Abstract
Platelet glycoprotein VI (GPVI) is a key receptor for collagens that mediates the propagation of platelet attachment and activation. Targeted disruption of the murine gene Gp6 on a mixed 129 × 1/SvJ × C57BL/6J background causes the expected defects in collagen-dependent platelet responses in vitro. The extent of this dysfunction in all Gp6−/− mice is uniform and is not affected by genetic background. However, the same Gp6−/− mice exhibit 2 diametrically opposed phenotypes in vivo. In some mice, tail bleeding times are extremely prolonged, and thrombus formation in an in vivo carotid artery ferric chloride-injury model is significantly impaired. In other littermates, tail bleeding times are within the range of wild-type mice, and in vivo thrombus formation is indistinguishable from that of control mice. Directed intercrosses revealed that these phenotypes are heritable, and a genome-wide single-nucleotide polymorphism scan revealed the most significant linkage to a single locus (8 megabases) on chromosome 4 (logarithm of the odds [LOD] score = 6.9, P < .0001) that we designate Modifier of hemostasis (Mh). Our results indicate that one or more modifier genes in Mh control the extent to which in vivo platelet thrombus formation is disrupted by the absence of platelet GPVI.
Introduction
Glycoprotein VI (GPVI), which is restricted in expression to megakaryocytes and platelets, is an important receptor for colla-gens. GPVI (60-65 kDa) is noncovalently associated with the fragment crystallizable receptor γ (FcRγ) subunit, an immunoreceptor tyrosine-based activation motif (ITAM)-based signaling coreceptor,1,2 and in the mouse, surface GPVI expression requires coexpression of FcRγ.3 Upon engagement of collagens, the GPVI-FcRγ complex transduces outside-in signals that involve spleen tyrosine kinase (Syk), linker for the activation of T cells (LAT), and Src homology 2 domain-containing leukocyte-specific phosphoprotein of 76 kDa (SLP-76), resulting in phospholipase γ2 and phosphatidylinositol 3-kinase activation, leading to release of granule contents, prothrombinase activity, and platelet aggregation.4-7
Targeted gene deletion in the mouse is one strategy to generate experimental models to assess the role of membrane receptors in hemostasis and thrombosis. We were the first to report the targeted disruption of murine Gp68 and demonstrated, using functional in vitro assays, that platelets from homozygous Gp6−/− mice fail to aggregate upon stimulation with type I fibrillar collagen and fail to form thrombi upon perfusion over collagen-coated surfaces. However, the in vivo tail bleeding times for the same Gp6−/− mice were more often within the range seen with wild-type littermates (45-140 seconds), and only a minority of the Gp6−/− F2 littermates (3/13) exhibited an abnormal, extremely prolonged tail bleeding time (> 600 seconds). Similar proportions of normal and abnormal mice have been observed in studies of genetically engineered FcRγ−/− mice, in which platelet surface GPVI expression is ablated, or wild-type mice made GPVI-deficient after 7 days by intraperitoneal injection of rat anti-mouse GPVI monoclonal antibody JAQ1.9-11
On repeated examination, we determined that these differences in bleeding phenotype of Gp6−/− littermates in vivo are not only reproducible,8 but also transmitted as a hereditary trait in mice with a mixed background, indicating that the in vivo bleeding tendency is being modified by underlying genetic differences. In this study, we have defined the location of a single modifier gene locus on chromosome 4 that exhibits the highest linkage to the bleeding phenotype in Gp6−/− mice.
Methods
Approval for these studies was obtained from the Scripps Research Institute Animal Care Review Board.
Reagents
Alexa Fluor 488 (Invitrogen, Carlsbad, CA)-conjugated hamster monoclonal anti–mouse integrin α2 (HM α2) and unconjugated hamster anti–mouse integrin α1 (Ha31/8) were purchased from BD Pharmingen (San Jose, CA). Fluorescein isothiocyanate (FITC)-conjugated rat anti–mouse GPVI (6.E10), FITC-conjugated anti–mouse integrin α5 (Tap.A12), FITC-conjugated anti–mouse GPV (Gon.C2), Phycoerythrin (PE)-conjugated rat anti–mouse αIIbβ3 (Leo.D2), and PE-conjugated anti–mouse GPIbα (Xia.G5) were purchased from Emfrets Analytics (Wurzburg, Germany). FITC-conjugated rat anti–mouse α6 was purchased from Serotec (Raleigh, NC). FITC-conjugated goat anti–hamster IgG heavy plus light chains (H + L) was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Mouse platelet preparation
Mouse blood (0.3 mL) was obtained from the retroorbital venous plexus of anesthetized mice through a heparinized glass capillary tube (Thermo Fisher Scientific, Waltham, MA). For ex vivo perfusion studies, the blood was transferred immediately into plastic tubes containing heparin at a final concentration of 40 U/mL (Sigma, St Louis, MO). Otherwise, 9 volumes of blood were admixed with 1 volume of 3.2% (wt/vol) sodium citrate (105 mmol/L). For the coagulation studies only, the blood was obtained through plain (nonheparinized) glass capillary tubes, and 9 volumes of blood was mixed with 1 volume of 120 mmol/L sodium citrate and 10 mmol/L HEPES pH 7.4. In certain assays in which larger volumes of blood are required, the blood from several phenotype-identical mice was pooled to perform an experiment.
Platelet aggregation
Aggregation assays required pooled blood drawn from 6 to 8 male mice with identical GPVI phenotype (LONG or SHORT; see “Bleeding time assays”). Platelet-rich plasma (PRP) was obtained by centrifugation of whole blood at 500g for 6 minutes, and platelet-poor plasma (PPP) was obtained by centrifuging PRP at 2000g for 7 minutes. Platelet count and mean platelet volume (MPV) were measured using a Coulter 9000 apparatus (Mallinckrodt Baker, Phillipsburg, NJ), and the platelet count was normalized to 2.4 × 105/μL with PPP. Four hundred microliters of PRP was prewarmed at 37°C. Horse tendon collagen (2 μg/mL) or adenosine diphosphate (ADP; 10 μmol/L) were the selected agonists. Aggregation profiles were generated in a Chrono-Log aggregometer (Havertown, PA), and PPP was used as a blank for the aggregation study.
Coagulation assays
For the activated partial thromboplastin time (APTT) test, the reagent used was Platelin LS from bioMerieux (Durham, NC). For the prothrombin time (PT) test, the reagent used was Innovin from DADE Behring (Newark, DE). Innovin contains recombinant human tissue factor and a mixture of synthetic phospholipids (20% phosphatidylserine and 80% phosphatidylcholine). The amount of tissue factor present in the undiluted solution is 4.1 nmol/L. When used at 1/300 dilution, the concentration of tissue factor added to the clotting mixture is 13.6 pmol/L.
Bleeding time assays
The bleeding phenotype of each Gp6−/− mouse was defined by the outcome of 2 tail bleeding times that had to be confirmatory. Bleeding times were performed on each Gp6−/− mouse (both male and female; 2-6 months in age) on 2 separate occasions. The mice were considered to bear a legitimate phenotype only if the initial bleeding time was confirmed on the second occasion, and only mice with confirmed bleeding times were used in subsequent genetic and functional experiments. Gp6−/− mice with confirmed bleeding times that fell within the upper limit of the range for wild-type mice (250 seconds) are referred to as Gp6−/− SHORT mice; those with confirmed, excessively prolonged bleeding times (> 600 seconds) that stopped only after physical intervention are referred to as Gp6−/− LONG mice.
Bleeding times were performed as described previously.8 In brief, nonanesthetized mice were immobilized in a cylindrical Plexiglas restrainer. A 2- to 3-mm distal section of the tail was removed with a sterile scalpel blade, and the tail was immediately immersed in a container of isotonic saline maintained at 37°C. A complete cessation of bleeding was defined as the end of the bleeding time. If, after 600 seconds, bleeding had not yet ceased, pressure was applied with a sterile gauze pad to the tail until bleeding ceased, and the bleeding time was recorded as 600 seconds. A second bleeding time was performed 2 to 3 weeks after the first occasion.
Carotid artery injury model
The carotid artery of anesthetized mice was injured with FeCl3 according to a standardized protocol.12 In brief, the common carotid artery was dissected free from the surrounding tissue and a miniature ultrasound flow probe (0.5 VB; Transonic Systems, Ithaca, NY) was positioned around the vessel. The flow probe was interfaced with a flow meter (T106; Transonic Systems) and a computer-based data acquisition program (WinDaq Lite; DATAQ Instruments, Akron, OH). After measuring baseline flow in the artery, a 0.5 × 1.0-mm strip of filter paper (No. 1; Whatman, Clifton, NJ) soaked in 10% FeCl3 was applied to the surface of the adventitia for 3 minutes. After removing the strip, the area surrounding the vessel was carefully washed with warm physiologic saline solution, and carotid blood flow monitoring was resumed and continued for 25 minutes after the injury. The artery was considered occluded when flow was below 0 plus or minus 0.2 mL/min, a range corresponding to the accuracy of the system (zero offset) as specified by the manufacturer.
Ex vivo perfusion studies
The blood from 5 to 8 mice was pooled to perform each experiment. Mepacrine (10 μmol/L) was added to render platelets fluorescent8 before perfusing the blood over immobilized fibrillar type I collagen (acid insoluble) coating a glass coverslip13 or extracellular matrices prepared from mouse fibroblasts cultured on glass coverslips. The coverslips were assembled at the bottom of a Hele-Shaw parallel plate flow chamber with linearly variable shear rate decreasing from the inlet to the outlet14 or a rectangular chamber with constant shear rate across the flow field. Events on the surface were visualized by epifluorescence and confocal video microscopy, and analyzed as described previously to obtain direct quantitative thrombus volume measurements.13,15,16
Flow cytometry
Mouse whole blood (2 μL) was mixed with 18 μL of Tyrode's buffer (12 mmol/L NaHCO3, 2 mmol/L MgCl2, 137.5 mmol/L NaCl, and 2.6 mmol/L KCl, pH 7.4) containing 1% (wt/vol) bovine serum albumin and incubated with 5 μL of FITC- or PE-conjugated anti–mouse antibody for 15 minutes. In the case of anti–mouse integrin α1 only, 10 μg/mL was added, and after 60-minute incubation at ambient temperature, FITC-conjugated secondary antibody (1:100 dilution) was added for an additional 30 minutes. Then, 400 μL of 0.008 mol/L Na2HPO4, 0.002 mol/L KH2PO4, pH 7.4, containing 0.138 mol/L NaCl and 0.0027 mol/L KCl, was added to all samples, and the amount of bound antibody was determined within 30 minutes using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Data were analyzed with BD CellQuestPro software, and the amount of bound antibody was reported as the geometric mean fluorescence intensity (GMFI).
Polymerase chain reaction
Genomic DNA was extracted from mouse tail or ear sections using the DNeasy tissue kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. An amplicon encompassing a 4.5-kilobbase pair (kb) segment of the wild-type sequence or a 6.2-kb sequence of the mutated gene8 was generated using LA Taq Polymerase according to manufacturer's instructions (Takara Bio USA, Madison, WI). The primer pair was GPVIF (5′-GACTGTGCTTTACCTGTTTCTG-3′), situated at nucleotides −4430 to −4407, relative to the transcription initiation site, and GPVIR (5′-CCAATACAGAAGAAAGTGGGTG-3′), situated at nucleotides +49 to +71.
Genome-wide linkage scan
Single-nucleotide polymorphisms (SNPs) were identified by direct sequencing of polymerase chain reaction (PCR) products generated for each of the SNP loci from 8 inbred mouse strains, including C57BL/6J and 129 × 1/SvJ.17 For the first low-resolution scan (roughly 10-Megabase [Mb] intervals), 355 SNPs were selected. SNP assays are performed using the Sequenom MassARRAY system, and primers for PCR and single base extension were designed with the use of the SPECTRODESIGNER software package (Sequenom, San Diego, CA).
Results
The Gp6−/− mice used in this study were either F2 or the progeny of multiple, consecutive intercrosses between F2 Gp6−/− mice, and all mice were bred to maintain a mixed genetic background. Wild-type mice are either F2 littermates that were determined to be genetically Gp6+/+ or the progeny of multiple, consecutive intercrosses between these Gp6+/+ mice. Platelets from female mice are more resistant to inhibitors of platelet function than platelets from male mice, regardless of strain.18 Because of this sex difference, we used only male mice for our in vitro functional studies, unless otherwise specified.
All Gp6−/− mice exhibit a single 6.2-kb amplicon, the increased size of which results from the insertion of the neomycin-resistance cassette (neo) into exon 18 ; the amplicon from wild-type littermates is 4.5 kb, the anticipated length of the native Gp6 genomic sequence.
Platelet surface expression of GPVI and other relevant receptors
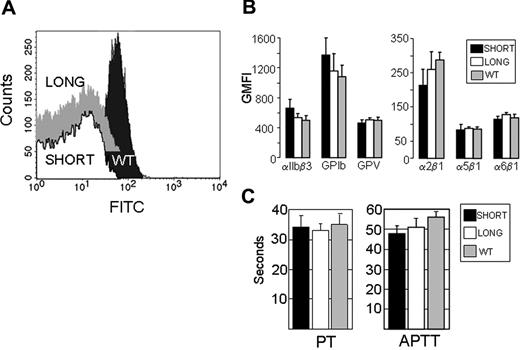
To measure platelet surface expression of GPVI protein by flow cytometry (Figure 1A), we used a monoclonal rat anti-mouse GPVI (6.E10) (Emfrets Analytics). Representative histograms are depicted. Wild-type mice express normal levels of GPVI (black peak), whereas background antibody binding is found with Gp6−/− mice, whether they are phenotyped as Gp6−/− SHORT (Figure 1A white peak) or Gp6−/− LONG (Figure 1A gray peak). In this figure, the Gp6−/− LONG peak is shifted vertically upward to enable it to be distinguished from the virtually identical Gp6−/− SHORT peak.
Characterization of Gp6−/− mice. (A) Platelet surface expression of GPVI. Platelet surface expression of GPVI was measured by flow cytometry of whole blood using monoclonal rat anti-mouse GPVI (6.E10). Representative histograms depict platelets from WT mice (dark gray peak), Gp6−/− SHORT mice (white peak), or Gp6−/− LONG mice (gray peak). The Gp6−/− LONG peak is shifted vertically upward to enable it to be distinguished from the virtually identical Gp6−/− SHORT peak. Values on the abscissa represent the FITC fluorescence intensity. These results are typical of measurements performed on more than 30 mice with each phenotype. (B) Platelet surface expression of other relevant receptors. In flow cytometry, rat monoclonal antibodies were used to quantitate integrins αIIbβ3, α2β1, α5β1, and α6β1, as well as platelet GPIb and GPV. In each case, the GMFI (mean ± SD for 5 mice) is depicted. None of the differences were statistically significant (P > .05). (C) Blood coagulation assays. Plasmas from 5 WT, 5 Gp6−/− LONG, and 5 Gp6−/− SHORT mice were individually analyzed, and the mean plus or minus SD for each group is depicted. To increase the sensitivity, the prothrombin time (PT) assay (left panel) was carried out with a 1/300 dilution of tissue factor reagent. The right panel depicts the results of the activated partial thromboplastin time (APTT) assay. The differences in PT or APTT between Gp6−/− LONG and Gp6−/− SHORT mice were not statistically different (P > .05).
Characterization of Gp6−/− mice. (A) Platelet surface expression of GPVI. Platelet surface expression of GPVI was measured by flow cytometry of whole blood using monoclonal rat anti-mouse GPVI (6.E10). Representative histograms depict platelets from WT mice (dark gray peak), Gp6−/− SHORT mice (white peak), or Gp6−/− LONG mice (gray peak). The Gp6−/− LONG peak is shifted vertically upward to enable it to be distinguished from the virtually identical Gp6−/− SHORT peak. Values on the abscissa represent the FITC fluorescence intensity. These results are typical of measurements performed on more than 30 mice with each phenotype. (B) Platelet surface expression of other relevant receptors. In flow cytometry, rat monoclonal antibodies were used to quantitate integrins αIIbβ3, α2β1, α5β1, and α6β1, as well as platelet GPIb and GPV. In each case, the GMFI (mean ± SD for 5 mice) is depicted. None of the differences were statistically significant (P > .05). (C) Blood coagulation assays. Plasmas from 5 WT, 5 Gp6−/− LONG, and 5 Gp6−/− SHORT mice were individually analyzed, and the mean plus or minus SD for each group is depicted. To increase the sensitivity, the prothrombin time (PT) assay (left panel) was carried out with a 1/300 dilution of tissue factor reagent. The right panel depicts the results of the activated partial thromboplastin time (APTT) assay. The differences in PT or APTT between Gp6−/− LONG and Gp6−/− SHORT mice were not statistically different (P > .05).
The surface expression of each relevant platelet receptor that was tested is essentially identical in Gp6−/− LONG or Gp6−/− SHORT mice (Figure 1B). For these assays, rat monoclonal antibodies (Emfrets) were used to quantitate integrins αIIbβ3, α2β1, α5β1, and α6β1, as well as platelet glycoproteins Ib and V. In each case, the GMFI (mean ± SD for 5 mice) is depicted. None of the differences were statistically significant (P > .05).
Platelet count and MPV
Platelet counts (× 103 per μL; mean ± SD) were 671 (± 112) for wild-type mice (n = 16); 692 (± 99) for Gp6−/− SHORT mice (n = 18); and 623 (± 53) for Gp6−/− LONG mice (n = 14). The corresponding MPV values (in femtoliters) were 5.98 (± 0.1), 5.84 (± 0.07), and 5.93 (± 1). Thus, there is no difference in mean platelet count or MPV between Gp6−/− LONG and Gp6−/− SHORT mice.
Coagulation tests
For coagulation assays, plasmas from 5 wild-type, 5 Gp6−/− LONG, and 5 Gp6−/− SHORT mice were individually analyzed, and the mean plus or minus SD for each group is depicted in Figure 1C. To increase the sensitivity, the PT assay (left panel) was carried out with a 1/300 dilution of tissue factor reagent. The right panel depicts the results of the APTT assay. There was no difference in the PT and APTT between Gp6−/− LONG and Gp6−/− SHORT mice. In addition, no significant difference was observed between the PT when undiluted tissue factor reagent was used (wild-type = 14.0 ± 0.4 seconds, Gp6−/− SHORT = 13.5 ± 0.6 seconds, and Gp6−/− LONG = 13.6 ± 0.5 seconds).
Tail bleeding time
The sequential bleeding times for each of 262 Gp6−/− mice and 28 wild-type littermates are depicted in Figure 2. For each mouse, the initial bleeding time (BT1) to the left of the panel is connected to the second bleeding time (BT2) to the right of the panel by a straight line. Based on the actual bleeding times and the reproducibility between BT1 and BT2, we established 4 categories among the 262 Gp6−/− mice (Figure 2A-D). For 120 mice, both bleeding times reached the maximal limit of 600 seconds and were deliberately stopped by physical pressure to the cut tail area (Figure 2A). In 2 mice, the second bleeding time was decreased by no more than 10% relative to the initial bleeding time. These 122 mice were designated Gp6−/− LONG. In 24 mice (Figure 2B), the initial bleeding time of 600 seconds was not confirmed because of a significantly shortened second bleeding time. In 50 mice (Figure 2C), the second bleeding time was 600 seconds and significantly higher than the first bleeding time. The mice in categories B and C were excluded from subsequent genetic linkage analyses because it was not possible to confidently assign a bleeding phenotype. Thus, 74 of 262 mice (28%) were excluded for this reason. In the remaining 66 Gp6−/− mice (Figure 2D), there was excellent reproducibility between the 2 bleeding times, and each mouse had an average time within the range of values obtained for wild-type (Gp6+/+) littermates. These mice were designated Gp6−/− SHORT. Among them, BT2 was shorter than BT1 in 37 mice (56%), and BT2 was longer than BT1 in 29 mice (44%). The consecutive bleeding times for the 28 wild-type littermates are depicted in Figure 2E (mean ± SD = 114 ± 27 seconds; range = 47-198 seconds).
Tail bleeding time. Sequential tail bleeding times (BT1 followed by BT2) are shown for 262 Gp6−/− mice and 28 Gp6+/+ littermates. In the case of Gp6−/− mice, 4 categories (A through D) were defined based on the reproducibility of the BT, depicted on the ordinate (in seconds). (A) In 120 mice, both bleeding times reached the maximal limit of 600 seconds and were deliberately stopped by physical pressure to the cut tail area. In 2 mice, the second bleeding time was decreased by no more than 10% relative to the initial bleeding time. These 122 mice were designated Gp6−/− LONG. (B) In 24 mice, the initial bleeding time of 600 seconds was not confirmed because of a significantly shortened second bleeding time. (C) In 50 mice, the second bleeding time was 600 seconds and significantly higher than the first bleeding time. The mice in categories B and C were excluded from subsequent genetic linkage analyses. (D) In 66 mice, there was excellent reproducibility between the 2 bleeding times, and each mouse had an average time within the range of values obtained for Gp6+/+ mice. These mice were designated Gp6−/− SHORT. (E) Consecutive bleeding times for 28 wild-type (Gp6+/+) littermates (mean ± SD = 114 ± 27 seconds; range = 47-198 seconds).
Tail bleeding time. Sequential tail bleeding times (BT1 followed by BT2) are shown for 262 Gp6−/− mice and 28 Gp6+/+ littermates. In the case of Gp6−/− mice, 4 categories (A through D) were defined based on the reproducibility of the BT, depicted on the ordinate (in seconds). (A) In 120 mice, both bleeding times reached the maximal limit of 600 seconds and were deliberately stopped by physical pressure to the cut tail area. In 2 mice, the second bleeding time was decreased by no more than 10% relative to the initial bleeding time. These 122 mice were designated Gp6−/− LONG. (B) In 24 mice, the initial bleeding time of 600 seconds was not confirmed because of a significantly shortened second bleeding time. (C) In 50 mice, the second bleeding time was 600 seconds and significantly higher than the first bleeding time. The mice in categories B and C were excluded from subsequent genetic linkage analyses. (D) In 66 mice, there was excellent reproducibility between the 2 bleeding times, and each mouse had an average time within the range of values obtained for Gp6+/+ mice. These mice were designated Gp6−/− SHORT. (E) Consecutive bleeding times for 28 wild-type (Gp6+/+) littermates (mean ± SD = 114 ± 27 seconds; range = 47-198 seconds).
Ex vivo perfusion studies
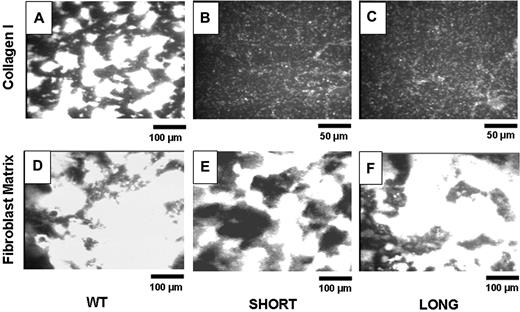
The blood from 5 to 8 mice was pooled to perform each experiment. Figure 3 depicts representative images of adhesion of platelets from wild-type mice (Figure 3A,D), Gp6−/− SHORT mice (Figure 3B,E) and Gp6−/− LONG mice (Figure 3C,F) to insoluble, fibrillar type I collagen (Figure 3A-C) or mouse fibroblast matrix (Figure 3D-F). The bar in the bottom right corner of each panel indicates distance, such that the magnification in Figure 3B,C is twice that of the remaining panels. It is evident that platelets from both Gp6−/− SHORT and Gp6−/− LONG mice fail to adhere to type 1 collagen (Figure 3B,C) under these conditions but adhere normally to and facilitate normal thrombus formation on fibroblast matrix (Figure 3E,F). At this time, the components of fibroblast matrix that support platelet adhesion and activation remain to be precisely determined. Nonetheless, these findings confirm that platelets in whole blood from Gp6−/− mice fail to adhere ex vivo to fibrillar type I collagen, regardless of the in vivo bleeding phenotype. On the other hand, their normal adhesion to fibroblast matrix attests to the specificity of the GPVI defect.
Ex vivo perfusion studies. Blood from 5 to 8 mice was pooled to perform each experiment. Representative images show platelet adhesion in WT mice (panels A and D), Gp6−/−SHORT mice (panels B and E), or Gp6−/− LONG mice (panels C and F) to insoluble, fibrillar Type I collagen (panels A-C) or mouse fibroblast matrix (panels D-F). These results are representative of more than 4 experiments using genetically similar mice. The bar in the bottom right corner of each panel indicates distance, such that the magnification in panels B and C is twice that of the remaining panels.
Ex vivo perfusion studies. Blood from 5 to 8 mice was pooled to perform each experiment. Representative images show platelet adhesion in WT mice (panels A and D), Gp6−/−SHORT mice (panels B and E), or Gp6−/− LONG mice (panels C and F) to insoluble, fibrillar Type I collagen (panels A-C) or mouse fibroblast matrix (panels D-F). These results are representative of more than 4 experiments using genetically similar mice. The bar in the bottom right corner of each panel indicates distance, such that the magnification in panels B and C is twice that of the remaining panels.
Platelet aggregation
The amount of PRP required to complete a set of aggregation assays necessitates the pooling of blood from 6 to 8 mice. Because platelets from female mice have an increased sensitivity to agonists compared with those from male mice, regardless of strain,18 we selected only male mice for the platelet aggregation tests.
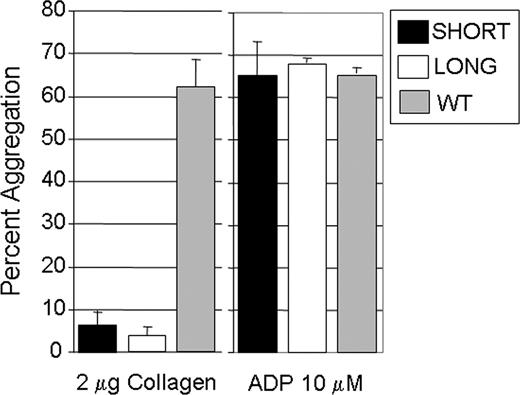
Platelet aggregation was induced by addition of collagen (2 μg/mL) or 10 μmol/L ADP to PRP (Figure 4). When exposed to ADP, no difference in the extent of aggregation was observed (Figure 4 right panel). When stimulated with collagen (2 μg/mL) (Figure 4 left panel), aggregation of platelets from Gp6−/− SHORT (17 ± 5%; mean ± SD) or Gp6−/− LONG (14 ± 2%) mice was comparable and significantly lower than that seen in wild-type littermates (62 ± 7%; P < .01 in both cases).
In vitro platelet aggregation. For these assays, blood was pooled from 6 to 8 male mice. Platelet aggregation was induced by addition of collagen (2 μg/mL) (left panel) or 10 μmol/L ADP (right panel). When exposed to ADP, no difference in the extent of aggregation was observed (right panel). When stimulated with collagen (2 μg/mL) (left panel), aggregation of platelets from Gp6−/− SHORT mice (17 ± 5%; mean ± SD) or Gp6−/− LONG mice (14 ± 2%) was comparable and significantly lower than that seen in wild-type littermates (62 ± 7%; P < .01 in both cases). The mean (± SD) for 3 separate assays is shown.
In vitro platelet aggregation. For these assays, blood was pooled from 6 to 8 male mice. Platelet aggregation was induced by addition of collagen (2 μg/mL) (left panel) or 10 μmol/L ADP (right panel). When exposed to ADP, no difference in the extent of aggregation was observed (right panel). When stimulated with collagen (2 μg/mL) (left panel), aggregation of platelets from Gp6−/− SHORT mice (17 ± 5%; mean ± SD) or Gp6−/− LONG mice (14 ± 2%) was comparable and significantly lower than that seen in wild-type littermates (62 ± 7%; P < .01 in both cases). The mean (± SD) for 3 separate assays is shown.
Inheritance
Five intercrosses between Gp6−/− LONG F2 parents generated 38 progeny, all of which were phenotyped Gp6−/− LONG. On the other hand, 3 intercrosses between Gp6−/− SHORT F2 parents generated 19 mice, 5 of which were phenotyped Gp6−/− LONG and 14 of which were phenotyped Gp6−/− SHORT. The most likely conclusion from these preliminary results based solely on the bleeding time would be that Gp6−/− LONG is an autosomal recessive trait. However, subsequent results from the in vivo carotid artery injury model are more consistent with a codominant mode of inheritance.
Linkage analyses
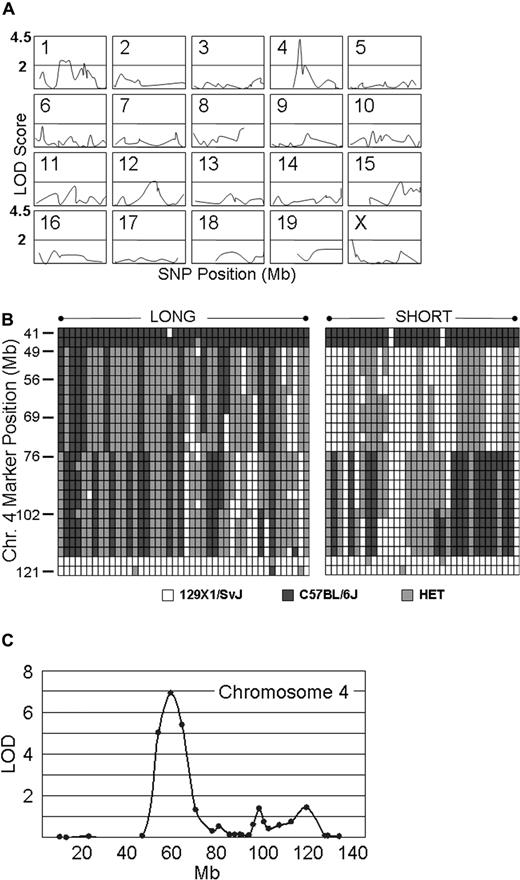
Genotyping assays were carried out on 34 Gp6−/− SHORT and 44 Gp6−/− LONG mice. The results were analyzed by MAPMANAGER QTX, which generates a linkage statistic that can be converted to a LOD score. Figure 5A summarizes schematically the output of the statistical analysis of linkage for the 274 most informative SNPs of the 355 selected for the first screen. Because of the extensive intercrosses conducted to generate the mice in this study, certain segments of chromosomes have already become congenic. These congenic segments are represented by the absence of a plotted line in any of the panels. For example, one of these DNA segments is located in the upstream region of Chr 4 (≤ roughly position 45 000 000), where the C57BL/6J haplotype was found in all mice tested. The most significant linkage was to a locus on chromosome 4, and the SNP with the highest LOD score of 4.2 (P < .001) was located at position chr4:63 714 000 (marker = rs13477745). It is noteworthy that, in the unaffected (SHORT) group, 34 of 34 mice were either homozygous or heterozygous for the 129 × 1/SvJ chromosome 4 haplotype represented by this SNP. For the remaining 18 mouse autosomes and the X chromosome, none of the SNPs that we analyzed generated LOD scores higher than 2.5, whereas 97% were less than 2.0, and 84% were less than 1.0. Stratification of male and female mice before statistical analysis did not affect selection of this locus (not shown). The haplotypes on chromosome 4 for each the 44 Gp6−/− LONG and 34 Gp6−/− SHORT mice used in this genome wide linkage scan are depicted schematically in Figure 5B.
Genome-wide linkage scan. (A) Linkage analysis on 20 murine chromosomes. Genotyping assays were performed on 34 Gp6−/− SHORT and 44 Gp6−/− LONG mice. The results were analyzed by MAPMANAGER QTX, generating a linkage statistic that was converted to a LOD score. The chromosome number is indicated to the top left of each panel, wherein the LOD score (ordinate) is plotted against the SNP position (abscissa) on that chromosome. Congenic segments are represented by the absence of a plotted line in any of the panels. A horizontal bar in each panel indicates a LOD score of 2.0, and the top of each panel represents a LOD score of 4.5. The LOD score obtained for each SNP tested as a function of position (Mb) on each chromosome is plotted. The SNP with the highest LOD score of 4.2 (P < .001) is located at position chr4: 63 714 000 (marker = rs13477745). For the remaining 18 mouse autosomes and the X chromosome, none of the SNPs generated LOD scores more than 2.5, and 97% were less than 2.0 and 84% were less than 1.0. (B) Haplotype diagram of chromosome 4. This diagram schematically depicts the haplotypes for the 44 Gp6−/− LONG (LEFT).and 34 Gp6−/− SHORT (RIGHT) mice used in the initial genome wide linkage scan (see A). Each column represents the results for a single mouse. Each row depicts the typing results at a particular SNP, and the position of a selected number of key SNPs is indicated on the ordinate as the position from the 5′ end (TOP) in Mb. White boxes indicate the 129 × 1/SvJ allele; black boxes, the C57BL/6J allele; and gray boxes, heterozygosity. (C) Fine mapping on chromosome 4. Additional fine mapping of chromosome 4 using 76 SNPs conducted with 129 mice. Linkage to markers encompassing the locus from chr4:48 163 000 to chr4:69 206 000 remained highly significant, and rs13477745 remains the SNP with the highest LOD score of 6.9 (P < .001).
Genome-wide linkage scan. (A) Linkage analysis on 20 murine chromosomes. Genotyping assays were performed on 34 Gp6−/− SHORT and 44 Gp6−/− LONG mice. The results were analyzed by MAPMANAGER QTX, generating a linkage statistic that was converted to a LOD score. The chromosome number is indicated to the top left of each panel, wherein the LOD score (ordinate) is plotted against the SNP position (abscissa) on that chromosome. Congenic segments are represented by the absence of a plotted line in any of the panels. A horizontal bar in each panel indicates a LOD score of 2.0, and the top of each panel represents a LOD score of 4.5. The LOD score obtained for each SNP tested as a function of position (Mb) on each chromosome is plotted. The SNP with the highest LOD score of 4.2 (P < .001) is located at position chr4: 63 714 000 (marker = rs13477745). For the remaining 18 mouse autosomes and the X chromosome, none of the SNPs generated LOD scores more than 2.5, and 97% were less than 2.0 and 84% were less than 1.0. (B) Haplotype diagram of chromosome 4. This diagram schematically depicts the haplotypes for the 44 Gp6−/− LONG (LEFT).and 34 Gp6−/− SHORT (RIGHT) mice used in the initial genome wide linkage scan (see A). Each column represents the results for a single mouse. Each row depicts the typing results at a particular SNP, and the position of a selected number of key SNPs is indicated on the ordinate as the position from the 5′ end (TOP) in Mb. White boxes indicate the 129 × 1/SvJ allele; black boxes, the C57BL/6J allele; and gray boxes, heterozygosity. (C) Fine mapping on chromosome 4. Additional fine mapping of chromosome 4 using 76 SNPs conducted with 129 mice. Linkage to markers encompassing the locus from chr4:48 163 000 to chr4:69 206 000 remained highly significant, and rs13477745 remains the SNP with the highest LOD score of 6.9 (P < .001).
A subsequent fine mapping of chromosome 4 using 76 SNPs was conducted on 129 mice that included the original 68 and the progeny of additional intercrosses. As shown in Figure 5C, linkage to markers encompassing the locus from chr4:48 163 000 to chr4:69 206 000 remained highly significant, and rs13477745 remained the SNP with the highest LOD score of 6.9 (P < .001). The majority of mice (121) are either completely C57BL/6J, completely 129 × 1/SvJ, or completely heterozygous across the original Mh locus. Thus, typing of additional SNPs within this locus was not informative.
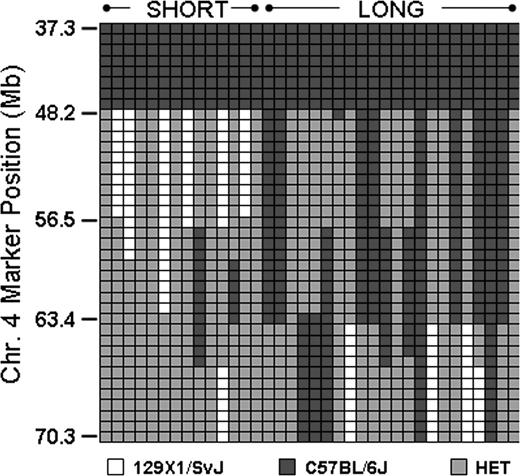
This obstacle was overcome through selected intercrosses between mice homozygous for one haplotype and mice heterozygous at this locus. In this manner, we identified several progeny with recombinations within Mh, and the results from 14 Gp6−/− SHORT and 22 Gp6−/− LONG progeny are depicted schematically in Figure 6. Each column represents a single mouse, and each row represents a SNP. Black indicates homozygous C57BL/6J, white is homozygous 129 × 1/SvJ, and gray is heterozygous. Mice with a SHORT phenotype are grouped to the left; those with a LONG phenotype are grouped to the right. A comparison of the bleeding phenotypes of these recombinant mice permitted us to further localize the Mh locus to an 8-Mb region between chr4:48 163 000 and chr4:56 497 000.
Analysis of mice with recombinant events occurring within Mh. In a selected number of progeny, recombinations occurred within Mh permitting a more precise resolution of the QTL. This diagram schematically depicts the haplotypes for representative number of mice: 14 Gp6−/− SHORT and 22 Gp6−/− LONG. Each column represents the results for a single mouse. Each row depicts the typing results at a particular SNP, and the position of a selected number of key SNPs is indicated on the ordinate. □ indicate the 129 × 1/SvJ allele; ■, the C57BL/6J allele; and  , heterozygosity. It is evident that the phenotype SHORT is segregating within a limited sequence ranging from 48 163 to 56 497.
, heterozygosity. It is evident that the phenotype SHORT is segregating within a limited sequence ranging from 48 163 to 56 497.
Analysis of mice with recombinant events occurring within Mh. In a selected number of progeny, recombinations occurred within Mh permitting a more precise resolution of the QTL. This diagram schematically depicts the haplotypes for representative number of mice: 14 Gp6−/− SHORT and 22 Gp6−/− LONG. Each column represents the results for a single mouse. Each row depicts the typing results at a particular SNP, and the position of a selected number of key SNPs is indicated on the ordinate. □ indicate the 129 × 1/SvJ allele; ■, the C57BL/6J allele; and  , heterozygosity. It is evident that the phenotype SHORT is segregating within a limited sequence ranging from 48 163 to 56 497.
, heterozygosity. It is evident that the phenotype SHORT is segregating within a limited sequence ranging from 48 163 to 56 497.
Carotid artery injury model
The determination of Mh haplotypes allowed us to evaluate more accurately the response of individual mice in the in vivo carotid artery injury model. In studies of 18 Gp6−/− mice, we observed a dichotomous response that roughly paralleled the findings made with the tail bleeding time but more precisely reflected Mh haplotypes (Figure 7).
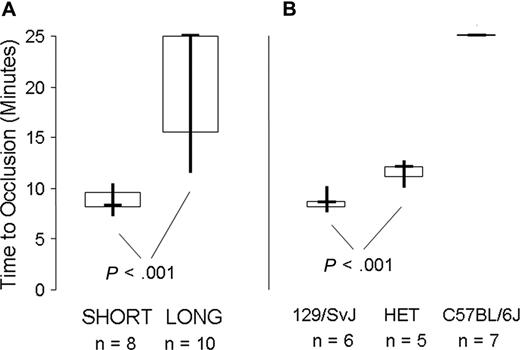
In vivo carotid artery injury model. (A) 8 Gp6−/− SHORT mice were compared with 10 Gp6−/− LONG mice, as indicated on the abscissa. The data are depicted as a box plot, where the bold horizontal line represents the median, the box represents the second and third quartiles, and the vertical lines indicate the limits of the data range. The time to occlusion (ordinate) for the 8 Gp6−/− SHORT mice (4 male mice; 4 female mice) was 8.6 ± 1.2 minutes (mean ± SD). In contrast, the time to occlusion of 10 Gp6−/− LONG mice (5 male mice; 5 female mice) ranged from 11.5 to at least 25 minutes, with a median of at least 25 minutes. The difference in time to occlusion between Gp6−/− LONG and Gp6−/− SHORT mice was statistically significant (P < .001). (B) Correlation between time to occlusion (ordinate) and haplotype at Mh (abscissa). For 6 mice that are homozygous 129 × 1/SvJ at Mh, the mean time to occlusion was 8.2 (± 0.9) minutes (mean ± SD). For 5 mice that are heterozygous 129 × 1/SvJ/C57BL/6J at Mh (HET), the time to occlusion was significantly increased to 11 (± 1) minute (mean ± SD; P < .001). In marked contrast, occlusion was not observed (time to occlusion ≥ 25 minutes) in any of the 7 mice that were homozygous C57BL/6J at Mh.
In vivo carotid artery injury model. (A) 8 Gp6−/− SHORT mice were compared with 10 Gp6−/− LONG mice, as indicated on the abscissa. The data are depicted as a box plot, where the bold horizontal line represents the median, the box represents the second and third quartiles, and the vertical lines indicate the limits of the data range. The time to occlusion (ordinate) for the 8 Gp6−/− SHORT mice (4 male mice; 4 female mice) was 8.6 ± 1.2 minutes (mean ± SD). In contrast, the time to occlusion of 10 Gp6−/− LONG mice (5 male mice; 5 female mice) ranged from 11.5 to at least 25 minutes, with a median of at least 25 minutes. The difference in time to occlusion between Gp6−/− LONG and Gp6−/− SHORT mice was statistically significant (P < .001). (B) Correlation between time to occlusion (ordinate) and haplotype at Mh (abscissa). For 6 mice that are homozygous 129 × 1/SvJ at Mh, the mean time to occlusion was 8.2 (± 0.9) minutes (mean ± SD). For 5 mice that are heterozygous 129 × 1/SvJ/C57BL/6J at Mh (HET), the time to occlusion was significantly increased to 11 (± 1) minute (mean ± SD; P < .001). In marked contrast, occlusion was not observed (time to occlusion ≥ 25 minutes) in any of the 7 mice that were homozygous C57BL/6J at Mh.
When the results are plotted as a function of the LONG versus SHORT phenotype (Figure 7A), the time to occlusion for the 8 Gp6−/− SHORT mice (4 male mice; 4 female mice) was 8.6 (± 1.2) minutes (mean ± SD) and not significantly different (P > .9) from that previously recorded for wild-type mice19 ). In contrast, the time to occlusion of 10 Gp6−/− LONG mice (5 male mice; 5 female mice) ranged from 11.5 to at least 25 minutes, with a median at of at least 25 minutes. In either case, there was no correlation of the platelet response with sex. The difference in time to occlusion between Gp6−/− LONG and Gp6−/− SHORT mice was statistically significant (P < .001).
A more precise correlation was observed between time to occlusion and haplotype at Mh (Figure 7B). For 6 mice that are homozygous 129 × 1/SvJ at Mh, the mean time to occlusion was 8.2 (± 0.9) minutes (mean ± SD). For 5 mice that are heterozygous 129 × 1/SvJ / C57BL/6J at Mh, the time to occlusion was significantly increased to 11 (± 1) minute (mean ± SD; P < .001). In marked contrast, occlusion was not observed (time to occlusion ≥ 25 minutes) in any of the 7 mice that were homozygous C57BL/6J at Mh.
Thus, the bleeding phenotype is reflected in both a prolonged in vivo bleeding time and delayed in vivo thrombus formation in this carotid artery injury model. These results indicate that the carotid artery injury model more accurately reflects the haplotype at Mh than does the tail bleeding time, because even a single 129 × 1/SvJ haplotype at Mh is sufficient to support thrombus formation, albeit with a slight delay to onset. Finally, sex does not significantly modify the in vivo phenotype attributable to Mh.
Discussion
GPVI is considered a major receptor that mediates collagen-dependent platelet function, an assertion that finds support in the complete lack of collagen-initiated platelet responses in vitro in mice that are genetically FcRγ deficient,9,20 that are temporarily GPVI-depleted by administration of JAQ1,10 and that are Gp6−/−.8
All of the Gp6−/− mice in our current study exhibit markedly deficient platelet responses in ex vivo perfusion studies on fibrillar type I collagen and a virtual absence of in vitro collagen-induced platelet aggregation, as expected. Nonetheless, the same mice can be distinguished by a significant and heritable difference in both the tail bleeding time and the rate of thrombus formation in vivo in response to ferric chloride-induced carotid artery injury. Those mice in our study with consistently prolonged tail bleeding times also exhibited delayed or impaired thrombosis in the in vivo model. These findings suggest that there is an underlying genetic difference between those mice that have markedly prolonged tail bleeding times (Gp6−/− LONG) and those mice that are otherwise normal (Gp6−/− SHORT). Nonetheless, the correlation between the tail bleeding time and performance in the carotid artery injury model was not always perfect, because the latter was able to distinguish mice that were heterozygous at Mh from those homozygous for either haplotype, whereas the former could not. This led to the appearance of an autosomal recessive effect when the actual influence of the Mh haplotypes was likely codominant. These 2 in vivo tests are presumably measuring 2 different processes, combined venous and arterial bleeding in the tail bleeding assay, but specifically arterial hemostasis in the carotid injury model. The ideal assay for our genetic analyses would have been a nonterminal test of arterial hemostasis. Unfortunately, the carotid artery injury model is a terminal assay, and the only other established test of arterial hemostasis that is nonterminal, the ear artery model, does not work in our mice, whose ears are too pigmented for noninvasive, optical analyses. Fortunately, despite its drawbacks, the tail bleeding time data did provide enough correlation to generate meaningful linkage results.
There is indirect support for our findings in the literature, where the effect of GPVI depletion on platelet function in vivo has not been consistent. Lockyer et al21 reported that some Gp6−/− mice (proportion not stipulated) exhibited prolonged bleeding times, whereas most had an average bleeding time not significantly different from Gp6+/+mice. Moreover, only 10 of 18 Gp6−/− mice (55%) survived an otherwise lethal thromboembolic challenge of intravenous collagen plus epinephrine infusion. Likewise, dichotomous bleeding times have been observed in FcRγ-deficient mice.11
Using a genome wide linkage analysis coupled with a recombination fine mapping approach, we have identified a single major locus Mh on chromosome 4 that is associated with the bleeding phenotype (LOD = 6.9; P < .001) of our Gp6−/− mice. At this locus, the 129 × 1/SvJ haplotype predominates in Gp6−/− SHORT mice, whereas the C57 BL/6J haplotype is more often associated with Gp6−/− LONG mice. An additional locus of interest was located on chromosome 1 (Mb 68-94), but based on the LOD score (2.4), this linkage was barely significant. Some would consider it an unexpected finding that a complex phenotype would so strongly linked to a single major locus, such as Mh. One explanation is that there are multiple, closely associated genes within Mh that control the phenotype. Another possible explanation is that the gene in question is a transcription factor that regulates the expression of several genes, each of which contributes to the complex phenotype. One can also not exclude the possibility that the gene in question is responsible for posttranslational modification and thus expression or activity of several proteins that modulate the phenotype.
The refined 8 Mb locus of chromosome 4 contains less than sixty known genes (NCBI, Build 37.1), none of which are known to be expressed by platelets. However, 9 of these genes are potential candidates because of their documented or implied association with thrombosis or their expression by cells of the vascular bed. These are the genes for: the α chain of type XV collagen (Cofa1; 47.2 Mb)22 ; transforming growth factor β receptor I (Tgfbr1; 47.4 Mb)23 ; ATP-binding cassette subfamily A member 1 (Abca1; 53.1 Mb)24 ; Klf4 (55.5 Mb), which encodes Kruppel-like factor 4 (Klf4), also known as gut-enriched Kruppel-like factor (GKLF) or epithelial zinc-finger protein (EZF)25-28 ; α-catulin (α-catenin-related protein) (Ctnnal1; 56.9 Mb)29 ; a sushi-, von Willebrand factor type A-, and EGF-domain bearing protein (Svep1; 58.2 Mb)30 ; the α chain of type XXVII collagen (Col27a1; 62.7 Mb)31 ; tenascin C (Tnc; 63.5 Mb)32 ; and toll-like receptor 4 (Tlr4; 66.3 Mb).33 To further refine the Mh locus, we are pursuing additional selected intercrosses to identify meaningful recombinant offspring. We are also characterizing the potential contributions of candidate genes within Mh to the in vivo phenotype through the study of inducible, conditional knockouts and conditional transgene overexpression.
There are obvious implications of our discovery of modifier genes in the context of our Gp6 knockout model. Clearly, future knockout models of any protein or cell receptor involved in hemostasis or thrombosis must be more rigorously defined with respect to genetic background. In the case of GPVI, the absence of this platelet receptor does not create an inherent bleeding phenotype in vivo, and defects in thrombus formation or prolonged bleeding in vivo are dependent upon the genetic background at Mh. Thus, conclusions from previously established murine models of GPVI deficiency in animals with mixed backgrounds, whether induced by immune-mediated down-regulation9,10 or targeted gene disruption of related molecules, such as FcRγ,11,20 or even direct targeted disruption of Gp6 itself,21 need to be reevaluated. Moreover, the indication of GPVI as a target for antithrombotic intervention must also be reconsidered, because its potential efficacy as well as side effects may be influenced by putative human equivalents of Mh genes.
Hemostasis and pathologic thrombosis are complex, redundant processes that require the synergistic effects of multiple gene products, including plasma coagulation factors, cell membrane receptors, and matrix components produced by endothelial cells, smooth muscle cells, and perhaps fibroblasts. The “sensitized” mouse model for gene discovery has been promoted by Ginsburg and colleagues34 and based on the premise that genetic modifiers modulate the phenotype expected of a single gene defect. This approach has led to an understanding of genetic factors that modify the predisposition for and severity of factor V Leiden thrombosis35 and the severity of the hypercoagulable state resulting from thrombomodulin mutations.36
In summary, modifier genes may indirectly influence the composition and thrombotic nature of the extracellular matrix through regulation of gene expression in endothelial cells, smooth muscle cells, and/or fibroblasts. A change in the basal gene activity could certainly alter cell protein profiles, extracellular matrix protein composition, and the responsiveness of vascular cells to proinflammatory or shear stress-induced stimuli. For the study of genetic regulation of hemostasis and thrombosis, the exploitation of sensitized mouse models, such as our Gp6−/− mice, provides a powerful tool to dissect and analyze the variety of genetic differences that can modulate this complex physiologic process. The interplay of heritable but subtle variations in expression of numerous genes likely dictates the hemostatic and/or thrombotic status of the vasculature in any single individual organism.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr David Ginsburg (University of Michigan, Ann Arbor) for his advice and encouragement and Dr John Griffin (Scripps Research Institute) for his expertise in the design and completion of coagulation factor assays.
This work was funded by National Heart, Lung and Blood Institute grants HL075821 to T.J.K. and HL031950 to Z.M.R.
National Institutes of Health
Authorship
Contribution: Y.C. performed research and wrote the manuscript. D.J., P.M., D.H., and J.A.F. performed research and analyzed data. T.W. and M.C. directed genomic scan research. J.W. and Z.M.R. contributed analytical tools, directed research, and analyzed data. T.J.K. designed and directed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas J. Kunicki, PhD, Scripps Research Institute, MEM150, 10550 North Torrey Pines Road, La Jolla, CA 92037; e-mail: tomk@scripps.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal