HLA-A2− LCLs (Table S2). The capacity of alloreactive TCRs to bind to different MHC-peptide complexes has been repeatedly shown. Specificity for the same peptide presented by different MHC alleles as well as for different peptides presented by the same or different MHC alleles has been described.51-56 It is intensively discussed if this capacity of alloreactive T cells represents a state of degeneracy or polyspecificity.43,57,58 However, it has been shown recently that the mechanism of cross-reactivity can occur independently of molecular mimicry.43,59 Moreover, cross-reactivity has been also observed for high-affinity TCRs and therefore might be a general property of T-cell recognition.60-62 This is certainly an important issue for the development of immunotherapies using allorestricted peptide-specific T cells. Although FMNL1-PP2 peptide–specific T cells are highly specific and cross-reactivity has been observed only exceptionally, further investigations in vitro and in vivo are necessary to clarify if other peptides and MHC molecules might be recognized by FMNL1-PP2–specific T cells. Knowledge of the TCR chains responsible for peptide-specific killing will allow TCR transfer studies and extensive investigation of the specific TCR using peptide libraries and multiple MHC alleles. For clinical use, patients would need to be carefully selected prior to therapeutic application.
In summary, we identified a novel antigenic peptide (FMNL1-PP2) derived from FMNL1 with high potential for the use as a target antigen for immunotherapeutic purposes. HLA-A2–allorestricted T cells with specificity for this peptide have been isolated and characterized. These T cells preferentially killed EBV-transformed B cells as well as tumor cells derived from lymphoma and renal cell carcinoma, suggesting that the specific TCR is a highly interesting tool for the development of adoptive immunotherapies in EBV-associated lymphoproliferative diseases, lymphoma, and renal cell carcinoma. We observed only minor killing against nontransformed cells, with the exception of healthy B cells that were better recognized. Future studies including additional testing of peptide libraries and different MHC alleles, mutational TCR analyses, and in vivo experiments evaluating the antitumor and autoimmune potential of transgenic TCR-expressing T cells, as well as functional studies of FMNL1 to clarify its functional role in healthy and malignant tissue, will provide the basis for the design of clinical studies to assess the role of this epitope and the corresponding TCR in immunotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Life Science–Stiftung and Deutsche Forschungsgemeinschaft (DFG) (KR-2305/2–1) to A.M.K. as well as DFG-SFB/TR36 (projects A1, A7, A8, and B3).
We thank D. J. Schendel, R. Mocikat, and T. Blankenstein for critical reading of the paper; S. Stevanovic, J. Mautner, R. Mocikat, and T. Wölfel for providing cell lines and T cells; A. Moosmann for providing the peptide IPS; and C. S. Falk and B. Mosetter for introduction into BioPlex analysis.
Authorship
Contribution: I.G.S. performed research and analyzed data; D.H.B. contributed new agents; E.E. performed research; E.K. contributed new agents; S.M., C.H., C.K., J.W.E., B.F., and E.N. contributed analytical tools; C.S., C.B., A.B., and H.-J.K. provided clinical data and samples; and A.M.K. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: I.G.S., D.H.B., and A.M.K. are holding a patent application about the peptide sequence of FMNL1-PP2 and the sequences of the FMNL1-PP2–specific TCR. All other authors declare no competing financial interests.
Correspondence: Angela M. Krackhardt, Institute of Molecular Immunology, GSF–National Research Center for Environment and Health, Marchioninistr. 25, 81377 Munich, Germany; e-mail: angela.krackhardt@gsf.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal