Abstract
The cAMP-responsive element binding protein (CREB) is a 43-kDa nuclear transcription factor that regulates cell growth, memory, and glucose homeostasis. We showed previously that CREB is amplified in myeloid leukemia blasts and expressed at higher levels in leukemia stem cells from patients with myeloid leukemia. CREB transgenic mice develop myeloproliferative disease after 1 year, but not leukemia, suggesting that CREB contributes to but is not sufficient for leukemogenesis. Here, we show that CREB is most highly expressed in lineage negative hematopoietic stem cells (HSCs). To understand the role of CREB in hematopoietic progenitors and leukemia cells, we examined the effects of RNA interference (RNAi) to knock down CREB expression in vitro and in vivo. Transduction of primary HSCs or myeloid leukemia cells with lentiviral CREB shRNAs resulted in decreased proliferation of stem cells, cell- cycle abnormalities, and inhibition of CREB transcription. Mice that received transplants of bone marrow transduced with CREB shRNA had decreased committed progenitors compared with control mice. Mice injected with Ba/F3 cells expressing either Bcr-Abl wild-type or T315I mutation with CREB shRNA had delayed leukemic infiltration by bioluminescence imaging and prolonged median survival. Our results suggest that CREB is critical for normal myelopoiesis and leukemia cell proliferation.
Introduction
Hematopoiesis is regulated by transcription factors that drive bone marrow progenitor cells to proliferate and differentiate. Among the nuclear factors that control gene transcription is a leucine zipper transcription factor, cAMP-responsive element binding protein (CREB), which activates genes that control metabolism, cell cycle, signal transduction, and cell survival. CREB is a member of the activating transcription factor (ATF)/CREB family of transcription factors and requires phosphorylation of serine 133 for function.1,2 We demonstrated previously that CREB is a downstream target of hematopoietic growth factor signaling activated by granulocyte-macrophage–colony stimulating factor and interleukin-3.3-5 A role for CREB in oncogenesis has been suggested by its overexpression in the majority of bone marrow samples from patients with acute leukemia.6 CREB is overexpressed at both the protein and mRNA levels in leukemic blasts and in leukemia stem cells.7-9 Furthermore, CREB is amplified in blast cells from CREB-overexpressing patients.6
We also demonstrated previously that CREB overexpression in myeloid cells increases cell proliferation and survival. CREB transgenic mice that overexpress CREB in the myeloid lineage develop myeloproliferative disease/myelodysplastic syndrome but not acute leukemia, suggesting that CREB contributes to myeloid cell proliferation but is not sufficient for development of acute leukemia. Bone marrow progenitors from CREB transgenic mice demonstrate increased stem-cell self-renewal in replating assays and increased sensitivity to hematopoietic growth factors.8 We demonstrated that CREB overexpressing myeloid cells also have increased expression of cyclin A associated with an increase in the number of cells in S phase. Therefore, CREB seems to play a role in hematopoietic stem cell (HSC) proliferation and survival through its effects on cell-cycle regulation.
To understand the requirement of CREB in hematopoietic stem cells and myeloid leukemia cells, we investigated the expression of CREB in normal mouse and human HSCs and studied the effects of CREB down-regulation on normal and leukemic cell proliferation and maturation. In this article, we report that CREB is highly expressed in normal lineage negative (lin−) or uncommitted hematopoietic progenitor cells and that inhibition of CREB expression using shRNAs resulted in decreased proliferation and differentiation of normal and neoplastic hematopoietic cells in vitro and in vivo, respectively. We also demonstrate by expression profiling, potential mechanisms by which CREB may influence HSC fate. Our results suggest that CREB plays a critical role in normal HSC proliferation and leukemia progression.
Methods
Stem cells and preparation
Murine hematopoietic stem cells and progenitors were isolated from adult C57BL6 mice as described previously.10-15 Mouse whole bone marrow cells were divided into lin− and lineage-positive (lin+) cells using the mouse lineage cell-depletion kit of the magnetic activated cell-separation system in combination with the auto MACS magnetic cell separator (Miltenyi Biotec, Auburn, CA). The lin− population was sorted either on a FACSDiVa or a BD FACSAria cell sorter (BD Biosciences, Rockville, MD) into hematopoietic stem cells and progenitors. The lin+ fraction was sorted into mature hematopoietic cells, including T cells, B cells, granulocytes, macrophages, and erythroid cells. Human cord blood cells were obtained from Cambrex Charles City (Charles City, IA). Human lin− cord blood cells were separated into CD34− and CD34+ cells using the human CD34 MicroBead kit in combination with the auto MACS separator. Human lin− CD34+ cord blood cells were sorted by FACS into hematopoietic stem cells and progenitors, and lin+ CD34− cord blood cells were sorted into mature hematopoietic cells. Human peripheral blood stem cells (PBSCs) and bone marrow cells were sorted by FACS into hematopoietic stem cells (CD34+ CD38−) and progenitors (CD34+ CD38+). Human lin+ PBSCs were sorted by FACS into matured hematopoietic cells, including T cells (CD3+), B cells (CD19+), NK cells (CD56+), granulocytes/macrophages (CD14+), and erythroid cells (glycophorin-A+). For cell-cycle experiments, samples were stained according to a hypotonic propidium iodide buffer–based protocol.16,17
Quantitative real-time and reverse transcription-polymerase chain reaction
Total RNA was extracted from sorted cells (2-5 × 104 cells per sample) using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was synthesized using the Omniscript Reverse Transcriptase Kit (Qiagen) or Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Indianapolis, IN). To detect the expression of transcription factors SCL, c-Mpl, Pu-1, Aiolos, and CEBPα in mouse HSC and progenitors, reverse transcription-polymerase chain reaction (RT-PCR) was performed using primers and PCR conditions described previously.18,19 Quantitative real-time PCR (q-PCR) was performed in triplicate using the TaqMan probe system with CREB- and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific probe and primers on an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) according to manufacturer's instructions. CREB expression data were standardized using GAPDH expression data. Primer sequences are available upon request. For shRNA experiments, cells (5 × 106) were lysed in TRIzol and stored at −80°C before RNA extraction. RNA extraction was performed according to a standard protocol supplied by the manufacturer (Invitrogen, Carlsbad, CA), and pellets were resuspended in RNAse-free water. The cDNA was transcribed with a Superscript RT III based-protocol. DNAse treatment was not performed as a result of the selection of intron-spanning primers. Quantitative real-time PCR was performed with the SyberGreen reagent (Bio-Rad Laboratories, Hercules, CA) in triplicate and analyzed by the standard curve method standardized to the housekeeping gene β-actin.20,21
shRNA sequence design and constructs
The CREB-specific shRNA sequences (CREB shRNA-1, CREB shRNA-2, CREB shRNA-3) were selected and validated based on accepted parameters established by Tuschl et al22-24 (http://www.rockefeller.edu/labheads/tuschl/sirna.html). Sequence 2 was chosen for in vivo experiments based on maximal gene inhibition. Controls included empty vector, luciferase shRNA, and scrambled shRNA. shRNA sequences are CREB shRNA-1 (5′-GCAAATGACAGTTCAAGCCC-3′), shRNA-2 (5′-GTACAGCTGGCTAACAATGG-3′), shRNA-3 (5′-GAGAGAGGTCCGTCTAATG-3′), Luciferase shRNA (5′-GCCATTCTATCCTCTAGAGGA-3′), and scramble shRNA (5′-GGACGAACCTGCTGAGATAT-3′). Short-hairpin sequences were synthesized as oligonucleotides and annealed according to standard protocol. Annealed shRNAs were then subcloned into pSICO-R shRNA vectors from the Jacks laboratory at MIT (http://web.mit.edu/ccr/labs/jacks/index.html).25 The second generation SIN vector HIV-CSCG was used to produce human shRNA vectors.26
Cell lines
The following human leukemia cell lines were transduced with shRNAs: K562 (Iscove medium + 10% fetal calf serum [FCS]), TF-1 (RPMI medium + 10% FCS + recombinant human granulocyte macrophage–colony-stimulating factor [rhGM-CSF]), and MV-411 (Iscove medium + 10% FCS). Murine leukemia cell lines included Ba/F3-Bcr/Abl wild type (RPMI medium + 10% FCS) and Ba/F3-Bcr/Abl T315I (RPMI medium + 10% FCS). All leukemia cell lines express CREB. Cells were cultured at 37°C, 5% CO2 and split every 3 to 4 days. Proliferation and viability assays in Figure 4 were performed in triplicate by the trypan blue exclusion method. Several transductions were performed for each shRNA sequence to avoid clonal effects or selection. Each experiment was performed within a week after transduction.
Western blot analysis
Boiling SDS-Laemmli method was used for all Western blot analyses. Protein lysates were separated on a 10% SDS-polyacrylamide gel. Immunoblot was performed with anti-CREB (UBI, New York, NY), or β-tubulin antisera (Santa Cruz Biotechnology, Santa Cruz, CA) as described previously.8 All experiments were performed 3 times.
Lentiviral transduction of primary cells
One day before transduction, 7 × 105 cells were cultured in retronectin-treated 96-well plates and pre-stimulated with appropriate cytokines; for murine bone marrow, interleukin (IL)-3 (6 ng/mL), human IL-6 (10 ng/mL), and stem-cell factor (50 ng/mL) were used; for human peripheral blood, IL-3 (10 ng/mL), IL-6 (10 ng/mL), stem-cell factor (50 ng/mL), Flt-3 ligand (6 ng/mL), and thrombopoietin (6 ng/mL) were used.27-30 All experiments were repeated at least 3 times. Human blood and bone marrow cells were obtained with institutional review board consent approved by the institution (UCLA IRB #98-09-036-21) in accordance with the Declaration of Helsinki.
Colony-forming assays
Transduced and GFP-sorted hematopoietic cells (2 × 104 murine bone marrow cells or 500 human CD34+ cells) were plated in methylcellulose (Stem Cells, Palo Alto, CA) containing a full complement of cytokines (GM-CSF, IL-3, IL-6, stem-cell factor, erythropoietin [EPO]) or EPO alone for blast forming unit–erythroid (BFU-E)/colony-forming unit–erythroid (CFU-E) assays, and cultured for 21 days. Colony counts, cytospins, flow cytometric analysis, and Western blot analysis were performed in triplicate as described previously.8 Fluorescence-activated cell sorting (FACS) antibodies used include CD150, CD135, Thy 1.1, CD117, Flk2, Sca-1, Mac-1, Gr-1, CD3e, B220, and Ter119.
In vivo bioluminescence
SCID mice (The Jackson Laboratory, Bar Harbor, ME) were tail vein-injected with 106 Ba/F3-Bcr/Abl T315I cells expressing luciferase that were transduced with either CREB shRNA or a control scrambled shRNA. Mice were followed over a period of approximately 4 weeks with serial examinations for disease. Luciferin (15 μg) was injected 10 to 15 minutes before imaging and repeated every 7 days for a total of 2 weeks. Relative intensity units for regions of interest were measured in triplicate and averaged,31 with antilogs of log transform statistics used to estimate the geometric mean ratio of intensities and its confidence interval. These experiments were repeated twice. A similar approach was taken with Ba/F3-Bcr/Abl wild-type cells expressing luciferase that were either not transduced or transduced with CREB shRNA or a control, scrambled shRNA. SCID mice were injected with 5 × 106 cells, monitored, and images were acquired over 4 weeks as described previously in this paragraph.
Apoptosis experiments
Standard Western blot analysis methods were applied to lysates made from mouse bone marrow cells (106) that were transduced with CREB shRNA or control shRNA lentivirus. Blots were probed with anti-PARP antisera (Cell Signaling Technology, Danvers, MA) to assess for cleavage fragments confirmatory for apoptosis. In addition, flow cytometic methods were performed using an allophycocyanin-tagged annexin-V monoclonal antibody and propidium iodide to assay for apoptotic cells.
Murine bone marrow transplantation
C57BL/6j CD45.2 mice were purchased from The Jackson Laboratory and treated with prophylactic trimethoprim-sulfa therapy from four days prior to transplant in water. Recipient mice were sublethally irradiated on day 0 with a single dose of 9.5 Gy (950 rads) and subsequently injected through the tail vein with 2 × 105 green fluorescent protein-positive (GFP+) bone marrow mononuclear cells from CD45.1 donors. Donor cells were transduced as described previously (“Lentiviral transduction of primary cells”) and positive transductants were FACS sorted. Engraftment was monitored by serial eye bleeds and analysis of bone marrow with complete blood counts and lineage-specific monoclonal antibodies using flow cytometry.32-34 Antibodies used included myeloid (Gr-1;Mac-1) and lymphoid (B220;CD3e). These experiments were performed twice.
Results
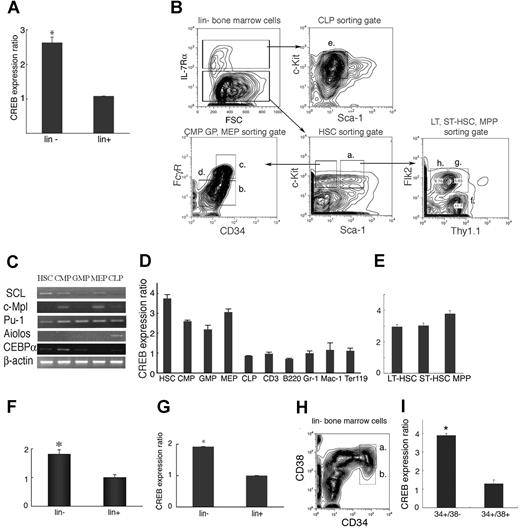
To characterize the expression of CREB during adult mouse hematopoiesis, we performed q-PCR on mRNA derived from lin− and lin+ murine bone marrow cells, and distinct lin− and lin+ subpopulations. CREB expression was higher (2.6-fold) in the lin− population than in the lin+ population (Figure 1A, P < .01). Next, lin− cells were further fractionated into HSCs (Figure 1Ba), common myeloid progenitor (CMP; Figure 1Bb), common lymphoid progenitor (CLP; Figure 1Be), granulocyte-macrophage progenitor (GMP; Figure 1Bc), and megakaryocytic-erythroid progenitor (MEP; Figure 1Bd) using their characteristic cell surface antigen expression pattern and the gating and cell-sorting strategy as outlined in Figure 1B. After sorting, cDNAs from HSC, CMPs, CLPs, GMPs, and MEPs were confirmed by expression of SCL, c-Mpl, PU.1, Aiolos, and CCAAT/enhancer-binding protein α (C/EBPα; Figure 1C). As expected from the data we had obtained from lin− cells, CREB expression was higher in HSC, CMP, GMP, and MEP compared with differentiated cells (Figure 1D). To further define CREB expression in HSC, we examined the expression of CREB in long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and multipotent progenitors (MPPs; Figure 1E). CREB was slightly higher in the MPP fraction compared with the LT-HSC and ST-HSC populations, but this was not found to be statistically significant (Figure 1E, P = .06).
CREB expression in mouse and human hematopoietic progenitors. (A) CREB expression levels in lin− and lin+ cells as measured by q-PCR. CREB expression was higher (2.6-fold) in the lin− population than the lin+ population (P = .01). (B) Gating strategy for cell sorting of lin− cells to isolate HSC (Ba), CMP (Bb), GMP (Bc), MEP (Bd), CLP (Be), LT-HSC (Bf), ST-HSC (Bg), and MPP (Bh). (C) cDNA from HSC CMPs, CLPs, GMPs, and MEPs were confirmed by expression of SCL, c-Mpl, Pu-1, Aiolos, and CEBPα. (D) CREB expression levels in HSC, CMP, GMP, MEP, and differentiated cells. Compared with mature cells, CREB was 2.5- to 4-fold higher in HSC, CMP, GMP, and MEP. (E) CREB expression in LT-HSC, ST-HSC, and MPP. CREB was slightly higher (4- vs 3-fold) in the MPP fraction compared with the LT-HSC and ST-HSC populations, but this was not found to be statistically significant (P = .06). CREB expression in human lin− and lin+ cells isolated from cord blood (F) or peripheral blood (G) stem cells. CREB expression was higher for both cell types in the lin− population than the lin+ population (P = .01). (H and I) CREB expression in human bone marrow cells. CREB was also expressed at higher levels in CD34+ CD38− cells (b) than CD34+ CD38+ cells (a). All experiments were performed in triplicate. Error bars represent SE.
CREB expression in mouse and human hematopoietic progenitors. (A) CREB expression levels in lin− and lin+ cells as measured by q-PCR. CREB expression was higher (2.6-fold) in the lin− population than the lin+ population (P = .01). (B) Gating strategy for cell sorting of lin− cells to isolate HSC (Ba), CMP (Bb), GMP (Bc), MEP (Bd), CLP (Be), LT-HSC (Bf), ST-HSC (Bg), and MPP (Bh). (C) cDNA from HSC CMPs, CLPs, GMPs, and MEPs were confirmed by expression of SCL, c-Mpl, Pu-1, Aiolos, and CEBPα. (D) CREB expression levels in HSC, CMP, GMP, MEP, and differentiated cells. Compared with mature cells, CREB was 2.5- to 4-fold higher in HSC, CMP, GMP, and MEP. (E) CREB expression in LT-HSC, ST-HSC, and MPP. CREB was slightly higher (4- vs 3-fold) in the MPP fraction compared with the LT-HSC and ST-HSC populations, but this was not found to be statistically significant (P = .06). CREB expression in human lin− and lin+ cells isolated from cord blood (F) or peripheral blood (G) stem cells. CREB expression was higher for both cell types in the lin− population than the lin+ population (P = .01). (H and I) CREB expression in human bone marrow cells. CREB was also expressed at higher levels in CD34+ CD38− cells (b) than CD34+ CD38+ cells (a). All experiments were performed in triplicate. Error bars represent SE.
To assess the mRNA CREB expression patterns in human hematopoiesis, lin− and lin+ cells were isolated from human cord blood and peripheral blood stem cells and subjected to q-PCR. CREB expression was higher (2-fold) in the lin− population than in the lin+ population for both cord and peripheral blood stem cells (Figure 1F,G, respectively, P < .01). CREB expression in human bone marrow cells was higher in CD34+ CD38− cells than in CD34+ CD38+ cells (Figure 1H,I). Our results demonstrate that the lineage negative population consistently expressed higher levels of CREB regardless of the stem-cell source.
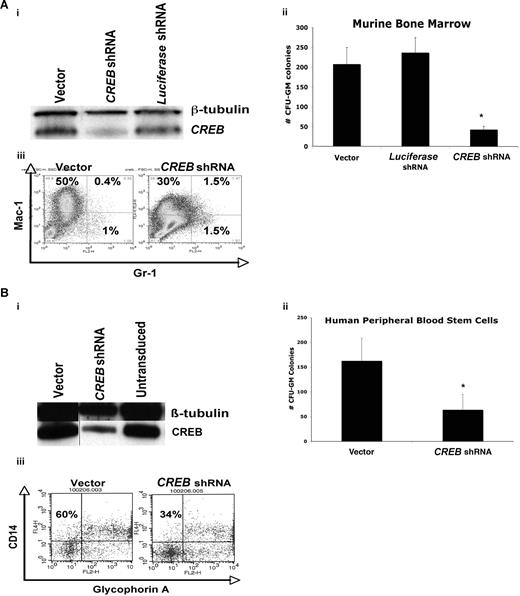
We examined the requirement of CREB for normal hematopoiesis by transducing normal murine bone marrow cells with CREB or control lentiviral shRNAs. Although 3 separate shRNAs were tested in cell lines initially (Figure 4 and data not shown), shRNA sequence 2 was used to transduce primary bone marrow cells because it was the most effective in down-regulating CREB. Methylcellulose colony assays were performed to assess qualitative and quantitative effects of CREB on normal hematopoiesis in vitro (Figure 2A,B). Transduction efficiency varied between 5% and 34%, as assessed by measuring the GFP+ fraction by flow cytometry (data not shown). We first analyzed CREB expression in primary mouse bone marrow cells transduced with CREB shRNA lentivirus by Western blot analysis and observed a significant inhibition of CREB expression (∼80%) in CREB shRNA–transduced cells, compared with control cells (Figure 2Ai). After 21 days, we detected a statistically significant decrease (up to 5-fold) in the number of GFP+ CFU-GM colonies with CREB shRNA-transduced mouse bone marrow cells compared with vector or luciferase control (Figure 2Aii). These colonies consisted of myeloid progenitor cells that were mostly Mac-1+ (Figure 2Aiii). The Mac-1+ Gr-1+ progenitor cell fraction, an immature population of cells, was noted to be up to 4-fold higher in the CREB shRNA–transduced cells compared with control cells.35 It is noteworthy that there were fewer differentiated Mac-1+ cells in the CREB shRNA–transduced cells (30%) compared with vector control (50%), suggesting that CREB is critical for both hematopoietic cell proliferation and possibly terminal differentiation of monocytes. There was no statistically significant difference in the numbers of BFU-E or CFU-granulocyte/erythrocyte/monocyte colonies in methylcellulose with CREB and scrambled shRNA-transduced murine bone marrow progenitor cells. Likewise, long-term culture-initiating cell assays showed no difference between CREB and scrambled shRNA-transduced cells, suggesting that although CREB is a critical regulator of early myelopoiesis, it does not appear to be necessary for hematopoietic stem-cell proliferation and differentiation (data not shown).
CREB is critical for normal myelopoiesis in vitro. (A) (i) Western blot analysis demonstrating knockdown of CREB approaching 80% compared with control cells. (ii) Total numbers of CFU-GM colonies after 21 days in methylcellulose for murine hematopoietic cells. (iii) Flow cytometric analysis of murine bone marrow transduced with CREB shRNA or control lentivirus and sorted for GFP+ fraction, cultured in methylcellulose over 21 days. CREB knockdown cells had a lower fraction of mature granulocyte and monocytes compared with control cells. (B) (i) Western blot analysis demonstrating knockdown of CREB up to 65% compared with control cells. (ii) Total number of CFU-GM colonies after 21 days in methylcellulose for human peripheral blood stem cells. (iii) Flow cytometry analysis of transduced CD34+ human peripheral blood stem cells cultured in methylcellulose over 21 days. All experiments were performed in triplicate. Error bars in Aii,Bii represent SE.
CREB is critical for normal myelopoiesis in vitro. (A) (i) Western blot analysis demonstrating knockdown of CREB approaching 80% compared with control cells. (ii) Total numbers of CFU-GM colonies after 21 days in methylcellulose for murine hematopoietic cells. (iii) Flow cytometric analysis of murine bone marrow transduced with CREB shRNA or control lentivirus and sorted for GFP+ fraction, cultured in methylcellulose over 21 days. CREB knockdown cells had a lower fraction of mature granulocyte and monocytes compared with control cells. (B) (i) Western blot analysis demonstrating knockdown of CREB up to 65% compared with control cells. (ii) Total number of CFU-GM colonies after 21 days in methylcellulose for human peripheral blood stem cells. (iii) Flow cytometry analysis of transduced CD34+ human peripheral blood stem cells cultured in methylcellulose over 21 days. All experiments were performed in triplicate. Error bars in Aii,Bii represent SE.
The role of CREB in normal human hematopoietic cells was studied by knocking down CREB through lentiviral transduction of mobilized, CD34+ normal peripheral blood mononuclear cells. Efficiency of transduction was as high as 34% by GFP expression (data not shown). CREB knockdown was 65% by Western blot analysis after 21 days in methylcellulose (Figure 2Bi). We observed a 3-fold decrease in CFU-GM colony formation in methylcellulose with CREB shRNA–transduced cells compared with control cells (P < .05; Figure 2Bii). Cytospin assays confirmed that CFU-GM were in the form of immature myeloid cells (data not shown). CREB shRNA–transduced stem cells had decreased numbers of mature CD14+ monocytes (Figure 2Biii). This suggests that CREB down-regulation could be inhibiting terminal differentiation in addition to proliferation and survival in vitro.
CREB has been previously demonstrated to enhance cell proliferation and survival. To investigate the role of CREB in apoptosis of primary bone marrow progenitor cells, we transduced human and mouse bone marrow cells with CREB, scrambled, or vector control shRNA lentivirus. Identical sequences were used as described in Figure 2. In colony assays after 5 days, CREB shRNA–transduced cells showed a decrease in the human CD34+ fractions and an undetectable CD34+ CD38− HSC population compared with controls (Figure 3A and data not shown). For mouse bone marrow transductions, a relatively higher proportion of apoptotic cells in GFP+CREB shRNA–transduced cells with an increase in cells stained with annexin-V and propidium iodide (PI) (Figure 2B; P < .05). However, the overall percentage of apoptotic cells was less than 2%, suggesting that apoptosis occurs in a small proportion of the total number of transduced cells. Primary mouse BM progenitor cells transduced with CREB shRNA had increased PARP cleavage compared with control cells (Figure 3C). Therefore, our results suggest that CREB down-regulation in HSCs inhibits proliferation and survival and to a lesser extent induces apoptosis in vitro.
CREB shRNA induces apoptosis in HSCs and bone marrow progenitor cells. (A) Transduced human peripheral blood cells plated in methylcellulose for 3 weeks and stained with monoclonal antibodies for CD34 and CD38 expression. Cells were stained with PI to assess cell death. (B) Murine bone marrow cells were transduced at a density of 106 cells/mL with lentivirus expressing CREB, scrambled, or vector shRNA at a multiplicity of infection (MOI) of 100. After 2 days of culturing in media containing cytokines (mIL-3, 10ng/mL; mSCF, 25 ng/mL; and hIL-6, 10 ng/mL), cells were sorted using flow cytometry for GFP expression. Sorted calls were cultured in cytokine containing media for 5 days and stained for annexin-V and PI. All experiments were performed in triplicate. (C) Western blot analysis with lysates from mouse BM cells (106) transduced with CREB shRNA, scrambled, luciferase, and vector control lentivirus. Immunoblots were probed with anti-PARP or β-tubulin antisera. Error bars in panels A and B represent SE.
CREB shRNA induces apoptosis in HSCs and bone marrow progenitor cells. (A) Transduced human peripheral blood cells plated in methylcellulose for 3 weeks and stained with monoclonal antibodies for CD34 and CD38 expression. Cells were stained with PI to assess cell death. (B) Murine bone marrow cells were transduced at a density of 106 cells/mL with lentivirus expressing CREB, scrambled, or vector shRNA at a multiplicity of infection (MOI) of 100. After 2 days of culturing in media containing cytokines (mIL-3, 10ng/mL; mSCF, 25 ng/mL; and hIL-6, 10 ng/mL), cells were sorted using flow cytometry for GFP expression. Sorted calls were cultured in cytokine containing media for 5 days and stained for annexin-V and PI. All experiments were performed in triplicate. (C) Western blot analysis with lysates from mouse BM cells (106) transduced with CREB shRNA, scrambled, luciferase, and vector control lentivirus. Immunoblots were probed with anti-PARP or β-tubulin antisera. Error bars in panels A and B represent SE.
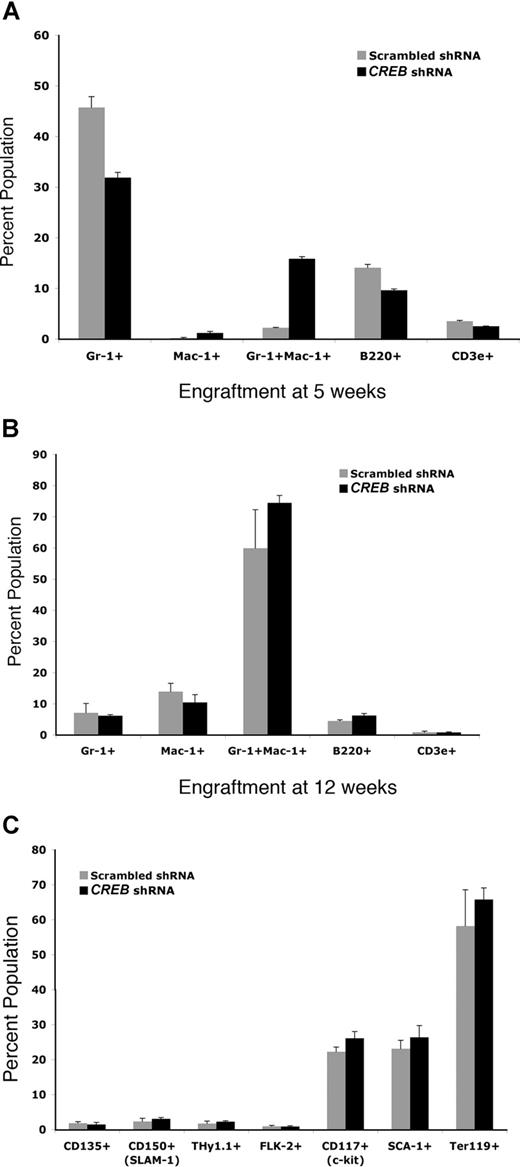
To study the requirement of CREB in normal hematopoiesis in vivo, we also analyzed hematopoietic reconstitution of sublethally irradiated B6/C7 CD45.2 mice at 5 and 12 weeks after infusion of marrow progenitor cells from B6/C7 CD45.1 transduced with CREB shRNA or scrambled shRNA lentivirus. We analyzed by flow cytometry those cells that were GFP+ and CD45.1+. Peripheral blood counts were comparable between both groups; however, there was a statistically significant increase (4-fold) in myeloid (Gr-1/Mac-1+) progenitors and less dramatic increase in Mac-1+ cells in the CREB shRNA group (Figure 4A and data not shown). At 12 weeks, very little difference was observed in lineage-specific cells (Gr-1/Mac-1+, Mac-1+, and Gr-1+, B220+, and CD3e+) or hematopoietic stem cells (CD135+, CD150+, Thy1.1+, Flk-2+, CD117+, Sca-1+, Ter119+) from bone marrow between CREB shRNA compared with scrambled shRNA (Figure 4B). These results suggest that CREB could also be playing a role in regulating myeloid differentiation of committed progenitor cells.
CREB is critical for myelopoiesis in vivo. Bone marrow from CD45.1 mice transplanted into CD45.2 mice were analyzed with lineage-specific and hematopoietic stem-cell markers using FACS analysis at 5 and 12 weeks after transplantation. (A) Myeloid engraftment as measured by staining of bone marrow cells from transplant-recipient mice at 5 weeks. (B) Myeloid engraftment as measured by FACs staining of bone marrow cells at 12 weeks. At least 5 mice in each group were analyzed. Experiments were performed in triplicate and repeated twice. Error bars represent SE.
CREB is critical for myelopoiesis in vivo. Bone marrow from CD45.1 mice transplanted into CD45.2 mice were analyzed with lineage-specific and hematopoietic stem-cell markers using FACS analysis at 5 and 12 weeks after transplantation. (A) Myeloid engraftment as measured by staining of bone marrow cells from transplant-recipient mice at 5 weeks. (B) Myeloid engraftment as measured by FACs staining of bone marrow cells at 12 weeks. At least 5 mice in each group were analyzed. Experiments were performed in triplicate and repeated twice. Error bars represent SE.
We next studied the effects of CREB down-regulation in proliferation of myeloid leukemia cells. All cell lines had strong basal CREB expression (Figure 5A). CREB was successfully knocked-down in human (TF-1 and K562) myeloid leukemia cell lines transduced with CREB shRNA at an efficiency of 45% to 92% confirmed by Western blot analysis and q-PCR (Figure 5A,B). In addition, phosphorylated CREB levels were lower in CREB shRNA–transduced cells (Figure 5A). The growth and viability of K562 and TF-1 leukemia cells were significantly decreased with CREB shRNA–transduced cells compared with control cells (Figure 5C,i-iii). Furthermore, we did not see off-target effects indicated by 2′,5′-oligoadenylate synthetase 1 (OAS-1) expression in cells treated with interferon (Figure 5D).36 Multiple transductions were performed with the different shRNA sequences to avoid the effects of clonogenicity. We showed previously that overexpression of CREB in myeloid leukemia cells resulted in increased cyclin A promoter activity.8 To examine the effects of CREB shRNA on CREB transcriptional activity, we transfected TF-1 cells with a construct containing the cyclin A promoter with the cAMP-responsive element and the luciferase reporter gene.8 A significant decrease in cyclin A promoter activity was observed in cells transduced with CREB shRNA compared with vector control shRNA or untransduced cells (Figure 5E). We also analyzed the effects of CREB shRNA on cell-cycle regulation in TF-1 cells stimulated with GM-CSF for 12 hours after growth factor starvation (Figure 5F). Our results demonstrated decreased percentage of cells in S-phase and increased percentage of cells in G1 and G2/M when CREB was down-regulated with 2 different CREB shRNA sequences.
CREB is essential for leukemia cell proliferation and survival. (A) Human (K562, TF-1) leukemia cells were transduced with a lentivirus expressing no shRNA,1 CREB shRNA-1,2 CREB shRNA-2,3 or luciferase shRNA4 at a multiplicity of infection (MOI) of approximately 100. Wild-type cells5 were also used as a control. Western blot analyses were performed with CREB, phospho-CREB, and β-tubulin antisera. (B) Five micrograms of total RNA were extracted from transduced leukemia cells, and q-PCR was performed to determine CREB expression. CREB was knocked down by up to 75% relative to control shRNA (vector) in human myeloid leukemia cells. (C) Trypan blue exclusion method was performed in triplicate to assess growth and survival of transduced leukemia cells. CREB knocked-down cells demonstrated diminished proliferation and viability 72 hours after transduction. (D) K562 cells were transduced and cultured for 48 hours before harvesting total RNA. Parental K562 cells were cultured in the presence of interferon-2α (100 units/mL) for 48 hours as a positive control. Quantitative reverse transcription-PCR was performed in triplicate with primers specific for CREB, actin, and OAS-1. (E) Luciferase reporter assays in human TF-1 leukemia cells transduced with CREB or control shRNAs. Decreased transcriptional activity was observed in CREB knocked-down cells and repeated in triplicate. (F) Cell-cycle analysis of CREB knocked-down TF-1 cells after synchronization by serum starvation overnight and stimulated for 12 hours with GM-CSF revealed decreased percentage of cells in S-phase. Experiment was performed in triplicate. Error bars in panels B-F represent SE.
CREB is essential for leukemia cell proliferation and survival. (A) Human (K562, TF-1) leukemia cells were transduced with a lentivirus expressing no shRNA,1 CREB shRNA-1,2 CREB shRNA-2,3 or luciferase shRNA4 at a multiplicity of infection (MOI) of approximately 100. Wild-type cells5 were also used as a control. Western blot analyses were performed with CREB, phospho-CREB, and β-tubulin antisera. (B) Five micrograms of total RNA were extracted from transduced leukemia cells, and q-PCR was performed to determine CREB expression. CREB was knocked down by up to 75% relative to control shRNA (vector) in human myeloid leukemia cells. (C) Trypan blue exclusion method was performed in triplicate to assess growth and survival of transduced leukemia cells. CREB knocked-down cells demonstrated diminished proliferation and viability 72 hours after transduction. (D) K562 cells were transduced and cultured for 48 hours before harvesting total RNA. Parental K562 cells were cultured in the presence of interferon-2α (100 units/mL) for 48 hours as a positive control. Quantitative reverse transcription-PCR was performed in triplicate with primers specific for CREB, actin, and OAS-1. (E) Luciferase reporter assays in human TF-1 leukemia cells transduced with CREB or control shRNAs. Decreased transcriptional activity was observed in CREB knocked-down cells and repeated in triplicate. (F) Cell-cycle analysis of CREB knocked-down TF-1 cells after synchronization by serum starvation overnight and stimulated for 12 hours with GM-CSF revealed decreased percentage of cells in S-phase. Experiment was performed in triplicate. Error bars in panels B-F represent SE.
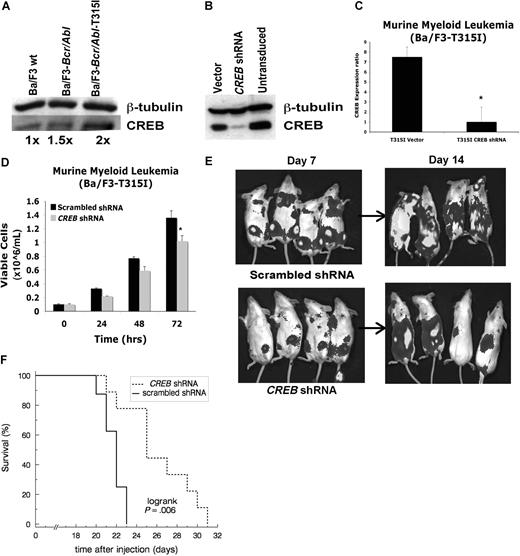
To study the requirement of CREB during progression of leukemia in vivo, Ba/F3 cells overexpressing Bcr-Abl wild type or Bcr-Abl with the imatinib-resistant T315I mutation were injected into SCID mice (Figures 6 and S1, available on the Blood website; see the Supplemental Materials link at the top of the online article.). These mice normally die within 21 days of injection from leukemic infiltration of liver and spleen. The Bcr-Abl–expressing Ba/F3 cells express CREB equal to the Ba/F3 wild-type cells. The T315I mutant cell line was chosen based on the 2-fold increased expression of CREB protein compared with Bcr-Abl or untransduced Ba/F3 cells (Figure 6A). Thus, we hypothesized that removing the CREB-dependent component of Bcr-Abl T315I-induced transformation pathways could affect leukemia progression. Transduction of Ba/F3 Bcr-Abl T315I cells resulted in an 80% decrease in CREB protein and mRNA expression (Figure 6B,C). Six-week-old SCID mice were injected with either 106 or 5 × 105 Ba/F3 Bcr-Abl wild-type or T315I transduced cells also expressing luciferase (kindly provided to us by Neil Shah, University of San Francisco, CA).37,38 Mice were analyzed after each luciferin injection 15 minutes before imaging at days 7 and 14. A modest effect on proliferation was also noted in vitro after CREB knockdown in Ba/F3 T315I cells and analysis using trypan blue exclusion (Figure 6D). Bioluminescence imaging performed on days 7 and 14 demonstrated a significant decrease in disease burden in CREB knockdown mice with Ba/F3 Bcr-Abl T315I cells compared with mice injected with scrambled shRNA construct or Ba/F3 Bcr-Abl wild-type cells transduced with control shRNA (Figures 6E,S1). This was confirmed by comparing quantified bioluminescence intensities between comparable regions of interest. For Bcr-Abl T315I-transduced cells, geometric mean bioluminescent intensity in control mice (scrambled shRNA) was 3.4-fold larger than the knockdown group at day 7 and 4.9-fold at day 14 (95% confidence intervals, 2.1- to 5.4-fold and 3.1- to 7.9-fold, respectively). Flow cytometric analysis of splenocytes from injected mice confirmed the presence of GFP+ cells (data not shown). Bioluminescence in control cells was similarly elevated above knockdown using unmutated Bcr-Abl transduced cells (Figure S1). Furthermore, Kaplan-Meier analysis demonstrated a 3-day increase in median survival time for mice injected with Ba/F3 T315I cells transduced with CREB shRNA compared with scrambled shRNA at both 5 × 105 cells injected (Figure 6F, P = .006) and 106 cells (data not shown, P = .014 by logrank test). For mice injected with 106 Ba/F3 Bcr-Abl wild-type cells, there was a 4-day increase in median survival using CREB shRNA transduction compared with scrambled shRNA (Figure S1, P = .09 by Wilcoxon-Mann-Whitney test).
CREB inhibits progression of leukemia in vivo. (A) Western blot analysis with CREB and tubulin antisera, demonstrating 2-fold increase in expression of CREB in T315I mutant of Bcr-Abl in murine pro-B lymphocyte line (Ba/F3) compared with wild-type Ba/F3 cells. (B) Western blot analysis after lentiviral transduction with CREB shRNAs demonstrating 90% inhibition. (C) Quantitative reverse transcription-PCR showing diminished CREB mRNA levels in transduced Ba/F3 T315I cells. (D) Trypan blue exclusion method performed in triplicate shows diminished growth after transduction with CREB shRNA compared with empty vector. (E) Bioluminescence imaging of SCID mice injected with 106 cells transduced with CREB shRNA or CREB scrambled shRNA lentivirus. Mice were imaged at days 7 and 14. Tumor burden is lower in CREB shRNA–injected mice. (F) Kaplan-Meier survival analysis of mice injected with 5 × 105 cells showing longer survival with CREB knockdown (n = 9) compared with scrambled shRNA (n = 9). All deaths were due to leukemia, except for a day 7 handling event in the scrambled group treated as a censored observation. Error bars in panels C and D represent SE.
CREB inhibits progression of leukemia in vivo. (A) Western blot analysis with CREB and tubulin antisera, demonstrating 2-fold increase in expression of CREB in T315I mutant of Bcr-Abl in murine pro-B lymphocyte line (Ba/F3) compared with wild-type Ba/F3 cells. (B) Western blot analysis after lentiviral transduction with CREB shRNAs demonstrating 90% inhibition. (C) Quantitative reverse transcription-PCR showing diminished CREB mRNA levels in transduced Ba/F3 T315I cells. (D) Trypan blue exclusion method performed in triplicate shows diminished growth after transduction with CREB shRNA compared with empty vector. (E) Bioluminescence imaging of SCID mice injected with 106 cells transduced with CREB shRNA or CREB scrambled shRNA lentivirus. Mice were imaged at days 7 and 14. Tumor burden is lower in CREB shRNA–injected mice. (F) Kaplan-Meier survival analysis of mice injected with 5 × 105 cells showing longer survival with CREB knockdown (n = 9) compared with scrambled shRNA (n = 9). All deaths were due to leukemia, except for a day 7 handling event in the scrambled group treated as a censored observation. Error bars in panels C and D represent SE.
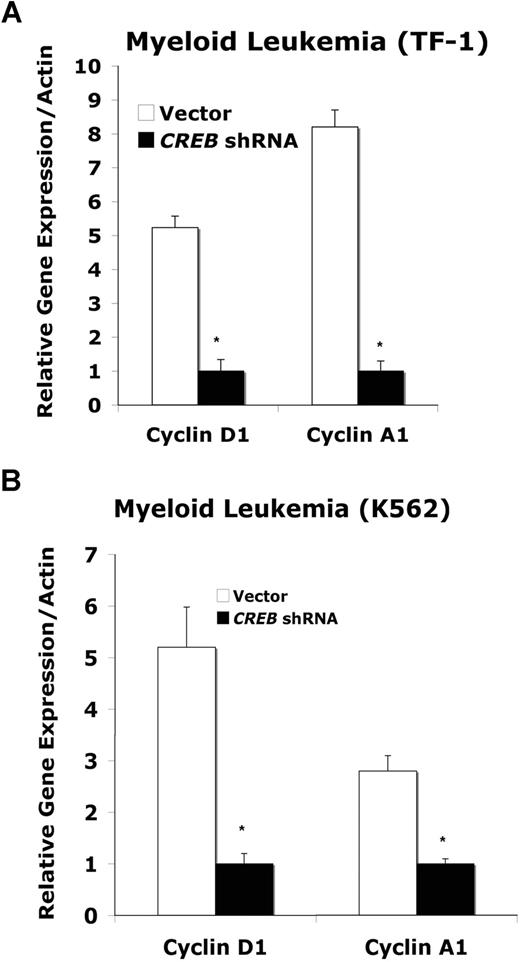
To understand possible downstream pathways mediating the effects of CREB down-regulation, including cell-cycle abnormalities, we sought to determine the levels of known CREB target genes, cyclins D and A, which both affect cycle progression from G1 to S. Cyclin A also regulates the G2/M transition.1 Cyclin D is known to be a direct target of CREB by serial analysis of gene expression (SAGE).39 Cyclin A has been reported to be induced by CREB and is a direct target gene of CREB.40 We observed a 5-fold decrease in cyclin D mRNA expression in TF-1 and K562 cells transduced with CREB shRNA compared with control cells (Figure 7A,B, P < .05).41 We also observed decreased cyclin A mRNA levels in CREB shRNA–transduced TF-1 and K562 cells (Figure 7A,B, P < .05). These results suggest that one possible reason for increased numbers of cells in G1 and G2/M in CREB shRNA–transduced leukemia cells is the inhibition of known target genes, such as cell-cycle proteins, cyclin D, and cyclin A.
Expression of cyclins A and D in leukemia cells. TF-1 (A) or K562 (B) myeloid leukemia cells were transduced with control and CREB shRNA lentivirus and synchronized. At 12 hours, 5 μg of total RNA was isolated for q-PCR by SyberGreen method. Cyclin A1- and D1-specific primers were used, and expression was normalized to the house keeping gene β-actin. Experiments were performed in triplicate. Error bars represent SE.
Expression of cyclins A and D in leukemia cells. TF-1 (A) or K562 (B) myeloid leukemia cells were transduced with control and CREB shRNA lentivirus and synchronized. At 12 hours, 5 μg of total RNA was isolated for q-PCR by SyberGreen method. Cyclin A1- and D1-specific primers were used, and expression was normalized to the house keeping gene β-actin. Experiments were performed in triplicate. Error bars represent SE.
Discussion
We report that CREB can be successfully down-regulated through lentiviral transduction in murine and human primary stem cells and leukemia cell lines in vitro and in vivo. Our work demonstrates that CREB is an important regulator of stem-cell proliferation, survival, and differentiation during normal hematopoiesis. These observations are consistent with previous work suggesting that CREB is critical for neuronal differentiation and survival.42 Our results are also consistent with previous observations that when overexpressed, CREB acts as a proto-oncogene, resulting in increased myeloid cell proliferation and myeloproliferative disease.8,43 In this article, we show that inhibition of CREB expression leads to abnormal proliferation, survival, and cell-cycle regulation of normal HSCs and leukemia cells.
Analysis of CREB expression in normal HSC populations support a role for CREB in uncommitted progenitors, because lineage-negative stem cells have higher levels of CREB expression compared with lineage-positive cells. In colony assays, CREB shRNA had a more dramatic effect on murine HSC proliferation than differentiation. Furthermore, there is an increase in less mature Gr-1+ /Mac-1+ cells compared with differentiated Gr+ 1 or Mac-1+ cells. It is noteworthy that the effects of CREB shRNA, as shown in colony assays, seemed to affect monocyte differentiation more than granulocyte differentiation. Reduced proliferation of human peripheral blood CD34+ CD38− cells transduced with CREB shRNA suggests the dependence of these cells on CREB. CREB shRNA–transduced human peripheral blood CD34+ CD38− HSCs also appeared to undergo apoptosis more readily than CD34+ CD38+ HSCs. Although CREB down-regulation resulted in increased apoptosis, this does not seem to be a major factor in the role of CREB in differentiation. Future studies will focus on analysis of target genes and pathways that regulate hematopoietic differentiation and proliferation downstream of CREB.
CREB also affected early engraftment of normal HSCs in lethally irradiated mice. There were more immature myeloid cells that were Gr-1/Mac-1+ at 5 and 12 weeks after transplantation (although more dramatic at 5 weeks), which is consistent with in vitro colony assays. One explanation for this discrepancy is that the CFU-GM colonies were counted after 2 weeks, whereas cells from mice were analyzed after 5 and 12 weeks. The numbers of terminally differentiated Gr-1+ or Mac-1+ cells were the same in CREB and control shRNA-transduced HSCs at 5 and 12 weeks after transplant. It is possible that other CREB family members (eg, ATF2) are able to compensate for the decreased levels of CREB in myeloid cells transduced with CREB shRNA. Furthermore, the overall survival of mice that received transplants was not affected by CREB shRNA. These results suggest that, in vivo, additional mechanisms or redundant pathways could be responsible for regulating HSCs under stress conditions and that CREB is not required for early hematopoietic reconstitution.
CREB seems to regulate leukemia cell proliferation in vitro and in vivo. Our ability to successfully suppress but not completely inhibit the growth of several leukemia cell lines suggests that CREB is one of several transcriptional factors or signal transduction pathways driving leukemic proliferation. CREB shRNA not only affected proliferation, but also phosphorylation and transcriptional activity of CREB in acute myeloid leukemia cells. Our results demonstrate that CREB is necessary for maximal proliferation of myeloid leukemia cells in vitro and that CREB is probably one but not the only critical target of signaling pathways regulating growth of these cells. CREB may ultimately function in a role analogous to C/EBPα and PU.1, which have been shown to be critical in early myeloid hematopoiesis and leukemia.44
We observed that CREB shRNA in vivo inhibited early leukemic progression but did not significantly prolong maximal survival. This may be due in part to the aggressive nature of Bcr-Abl wild type and T315I mutation expressed in Ba/F3 cells injected into SCID mice. Our results also suggest that CREB-dependent signaling pathways are critical for Bcr-Abl T315I cell proliferation. The time course of splenic and bone marrow infiltration was delayed with CREB shRNA–transduced cells compared with control cells. This suggests the possibility that CREB may be affecting initial homing or engraftment of leukemia cells in the spleen and bone marrow. However, the pattern of progression (ie, organs infiltrated) appeared to be the same between CREB shRNA and control cells. Alternatively, CREB may be slowing down the initial growth of cells in mice, resulting in a longer latency period for the leukemia to kill the mice. Experiments are in progress to identify the pathways regulating leukemic progression of Bcr-Abl–expressing cells.
Our results demonstrated that CFU-GM was decreased in mouse bone marrow progenitor colony assays transduced with CREB shRNA. In the transplantation experiments, the mice had increased Gr-1+ /Mac-1+ cells with CREB shRNA. Except for the Mac-1 population in colony assays, CREB shRNA had minimal effect on terminal differentiation of myeloid and lymphoid populations in vitro and in vivo (Figure 4 and data not shown). This could reflect the contribution of CREB in monocyte differentiation, under the specific culture conditions. The differences in the mouse bone marrow colony assays at day 14 and knockdown of CREB did not seem to affect hematopoietic reconstitution after 12 weeks and only minimally induced apoptosis in progenitor cells. In contrast, myeloid leukemia cells transduced with CREB shRNA had significant inhibition in growth and increased apoptosis. Taken together, our data suggest that CREB may be a viable target for leukemia therapy.
CREB knockdown resulted in aberrant progression from G1 to S and exit from G2/M to G1, suggesting a critical link between CREB and the cell-cycle machinery in normal HSCs and leukemia cells. Known CREB target genes include cyclin D1, which activates G1/S cyclin-dependent kinases Cdk4 and Cdk6.39 Indeed, our results demonstrated that CREB knockdown inhibited cyclin D1 expression, which could provide a mechanism by which cells arrest in G1 and fail to progress to S phase. Likewise, cyclin A1 has a CREB binding site in its promoter, suggesting that cells could fail to progress to S phase or exit from G2/M as a result of decreased expression of cyclin A and failure to activate Cdk1 and Cdk2. It is noteworthy that CREB transgenic mice develop myeloproliferative disease with dysplastic myeloid cells in the spleen. CREB overexpression in myeloid leukemia cell lines and mouse spleens was associated with increased cyclin A expression and increased percentage of cells in S phase.9 Thus, it is possible that by being a positive regulator of cyclins D and A, CREB overexpression could lead to genomic instability and ultimately transformation. In this article, we have shown that CREB down-regulation led to cell-cycle arrest and effects opposite of those observed with CREB overexpression. The key to understanding the role of CREB is to elucidate the target genes that mediate its effects in proliferation and survival. Future work will focus on the downstream pathways regulating CREB function during normal and aberrant hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health grants HL75826 (K.M.S.), HL83077 (K.M.S.), CA16042 (I.S. and E.L.), AI28697 (I.S.), 2P30-DK041301 (N.K.), and F32-HL085013 (J.C.); American Cancer Society grant RSG-99-081-01-LIB (K.M.S.); and Department of Defense grant CM050077 (K.M.S.). K.M.S. is a scholar of the Leukemia and Lymphoma Society.
Authorship
Contribution: J.C. wrote the manuscript and designed and performed experiments. K.K., W.S.W., D.J., and J.C. performed experiments. I.S. helped analyze data, supervised experiments, and edited the manuscript. D.B.S. helped design experiments. N.K. contributed reagents. R.B. edited the manuscript and consulted on project. E.M.L. performed statistical analysis and wrote the manuscript. K.M.S. supervised the research, designed experiments, contributed to analysis, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Kathleen M. Sakamoto, Division of Hematology/Oncology, Mattel Children's Hospital, David Geffen School of Medicine at UCLA, Los Angeles, CA 90095-1752; e-mail: kms@ucla.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal