Cancer cells acquire disruptions in normal signal transduction pathways and homeostatic mechanisms that would trigger apoptosis in normal cells. These abnormalities include genomic instability, oncogene activation, and growth factor independent proliferation. Therefore, cancer cells likely require a block in apoptosis in order to survive. Overexpression of the antiapoptotic protein BCL-2 provides a block in apoptosis that is frequently observed in cancer cells. We have developed methods for the detection and analysis of BCL-2 dependence and here apply them to acute lymphoblastic leukemia (ALL). BH3 profiling, a mitochondrial assay that classifies blocks in the intrinsic apoptotic pathway, indicated a dependence on BCL-2 of both ALL cell lines and primary samples. This dependence predicted that BCL-2 would be complexed with select pro-death BH3 family proteins, a prediction confirmed by the isolation of BCL-2 complexes with BIM. Furthermore, the BH3 profiling and protein analysis predicted that ALL cell lines and primary cells would be sensitive to ABT-737 as a single agent. Finally, BH3 profiling and protein studies accurately predicted a relative degree of sensitivity to BCL-2 antagonism in cell lines. The ALL cells studied exhibit BCL-2 dependence, supporting clinical trials of BCL-2 antagonists in ALL as single agents or combination therapies.
Introduction
Although pediatric acute lymphoblastic leukemia (ALL) has a cure rate of 80%, the remaining 20% of cases are refractory to currently available therapies. Furthermore, the responsive pediatric ALL patients require intensive therapy, which can have serious acute and long-term side effects.1 Additionally, adult ALL survival rates are currently below 40%, further indicating that more effective therapies for ALL are required.2
Cancer cells acquire disruptions in normal signal transduction pathways and homeostatic mechanisms that result in abnormalities that would trigger apoptosis in normal cells. These abnormalities include genomic instability, oncogene activation, and growth factor independent proliferation. Therefore, it is likely that cancer cells require a block in apoptosis in order to survive.3,,,,,–9 Overexpression of the antiapoptotic protein BCL-2 provides a block in apoptosis that can be frequently observed in cancer cells.10,,,,–15 BCL-2 was originally identified at the breakpoint of t(14;18) in follicular lymphoma,16,–18 and its discovery has led to identification of a family of proteins that control commitment to apoptosis using the mitochondrial, or intrinsic, pathway. Several other antiapoptotic proteins such as MCL-1, BCL-XL, BCL-w, and BFL-1 have been identified, as well as proapoptotic proteins such as BAX and BAK, all of which share homology in several BCL-2 homology (BH) domains. The BH3-only proteins represent a third group within the BCL-2 family. These proteins have homology in only one domain, the BH3 domain, which is essential to their proapoptotic function. BH3-only proteins are distinguishable into 2 categories, activators and sensitizers.13,19 Activators, which include at least BID and BIM, induce activation and oligomerization of BAX and BAK. Oligomerized BAX and BAK cooperate in the formation of a pore that permeabilizes the outer mitochondrial membrane, releasing proapoptotic molecules such as cytochrome c, SMAC, and AIF to the cytosol.
Proteins like BCL-2 perform their antiapoptotic function by binding and sequestering activators, prohibiting their interaction with BAX and BAK. Additionally, antiapoptotic proteins may sequester monomeric BAX or BAK, particularly following activation. Sensitizers, meanwhile, lack the ability to activate BAX or BAK. They exert their proapoptotic function by binding antiapoptotic proteins and inhibiting their function. The interaction of BH3-only sensitizers and antiapoptotic proteins is selective: each antiapoptotic protein binds its own subset of sensitizer BH3 domains13,20 (Figure 1A). In cancer cells, therefore, overexpression of antiapoptotic BCL-2 allows survival in the face of activator protein expression, likely generated by phenotypic abnormalities acquired during oncogenesis. For instance, we have shown that chronic lymphocytic leukemia (CLL) cells depend on the function of BCL-2 to tonically sequester the BIM present in CLL cells. When BCL-2 function is abrogated, BIM is released and the CLL cells undergo apoptosis.
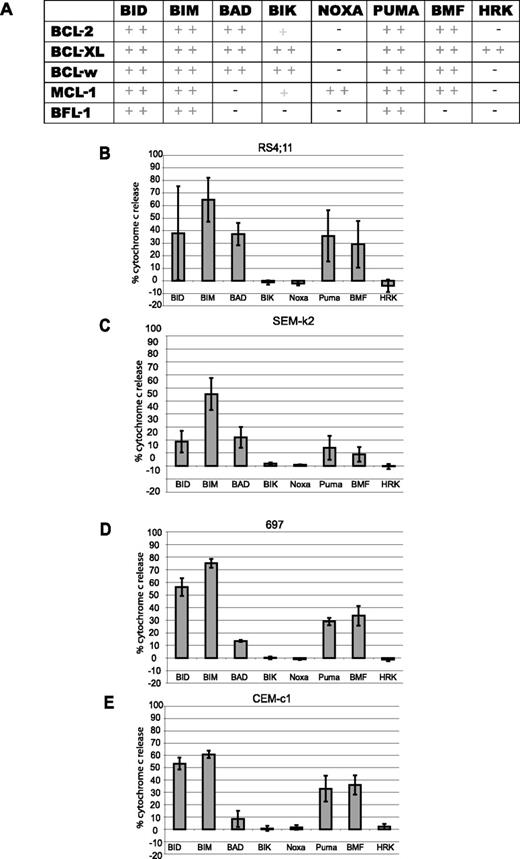
BH3 profiling determines BCL-2 dependence of ALL cell lines. (A) Summary of binding chart between BH3-only peptides and different antiapoptotic BCL-2 family members, adapted from Certo et al.13 (B-E) For BH3 profiling, at least 5 × 108 cells were used to isolate mitochondria by differential centrifugation for each cell line. Mitochondria were then treated with 100 μM BH3 peptides for 35 minutes at room temperature and cytochrome c release assessed by ELISA. Profiles were put in order of most to least primed. (B) RS4;11 cells. (C) SEM-k2 cells. (D) 697 cells. (E) CEM-c1 cells. Values are representative of the mean of 3 independent experiments (+ SD).
BH3 profiling determines BCL-2 dependence of ALL cell lines. (A) Summary of binding chart between BH3-only peptides and different antiapoptotic BCL-2 family members, adapted from Certo et al.13 (B-E) For BH3 profiling, at least 5 × 108 cells were used to isolate mitochondria by differential centrifugation for each cell line. Mitochondria were then treated with 100 μM BH3 peptides for 35 minutes at room temperature and cytochrome c release assessed by ELISA. Profiles were put in order of most to least primed. (B) RS4;11 cells. (C) SEM-k2 cells. (D) 697 cells. (E) CEM-c1 cells. Values are representative of the mean of 3 independent experiments (+ SD).
BH3 profiling is a novel method we have developed to identify blocks in the apoptotic pathway. We have demonstrated, for example, that BH3 profiling can accurately identify cells that are dependent on BCL-2 or MCL-1. As such, the strategy can also predict response to BCL-2 antagonists like ABT-737. To perform the assay, mitochondria are isolated from the cell in question. A panel of BH3 peptides is then applied to the cells, and permeabilization of the mitochondrial outer membrane is measured, for instance, by measuring release of cytochrome c. If sensitizer BH3 peptides induce permeabilization, that implies dependence on an antiapoptotic protein. Furthermore, the particular pattern of BH3 sensitizer peptides that induce permeabilization allows us to deduce the identity of the critical antiapoptotic proteins(s) involved. A particular advantage to BH3 profiling is that no ex vivo cell culture is required. Analysis can be directly performed on samples freshly obtained from the patient. Thus, the assay provides a realistic appraisal of the arrangement of the apoptotic machinery in vivo.
In this report, we turn our attention to the investigation of whether acute lymphoblastic leukemias are often dependent on BCL-2. Using BH3 profiling, analysis of BCL-2 family proteins, and in vitro response to ABT-737, we find that BCL-2 dependence is common in ALL cell lines as well as primary samples. These results provide fundamental information about the type of apoptotic blocks likely to be found in ALL. They furthermore provide support for the clinical testing of BCL-2 antagonism in ALL.
Methods
Collection of blood samples was obtained after Dana-Farber Cancer Institute Institutional Review Board approval. Informed consent was obtained in accordance with the Declaration of Helsinki.
Peptides
Peptides were synthesized by Tufts University Core Facility and purified by high-performance liquid chromatography (HPLC). Identity was confirmed by mass spectrometry. Stock solutions were made in dimethyl sulfoxide (DMSO). Sequences were taken from published sequences of BAD(LWAAQRYGRELRRMSDEFEGSFKGL),21 NOXA A (AELPPEFAAQLRKIGDKVYC),22 BMF (HQAEVQIARKLQLIADQFHR)23 BID(EDIIRNIARHLAQVGDSMDR),24 BIM (MRPEIWIAQELRRIGDEFNA),25 BIK(MEGSDALALRLACIGDEMDV),26 BNIP3-α (VVEGEKEVEALKKSADWVSD),27 HRK (SSAAQLTAARLKALGDELHQ),28 and PUMA (EQWAREIGAQLRRMADDLNA).29
ALL cell line culture
697, RS4;11, CEM-c1, and SEM-k2 cells were cultured in RPMI medium with 10% fetal bovine serum.
Isolation and short-term culture of human cells
Three to 15 mL of blood in heparin-treated tubes was obtained from each anonymous ALL patient and processed without freezing. An exception was sample L8, which was processed after freezing and storage in 10% DMSO. Most samples were pediatric except L1, L8, and the sample co-incubated with stromal cells, which were obtained from adults. Equal volume of media (RPMI medium, 10% human serum, supplemented with 10 ng/mL human recombinant interleukin-3 (IL-3) (R&D Systems, Minneapolis, MN), 50 ng/mL human stem-cell factor (cytokines and signaling, Invitrogen, Carlsbad, CA), and 1% ITS (insulin/transferrin/selenium; Sigma-Aldrich, St Louis, MO) was mixed with each sample and ALL cells isolated by centrifugation through Ficoll-PAQUE Plus (Amersham, Arlington Heights, IL). Cells were washed twice in media and cultured at a density of 1.0 × 106 cells/mL for up to 48 hours. For the primary ALL cells cocultured with stroma, immortalized human bone marrow mesenchymal cells30 were plated 24 hours prior to isolation and co-incubation with ALL cells. We conformed to all guidelines and regulations in accordance with international review board protocols #01-206 (Dana-Farber Cancer Institute and Brigham and Women's Hospital) and #05-001 (Dana-Farber Cancer Institute and Children's Hospital, Boston).
Cytochrome c release
Mitochondria were purified from freshly isolated ALL cells and cell lines by mechanical disruption followed by differential centrifugation, as previously described.19 Mitochondrial suspensions were made 0.5 mg protein/mL. Release of cytochrome c was determined by a comparison of cytochrome c in the pellet and supernatant quantitated by ELISA (R&D Systems).
Immunoblots
ALL protein lysates were obtained by cell lysis in CHAPS (100 mM NaCl, 5 mM NaPO4, 2.5 mM EDTA, 1% CHAPS [Sigma]) buffer supplemented with a complete protease inhibitor cocktail tablet (Roche, Indianapolis, IN). C-terminal truncated glutathione-S transferase (GST)–tagged BCL-XL and BCL-2 were prepared from bacterial lysates as previously described.13 Protein samples were electrophoretically separated on NuPAGE 10% Bis-Tris polyacrylamide gels (Invitrogen). Antibodies were used to detect the following proteins on membrane: BIM (Calbiochem 22-40 [San Diego, CA] or Abgent BH3 domain [San Diego, CA]); BCL-2 (Pharmingen [San Diego, CA], /100); MCL-1 (Chemicon [Temecula, CA], RC-13 or Santa Cruz [Santa Cruz, CA], S-19); BID (Santa Cruz, FL-195, full and cleaved), BCL-XL (kind gift from Larry Boise), actin (Chemicon, MAB 1501), BAX (Santa Cruz, N-20), BAK (Santa Cruz, G-23), and poly(ADP-ribose) polymerase (PARP) (BioVision [Mountain View, CA], c-2-10, full and cleaved).
Annexin-V assay
Cells were stained with fluorescent conjugates of annexin-V (BioVision) and analyzed on a FACSCalibur machine (Becton Dickinson, San Jose, CA).
Western blot protein quantification
Densitometry of protein bands were acquired using an AlphaImager EC gel documentation system (Alpha Innotec, Kasendorf, Germany), and bands analyzed with the spot densitometry analysis tool (Alpha Ease FC software, version, 4.1.0). Quantities of relative BCL-2, MCL-1, and BIM per ALL cell line or primary sample were obtained by dividing total values of each protein respectively by the corresponding actin value.
Statistical analyses
At least 3 experimental replicates were performed in experiments using lysates, whole cells, or isolated mitochondria from different cell lines or primary ALL samples. A P value for comparison of cytochrome c release from the BAD BH3 peptide for RS4;11 and CEM-c1 mitochondria was obtained using a 2-tailed Student t test, and P < .05 was considered statistically significant. GraphPad Prism 4 software was used to determine EC50 values by nonlinear dose response curve fitting using the mean of 3 independent experiments. Correlation of logEC50 with percent cytochrome c release due to the BAD BH3 peptide was obtained using a linear regression model.
Caspase-9 activation assay
ALL cells were treated with 100 nM ABT-737 or DMSO for 24 hours. Cells were counted and an equal number of each set aside for analysis using ApoAlert Caspase 9 Fluorescent Assay Kit (Clontech, Palo Alto, CA) according to the users' manual.
Results
BH3 profiling reveals varying degrees of BCL-2 dependence in ALL cell lines
We performed BH3 profiling on mitochondria isolated from a selection of ALL cell lines described in Table 1. Peptides derived from BH3-only activators like BIM and BID should cause cytochrome c release in any cell that has intact BAX and/or BAK, while peptides derived from the sensitizer BH3-only proteins yield cytochrome c release only if the cell is dependent on one or more antiapoptotic proteins and the antiapoptotic protein(s) are bound by activators. We have previously demonstrated in cell lines as well as mouse and human primary cancer cells that a BCL-2–dependent cell can be readily distinguished from an MCL-1–dependent cell, or from a cell not dependent on either.13,31 The BH3 profiles of all 4 cell lines generated a BCL-2–dependent pattern (Figure 1A,B-E). However, despite the qualitative similarities in the pattern of permeabilization, quantitative differences can be discerned. Examination of the response to the BAD BH3 peptide by each line reveals that RS4;11 cells have distinctly greater cytochrome c release compared with CEM-c1 cells (P = .01). The BAD BH3 cytochrome c release patterns suggest the following ranking according to dependence on BCL-2: RS11:14 > SEM-k2 > 697 > CEM-c1.
Characteristics and EC50 values for 24 hours' treatment of ALL cell lines with ABT-737
| Cell line . | ABT-737 EC50 (nM) . | Enant. EC50 (nM) . | Phenotype . | MLL +/− . |
|---|---|---|---|---|
| 697 | 506 | 10421 | Pre-B | Absent |
| CEM-c1 | 2400 | 50100 | T | Absent |
| SEM-k2 | 235 | 8200 | Pro-B | Present |
| RS11;14 | 24 | 3200 | Pro-B | Present |
| Cell line . | ABT-737 EC50 (nM) . | Enant. EC50 (nM) . | Phenotype . | MLL +/− . |
|---|---|---|---|---|
| 697 | 506 | 10421 | Pre-B | Absent |
| CEM-c1 | 2400 | 50100 | T | Absent |
| SEM-k2 | 235 | 8200 | Pro-B | Present |
| RS11;14 | 24 | 3200 | Pro-B | Present |
ALL cell lines demonstrate varying sensitivity to ABT-737 as a single agent
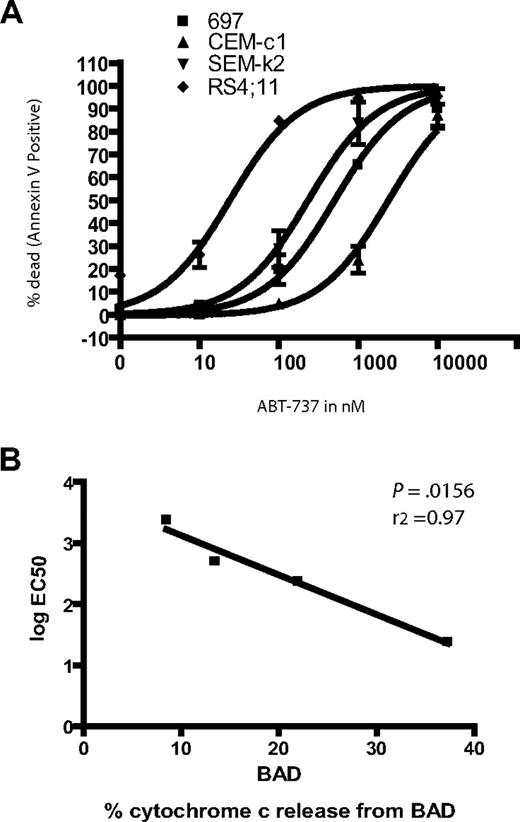
ABT-737 is a potent small molecule antagonist of BCL-2 (as well as BCL-w and BCL-XL).14 Notably, BAD BH3 has a similar interaction pattern to ABT-737, and we have found that ability of BAD BH3 to induce permeabilization in mitochondria correlates well with the toxicity of ABT-737 to cells. In our and others' hands, ABT-737 has shown single-agent toxicity against small-cell lung cancer cell lines, primary CLL cells, primary and established cell lines of acute myeloid leukemia, and multiple myeloma cell lines.13,14,31,,,–35 Our BH3 profiling suggested that in these ALL lines, there is tonic death signaling, conducted by pro-death BCL-2 family proteins, which are being held at bay by BCL-2 to varying degrees. Thus, certain cells are rendered quite dependent on BCL-2, so that BCL-2's antagonism is predicted to be fatal to the cells. Furthermore, we would predict that the cell lines would vary in their sensitivity to BCL-2 antagonism according to the ranking above. To test this prediction, the 4 ALL cell lines were treated with ABT-737 for 24 hours. The different cell lines gave a wide range of response to ABT-737 treatment (Figure 2) varying 2 logs from the most to least sensitive (Table 1).
Response of ALL cell lines demonstrates differential sensitivity to ABT-737 treatment. (A) 106 cells were treated in triplicate with increasing amounts of ABT-737. Apoptosis was assessed by counting annexin-V–positive cells by FACS analysis. (B) EC50 values were plotted against the BAD BH3 peptide cytochrome c release values for each line.
Response of ALL cell lines demonstrates differential sensitivity to ABT-737 treatment. (A) 106 cells were treated in triplicate with increasing amounts of ABT-737. Apoptosis was assessed by counting annexin-V–positive cells by FACS analysis. (B) EC50 values were plotted against the BAD BH3 peptide cytochrome c release values for each line.
ABT-737 sensitivity did support our hypothesis that the ALL cell lines have different degrees of BCL-2 dependence. For example, RS4;11 cells, which have an EC50 of 24 nM, give a 38% cytochrome c release when treated with BAD, while the relatively resistant CEM-c1 cells, which are 2 logs less responsive to ABT-737 (EC50 2400), gave only a 9% cytochrome c release when treated with the BAD peptide. In fact, sensitivity of cells to ABT-737 treatment followed the same order as the percent of cytochrome c release due to the BAD peptide: RS11:14 > SEM-k2 > 697 > CEM-c1 (Figures 1,2). This striking correlation is demonstrated graphically when the log EC50 is plotted against the cytochrome c release by BAD BH3 for each cell line, which yields a tightly correlated linear fit (Figure 2B).
Analysis of the BCL-2 family of proteins in ALL cells lines demonstrates further evidence of BCL-2 dependence
It is generally accepted that changes in apoptotic machinery are required to maintain survival of some, if not most, cancer cells.4 A variety of derangements exhibited by cancer cells can cause BH3-only proteins to become activated.36,,,,–41 Antiapoptotic proteins like BCL-2 or MCL-1 can sequester these pro-death proteins to permit cancer cell survival following up-regulation of BH3-only activators like BIM or BID.13,31 One telling example is CLL, in which up-regulated BIM is tolerated by CLL cells due to the concomitant expression of BCL-2. CLL cells survive because BIM is tonically sequestered in a complex with BCL-2.31 We call the cellular state where antiapoptotic proteins are complexed with activator BH3-only proteins “primed for death.”13 Primed cells are susceptible to apoptotic induction by antagonism of the antiapoptotic protein on which they depend. An example is the exquisite sensitivity of CLL cells to antagonism of BCL-2 function by ABT-737. Our BH3 profiling assay can not only readily detect if the cells are primed for death, but also uncover which antiapoptotic protein(s) a cell is dependent on.
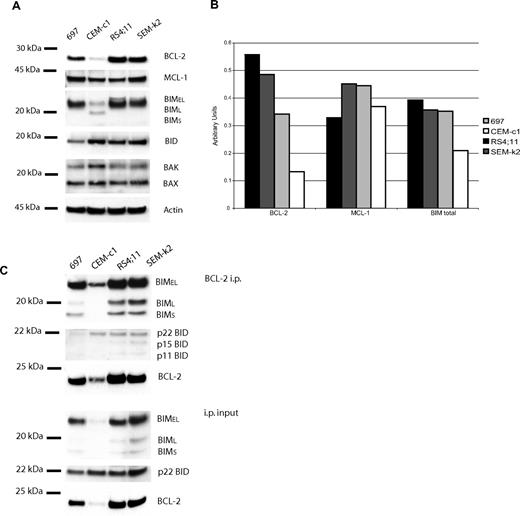
The BH3 profiling patterns that emerged from analysis of the ALL cell lines suggested to us that all of the ALL cell lines would be primed, but to different extents. We hypothesized that RS4;11 cells would be the most primed since they are the most responsive to ABT-737 and had the largest cytochrome c release from the BAD peptide. Indeed, when we analyzed the pro- and antiapoptotic proteins among the 4 cell lines, we found that the BCL-2 and BIM protein levels followed the same pattern as the whole-cell ABT-737 response and mitochondrial BH3 profiles, RS11:14 > SEM-k2 > 697 > CEM-c1 (Figure 3A,B). The BH3-only activator BID also is present in all 4 cell lines, as are BAX, BAK, and MCL-1, but at levels that do not correlate with response to BCL-2 inhibition. However, not only are BCL-2 and BIM present in each ALL cell line in a pattern consistent with the mitochondrial and whole-cell data, but they also are found in complex with each other as suggested by our “priming” model (Figure 3C). The levels of BCL-2:BIM complex also track with sensitivity to ABT-737. The activator BID was also found to be complexed with BCL-2, but there was no obvious correlation with the whole-cell or mitochondrial data. This data supports the notion that these cells are primed for death, and the amount of priming corresponds to ABT-737 sensitivity as well as to BH3 profile results.
Protein analysis in ALL cell lines reveals BCL-2, BIM, and BID are complexed. CHAPS lysates of the 4 ALL cell lines were used for protein experiments. (A) Western blot analysis of lysate (20 μg) reveals different expression of BCL-2, MCL-1, BIM, and BID among the cell lines. (B) Quantification of proteins bands by densitometry verifies that BCL-2 and BIM protein expression levels follow the rank order of whole-cell ABT-737 sensitivity and BAD-induced cytochrome c release: RS4;11 > SEM-k2 > 697 > CEMc-1. (C) Anti–BCL-2 immunoprecipitation of ALL cells lines (50 μg). Immunoprecipitation pellet (i.p.) and 10% of total lysate used for immunoprecipitation (input) were loaded. BIM and BID are complexed to BCL-2 in amounts proportional to whole-cell amounts.
Protein analysis in ALL cell lines reveals BCL-2, BIM, and BID are complexed. CHAPS lysates of the 4 ALL cell lines were used for protein experiments. (A) Western blot analysis of lysate (20 μg) reveals different expression of BCL-2, MCL-1, BIM, and BID among the cell lines. (B) Quantification of proteins bands by densitometry verifies that BCL-2 and BIM protein expression levels follow the rank order of whole-cell ABT-737 sensitivity and BAD-induced cytochrome c release: RS4;11 > SEM-k2 > 697 > CEMc-1. (C) Anti–BCL-2 immunoprecipitation of ALL cells lines (50 μg). Immunoprecipitation pellet (i.p.) and 10% of total lysate used for immunoprecipitation (input) were loaded. BIM and BID are complexed to BCL-2 in amounts proportional to whole-cell amounts.
Apoptosis induced by ABT-737 is through the intrinsic mitochondrial pathway
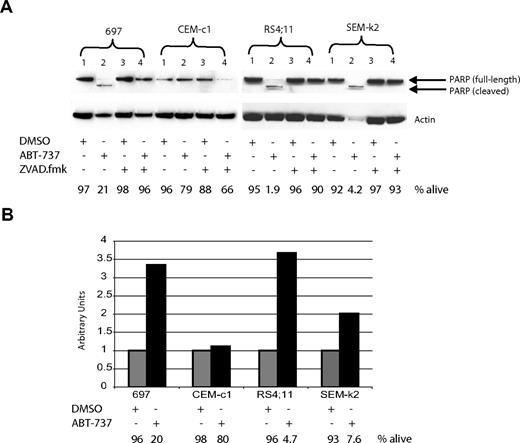
Apoptosis can be initiated either through the death-receptor–mediated extrinsic pathway or the intrinsic mitochondrial pathway. We have previously established that ABT-737 induces apoptosis through the mitochondrial pathway in CLL cells, through displacement of BIM from BCL-2.31 To distinguish which pathway is being engaged in ALL cell lines upon treatment with ABT-737, we first investigated if hallmarks of the mitochondrial pathway were present. Pretreatment of ALL cells with the pan caspase-inhibitor ZVAD.fmk prevented ABT-737–induced death at 24 hours as detected by annexin-V staining (Figure 4A bottom). Furthermore, PARP cleavage was detectable in ALL cells treated with ABT-737 but not with DMSO, which also was prevented by ZVAD.fmk (Figure 4A top). Next we examined the status of caspase-9, since only during apoptosis initiated by the intrinsic mitochondrial pathway, but not the extrinsic death-receptor–mediated pathway, is caspase-9 activated via cleavage following the mitochondrial permeabilization. Caspase-9 activity was detectable in ABT-737 but not in DMSO-treated cells (Figure 4B). Together these data support the notion that ALL cells treated with ABT-737 engage the mitochondrial pathway of apoptosis.
ABT-737 induces apoptosis through the mitochondrial pathway in ALL cells. Cells were treated for 24 hours with 100 nM ABT-737 or DMSO, or pretreated for 1 hour with 200 μM ZVAD.fmk prior to addition of ABT-737. (A) PARP cleavage correlates with cell death, as analyzed by annexin-V staining and reported as percent alive. (B) Caspase-9 activation was measurable by a fluorometric assay. The amount of annexin-V–negative cells was recorded just prior to harvesting cells (percent alive). Note that in both panels A and B the amount of ABT-737 used was not sufficient to cause significant apoptosis in CEM-c1 cells, which is consistent with the results reported in Figure 2.
ABT-737 induces apoptosis through the mitochondrial pathway in ALL cells. Cells were treated for 24 hours with 100 nM ABT-737 or DMSO, or pretreated for 1 hour with 200 μM ZVAD.fmk prior to addition of ABT-737. (A) PARP cleavage correlates with cell death, as analyzed by annexin-V staining and reported as percent alive. (B) Caspase-9 activation was measurable by a fluorometric assay. The amount of annexin-V–negative cells was recorded just prior to harvesting cells (percent alive). Note that in both panels A and B the amount of ABT-737 used was not sufficient to cause significant apoptosis in CEM-c1 cells, which is consistent with the results reported in Figure 2.
Primary ALL cells are sensitive to ABT-737 as a single agent in the low- to mid-nM range
Our results in the 4 ALL cell lines suggested that primary ALL cells may be primed and BCL-2 dependent, and therefore responsive to ABT-737 treatment in a reasonable therapeutic range. To identify if primary ALL cells are indeed sensitive to ABT-737, 6 primary ALL samples (Table 2) were collected and treated for 24 and 48 hours with increasing concentrations of either ABT-737 or to an enantiomer of much lower affinity to BCL-2. The resulting EC50 values illustrate that primary ALL cells are indeed sensitive to ABT-737 in the low- to mid-nM range (Table 3). It is notable that the range of EC50 values is narrower than that found for the cell lines. While these values suggest that ALL may be a more heterogeneous leukemia than CLL, the 6 primary ALL samples tested nonetheless exhibit fairly consistent single-agent toxicity.
Clinical characteristics of primary ALL samples studied
| Sample no. . | WBC . | Blast % . | Phenotype . | MLL +/− . |
|---|---|---|---|---|
| L1 | 85 | 83 | B cell | Present |
| L2 | 40.1 | 95 | Pre-B | Absent |
| L5 | 89 | 69 | Pre-B | Absent |
| L7 | 34.8 | 86 | Pre-B | Absent |
| L8 | >100 | >90 | Pre-B | Present |
| L9 | 130 | 78 | T | Absent |
| Sample no. . | WBC . | Blast % . | Phenotype . | MLL +/− . |
|---|---|---|---|---|
| L1 | 85 | 83 | B cell | Present |
| L2 | 40.1 | 95 | Pre-B | Absent |
| L5 | 89 | 69 | Pre-B | Absent |
| L7 | 34.8 | 86 | Pre-B | Absent |
| L8 | >100 | >90 | Pre-B | Present |
| L9 | 130 | 78 | T | Absent |
EC50 values for 24 hours' treatment of primary ALL cells with ABT-737
| Sample no. . | ABT-737 EC50, nM . | Enant. EC50, nM . |
|---|---|---|
| L1 | 47 | 757 |
| L2 | 361 | 2850 |
| L5 | 106 | 6710 |
| L7 | 16 | 1860 |
| L8 | 147 | 4900 |
| L9 | 207 | 4600 |
| Sample no. . | ABT-737 EC50, nM . | Enant. EC50, nM . |
|---|---|---|
| L1 | 47 | 757 |
| L2 | 361 | 2850 |
| L5 | 106 | 6710 |
| L7 | 16 | 1860 |
| L8 | 147 | 4900 |
| L9 | 207 | 4600 |
BH3 profiling and protein analysis of primary ALL cells reveal BCL-2 dependence
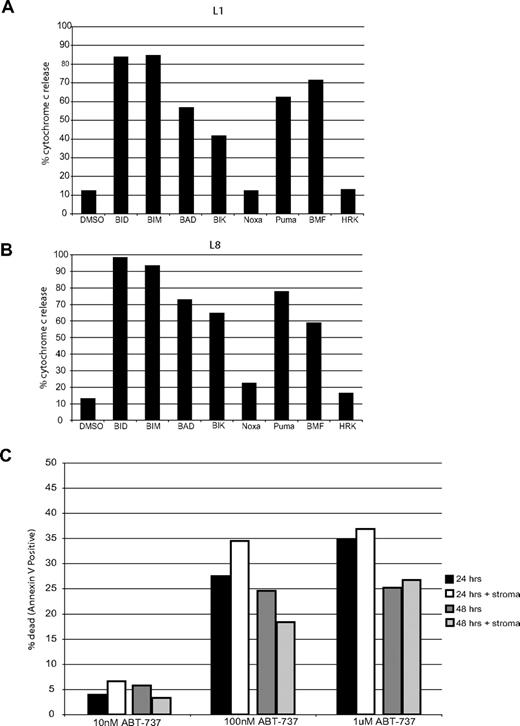
BH3 profiling requires a large number of cells in order to isolate mitochondria. Since the majority of the primary ALL samples we tested were from pediatric patients, a limited number of leukemia cells were available. However, cell amounts of 2 samples, L1 and L8, were adequate to perform BH3 profiling. Both leukemias demonstrated a BCL-2 profile (Figure 5A,B). Both samples also are very sensitive to ABT-737 treatment, with EC50's under 150 nM (Table 3), suggesting that BCL-2 is primed in each. These results suggest that BH3 profiling accurately assesses sensitivity of primary cells to ABT-737 without the need for ex vivo culturing.
BH3 profiling of primary ALL cells also shows a BCL-2 pattern. 109 cells from primary sample L1 (A) or 3.5 × 108 cells from primary sample L8 (B) were subjected to mechanical disruption and differential centrifugation. Isolated mitochondria were brought to a final concentration of 0.5 mg/mL and incubated with 100 μM peptides for 35 minutes at room temperature. Mitochondria pellet and supernatant were separated and analyzed using a cytochrome c ELISA kit. (C) Primary ALL cells were plated with or without stromal cells (Mihara, 2003 #654) with increasing concentrations of ABT-737. Apoptosis was assessed at 24 and 48 hours by counting annexin-V–positive cells by FACS analysis.
BH3 profiling of primary ALL cells also shows a BCL-2 pattern. 109 cells from primary sample L1 (A) or 3.5 × 108 cells from primary sample L8 (B) were subjected to mechanical disruption and differential centrifugation. Isolated mitochondria were brought to a final concentration of 0.5 mg/mL and incubated with 100 μM peptides for 35 minutes at room temperature. Mitochondria pellet and supernatant were separated and analyzed using a cytochrome c ELISA kit. (C) Primary ALL cells were plated with or without stromal cells (Mihara, 2003 #654) with increasing concentrations of ABT-737. Apoptosis was assessed at 24 and 48 hours by counting annexin-V–positive cells by FACS analysis.
It has been suggested that co-incubation of leukemia cells with stroma up-regulates antiapoptotic proteins42,43 that might potentially effect ABT-737 sensitivity. To verify that the absence of stroma did not affect the ABT-737 response we observed (Table 3), we cultured a primary ALL sample with and without stroma. We found that stromal cells had no effect on ABT-737 response within the first 24 and 48 hours of treatment (Figure 5C).
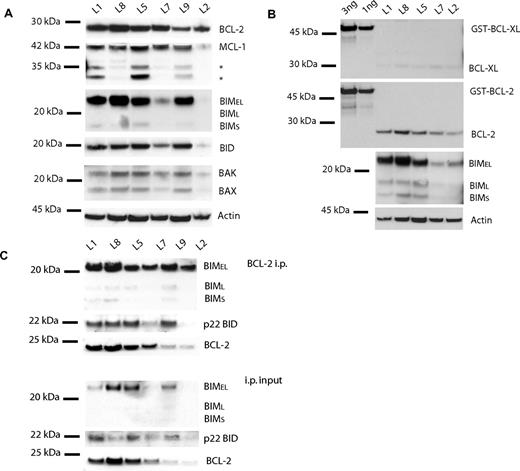
To verify the presence of BCL-2 as well as activators BIM and BID, Western blot analysis of the BCL-2 family of proteins was performed with 6 primary ALL samples. Indeed BCL-2, BIM, and BID were present in all samples, although at different levels, as well as BAK, BAX, and MCL-1 (Figure 6A,B). Our finding that BCL-2 protein levels vary among primary ALL samples is in agreement with an earlier report where BCL-2 protein expression levels were found to vary by as much as 10-fold in primary samples.44 Samples L1 and L8 do have a large amount of BCL-2, BIM, and BID (Figure 6A), consistent with their whole-cell response to ABT-737 as well as their BH3 profiles. Note that levels of BCL-XL, another protein that is targeted by ABT-737, are much lower (at least 5 times lower, based on densitometry; data not shown) than those for BCL-2. This finding is consistent with the low signal from the BCL-XL–specific HRK peptide in the BH3 profiles (Figures 1A,5).13 These data suggest that BCL-2 plays a more important role than BCL-XL in these primary ALL cells. Supporting the hypothesis that BCL-2 is primed with activator proteins, BIM and BID are complexed to BCL-2 as detected via immunoprecipitation (Figure 6C). Given the fairly narrow range of response to ABT-737 observed in these primary samples, abundance of no single protein or complex was able to quantitatively predict EC50. In a separate study, using a larger sample18 of lymphoma cell lines with a much broader range of response, we could show that both BCL-2 levels and BCL-2:BIM complex levels were predictive of EC50.45
Protein analysis of primary ALL cells is consistent with whole-cell ABT-737 response and BH3 profiles. (A) Western blot analysis of CHAPS lysates (20 μg) shows detectable expression of BCL-2, MCL-1, BIM, and BID among primary ALL cells. (B) Purified GST-BCL-XL and GST-BCL-2 (1 and 3 ng) were run next to 5 independent ALL lysates (10 μg) to quantitate BCL-XL (top panel) and BCL-2 (middle panel). Primary ALL lysates with better equal loading reveal clearer BIM levels among samples (bottom panel). (C) BCL-2, BIM, and BID are also complexed in primary ALL cells. Anti–BCL-2 immunoprecipitation of ALL primary cells (50 μg). Immunoprecipitation pellet (i.p.) and 10% of total lysate used for immunoprecipitation (input) were loaded. BIM and BID are complexed to BCL-2 in amounts proportional to whole-cell amounts.
Protein analysis of primary ALL cells is consistent with whole-cell ABT-737 response and BH3 profiles. (A) Western blot analysis of CHAPS lysates (20 μg) shows detectable expression of BCL-2, MCL-1, BIM, and BID among primary ALL cells. (B) Purified GST-BCL-XL and GST-BCL-2 (1 and 3 ng) were run next to 5 independent ALL lysates (10 μg) to quantitate BCL-XL (top panel) and BCL-2 (middle panel). Primary ALL lysates with better equal loading reveal clearer BIM levels among samples (bottom panel). (C) BCL-2, BIM, and BID are also complexed in primary ALL cells. Anti–BCL-2 immunoprecipitation of ALL primary cells (50 μg). Immunoprecipitation pellet (i.p.) and 10% of total lysate used for immunoprecipitation (input) were loaded. BIM and BID are complexed to BCL-2 in amounts proportional to whole-cell amounts.
Discussion
In this study we used ALL cell lines as well as primary cells to determine if ALL is generally dependent on the apoptotic block provided by BCL-2 expression. Certainly, it has been shown that BCL-2 is widely expressed in ALL.46,47 However, previous studies that have focused on the correlation of BCL-2 levels and clinical outcome48,–50 or BCL-2 gene expression and drug resistance51 concluded that there is no significant prognostic value to BCL-2 levels. However, expression of BCL-2 and dependence on BCL-2 are 2 very distinct concepts. We expect that BCL-2 dependence, rather than mere expression, might be a more clinically significant parameter. Therefore, we sought to investigate the functionally critical question of whether ALL cells are dependent on BCL-2.
BH3 profiling predicted which ALL cell lines and primary cells would be sensitive to BCL-2 antagonism, a prediction that was validated by in vitro testing. Since BH3 profiling is performed on fresh cells without ex vivo subculture, it describes the biology of the tumor in vivo. Thus, our results support the hope that patients' ALL cells would also respond to BCL-2 antagonists in vivo. BH3 profiling suggested that the cells in question were “primed,” a model subsequently supported by our protein studies, in which BIM:BCL-2 complexes were easily identified in all cells sensitive to ABT-737. Furthermore, we found that the 4 cell lines displayed varying sensitivity to ABT-737 (Figure 2) and that the degree of sensitivity correlated with both the BH3 profile (Figure 1A-E) and amounts of BCL-2 proteins present (Figure 3A,B) in each line. These results all correlate priming of BCL-2 to sensitivity to ABT-737 in ALL cells, just as was the case for CLL.31
It is worth noting that it is not expected that normal cells will exhibit “priming” to the same extent that cancer cells do. Normal vegetative cells performing their physiologically appropriate function are unlikely to bear a large quantity of death signaling, conducted by pro-death activators, that would require buffering by antiapoptotic proteins. To put it another way, what antiapoptotic proteins are present on normal cells are expected to be largely empty, while those in cancer cells may well be fuller. While definitive support for this model awaits a more exhaustive and broad study of normal and malignant tissues, our work so far supports the concept that malignant cells are more likely to be primed than nonmalignant cells.13,31 We feel this model is particularly enticing, as it predicts a dependence of many cancer cells on antiapoptotic proteins that would not be shared by nonmalignant cells, offering the possibility of a clinically useful therapeutic window.
The concept of identifying tumor-specific targets and antagonizing them with rationally designed pharmacologic agents has become the paradigm for much translational cancer research. However, as targets become more specific, the proportion of cancers bearing the target may also become more limited. An excellent example is that of inhibition of the epidermal growth factor receptor in non–small cell lung cancer with either gefitinib or erlotinib. While effective when used on cells bearing the target, cells bearing the appropriate EGFR mutation represent only approximately one-tenth of all non-small cell lung cancers. Therefore, use of an appropriate genetic screening mechanism was critical to their clinical implementation.52,53 In the case of targeting BCL-2 in cancers like ALL, it appears that BH3 profiling may be an effective tool in identifying leukemias most likely to be sensitive to BCL-2 antagonism.
In these studies, we did not directly address the possibility that differential sensitivity to BCL-2 antagonism might be exhibited by leukemia stem cells. Such a difference might well affect the ability of this new therapy to improve cure rates. However, the relevance of the leukemia stem cell concept to lymphoid malignancies remains unclear.54 Here we provide evidence that at least the bulk of cells in the ALL samples we tested exhibit dependence on BCL-2 by in vitro drug testing and BH3 profiling of fresh samples. Investigation of the ability to improve cure or the influence of environmental factors will likely best be addressed in the setting of clinical trials of BCL-2 antagonists in ALL. We believe our results provide support for the initiation of such clinical trials.
While many times the most effective chemotherapeutic regiments involve the combination of several different drugs, it is mechanistically reassuring that a BCL-2 inhibitor can be effective as a single agent. Our initial study suggests that BCL-2 antagonists such as ABT-737 may be worth considering as either single agents or in combination for the treatment of ALL. Furthermore, our studies suggest that BH3 profiling and BCL-2 protein analysis may be useful in identifying leukemias that are most likely to benefit from BCL-2 antagonism.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors wish to acknowledge technical assistance from Nicole Carlson, Jeremy Ryan, and Joerg Faber, and the kind gift of ABT-737 and negative control enantiomer compounds from Saul Rosenberg of Abbott Laboratories, Dan DeAngelo for adult ALL samples, and the DFCI Pediatric Hematology/Oncology Research Consortium core for providing clinical data.
This work was supported by funding from the American Cancer Society Steven Stanley and Edward Albert Bielfelt Postdoctoral Fellowship (V.D.G.M.); National Institutes of Health (NIH) grant #K08 CA10254; the Smith Family Foundation, Chestnut Hill, MA; the Sidney Kimmel Foundation for Cancer Research (A.L.); and NIH grant P01 CA068484 (A.L., S.A.).
National Institutes of Health
Authorship
Contribution: V.D.G.M. and A.L. contributed to the design and analysis of experiments and wrote the manuscript. V.D.G.M. conducted all of the experiments, performed statistical analysis, and made the figures. A.L. and S.A.A. initiated and supervised the study. S.E.S. provided essential material. K.D.S. coordinated primary pediatric ALL sample collection and prepared samples.
Conflict-of-interest disclosure: A.L. is a founder of Eutropics Pharmaceuticals and a member of its scientific advisory board. A patent application covering the BH3 profiling technology has been filed with the United States Patent Office. All other authors declare no competing financial interests.
Correspondence: Anthony Letai, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115; e-mail: anthony_letai@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal