The oncogene latent membrane protein 1 (LMP1) of Epstein-Barr virus (EBV) without a ligand drives proliferation of EBV-infected B cells. Its levels vary in cells of clonal populations by more than 100-fold, which leads to multiple distinct activities of the oncogene. At intermediate levels it drives proliferation, and at high levels it inhibits general protein synthesis by inducing phosphorylation of eukaryotic initiation factor 2α (eIF2α). We have found that LMP1 activates PERK to induce phosphorylation of eIF2α, which upregulates activating transcription factor 4 (ATF4) expression. ATF4, in turn, transactivates LMP1's own promoter. LMP1 activates not only PERK but also inositol requiring kinase 1 (IRE1) and ATF6, 3 pathways of the unfolded protein response (UPR). Increasing expression levels of LMP1 induced a dose-dependent increase in IRE1 activity, as measured by its “splicing” of XBP-1. These infected B cells secrete immunoglobins independent of the levels of LMP1, indicating that only a threshold level of XBP-1 is required for the secretion. These findings indicate that LMP1's activation of the UPR is a normal event in a continuum of LMP1's expression that leads both to stimulatory and inhibitory functions and regulates the physiology of EBV-infected B cells in multiple, unexpected modes.
Introduction
Oncogenes are dominantly acting regulators of cell proliferation and survival. They can be derived from proto-oncogenes by mutations that alter their enzymatic activity, as in the case of Ha-ras in carcinomas, or their levels of expression, as in the case of Bcl-2 in diffuse, large B-cell lymphomas.1,2 The latent membrane protein 1 (LMP1) oncogene of Epstein-Barr virus (EBV) stands out among the regulators of cell proliferation because it has opposing activities dependent on its own level of expression.3 These levels vary naturally in individual cells of clonal populations by more than 100-fold such that in any untreated population, some cells are proliferating while others are not
EBV contributes causally to multiple lymphomas and carcinomas. An important part of its oncogenicity in lymphoid cells lies in its ability to infect, induce, and maintain proliferation in B lymphocytes. This proliferation requires signaling by the LMP1 oncogene.4,–6 LMP1's signaling in B cells is in one way similar to that of the cellular CD40 receptor: both activate nuclear factor-κB (NF-κB), AP-1, and Stat-1 by associating with molecules such as TRAFs and JAK3.7,,,,,,,–15 LMP1's signaling, however, differs fundamentally from CD40, because LMP1 regulates these signaling pathways without itself being regulated by a ligand, as is CD40. When LMP1 is expressed at intermediate levels, it drives signaling through NF-κB, for example, to promote cell proliferation.6,16 When it is expressed at the high end of physiologic levels, it has been shown to induce the phosphorylation of eukaryotic initiation factor 2α (eIF2α), an inhibition of general protein synthesis and cellular cytostasis.3,17,–19 It is thus essential to elucidate the mechanism by which the levels of LMP1 are regulated to understand its functioning as an oncogene.
We examined the path by which LMP1 induces the phosphory-lation of eIF2α and have found that LMP1 activates the kinase, PERK, which phosphorylates eIF2α. LMP1's activation of PERK is accompanied by its activation of the inositol requiring kinase 1 (IRE1), and activating transcription factor 6 (ATF6), indicating that LMP1 induces the unfolded protein response (UPR) pathway. LMP1's induction of the UPR is dependent on the expression level of LMP1; intermediate levels of LMP1 activate IRE1 to intermediate levels, as measured by IRE1's “splicing” of its substrate, XBP-1. The spliced form of XBP-1 has been shown to play a role in differentiation of B cells to plasma cells, which leads to immunoglobulin (Ig) secretion.20,–22 EBV-infected cells secrete Ig independently of the level of spliced XBP-1 product present. LMP1's dose-dependent activation of PERK leads to a proportional phosphorylation of eIF2α and also to a proportional induction of expression of the bZIP transcription factor ATF4. We have found that ATF4 drives LMP1 transcription through an ATF/CRE-binding site. These observations together support a model in which EBNA2 first induces LMP1 transcription. As the levels of LMP1 accumulate, LMP1 induces phosphorylation of eIF2α to induce low levels of ATF4, leading to increased expression of LMP1. Increasing levels of LMP1 lead both to more LMP1 and to its signaling required for B-cell proliferation. High physiologic levels of LMP1 lead to cytostasis and to cell survival. We have engineered EBV to express LMP1 tagged with mRFP so that the fate of EBV-infected B cells expressing high levels of LMP1 could be monitored. These cells proliferate clonally as well as those with low levels of LMP1 and regenerate populations of cells with the natural wide range of levels of LMP1, illustrating that LMP1's induction of the UPR is tied to its own expression and its regulation of its host cell's proliferation.
Methods
Cell culture
The cell lines BJAB/HA-LMP1 and BJAB/HA6MLMP1-GFP have been described previously.3 The 721 cell is an EBV-positive lymphoblastoid cell line.23 BJAB is an EBV-negative Burkitt lymphoma cell line.24 The PKR−/− cell line was kindly provided by Dr Ian Mohr, New York University School of Medicine. PERK−/− mouse embryonic fibroblasts (MEFs) were immortalized using a retrovirus expressing human telomerase IRES puromycin. Wild-type (wt) PERK cells were made by cotransfecting PERK−/− cells with mouse PERK, wild type, 9E10 tagged at the C-terminus (kindly provided by Dr David Ron, New York University School of Medicine) and expressed from a plasmid also expressing resistance to hygromycin. BJAB and 721 cells were grown in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. BJAB/HA-LMP1 and BJAB/HA6MLMP1-GFP cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1 mg/mL G418, and 1 μg/mL puromycin. The PKR−/− cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, and 10 mM HEPES. The PERK−/− cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 1 μg/mL puromycin. The wt PERK cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 1 μg/mL puromycin, and 200 μg/mL hygromycin. All cell culture media were supplemented with 200 U/mL penicillin and 200 μg/mL streptomycin, and cells were grown at 37°C in a 5% CO2 humidified atmosphere.
Plasmids
To construct a retrovirus expressing dominant-negative (dn) PERK, the parent retroviral plasmid pCMMP-MCS-ires-eGFP, p3051,25 was digested with HpaI and XhoI and ligated to an insert containing dn PERK that was derived from digesting a plasmid encoding dn PERK (kindly provided by Dr Ron Wek, Indiana University School of Medicine, Indianapolis) with BamHI, blunted with T4 DNA polymerase, and XhoI. The retrovirus expressing dn PKR was constructed by inserting a polymerase chain reaction (PCR) fragment amplified from a plasmid encoding dn PKR (kindly provided by Dr Kenzo Takada, Hokkaido University, Sapporo, Japan) into p3051 digested with MluI and XhoI. The retrovirus expressing LMP1 was constructed by ligating p3051 digested with MfeI and MluI with the insert encoding LMP1 that was derived from digesting a plasmid encoding cDNA of LMP1, p1990,26 with EcoRI and MluI. The retrovirus expressing human telomerase, Htert, was constructed by digesting the parent retroviral plasmid, pCMMP-MCS-ires-puro, p3048, with AgeI and AvrII. The Htert fragment was derived from digesting a plasmid encoding Htert, p3075, with AgeI and AvrII. Mouse PERK, wild type, 9E10 tagged at C-terminus, mammalian expression vector27 and CHOP:GFP reporter plasmid28 were kindly provided by Dr David Ron. The pgLRS (−106) CAT and pgLRS (−106)(ATF/CREmut.) CAT were kindly provided by Dr Lars Rymo (Sahlgrenska University Hospital, Sweden) and have been described previously.29 The p3xFLAG-ATF6 was kindly provided by Dr Ron Prywes (Columbia University, New York, NY) and has been described previously.30
Retroviral production and infection
Retrovirus was generated by cotransfecting 293T in a 70% confluent 10-cm dish with 3 μg of a plasmid encoding the Gag-Pol element, 1 μg of a plasmid encoding the vesicular stomatitis virus G protein, 1 μg of a plasmid encoding a derivative of NF-κB, and 10 μg of a plasmid carrying the retroviral backbone encoding either dn PERK, dn PKR, or LMP1 using polyethylenimine (PEI; Sigma-Aldrich, St Louis, MO). A total of 40μg PEI per 10-cm dish was used. Twenty-four hours after transfection, the culture medium was replaced with Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 50 mM HEPES. On days 3 and 4 after transfection, the media was collected and filtered through a 0.8-μm pore size filter. The retrovirus was concentrated via sedimentation at 36 000 rpm (77 000g) for 2.5 hours, resuspended in RPMI 1640 medium supplemented with 10% fetal bovine serum and 50 mM HEPES, and stored at −80°C. The titers of the viral stocks were measured by infecting BJAB cells plated at 5 × 104 cells per well of a 24-well plate with 1:5 serial dilutions of viral stocks. Infected cells were identified by the expression of enhanced green fluorescent protein (eGFP), and titers were calculated according to the Poisson distribution.
BJAB/HA-LMP1, BJAB/HA6MLMP1-GFP, PERK−/−, PKR−/−, and wt PERK cells growing exponentially were infected with 10 GBU (green BJAB units) by incubating the suspension in complete medium with 50 mM HEPES for 1 hour at 4°C with gentle rocking. After the incubation, cells were resuspended in 10 mL complete medium and incubated at 37°C. After 48 hours of infection, cells were sorted.
Cell sorting
Single cells were sorted on FACS-Vantage SE with the FACS-DIVA option (Becton Dickinson, San Jose, CA). Cells infected with control or dominant-negative retrovirus were sorted for those expressing the highest 10% GFP. Infected cells were sorted by fluorescent-activated cell sorting (FACS) based on the intensity of eGFP encoded by the retrovirus, because the intensity of eGFP correlates with expressed levels of proteins upstream of the IRES.31 EBV-positive strains were sorted for low, medium, and high levels of mRFP. The cells were analyzed on FACS-Vantage SE with the FACS-DIVA option. Data were analyzed with FlowJo software (Tree Star, Ashland, OR).
Western blotting
Cells were harvested at a density of 2 to 3 × 105/mL. The cells then were solubilized with 1× sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer made from a dilution of a 2× stock solution with 1× RIPA buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 0.5% deoxycholate; and 0.5% SDS). Samples were briefly sonicated. Cell lysates (105 cells per sample) were separated by SDS-10% PAGE for eIF2α, LMP1, ATF4, 3xFLAG-ATF6, and tubulin, and SDS-12% PAGE for human Ig and GFP, and were transferred to nitrocellulose membranes. The blots were blocked with 5% nonfat milk and probed with primary antibodies followed by alkaline phosphatase–labeled secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). The signals were quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The following primary antibodies were used: mouse monoclonal anti-LMP1 antibody CS1-4 (Dako, Carpinteria, CA) at 1:500, mouse monoclonal anti–total eIF2α (Biosource, Carlsbad, CA) at 1:2000, rabbit polyclonal anti–phosphorylated eIF2α (Biosource) at 1:500, mouse monoclonal anti-myc (Cell Signaling Technology, Danvers, MA) at 1:1000, rabbit polyclonal anti-GFP (Clontech, Mountain View, CA) at 1:400, mouse monoclonal anti–α-tubulin (Sigma-Aldrich, St Louis, MO) at 1:2000, rabbit polyclonal anti-ATF4 (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:100, goat anti–human IgA, IgG, and IgM (KPL, Gaithersburg, MD) at 1:500, and mouse monoclonal anti–Flag M2 (Sigma) at 1:1000.
Reverse transcription PCR
RNA was prepared using the RNeasy Mini Kit from Qiagen (Valencia, CA). For GAPDH detection, each reaction contained 100 ng RNA, 1× EZ buffer (GeneAmp; Applied Biosystems, Foster City, CA), 0.4 M Betaine (Sigma), 75 ng primers, 300 mM dinucleotide triphosphates, 2.5 Mn(OAc)2, and 5 units of rTth polymerase (GeneAmp). A forward primer, gAgCCAAAAgggTCATC, and a reverse primer, gTggTCATgAgTCCTTC, were used to amplify GAPDH. The conditions for the reaction are as follows: 60°C for 30 minutes, 95°C for 5 minutes, 25 cycles of 95°C for 1 minute, 47°C for 2 minutes, 60°C for 1 minute, and 10 minutes at 60°C. For XBP-1 detection, a protocol available in David Ron's laboratory was used.32 RNA was reverse transcribed using the primer gATgACgTCCCCACTgACAgAgAAAgggAg. A PCR reaction was done using as a forward primer, AAACAgAgTAgCAgCTCAgACTgC, and a reverse primer TCCTTCTgggTAgACCTCTgggAg. PCR with the XBP-1 primer sets gives rise to a 473-bp product. This RT-PCR product was digested with PstI to distinguish the unspliced product from the spliced product. PstI cuts only the unspliced cDNA product and gives rise to 2 fragments, a 290-bp and a 183-bp fragment.
Electroporation
BJAB, BJAB/HA-LMP1, and BJAB/HA6MLMP1-GFP cells were transfected by electroporation. A total of 5 × 106 cells were electroporated in 500 μL RPMI 1640 medium supplemented with 10% fetal bovine serum and 50 mM Hepes with a custom-built electroporator at 1500 V, 3 capacitor banks with 1540 microfarads of capacitance, R-adjust set at 6 o'clock, and a rise time set at 10 o'clock, as described previously.33
CAT activity in B cells
A total of 106 cells were assayed for chloramphenicol acetyl transferase (CAT) activity. The cells were washed once with PBS and resuspended in 50 μL of reporter lysis buffer (Promega, Madison, WI). The lysate was incubated at room temperature for 15 minutes. The nuclei were pelleted by centrifugation, and the supernatant was incubated at 65°C for 10 minutes. A total of 5 μL of 14C-labeled chloramphenicol at 0.05 mCi/mL (Perkin-Elmer Life and Analytical Sciences, Waltham, MA), 10 μL n-Butyryl CoA at 5 mg/mL, and 60 μL of water were added to each reaction and incubated at 37°C for 2 hours. A total of 500 μL ethyl acetate was added to each reaction, vortexed, and centrifuged for 3 minutes. A total of 450 μL organic layer was removed and evaporated in a SpeedVac for 30 minutes. The solute was resuspended in 10 μL ethyl acetate and spotted onto a silica gel TLC plate (J.T. Baker/Mallinckrodt Baker, Phillipsburg, NJ). The silica gel was placed in a developing chamber that was pre-equilibrated with chloroform-methanol in a 19:1 ratio. The separated products were detected and quantified by PhosphoImager analysis (Molecular Dynamics).
Recombinant EBV
Generation of recombinant EBV has been described previously.34 The p2167.1 plasmid4 was modified to express a fusion protein of LMP1-mRFP. The stop codon of LMP1 was replaced with a linker GGC CAG AGT GGT CCC GGT GGT, followed by the open reading frame of mRFP. The resulting maxi-EBV plasmid was checked by restriction analyses (data not shown) and then used to establish virus-producing 293 cell lines.4,35,–37 Primary B cells were isolated from human blood according to the protocol provided in the Miltenyi Biotech Human B cell Isolation Kit II (MACS). Primary human B lymphocytes purified from blood were infected with the recombinant EBV virus with an MOI of 1. One to 2 weeks after infection, 1, 3, or 10 infected cells/well were plated in 96-well plates with γ-irradiated human fibroblasts as feeder layer. A total of 1, 3, or 10 cells of each population were deposited into multiple wells of a 96-well plate. Wells containing surviving and proliferating cells were identified visually after 4 weeks. The cloning efficiency was determined using the Poisson distribution. Proliferating cell clones were expanded in suspension for 4 to 6 weeks.
Results
LMP1 induces phosphorylation of eIF2α through PERK
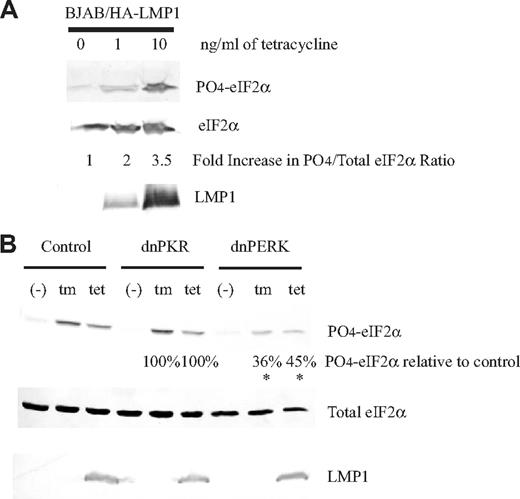
We have previously shown that induction of phosphorylation of eIF2α by the N-terminus and the 6 membrane-spanning domains of LMP1 occurs normally in EBV-infected cells.3,18 There are currently 4 kinases known to phosphorylate eIF2α: general control nonderepressible 2 (GCN2),38 interferon-inducible RNA-dependent protein kinase (PKR),39 heme-regulated inhibitor kinase (HRI),40 and PKR-like ER kinase (PERK).27 We tested whether LMP1 activates PERK because both proteins associate with membranes, and whether LMP1 activates PKR because several herpes viruses have been shown to regulate it.41,–43 We used an EBV-negative B-cell line termed BJAB/HALMP1 that was constructed to express LMP1 inducibly following treatment with tetracycline.3 These cells express 5-fold and 50-fold more LMP1 than does a normal EBV-infected cell line, 721,23 when they are treated with 1 and 10 ng/mL tetracycline, respectively. This treatment leads to a dose-dependent increase in phosphorylation of eIF2α (Figure 1A). The BJAB/HALMP1 cells were infected with a control retrovirus, one expressing a dominant-negative derivative of PKR, or one expressing a dominant-negative derivative of PERK. They were treated with tunicamycin as a positive control or with tetracycline to induce the expression of LMP1. Both treatments for the cells infected with the control retrovirus or those expressing the dominant-negative derivative of PKR induced phosphorylation of eIF2α efficiently (Figure 1B). This phosphorylation was significantly inhibited, however, in the cells expressing the dominant-negative derivative of PERK (Figure 1B).
Expression of dominant-negative PERK, but not dominant-negative PKR, inhibits phosphorylation of eIF2α induced by LMP1 expression. (A) The EBV-negative B cell, BJAB, constructed to express LMP1 inducible by tetracycline, BJAB/HA-LMP1, was untreated or treated with 1 ng/mL or 10 ng/mL tetracycline for 48 hours. The cells were harvested and analyzed by Western blotting using anti–phosphorylated eIF2α, anti–total eIF2α, and anti-LMP1 antibodies. (B) BJAB/HA-LMP1 cells were infected with a control retrovirus and either dominant-negative PERK or dominant-negative PKR. After 48 hours of infection, the cells were sorted for the highest levels of expression of GFP encoded by the retroviruses. Each population was either left untreated, treated with 10 ng/mL tetracycline for 48 hours to induce LMP1, or treated with 2.5 μg/mL tunicamycin for 4 hours to activate PERK. The cells were harvested and analyzed as described in panel A. The ratios of phosphorylated eIF2α to total eIF2α were calculated. The numbers represent the percentage of phosphorylation of eIF2α relative to cells infected with control retrovirus. *P (2-sided) <.05, Wilcoxon signed rank test. A representative example of 4 independent experiments is shown.
Expression of dominant-negative PERK, but not dominant-negative PKR, inhibits phosphorylation of eIF2α induced by LMP1 expression. (A) The EBV-negative B cell, BJAB, constructed to express LMP1 inducible by tetracycline, BJAB/HA-LMP1, was untreated or treated with 1 ng/mL or 10 ng/mL tetracycline for 48 hours. The cells were harvested and analyzed by Western blotting using anti–phosphorylated eIF2α, anti–total eIF2α, and anti-LMP1 antibodies. (B) BJAB/HA-LMP1 cells were infected with a control retrovirus and either dominant-negative PERK or dominant-negative PKR. After 48 hours of infection, the cells were sorted for the highest levels of expression of GFP encoded by the retroviruses. Each population was either left untreated, treated with 10 ng/mL tetracycline for 48 hours to induce LMP1, or treated with 2.5 μg/mL tunicamycin for 4 hours to activate PERK. The cells were harvested and analyzed as described in panel A. The ratios of phosphorylated eIF2α to total eIF2α were calculated. The numbers represent the percentage of phosphorylation of eIF2α relative to cells infected with control retrovirus. *P (2-sided) <.05, Wilcoxon signed rank test. A representative example of 4 independent experiments is shown.
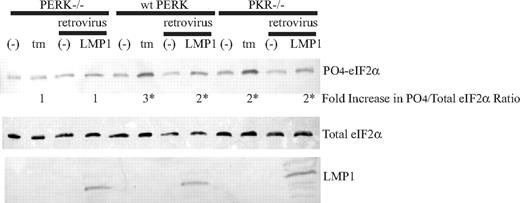
To assess further whether PERK is the predominant eIF2α kinase that is activated in response to LMP1, we measured eIF2α phosphorylation in mouse embryo fibroblasts (MEF) null for PERK, PERK−/− or null for PKR, PKR−/−. Expression of LMP1 in PKR−/− cells clearly induced phosphorylation of eIF2α, but this activity was abrogated in PERK−/− cells (Figure 2). This finding indicates that PERK is likely required for the induction of phosphorylation of eIF2α by LMP1. To confirm the role of PERK in LMP1 activity, we reconstituted PERK−/− MEFs with full-length wild type PERK, termed “wt PERK.” Treatment of wt PERK cells with tunicamycin led to an increase in phosphorylation of eIF2α, confirming the expression of a functional PERK protein in these cells (Figure 2). Efficient expression of LMP1 in these reconstituted wt PERK cells induced phosphorylation of eIF2α (Figure 2). In summary, these results in combination indicate that LMP1 induces phosphorylation of eIF2α by activating PERK.
Expression of LMP1 results in phosphorylation of eIF2α in PKR−/− and wt PERK but not in PERK−/− mouse embryo fibroblasts (MEF). PERK−/− MEFs were transfected with wild-type mouse PERK-myc; a clone that stably expressed PERK-myc was selected and termed wt PERK. PERK−/−, PKR−/−, and wt PERK MEFs were infected with a control retrovirus or a retrovirus that expresses LMP1 for 48 hours, and they were untreated or treated with 2.5 μg/mL tunicamycin for 4 hours. The cells were harvested and analyzed by Western blotting using anti–phosphorylated eIF2α, anti–total eIF2α, and anti-LMP1 antibodies. The ratios of phosphorylated eIF2α to total eIF2α were calculated. The numbers represent the fold difference of induced ratios relative to the uninduced ratios. *P (2-sided) <.05, Wilcoxon signed rank test. A representative example of 4 independent experiments is shown.
Expression of LMP1 results in phosphorylation of eIF2α in PKR−/− and wt PERK but not in PERK−/− mouse embryo fibroblasts (MEF). PERK−/− MEFs were transfected with wild-type mouse PERK-myc; a clone that stably expressed PERK-myc was selected and termed wt PERK. PERK−/−, PKR−/−, and wt PERK MEFs were infected with a control retrovirus or a retrovirus that expresses LMP1 for 48 hours, and they were untreated or treated with 2.5 μg/mL tunicamycin for 4 hours. The cells were harvested and analyzed by Western blotting using anti–phosphorylated eIF2α, anti–total eIF2α, and anti-LMP1 antibodies. The ratios of phosphorylated eIF2α to total eIF2α were calculated. The numbers represent the fold difference of induced ratios relative to the uninduced ratios. *P (2-sided) <.05, Wilcoxon signed rank test. A representative example of 4 independent experiments is shown.
LMP1's promoter is a novel downstream target of the PERK/ATF4 pathway
Activation of PERK and phosphorylation of eIF2α by high physiologic levels of LMP1 result in attenuation of global protein synthesis and cytostasis. Cells expressing average levels of LMP1 proliferate even in the presence of some phosphorylation of eIF2α, albeit the levels of this phosphorylation are lower than those found in cells with high levels of LMP1.3 To determine the biologic consequence of activating PERK and phosphorylation of eIF2α in a dose-dependent manner, we assayed the downstream effects of this pathway. Translation of ATF4 becomes upregulated only in the presence of phosphorylation of eIF2 because of the presence of 2 regulatory, upstream ORFs.44,45 The promoter-proximal part of the LMP1 promoter contains an ATF/CRE site at position −41 relative to the transcription initiation site.29,46 We tested whether the LMP1 promoter constitutes a novel downstream target of PERK/ATF4 pathway. EBV-negative B cells were electroporated with reporter plasmids containing a wild-type or mutated ATF/CRE site in LMP1's promoter driving chloramphenicol acetyl transferase (CAT) gene expression. In this experiment, expression of ATF4 induced a 50% increase in CAT activity in the presence of a wild-type ATF/CRE site but not in the presence of a mutated site (data not shown). The basal level of CAT activity depended on a wild-type ATF/CRE site because B cells express other ATF proteins that can activate it.29
Synthesis of LMP1 is regulated by an autocatalytic process.
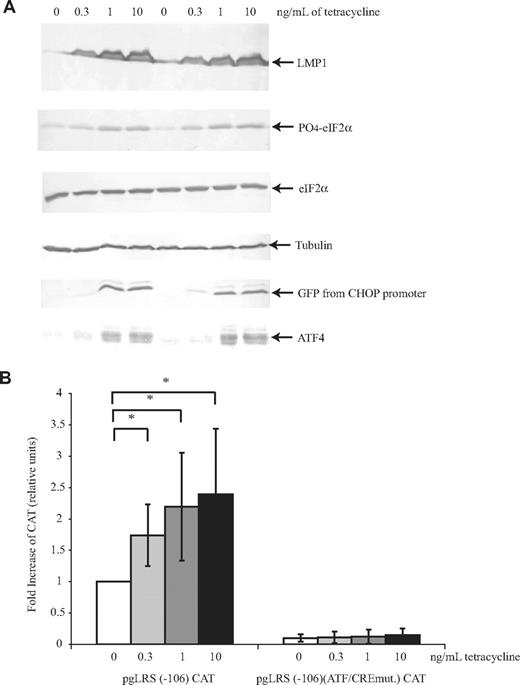
LMP1 induces PERK activation and its phosphorylation of eIF2α. Phosphorylation of eIF2α induces the expression of ATF4, which can transactivate LMP1's promoter. We tested whether expression of LMP1 will induce transactivation of its own promoter through PERK, eIF2α, and ATF4. Treatment of BJAB/HALMP1 cells with increasing amounts of tetracycline did induce a dose-dependent increase in LMP1's expression and phosphorylation of eIF2α (Figure 3A). In those same cells, treatment with tetracycline also resulted in a dose-dependent increase in the levels of expression of ATF4 (Figure 3A). To determine ATF4's level of activity, we measured ATF4's ability to transactivate a known target, C/EBP-homologous transcription factor (CHOP).47 Expression of ATF4 induced by LMP1's expression activated CHOP's promoter dose dependently. Similarly to its activation of the CHOP promoter, ATF4 also transactivated LMP1's promoter in a dose-dependent manner (Figure 3B). The increase in activity did not reflect a difference in the efficiency of transfection of the reporter plasmid, because the transfection efficiency as measured by mRFP-positive cells was similar (ranging from 7% to 9%).
LMP1 induces phosphorylation of eIF2α and expression of ATF4, which in turn leads to transactivation of LMP1's promoter dose-dependently. BJAB/HALMP1 cells were electroporated with an mRFP expression plasmid, a reporter plasmid that contains the CHOP promoter driving GFP gene expression, CHOP:GFP, and pgLRS (−106) CAT or pgLRS (−106)(ATF/CREmut.) CAT. pgLRS (−106) CAT has the wild-type ATF/CRE site, and pgLRS (−106)(ATF/CREmut.) CAT has a mutated ATF/CRE site in the −106 to +40 portion of the LMP1 promoter fused to chloramphenicol acetyl transferase (CAT). After 24 hours, electroporated cells were divided into 4 different plates and were either left untreated or treated with 300 pg/mL, 1 ng/mL, or 10/ng mL tetracycline for 48 hours to induce LMP1. (A) The cells were harvested and analyzed by Western blotting using antibodies to detect LMP1, phosphorylated eIF2α, total eIF2α, tubulin, ATF4, and GFP. A representative gel of 3 independent experiments is shown. (B) The cells were harvested and analyzed by CAT assays. The activity is given as the fold difference relative to the activity of pgLRS (−106) CAT in the absence of tetracycline, which was set at 1. Averages and standard deviations of 3 independent experiments are shown. *P (2-sided) <.05 (Wilcoxon rank sum test).
LMP1 induces phosphorylation of eIF2α and expression of ATF4, which in turn leads to transactivation of LMP1's promoter dose-dependently. BJAB/HALMP1 cells were electroporated with an mRFP expression plasmid, a reporter plasmid that contains the CHOP promoter driving GFP gene expression, CHOP:GFP, and pgLRS (−106) CAT or pgLRS (−106)(ATF/CREmut.) CAT. pgLRS (−106) CAT has the wild-type ATF/CRE site, and pgLRS (−106)(ATF/CREmut.) CAT has a mutated ATF/CRE site in the −106 to +40 portion of the LMP1 promoter fused to chloramphenicol acetyl transferase (CAT). After 24 hours, electroporated cells were divided into 4 different plates and were either left untreated or treated with 300 pg/mL, 1 ng/mL, or 10/ng mL tetracycline for 48 hours to induce LMP1. (A) The cells were harvested and analyzed by Western blotting using antibodies to detect LMP1, phosphorylated eIF2α, total eIF2α, tubulin, ATF4, and GFP. A representative gel of 3 independent experiments is shown. (B) The cells were harvested and analyzed by CAT assays. The activity is given as the fold difference relative to the activity of pgLRS (−106) CAT in the absence of tetracycline, which was set at 1. Averages and standard deviations of 3 independent experiments are shown. *P (2-sided) <.05 (Wilcoxon rank sum test).
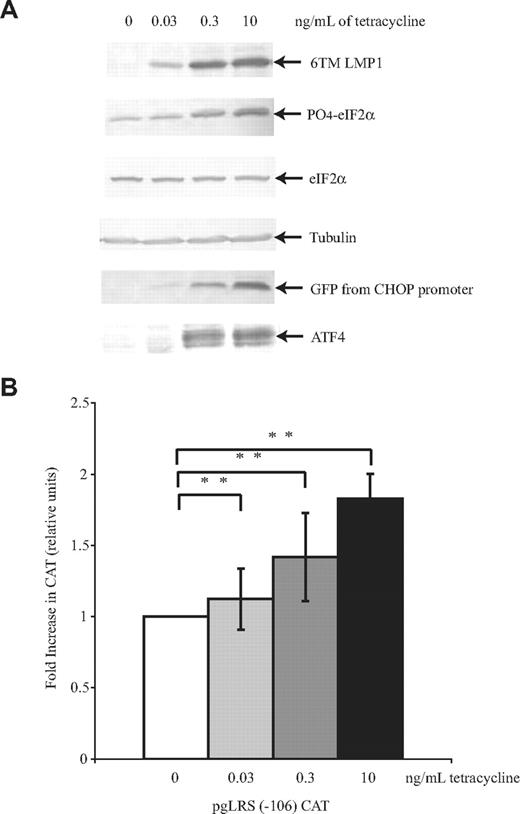
It has been shown that LMP1 induces phosphorylation of eIF2α through its transmembrane domains.3,17,19 To confirm that phosphorylation of eIF2α induced by the transmembrane domains drives ATF4's expression and transactivation of LMP1's promoter, a comparable study to the one described above was done using BJAB/HA6MLMP1-GFP cells. BJAB/HA6MLMP1-GFP is an EBV-negative B-cell line engineered to express a derivative of LMP1 that expresses only the N-terminus and 6 transmembrane domains of LMP1 conditionally upon treatment with tetracycline.3 LMP1's promoter was responsive to ATF4 induced by the 6 transmembrane domains of LMP1 (Figure 4). These data are consistent with the idea that LMP1's activation of PERK's, PERK's phosphorylation of eIF2α, and the subsequent upregulation of ATF4 support transactivation of LMP1's promoter.
Only the 6 transmembrane domains of LMP1 are necessary for induction of ATF4's expression and transactivation of LMP1's promoter. BJAB/HA6MLMP1-GFP cells were electroporated with mRFP, CHOP:GFP, and pgLRS (−106) and then divided into 4 different plates. After 24 hours, cells were either left untreated or treated with 30 pg/mL, 300 pg/mL, or 10 ng/mL tetracycline for 48 hours to induce LMP1. The cells were harvested and analyzed by Western blotting (A) using antibodies to detect LMP1, phosphorylated eIF2α, total eIF2α, tubulin, GFP, and ATF4. A representative gel of 3 independent experiments is shown. (B) The cells were harvested and analyzed by CAT assays. The activity is given as the fold-difference relative to the activity of pgLRS (−106) CAT in the absence of tetracycline, which was set at 1. Averages and standard deviations of 3 independent experiments are shown. **P (2-sided) <.01 (Wilcoxon rank sum test).
Only the 6 transmembrane domains of LMP1 are necessary for induction of ATF4's expression and transactivation of LMP1's promoter. BJAB/HA6MLMP1-GFP cells were electroporated with mRFP, CHOP:GFP, and pgLRS (−106) and then divided into 4 different plates. After 24 hours, cells were either left untreated or treated with 30 pg/mL, 300 pg/mL, or 10 ng/mL tetracycline for 48 hours to induce LMP1. The cells were harvested and analyzed by Western blotting (A) using antibodies to detect LMP1, phosphorylated eIF2α, total eIF2α, tubulin, GFP, and ATF4. A representative gel of 3 independent experiments is shown. (B) The cells were harvested and analyzed by CAT assays. The activity is given as the fold-difference relative to the activity of pgLRS (−106) CAT in the absence of tetracycline, which was set at 1. Averages and standard deviations of 3 independent experiments are shown. **P (2-sided) <.01 (Wilcoxon rank sum test).
Induction of phosphorylation of eIF2α in EBV-positive cells leads to an increase in LMP1's expression
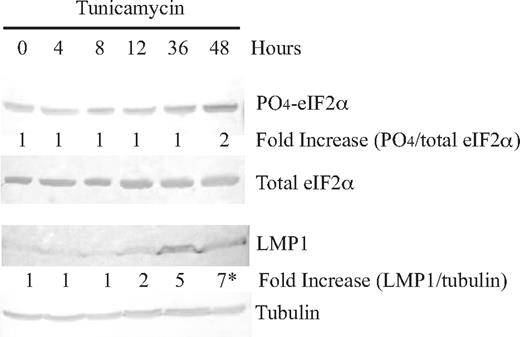
eIF2α is not phosphorylated in every cell within a clonal population of EBV-infected B cells because the phosphorylation of eIF2α is dependent on the levels of LMP1, which varies and ranges more than 100-fold.3 Knowing that LMP1's promoter is responsive to ATF4, we hypothesized that if the levels of phosphorylation of eIF2α were induced exogenously with, for example, tunicamycin in normal EBV-positive cells, the average levels of LMP1 would increase. This hypothesis proved correct; induction of phosphorylation of eIF2 by tunicamycin led to a 7-fold increase in the levels of expression of LMP1 48 hours after treatment (Figure 5). This finding confirms our studies with reporter assays that LMP1's synthesis is regulated by the PERK/ATF4 pathway.
Induction of phosphorylation of eIF2α with tunicamycin in an EBV-positive cell line, 721, leads to an increase in LMP1 protein levels. Cells of an EBV-positive cell line, 721, were treated with 2.5 μg/mL tunicamycin, harvested at 0, 4, 8, 12, 36, and 48 hours, and analyzed by Western blotting using antibodies to detect LMP1, phosphorylated eIF2α, total eIF2α, and tubulin. The top numbers represent the fold-difference of ratios of phosphorylation of eIF2α after tunicamycin treatment relative to 0 hours. The bottom numbers represent the fold-difference of ratios of LMP1 to tubulin after tunicamycin treatment relative to 0 hours. *P (2-sided) <.05, Wilcoxon signed rank test. A representative gel of 3 independent experiments is shown.
Induction of phosphorylation of eIF2α with tunicamycin in an EBV-positive cell line, 721, leads to an increase in LMP1 protein levels. Cells of an EBV-positive cell line, 721, were treated with 2.5 μg/mL tunicamycin, harvested at 0, 4, 8, 12, 36, and 48 hours, and analyzed by Western blotting using antibodies to detect LMP1, phosphorylated eIF2α, total eIF2α, and tubulin. The top numbers represent the fold-difference of ratios of phosphorylation of eIF2α after tunicamycin treatment relative to 0 hours. The bottom numbers represent the fold-difference of ratios of LMP1 to tubulin after tunicamycin treatment relative to 0 hours. *P (2-sided) <.05, Wilcoxon signed rank test. A representative gel of 3 independent experiments is shown.
LMP1 induces PERK's activation as part of the unfolded protein response
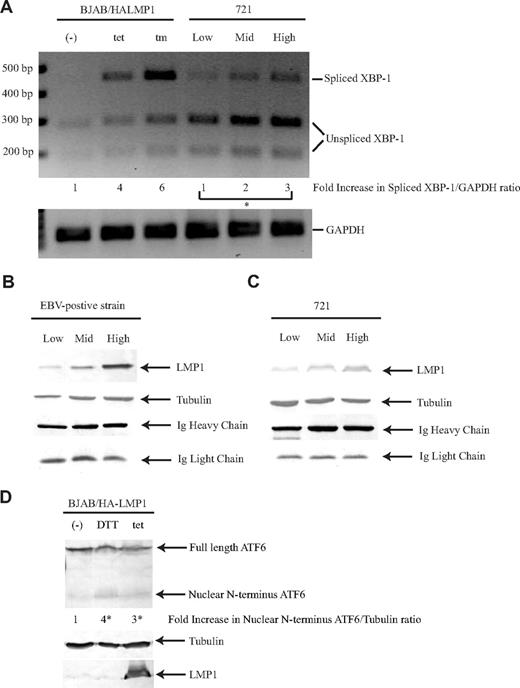
We tested whether LMP1 activates PERK by activating the UPR. During the UPR, both inositol requiring kinase 1 (IRE1) and PERK, which are transmembrane proteins that reside in the endoplasmic reticulum (ER), become activated. The luminal domains of IRE1 and PERK, which are the sensors for ER stress, share some homology and have been shown to be interchangeable.48 Oligomerization of IRE1 and PERK induces autophosphorylation and activates the RNase domain and kinase domain, respectively. Activated IRE1 cleaves XBP-1 mRNA, and activated PERK phosphorylates eIF2α.49 Recent studies have shown that the XBP-1 mRNA is spliced when the CD40 receptor is activated in B cells.20,21 We tested whether expression of LMP1 induces the “splicing” of XBP-1 mRNA. Expression of LMP1 in BJAB/HALMP1 cells induced an increase in spliced XBP-1 product (Figure 6A). We next tested whether LMP1 in the context of EBV also induces the splicing of XBP-1. In 721 cells, an EBV-positive cell line, the levels of spliced XBP-1 product correlated with the level of expression of LMP1; increasing amounts of LMP1 induced a dose-dependent increase of spliced XBP-1 (Figure 6A). LMP1 induces the activation of PERK and IRE1 in a dose-dependent manner, indicating that LMP1 induces the UPR pathway. We tested whether LMP1 activates PERK directly by asking whether it coimmunoprecipitated with PERK. When cell lysates were precipitated with antibodies against LMP1, PERK-myc was not coprecipitated detectably (data not shown). If 5% to 10% of PERK were to be associated with LMP1, a signal would have been detected in these experiments.
LMP1 induces all 3 pathways of the UPR. (A) BJAB/HA-LMP1 cells were untreated, treated with 10 ng/mL tetracycline for 48 hours, or treated with 2.5 μg/mL tunicamycin for 4 hours. The 721 cell line was sorted for low, medium, and high levels of LMP1. RNA was isolated from these cells and analyzed by RT-PCR using primers specific for both XBP-1 and GAPDH. To determine whether splicing of XBP-1 had occurred, the PCR fragments were digested with PstI. The ratios of spliced XBP-1 to GAPDH were calculated. The numbers represent the fold-difference relative to the ratio of untreated BJAB/HA-LMP1 cells, which was set at 1. *P (1-sided) <.05, 1-sided alternative: low, mid, high in increasing order, Jonckheere-Terpstra test. A gel representative of 3 independent experiments is shown. Production of Ig was measured by Western blotting the culture supernatant of EBV-positive strain cells (B) and 721 cells (C) that were sorted for low, medium, and high levels of LMP1 by staining with lysotracker red as described.60 For Ig heavy chain detection, 1 μL supernatant was analyzed, and for Ig light chain detection, 10 μL supernatant was analyzed, because the antibody preferentially detects Ig heavy chain. The cells were harvested and analyzed by Western blotting using antibodies to detect LMP1 and tubulin. A representative gel of duplicates of 2 independent experiments is shown. (D) BJAB/HALMP1 was electroporated with p3xFLAG-ATF6. Cells were either left untreated or treated with 10 mM DTT for 1.5 hours and 10 ng/mL tetracycline for 48 hours to induce LMP1. The cells were harvested and analyzed by Western blotting using antibodies to detect LMP1, tubulin, and Flag. The numbers represent the fold-difference of ratios of nuclear N-terminus ATF6 to tubulin. *P (2-sided) less than .05 (Wilcoxon rank sum test). A representative example of 3 independent experiments is shown.
LMP1 induces all 3 pathways of the UPR. (A) BJAB/HA-LMP1 cells were untreated, treated with 10 ng/mL tetracycline for 48 hours, or treated with 2.5 μg/mL tunicamycin for 4 hours. The 721 cell line was sorted for low, medium, and high levels of LMP1. RNA was isolated from these cells and analyzed by RT-PCR using primers specific for both XBP-1 and GAPDH. To determine whether splicing of XBP-1 had occurred, the PCR fragments were digested with PstI. The ratios of spliced XBP-1 to GAPDH were calculated. The numbers represent the fold-difference relative to the ratio of untreated BJAB/HA-LMP1 cells, which was set at 1. *P (1-sided) <.05, 1-sided alternative: low, mid, high in increasing order, Jonckheere-Terpstra test. A gel representative of 3 independent experiments is shown. Production of Ig was measured by Western blotting the culture supernatant of EBV-positive strain cells (B) and 721 cells (C) that were sorted for low, medium, and high levels of LMP1 by staining with lysotracker red as described.60 For Ig heavy chain detection, 1 μL supernatant was analyzed, and for Ig light chain detection, 10 μL supernatant was analyzed, because the antibody preferentially detects Ig heavy chain. The cells were harvested and analyzed by Western blotting using antibodies to detect LMP1 and tubulin. A representative gel of duplicates of 2 independent experiments is shown. (D) BJAB/HALMP1 was electroporated with p3xFLAG-ATF6. Cells were either left untreated or treated with 10 mM DTT for 1.5 hours and 10 ng/mL tetracycline for 48 hours to induce LMP1. The cells were harvested and analyzed by Western blotting using antibodies to detect LMP1, tubulin, and Flag. The numbers represent the fold-difference of ratios of nuclear N-terminus ATF6 to tubulin. *P (2-sided) less than .05 (Wilcoxon rank sum test). A representative example of 3 independent experiments is shown.
The spliced form of XBP-1 has been shown to play a role in differentiation of B cells to plasma cells.20,–22 This differentiation can be measured by an increase in the production of Ig. We have shown that LMP1 induces the production of spliced XBP-1 and tested whether increasing amounts of spliced XBP-1 would lead to increasing levels of Ig secretion. Secretion of Ig was observed in cells expressing spliced XBP-1 product independent of the level of spliced XBP-1 (Figure 6B,C). On average, 721 cells secreted 1.3 × 107 molecules of Ig/cell per hour, and cells of an EBV-positive strain secreted 3.2 × 107 molecules of Ig/cell per hour. Secretion of Ig from 721 cells and from those of the EBV-positive strain was inhibited in the presence of cycloheximide (data not shown).
LMP1 induces the activation of 2 branches of the UPR, PERK and IRE1. To determine whether LMP1 induces the third UPR pathway, we tested whether LMP1 induces ATF6 activation. Upon activation of the UPR, the 90-kDa full-length ATF6 is cleaved to release a 50-kDa N-terminal protein that migrates to the nucleus to activate transcription of genes.50,51 In untreated B cells, ATF6 was constitutively expressed and migrated as a 90-kDa protein. In response to ER stress inducers, such as DTT, a 50-kDa protein was visible (Figure 6D). Expression of LMP1 also induced the expression of the 50-kDa protein (Figure 6D). This findings show that LMP1 induces all 3 pathways of the UPR, PERK, IRE1, and ATF6.
Cells that express high levels of LMP1 regenerate a daughter population with a broad distribution of levels of expression of LMP1
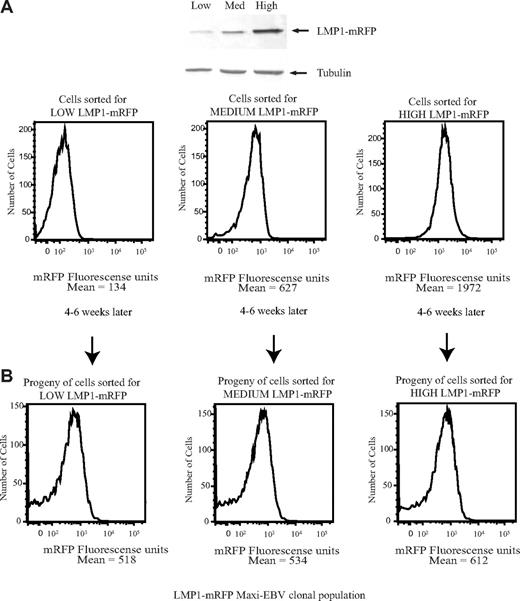
By inducing the UPR, LMP1 drives its own synthesis and at its highest levels leads to cytostasis. It is important to know whether cells expressing high physiologic levels of LMP1 can recover from the escalating levels of the UPR to resume protein synthesis and proliferation. To test whether cells can recover from their cytostatic state, a recombinant EBV was developed to express LMP1 fused to mRFP, termed LMP1-mRFP, so that live cells with differing levels of LMP1 could be sorted. We first confirmed that the level of LMP1 expression correlates with the levels of mRFP expression (Figure 7A). The cloning efficiencies of cells expressing low, medium, and high levels of LMP1 were statistically identical with 2% to 4% of the cells forming colonies. The same parental, broad distribution of LMP1 was observed from clones derived from cells expressing low, medium, or high levels of LMP1-mRFP (Figure 7B). Thus, cells that express high levels of LMP1 recover from their cytostatic state to proliferate and regenerate a population of daughter cells expressing a broad spectrum of levels of LMP1.
Cells that express high levels of LMP1 regenerate a daughter population with a broad distribution of levels of expression of LMP1. (A) EBV-positive strains expressing LMP1-mRFP were sorted for cells expressing low, medium, or high levels of mRFP and analyzed by FACS analysis and Western blotting using anti-LMP1 and antitubulin antibodies. (B) Cells sorted for low, medium, or high levels of mRFP were plated in 96-well plates. The clones were allowed to grow for 4 to 6 weeks. Proliferating cell clones were expanded and analyzed on FACS Vantage. Data were analyzed with FlowJo software. The broad distribution of LMP1 observed in EBV-positive strain cells expressing LMP1-mRFP was confirmed to be comparable with that observed in 721 cells by fixing the cells and staining with anti-LMP1 antibodies (data not shown).
Cells that express high levels of LMP1 regenerate a daughter population with a broad distribution of levels of expression of LMP1. (A) EBV-positive strains expressing LMP1-mRFP were sorted for cells expressing low, medium, or high levels of mRFP and analyzed by FACS analysis and Western blotting using anti-LMP1 and antitubulin antibodies. (B) Cells sorted for low, medium, or high levels of mRFP were plated in 96-well plates. The clones were allowed to grow for 4 to 6 weeks. Proliferating cell clones were expanded and analyzed on FACS Vantage. Data were analyzed with FlowJo software. The broad distribution of LMP1 observed in EBV-positive strain cells expressing LMP1-mRFP was confirmed to be comparable with that observed in 721 cells by fixing the cells and staining with anti-LMP1 antibodies (data not shown).
Discussion
In B cells, LMP1 is expressed mainly from the EDL1 promoter in the EBV genome.52,53 Both viral and cellular factors can transactivate the EDL1 promoter. The EBV nuclear antigen 2 (EBNA2) protein transactivates LMP1's promoter when bound to cellular proteins, such as RBP-Jκ54,,,–58 and PU.1 factor.59 EBNA2 does not have any specific DNA binding domain, and thus associates with cellular proteins to recognize the EDL1 promoter. When the levels of EBNA2 are compared in cells that express low levels of LMP1 and high levels of LMP1, a 2-fold difference is observed. When either levels of LMP1 RNA or protein are compared, a 30-fold or greater difference is observed.3 The 2-fold difference observed for the levels of EBNA2 cannot explain the 30-fold difference that we observe for the levels of LMP1 RNA and protein. These measurements indicate the presence of another mechanism that leads to the various levels of LMP1 among cells of clones of EBV-positive B cells. We have now found that LMP1 can regulate its own expression through an autocatalytic loop that likely accounts for its high levels of RNA and protein found in a subset of the cells of clonal populations. We have found that the LMP1 oncogene induces the UPR dose dependently in B cells (Figure 6), which leads to increasing activation of PERK, increasing phosphorylation of eIF2α, increasing expression of ATF4, and increasing expression of LMP1 (Figures 3,4).
This autocatalytic regulation is surprising for multiple reasons. First, it is autocatalytic. Autocatalytic loops are usually described as being uncontrolled and destructive, but LMP1 eventually limits its autocatalysis by inducing both autophagy and the inhibition of global protein synthesis efficiently.60 B cells showing the most advanced stages of autophagy, which are the cells that express high levels of LMP1, support rapid degradation of LMP1.60 Cells survive the high levels of LMP1 and regenerate daughter populations with a wide range of levels of LMP1 (Figure 7). This autocatalytic regulation is also unexpected because it reflects an oncogene's having evolved to regulate the UPR conditionally to regulate the oncogene's activity. Intermediate levels of LMP1 drive signaling through NF-κB efficiently, whereas high levels of LMP1 drive this signaling at reduced levels.19 This autocatalytic regulation can thus be likened to the regulation of a ligand for a receptor. Ligand-binding induces signaling; prolonged binding induces downregulation of the receptor and a decrease in its signaling. This autocatalytic regulation is intriguing for a third reason: it is mediated by the transmembrane domains of LMP1 and not by its carboxy signaling domain (Figure 4). LMP1 binds TRAFs, TRADD, and JAK3 through its carboxy terminus. Fusion of this terminus to the external and membrane-spanning moiety of the nerve growth factor-receptor (NGFR) leads to signaling through NF-κB, AP1, and STAT-1 being dependent on NGF.34 It is now clear that for intact LMP1, its membrane-spanning domains are crucial for its trafficking, for its oligomerization, and for the regulation of its levels of expression. These 3 properties determine its signaling. LMP1 can thus now be described as a fusion of a regulatory element, its membrane-spanning domain, to an effector element, its carboxy terminal signaling domain.
The wide range of expression of LMP1 in clonal cell populations reflects the combination of its regulation by its promoter and by its membrane-spanning domain. EBNA2 can activate this promoter robustly, and ATF4 more modestly. For example, LMP1's promoter driving luciferase in EBV-negative cells can be stimulated 10-fold,61 whereas our studies indicate that exogenous induction of the UPR induces LMP1 7-fold (Figure 5). These 2 modes of regulation acting multiplicatively likely account for the increases in LMP1's cycle of expression, whereas LMP1's eventual induction of an inhibition of protein synthesis through the UPR and its induction of autophagy account for its descending phase of this cycle.
We have also found that LMP1's induction of the UPR in B cells leads to increasing levels of expression of spliced XBP-1 (Figure 6A). The spliced form of XBP-1 has been shown to play a role in differentiation of B cells to plasma cells.20,–22 We have shown that secretion of Ig in EBV-positive cells is independent of the levels of XBP-1, indicating that this secretion requires only a threshold level of XBP-1 (Figure 6B,C). LMP1 may play a role in driving EBV-infected B cells toward plasma cell differentiation by inducing the expression of spliced XBP-1. To test this hypothesis, the ability to secrete Ig could be compared between a recombinant EBV that expresses full-length LMP1 to one that expresses the chimeric NGF-R:LMP1 protein.34 If LMP1 drives EBV-infected B cells toward plasma cell differentiation by inducing the expression of spliced XBP-1, the recombinant EBV expressing NGF-R:LMP1 protein would not secrete Ig because it lacks the 6-membrane spanning domains required to induce the UPR.
LMP1 induces the activation of all 3 pathways of the UPR: PERK, IRE1, and ATF6. Existing studies corroborate the idea of Bip being the master regulator of the UPR by binding to PERK, IRE1, and ATF6 and preventing their signaling during nonstressed conditions.62,63 Does LMP1 regulate Bip to induce the UPR and activate PERK, IRE1, and ATF6? Our studies indicate that LMP1 induces the UPR; the mechanism by which LMP1 does so still remains to be uncovered.
It is worth considering why EBV and its LMP1 oncogene have evolved such an apparently arcane mechanism to regulate expression of LMP1. The mechanism obviously works well for the virus; it now infects more than 90% of humanity. One satisfying explanation is that this mechanism leads to the daughter cells of any initially infected cell having a range of properties, allowing some to survive the human host's antagonistic response to infection. Infected cells with low levels of LMP1, for example, would be more likely to evade a host's cytotoxic response against LMP1 than those expressing higher levels. The survivors do rapidly yield daughter populations with the full range of levels of LMP1, allowing them to proliferate and eventually yield in vivo the proliferating memory B cells in which EBV resides latently64 or the proliferating B cells that can rarely evolve into lymphomas.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Ian Mohr for providing the PKR−/− MEF cells; Dr David Ron for providing the plasmid expressing mouse PERK, 9E10-tagged at C-terminus, and CHOP:GFP reporter plasmid; Dr Ron Wek for providing the plasmid encoding dn PERK, Dr Kenzo Takada for providing the plasmid encoding dn PKR; Dr Lars Rymo for providing the plasmids pgLRS (−106) CAT and pgLRS (−106)(ATF/CREmut.) CAT; and Dr Ron Prywes for providing the plasmid p3xFLAG-ATF6. We also thank Dr Jennifer Martin and Dr Ngan Lam for valuable discussions and comments on the manuscript. We are grateful to the Flow Cytometry Facility of the University of Wisconsin Paul P. Carbone Comprehensive Cancer Center for excellent technical support with FACS analysis.
This work was supported by National Institutes of Health grants CA070723 and P30 CA014520. B.S. is an American Cancer Society Research Professor.
National Institutes of Health
Authorship
Contribution: D.Y.L. designed and performed research, analyzed data, and wrote the paper. B.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bill Sugden, McArdle Laboratory for Cancer Research, University of Wisconsin-Madison, 1400 University Ave, Madison, WI 53706; e-mail: sugden@oncology.wisc.edu.