Abstract
Junctional adhesion molecule-A (JAM-A/JAM-1/F11R) is a cell adhesion molecule expressed in epithelial and endothelial cells, and also hematopoietic cells, such as leukocytes, platelets, and erythrocytes. Here, we show that JAM-A is expressed at a high level in the enriched hematopoietic stem cell (HSC) fraction; that is, CD34+c-Kit+ cells in embryonic day 11.5 (E11.5) aorta-gonod-mesonephros (AGM) and E11.5 fetal liver (FL), as well as c-Kit+Sca-1+Lineage− (KSL) cells in E14.5 FL, E18.5FL, and adult bone marrow (BM). Although the percentage of JAM-A+ cells in those tissues decreases during development, the expression in the HSC fraction is maintained throughout life. Colony-forming assays reveal that multilineage colony-forming activity in JAM-A+ cells is higher than that in JAM-A− cells in the enriched HSC fraction in all of those tissues. Transplantation assays show that long-term reconstituting HSC (LTR-HSC) activity is exclusively in the JAM-A+ population and is highly enriched in the JAM-A+ cells sorted directly from whole BM cells by anti–JAM-A antibody alone. Together, these results indicate that JAM-A is expressed on hematopoietic precursors in various hematopoietic tissues and is an excellent marker to isolate LTR-HSCs.
Introduction
Expression profiles of cell-surface molecules have been used to characterize lineages and differentiation stages of hematopoietic cells. A pioneering work by Osawa et al has shown that a single CD34− cell in the c-Kit+Sca-1+Lineage− (KSL) population is able to reconstitute the entire hematopoietic system for long term, establishing that CD34− KSL cells are long-term repopulating hematopoietic stem cells (LTR-HSCs) in adult bone marrow (BM).1 More recently, Matsuzaki et al. reported that a tip of the Hoechst 33342 side population2,3 in CD34− KSL cells is the most purified LTR-HSC.4 However, as these purification procedures require extensive cell sorting by a combination of multiple antibodies,1-6 it is desirable to find a novel HSC marker that can be used to purify HSC more efficiently.7-10 In the process of searching for the antigens expressed in murine fetal liver (FL) cells, we found that the junctional adhesion molecule A, JAM-A, also known as JAM-1 and F11R,11,12 is expressed in some FL cells (data not shown). JAM-A together with JAM-B and JAM-C forms a family of cell adhesion molecules with two immunoglobulin-like domains and is localized at a tight junction of epithelial and endothelial cells and also hematopoietic cells, such as leukocytes, platelets and erythrocytes.11-19
In this paper, we show that JAM-A is highly expressed in the enriched HSC fraction in the aorta-gonod-mesonephros (AGM), FL, and BM. JAM-A+ cells exhibit higher progenitor activity than JAM-A− cells, and transplantation assays reveal that the LTR-HSC activity is present in JAM-A+ cells but not in JAM-A− cells, and that LTR-HSCs in BM are highly enriched in JAM-A+ cells. These results indicate that JAM-A is expressed on HSCs, and anti–JAM-A antibody can be used to enrich LTR-HSC from BM cells efficiently.
Methods
Mice
C57BL/6(B6-Ly5.2) mice of 8 to 12 weeks of age were purchased from CLEA Japan (Tokyo, Japan). C57BL/6 mice congenic for the Ly5 locus (B6-Ly5.1) of 8 to 12 weeks of age were purchased from Sankyo-Lab Service (Tsukuba, Japan). Mice were kept under specific pathogen-free conditions in our animal facility. The day of appearance of vaginal plug was designated as day 0.5 of gestation. To obtain B6-F1 mice, B6-Ly5.1 male and B6-Ly5.2 female mice were mated. All experiments using mice received approval from the University of Tokyo Administrative Panel for Animal Care.
Isolation of cells and depletion of lineage-positive cells
AGM, FL, and BM cells were prepared as described previously.20-23 CD4, CD8, B220, Ter119, Mac-1, and Gr-1 were used as lineage markers for BM cells. Since it was shown that Mac-1 is a hematopoietic stem-cell marker at the fetal stage, Mac-1 was excluded from the lineage markers in the FL cells.24 Depletion of lineage marker–positive cells was performed as described previously.25 In brief, FL and BM cells were incubated at 4°C for 30 minutes in a cocktail of H129.19 (CD4), 53-6.7 (CD8a), RA-3-6B2 (B220), Ly-76 (Ter-119), and RB6-8C5 (Gr-1) antibodies (PharMingen, San Diego, CA). M1/70 (Mac-1) antibody was also added to BM cells.24 Cells (250 × 106 cells/mL) were incubated with M-450 sheep anti-rat IgG–conjugated immunomagnetic beads (Dynal, Oslo, Norway) at 4 beads per target cell according to the manufacturer's recommendations. Beads were removed with a magnetic particle concentrator (MPC-1; Dynal), and unattached cells were collected.
FACS analysis and cell sorting
KSL cells were prepared by sorting Sca1+c-Kit+ cells from lineage-depleted FL or BM cells as described above. CD34+c-Kit+ cells in embryonic day 11.5 (E11.5) AGM and E11.5 FL were considered the enriched HSC fraction,26 and lineage depletion was not performed because of the low abundance of lineage-positive cells.
After staining with a monoclonal antibody according to the manufacturer's protocol, cells were analyzed by fluorescence-activated cell sorting (FACS) with the FACSCalibur system or sorted by the FACS-Vantage system (Becton Dickinson Biosciences, San Jose, CA) for further analysis. E13-161.7 (FITC-Sca-1), RAM34 (FITC-CD34), 2B8 (PE–c-Kit), and 7-ADD antibodies were obtained from BD Biosciences PharMingen (San Diego, CA); 9D1 (APC-CD150) and eBio244F4 (PE-CD48) were from eBioscience (San Diego, CA); HM48-1 (FITC-CD48) was from Abcam (Cambridge, MA); and streptavidin-allophycocyanin (SA-APC) was from Molecular Probes (Eugene, OR). The anti–JAM-A monoclonal antibody was generated by immunization of a rat with E14.5 fetal liver cells. Among the panel of rat monoclonal antibodies, we found one antibody that recognized megakaryocytes.27 We identified the antigen as JAM-A by expression cloning of cDNA, as described previously.27 The specificity of the antibody was confirmed by COS7 cells transfected with JAM-A cDNA by flow cytometry. The monoclonal antibody was biotinylated and was used for flow cytometry.
Colony formation assays
To evaluate hematopoietic potentials, sorted cells were cultured in semisolid medium (MethoCult GF M3434; StemCell Technologies, Vancouver, BC) according to the manufacturer's protocols, and colonies were scored after 10 to 14 days.
Histologic analysis
After sorting of JAM-A+ and JAM-A− cells, cells were collected onto slides by Cytospin at 106g for 3 minutes (Cytospin 3; Thermo Shandon, Pittsburgh, PA). Cells were fixed in methanol for 2 minutes and stained with May-Grunwald Giemsa stains. The percentages of progenitors, mature myeloid (eg, neutrophil and monocyte), lymphoid, and erythroid/megakaryocyte lineage cells were evaluated by examining stained cells under light microscopy.
Transplantation
Test cells (25, 100, 300, 400, and 1000 cells) from a B6-Ly5.1 mouse were mixed with unfractionated adult BM cells (2 × 105) from a B6-F1 mouse and intravenously injected into the retro-orbital plexus of etherized adult recipients (B6-Ly5.2) that had been lethally irradiated at 10 Gy from a 137Cs source. For transplantation assays, 8- to 12-week-old mice were used as test, competitor, and donor. Peripheral blood (PB) samples were collected from the recipients after transplantation. After lysis of erythrocytes, the remaining cells were stained with biotinylated anti-Ly5.2, FITC–anti-Ly5.1, a mixture of PE–anti-Mac-1 and PE–anti-Gr-1, a mixture of PE–anti-CD4 and PE–anti-CD8, or PE–anti-B220, and followed by addition of SA-APC, and were analyzed with FACSCalibur. More than 1% of donor cell contribution in recipient PB were considered LTR-HSCs. Donor contribution was calculated as follows: The percentage of donor chimerism = (100 × %Ly-5.1 cells)/(%Ly-5.1 cells + %F1 cells).
CRU
The limiting dilution assay for competitive repopulating cells (CRUs) has been described previously.28 Cell number ranged from 25 to 300 cells in JAM-A+ KSL cells and from 25 to 400 cells in KSL cells. Hematopoietic repopulation was evaluated by taking PB at 8 weeks after transplantation. After lysis of erythrocytes, donor contribution was calculated. Donor contribution less than 1% was considered negative. The frequency of CRUs was calculated by applying Poisson statistics to the proportion of negative recipients at different dilution with using L-Calc software (StemCell Technologies).
Statistics
Data are presented as the means plus or minus SD, unless otherwise stated. Statistical significance was assessed by means of a Student t test. A P value less than .05 was considered statistically significant.
Results
Expression of JAM-A in hematopoietic tissues
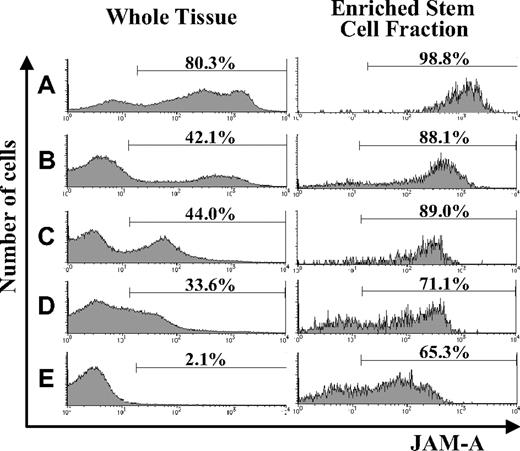
Using flow cytometry, we examined the expression of JAM-A in hematopoietic tissues during development and found that it was expressed in all of the hematopoietic tissues we examined. JAM-A was highly expressed in AGM and FL; however, the percentage of JAM-A+ cells was reduced dramatically in adult BM (Figure 1A-E). JAM-A was expressed not only on hematopoietic cells but also on nonhematopoietic cells in AGM and FL (eg, endothelial cells in AGM and hepatocytes in FL). Hematopoietic cells are a major cell population in BM, and the percentage of JAM-A+ cells is not significantly influenced by nonhematopoietic cells in BM. Furthermore, immature hematopoietic cells with JAM-A decreased along with development.
Expression of JAM-A in AGM, FL, and adult BM. Expression of JAM-A in whole tissues (left) and enriched stem cell fractions (right) in E11.5 AGM (A), E11.5 FL (B), E14.5 FL (C), E18.5 FL (D), and adult (8-12 weeks) BM (E). Representative data from more than 3 independent experiments are shown. Numbers on the graphs are percentages of JAM-A+ cells in whole tissue and on stem cell fractions.
Expression of JAM-A in AGM, FL, and adult BM. Expression of JAM-A in whole tissues (left) and enriched stem cell fractions (right) in E11.5 AGM (A), E11.5 FL (B), E14.5 FL (C), E18.5 FL (D), and adult (8-12 weeks) BM (E). Representative data from more than 3 independent experiments are shown. Numbers on the graphs are percentages of JAM-A+ cells in whole tissue and on stem cell fractions.
We then examined the expression of JAM-A in the enriched stem-cell fractions. It is known that HSCs are highly enriched in CD34+c-Kit+ cells in early embryonic hematopoietic tissues, such as E11.5 AGM and FL,26 whereas HSCs are in KSL cells from FL after E14.5 and adult BM. Most of the CD34+c-Kit+ cells in E11.5 AGM and E11.5 FL expressed JAM-A (Figure 1A,B), and most of the KSL cells in the E14.5 FL, E18.5FL, and adult BM cells were also JAM-A+ (Figure 1C-E). These results indicate that a majority of the enriched stem-cell fractions express JAM-A.
CD34+c-Kit+ and KSL cells are enriched in JAM-A+ cells
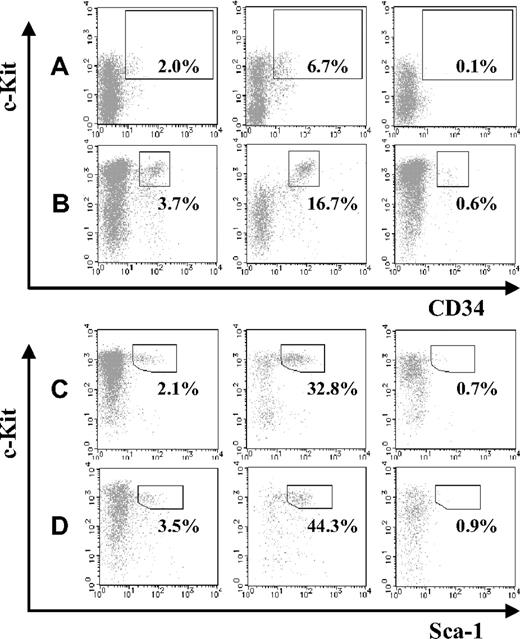
Next, we compared the abundance of HSCs in JAM-A+ and JAM-A− cells in the AGM and FL. JAM-A+ and JAM-A− cells were isolated from E11.5 AGM, E11.5 FL, E14.5 FL, and E18.5 FL cells using anti–JAM-A antibody, and the HSC fraction in each population was compared. CD34+c-Kit+ cells were significantly enriched in the JAM-A+ population of E11.5 AGM and FL as well as KSL cells in E14.5 FL and E18.5 FL (Figure 2A-D). These findings suggest that HSCs are highly enriched in JAM-A+ cells in all of those hematopoietic tissues.
Abundance of stem cells in JAM-A+ and JAM-A− fractions. CD34+c-Kit+ cells in whole cells (left), JAM-A+ cells (middle), and JAM-A− cells (right) were examined by flow cytometry. Enclosures represent percentages of enriched stem-cell fractions that are CD34+c-Kit+ cells in E11.5 AGM (A), E11.5 FL (B), Sca1+ c-Kit+ Lin− cells in E14.5 FL (C), and E18.5 FL (D). Representative data from more than 3 independent experiments are shown. Since these data are dot-plot, there seem to be no CD34+c-Kit− cells. However, the percentage of CD34+c-Kit− cells were 0.89% (± 0.26%) in whole 11.5 AGM and 0.58% (± 0.12%) in whole 11.5 FL, respectively.
Abundance of stem cells in JAM-A+ and JAM-A− fractions. CD34+c-Kit+ cells in whole cells (left), JAM-A+ cells (middle), and JAM-A− cells (right) were examined by flow cytometry. Enclosures represent percentages of enriched stem-cell fractions that are CD34+c-Kit+ cells in E11.5 AGM (A), E11.5 FL (B), Sca1+ c-Kit+ Lin− cells in E14.5 FL (C), and E18.5 FL (D). Representative data from more than 3 independent experiments are shown. Since these data are dot-plot, there seem to be no CD34+c-Kit− cells. However, the percentage of CD34+c-Kit− cells were 0.89% (± 0.26%) in whole 11.5 AGM and 0.58% (± 0.12%) in whole 11.5 FL, respectively.
Most of KSL cells in BM are JAM-A+
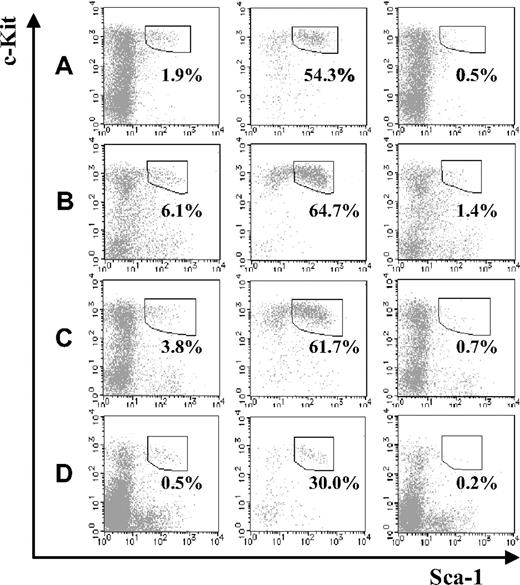
Next, we compared the fraction of KSL cells in JAM-A+ and JAM-A− cells from BM. Compared with the fetal stage, KSL cells in the JAMA− cell population were dramatically reduced in the BM, regardless of age (ie, from a 1-week-old to a 36- to 40-week-old mouse; Figure 3A-C). Moreover, even without depletion of lineage marker–positive cells, the percentage of JAM-A+ in the c-Kit+Sca-1+ population of BM was much higher than JAM-A−c-Kit+Sca-1+ cells (ie, 29.99% ± 8.72% vs 0.15% ± 0.08%; P < .001; Figure 3D). While it was shown that JAM-A is expressed in some leukocytes, platelets, and erythrocytes,12,14-19 JAM-A+ cells in adult BM were only 2.1%. Thus, JAM-A+ cells were enriched in KSL cells, suggesting that JAM-A is an excellent marker for HSCs.
Stem-cell fractions in JAM-A+ and JAM-A− cells in BM. Stem-cell fractions (Sca-1+c-Kit+) in whole lineage-negative cells (left), lineage-negative JAM-A+ cells (middle), and lineage-negative JAM-A− cells (right) in 1-week-old BM (A), 8- to 12-week-old BM (B), and 36- to 40-week-old BM (C). Each enclosure represents the percentage of enriched stem-cell fraction that is Sca-1+c-Kit+ in lineage-negative cells (A-C). Sca-1+c-Kit+ in whole cells (left), JAM-A+ cells (middle), and JAM-A− cells (right) in 8- to 12-week-old BM. Enclosures represent the percentages of Sca-1+c-Kit+ in cells that are without lineage depletion procedure (D). Representative data from more than 3 independent experiments are shown.
Stem-cell fractions in JAM-A+ and JAM-A− cells in BM. Stem-cell fractions (Sca-1+c-Kit+) in whole lineage-negative cells (left), lineage-negative JAM-A+ cells (middle), and lineage-negative JAM-A− cells (right) in 1-week-old BM (A), 8- to 12-week-old BM (B), and 36- to 40-week-old BM (C). Each enclosure represents the percentage of enriched stem-cell fraction that is Sca-1+c-Kit+ in lineage-negative cells (A-C). Sca-1+c-Kit+ in whole cells (left), JAM-A+ cells (middle), and JAM-A− cells (right) in 8- to 12-week-old BM. Enclosures represent the percentages of Sca-1+c-Kit+ in cells that are without lineage depletion procedure (D). Representative data from more than 3 independent experiments are shown.
May-Giemsa staining of JAM-A+ and JAM-A− cells
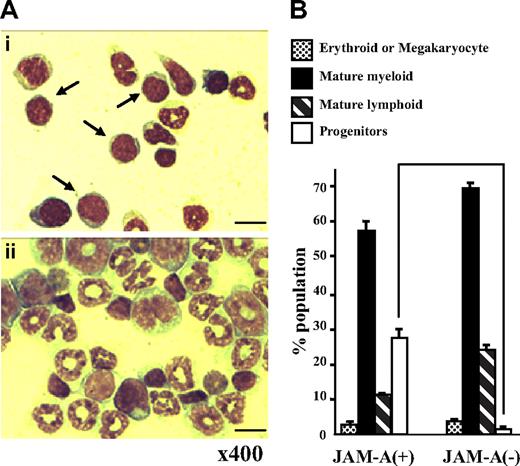
JAM-A+ and JAM-A− cells were sorted from the whole adult BM with anti–JAM-A antibody alone and were stained with May-Giemsa solution. Stained cells were classified into 4 phenotypically distinct groups: progenitors (more immature than myelocytes), mature myeloid, lymphoid, and erythroid/megakaryocyte lineage cells. Although both JAM-A+ and JAM-A− cells were heterogeneous, the progenitor population in JAM-A+ cells was larger than that in JAM-A− cells (27.75% ± 0.25% vs 1.50% ± 1.25%, P < .008; Figure 4A,B). In contrast, there was not much difference between JAM-A+ and JAM-A− cells in the erythroid (P > .05), neutrophil (P > .05), and lymphoid lineages (P > .009). These findings indicate that hematopoietic progenitor cells can be enriched from whole BM cells by anti–JAM-A antibody alone.
May-Giemsa staining of cells sorted with anti–JAM-A antibody. (A) May-Giemsa staining of JAM-A+ (i) and JAM-A− (ii) cells from BM of a 9-week-old mouse. Samples were examined using a Nikon Optiphot-2 microscope with a Nikon E plan 40×/0.65 numeric aperture objective and 10× eyepiece lens (Nikon, Tokyo, Japan). Images were acquired with an Evolution MP color camera (Media Cybernetics, Silver Spring, MD) and Image Pro Plus software (v4.5, Media Cybernetics). Arrows show progenitors. Scale bar = 10 μm. (B) The percentage of 4 phenotypically distinct groups in JAM-A+ and JAM-A− cells from BM of a 9-week-old mouse: progenitors (immature blastic cells), mature myeloid, mature lymphoid and erythroid, or megakaryocyte lineage cells. Results from 2 independent experiments are shown. Error bars represent SD.
May-Giemsa staining of cells sorted with anti–JAM-A antibody. (A) May-Giemsa staining of JAM-A+ (i) and JAM-A− (ii) cells from BM of a 9-week-old mouse. Samples were examined using a Nikon Optiphot-2 microscope with a Nikon E plan 40×/0.65 numeric aperture objective and 10× eyepiece lens (Nikon, Tokyo, Japan). Images were acquired with an Evolution MP color camera (Media Cybernetics, Silver Spring, MD) and Image Pro Plus software (v4.5, Media Cybernetics). Arrows show progenitors. Scale bar = 10 μm. (B) The percentage of 4 phenotypically distinct groups in JAM-A+ and JAM-A− cells from BM of a 9-week-old mouse: progenitors (immature blastic cells), mature myeloid, mature lymphoid and erythroid, or megakaryocyte lineage cells. Results from 2 independent experiments are shown. Error bars represent SD.
Colony-forming activity of JAM-A+ cells
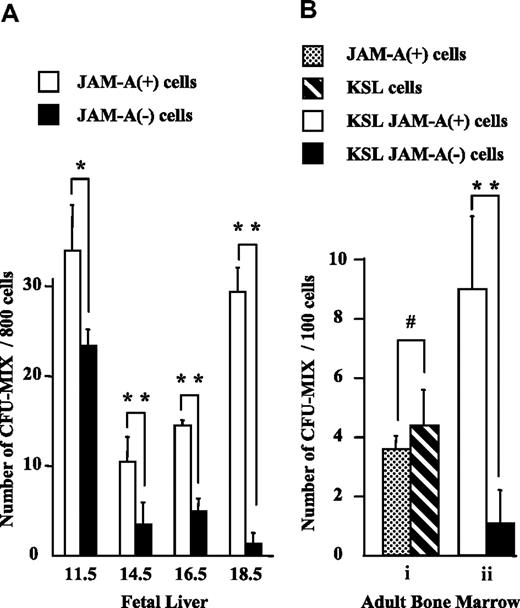
To evaluate the progenitor activity of JAM-A+ cells, we isolated JAM-A+ and JAM-A− cells from E11.5 FL CD34+c-Kit+ cells, and KSL cells from FL after E14.5 and from adult BM, and compared their colony-forming activities. Statistically similar levels of multilineage colony-forming activities (CFU-mix)29 were found in JAM-A+ and JAM-A− cells in E11.5 FL (Figure 5A). More CFU-mix29 was found in JAM-A+ cells than in JAM-A− cells in E14.5 and E16.5 FL (Figure 5A). Moreover, CFU-mix29 was almost exclusively in JAM-A+ cells in E18.5 FL and also in BM (Figure 5A,Bii). The results showed that CFU-mix29 is highly enriched in JAM-A+ KSL cells, especially in late FL and adult BM. Granulocyte-macrophage (GM)-type (CFU-GM, CFU-G, and CFU-M) colony-forming activity also was positively correlated with JAM-A expression among lineage-negative cells (data not shown); however, no such correlation was found in KSL cells. JAM-A+ cells were also sorted from the whole BM cells by anti–JAM-A antibody alone, and their potential to form colonies was examined. They contained CFU-mix29 at a level similar to that of KSL cells (Figure 5A). These results indicate that immature progenitors are highly enriched in JAM-A+ cells in BM.
Colony-forming activity of JAM-A+ and JAM-A− cells. Colony-forming activity of sorted cells from fetal and adult tissues was examined. (A) CFU-Mix activity of 800 JAM-A+ cells versus 800 JAM-A− cells in CD34+c-Kit+ cells in E11.5 FL and KSL cells in E14.5, E16.5, and E18.5 FL. (B) CFU-mix activity of 100 JAM-A+ cells without lineage depletion versus 100 KSL cells (i). CFU-mix activity of 100 JAM-A+ cells versus 100 JAM-A− cells in KSL cells in 8- to 12-week-old BM (ii). Results from more than 3 independent experiments are shown. Error bars indicate SEM; *P < .05, **P < .001, #P > .2.
Colony-forming activity of JAM-A+ and JAM-A− cells. Colony-forming activity of sorted cells from fetal and adult tissues was examined. (A) CFU-Mix activity of 800 JAM-A+ cells versus 800 JAM-A− cells in CD34+c-Kit+ cells in E11.5 FL and KSL cells in E14.5, E16.5, and E18.5 FL. (B) CFU-mix activity of 100 JAM-A+ cells without lineage depletion versus 100 KSL cells (i). CFU-mix activity of 100 JAM-A+ cells versus 100 JAM-A− cells in KSL cells in 8- to 12-week-old BM (ii). Results from more than 3 independent experiments are shown. Error bars indicate SEM; *P < .05, **P < .001, #P > .2.
LTR-HSC activity of JAM-A+ cells
To evaluate the LTR-HSC potential of JAM-A+ cells, we examined LTR-HSC activity by transplantation of sorted cells into irradiated mice. JAM-A+ KSL, JAM-A− KSL, KSL, and JAM-A+ cells were isolated from adult B6-Ly5.1 mice and were transplanted together with 2 × 105 unfractionated B6-F1 BM cells into the B6-Ly5.2 recipients. Donor contribution in PB was evaluated by flow cytometry 20 weeks after transplantation. High levels of donor contribution (>1%) were found in 7 of the 9 and 4 of the 4 mice that received 100 and 300 JAM-A+ KSL, respectively (Table 1). In contrast, none of the mice receiving a transplantation of 1000 JAM-A−KSL cells showed donor contribution (>1%) in PB (Table 1). Donor contribution was detected in 5 of the 6 mice receiving 300 KSL cells as a positive control (Table 1). Furthermore, donor contribution was detected in 4 of the 7 mice that received a transplantation of 100 JAM-A+ cells that were sorted from whole BM by JAM-A expression alone (Table 1). Consistent with the results of CFU assays, these results indicate that LTR-HSC activity in KSL cells is present in JAM-A+ cells but not in JAM-A− cells, and that LTR-HSCs in BM are highly enriched in JAM-A+ cells.
JAM-A marks LTR-HSCs in BM
| Cell population . | Cell no. . | Engrafted mice . | Total . | Myeloid cells . | T cells . | B cells . |
|---|---|---|---|---|---|---|
| KSL | 300 | 5/6 | 16.7 ± 10.6 | 15.6 ± 17.2 | 32.1 ± 19.8 | 28.0 ± 17.0 |
| KSL JAM-A− | 100 | 0/4 | — | — | — | — |
| KSL JAM-A− | 300 | 0/6 | — | — | — | — |
| KSL JAM-A− | 1000 | 0/6 | — | — | — | — |
| KSL JAM-A+ | 100 | 7/9 | 32.4 ± 26.4 | 29.7 ± 25.7 | 38.7 ± 25.3 | 24.6 ± 29.2 |
| KSL JAM-A+ | 300 | 4/4 | 77.4 ± 29.6 | 82.3 ± 20.8 | 73.8 ± 31.5 | 83.4 ± 17.1 |
| JAM-A+ | 100 | 4/7 | 15.4 ± 24.4 | 13.8 ± 21.9 | 10.4 ± 13.6 | 14.5 ± 24.4 |
| Cell population . | Cell no. . | Engrafted mice . | Total . | Myeloid cells . | T cells . | B cells . |
|---|---|---|---|---|---|---|
| KSL | 300 | 5/6 | 16.7 ± 10.6 | 15.6 ± 17.2 | 32.1 ± 19.8 | 28.0 ± 17.0 |
| KSL JAM-A− | 100 | 0/4 | — | — | — | — |
| KSL JAM-A− | 300 | 0/6 | — | — | — | — |
| KSL JAM-A− | 1000 | 0/6 | — | — | — | — |
| KSL JAM-A+ | 100 | 7/9 | 32.4 ± 26.4 | 29.7 ± 25.7 | 38.7 ± 25.3 | 24.6 ± 29.2 |
| KSL JAM-A+ | 300 | 4/4 | 77.4 ± 29.6 | 82.3 ± 20.8 | 73.8 ± 31.5 | 83.4 ± 17.1 |
| JAM-A+ | 100 | 4/7 | 15.4 ± 24.4 | 13.8 ± 21.9 | 10.4 ± 13.6 | 14.5 ± 24.4 |
Results of competitive hematopoietic stem-cell transplantation assay. Percentages of donor-derived cells of recipient mice were analyzed after 20 weeks of transplantation. Cell number indicates the test cells (B6-Ly5.1) that were transplanted with 2 × 105 competitive adult BM cells (B6-F1).
CRU frequency of JAM-A+ KSL and KSL cells
To further examine the effectiveness of JAM-A antibody, we performed limiting dilution assays to determine the CRUs of JAM-A+ KSL cells and KSL cells. The frequency of CRUs was estimated by assessing donor contribution in PB of recipient mice 8 weeks after transplantation. The frequency of CRUs in the fraction of JAM-A+ KSL and KSL was 1 in 64 (95% confidence interval for mean, 1 in 45 to 1 in 92, n = 16) and 1 in 109 (95% confidence interval for mean, 1 in 77 to 1 in 154, n = 18), respectively (Table 2). Importantly, selection of JAM-A+ cells from KSL cells resulted in the increase of HSC frequency more than 1.7-fold. This finding is consistent with colony assay data, showing that more primitive KSL cells, including HSCs, can be enriched using anti–JAM-A antibody (Figure 5 and Table 2).
CRU of JAM-A+ KSL and KSL
| Cell dose injected . | No. negative . | No. analyzed . |
|---|---|---|
| JAM-A+KSL | ||
| 25 | 2 | 3 |
| 100 | 2 | 9 |
| 300 | 0 | 4 |
| CRU frequency | 1 in 64 (95% confidence interval: 1 in 45 to 1 in 92) | |
| KSL | ||
| 25 | 2 | 3 |
| 100 | 2 | 7 |
| 300 | 1 | 6 |
| 400 | 0 | 2 |
| CRU frequency | 1 in 109 (95% confidence interval: 1 in 77 to 1 in 154) | |
| Cell dose injected . | No. negative . | No. analyzed . |
|---|---|---|
| JAM-A+KSL | ||
| 25 | 2 | 3 |
| 100 | 2 | 9 |
| 300 | 0 | 4 |
| CRU frequency | 1 in 64 (95% confidence interval: 1 in 45 to 1 in 92) | |
| KSL | ||
| 25 | 2 | 3 |
| 100 | 2 | 7 |
| 300 | 1 | 6 |
| 400 | 0 | 2 |
| CRU frequency | 1 in 109 (95% confidence interval: 1 in 77 to 1 in 154) | |
To determine the frequency of CRUs, recipient mice were analyzed at 8 weeks after transplantation. Negative mice were defined as animals with less than 1% of donor cell contribution in recipient PB. The frequency of CRUs was calculated by applying Poisson statistics with L-calc software.
Discussion
JAM proteins are expressed in various tissues in the human and murine body and are important for tight junction assembly, leukocyte transmigration, platelet activation, angiogenesis, and virus binding.16,30-35 It was reported recently that two of the JAM family members, JAM-B and JAM-C, are expressed in HSCs.36 Sakaguchi et al. showed that JAM-B is highly expressed in undifferentiated embryonic stem cells (ESCs), neural stem cells (NSCs), and HSCs, although it is not required for the maintenance of HSCs.37 JAM-A is expressed in a variety of hematopoietic cells12,14-19 ; however, the expression of JAM-A in HSCs had not been known. In this paper, we show that JAM-A is highly expressed in fetal hematopoietic tissues, AGM, and FL. Immunohistochemical staining revealed that JAM-A is detected in vascular networks with PECAM-1 from E9.5 to E12.5.38 Consistently, we also found that JAM-A is an endothelial marker, because almost all PECAM-1+ and Flk-1+ cells39 in E11.5 AGM were JAM-A+ (>99%, data not shown). Because of the increase of cells that do not express JAM-A during development, apparent JAM-A expression in whole fetal hematopoietic tissues decreases accordingly. However, the enriched HSC fraction maintains JAM-A expression during development.
In adults, JAM-A has been shown to be expressed in various hematopoietic cells in human PB.16 We also found that although as much as 25% of mouse PB mononuclear cells expressed JAM-A (data not shown), only about 2% of BM cells expressed JAM-A (Figure 1E). The KSL cells in BM showed high-level expression of JAM-A (Figure 1E), and Sca-1+c-Kit+ cells were more abundant in the JAM-A+ than JAM-A− cells in BM, regardless of age (Figure 3A-C). Compared with the fetal stage, the HSC fraction in JAM-A+ BM cells was much higher than that in JAM-A− cells throughout life. As only 2% of BM cells expressed JAM-A, the abundance of JAM-A+ cells in the enriched HSC fraction was quite high. In fact, c-Kit+Sca-1+ cells were enriched approximately 200-fold from whole BM cells by anti–JAM-A antibody alone; that is, the c-Kit+Sca-1+ cells in whole BM were only 0.15% (Figure 3D) and in JAM-A+ cells were 30% (Figure 3D). CFU-mix29 activity in the KSL population was almost exclusively in JAM-A+ cells from FL to BM (Figure 5Bii). Furthermore, CFU-mix29 activity of cells sorted by anti–JAM-A antibody alone was similar to that of KSL cells (Figure 5Bi), whereas CFU-GM and CFU-MegE were not different between JAM-A+ KSL and JAM-A− KSL cells (data not shown). Morphologic observation also supports that JAM-A+ cells contain more immature cells (Figure 4Ai). These results also indicate that most of the immature progenitors in FL and BM are highly enriched in JAM-A+ cells. Finally, transplantation of JAM-A+ cells demonstrates their LTR-HSC activity. Most of the mice received only 100 JAM-A+ KSL cells showed long-term engraftment (Table 1). More interestingly, donor contribution was detected in 4 of the 7 mice transplanted with only 100 JAM-A+ cells that were sorted directly from whole BM by JAM-A expression alone (Table 1). Since the ratio of JAM-A+ cells to JAM-A− cells in the KSL fraction was about 6.5:3.5 (Figure 1E), 100 JAM-A− KSL cells are equivalent to 285 total KSL cells. Consistently, most of the mice (5 of 6) receiving 300 KSL cells were engrafted for the long term. In contrast to JAM-A+ cells, 1000 JAM-A− KSL cells, which are equivalent to more than 2850 total KSL cells, failed to engraft for the long term. Taking these results and consideration together, HSCs in the KSL population are exclusively in JAM-A+ cells, and LTR-HSCs in BM can be highly enriched in JAM-A+ cells. The frequency of CRUs in the fraction of JAM-A+ KSL and KSL was 1 in 64 and 1 in 109, respectively (Table 2). Importantly, after selection of JAM-A expression, the frequency of CRUs increased approximately 1.7-fold. These results indicate that JAM-A is a useful marker for further purification of KSL cells. It was previously shown that CD34− KSL cells are a highly enriched HSC population, and we found that about 24% of CD34− KSL cells expressed JAM-A (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), suggesting that JAM-A+CD34− KSL may be the more purified HSCs. Yilmaz et al showed that SLAM family receptors CD150 and CD48 are expressed in HSCs and that CD150+CD48−CD244− KSL cells in BM are the most purified stem cells (1 in 2.1).8,40 They also showed that 21% of CD150+CD48−CD244− cells engrafted for the long term in recipient mice. We found that about 25% of CD150+CD48− BM cells were JAM-A+CD244−. Therefore, selection of JAM-A+ cells from CD34− KSL or CD150+CD48−CD244− may enhance the purity of HSCs.
Several lines of evidence suggest that JAM-A interacts with leukocyte via LFA-141 and αvβ333 in transendothelial migration of leukocytes. Leukocytic LFA-1 is a receptor for JAM-A and is involved in transendothelial migration of leukocytes.41 JAM-A is known to play a role in platelet activation and aggregation.15,16,42,43 JAM-A−/− polymorphonuclear leukocytes (PMNs) were shown to adhere more strongly and transmigrate less efficiently through endothelial monolayers, suggesting that the absence of JAM-A impairs PMN movement.44 JAM-A–deficient mice also revealed that JAM-A expression prevents spontaneous and random motility.45 By contrast, a role for JAM-A in hematopoiesis has not been elucidated. Considering the role of JAM-A in the adhesion of blood cells to endothelial cells, an interesting possibility may be that JAM-A is involved in the interaction between HSCs and bone marrow sinusoidal endothelial cells, the first step for hematopoietic stem-cell homing.46-48 Although we have not precisely located the JAM-A+ cells in AGM, we found that JAM-A was expressed in CD31+Flk+ cells in AGM. We previously showed that the FL cells support hematopoietic cells,23 and further study shows that immature hepatocytes support HSCs from AGM, and fetal hepatocytes also express JAM-A (data not shown). Thus, it is tempting to speculate that JAM-A may involve in the interaction of HSCs and their niche. In conclusion, the results in this paper show that JAM-A is an excellent marker for HSCs in BM and can be used to prepare a highly enriched HSC population by using anti–JAM-A antibody.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.S. designed, performed, and interpreted the experiments, and wrote the paper. M. Takeuchi designed the experiments and analyzed the data. A.H. and H.M. performed the experiments. M. Tanaka and T.K. interpreted the data and edited the paper. A.M. coordinated the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Atsushi Miyajima, Institute of Molecular and Cellular Biosciences, University of Tokyo. 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan; e-mail: miyajima@iam.u-tokyo.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal