Abstract
MMSET, identified by its fusion to the IgH locus in t(4;14)-associated multiple myeloma, possesses domains found within chromatin regulators, including the SET domain. MMSET protein is overexpressed and highly associated with chromatin in myeloma cell lines carrying t(4;14). MMSET possesses methyltransferase activity for core histone H3 lysine 4 and histone 4 lysine 20, whereas MMSET made in cells only modified H4. Segments of MMSET fused to the Gal4 DNA binding domain repressed transcription of a chromatin-embedded Gal4 reporter gene. MMSET-mediated repression was associated with increased H4K20 methylation gene and loss of histone acetylation. Consistent with this repressive activity, MMSET could form a complex with HDAC1 and HDAC2, mSin3a, and the histone demethylase LSD1, suggesting that it is a component of corepressor complexes. Furthermore, MMSET coexpression enhances HDAC1- and HDAC2-mediated repression in transcriptional reporter assays. Finally, shRNA-mediated knockdown of MMSET compromised viability of a myeloma cell line, suggesting a biologic role for the protein in malignant cell growth. Collectively, these data suggest that, by acting directly as a modifier of chromatin as well as through binding of other chromatin-modifying enzymes, MMSET influences gene expression and potentially acts as a pathogenic agent in multiple myeloma.
Introduction
Multiple myeloma (MM) is associated with recurrent chromosomal translocations that link the immunoglobulin promoter/enhancer with several partner genes, deregulation of which likely plays a key role in disease pathogenesis. MMSET (multiple myeloma SET domain) was identified as a gene involved in the t(4;14)(p16;q32) translocation present in approximately 15% to 20% of MM.1,2 This subtype of myeloma has a poor prognosis with frequent relapse after autologous stem-cell transplantation.3-5
The breakpoint within 14q32.3 occurs in the immunoglobulin switch region and dissociates the intronic enhancer (Eμ) from the 3′ enhancer (Eμ). The 4p16.3 breakpoint falls centromeric to the fibroblast growth factor receptor 3 (FGFR3) gene, placing it adjacent to the strong 3′ enhancer on the derivative chromosome 14. Concurrently, the intronic enhancer on derivative chromosome 4 is juxtaposed to the MMSET gene. The breakpoint on chromosome 4 can vary in different cases and can yield transcripts that can exclude up to 6 of the initial exons of MMSET.1 As a result of the translocation, both FGFR3 and MMSET can be deregulated; however, microarray analysis of t(4;14)-associated myeloma showed that all cases overexpress MMSET, but up to 25% of these cases do not overexpress FGFR3, implying a critical role for MMSET in this subset of myeloma.4,6
The MMSET gene, also known as Wolf-Hirschhorn Syndrome Candidate 1 (WHSC1)2 or Nuclear Receptor-binding SET Domain 2 (NSD2),7 spans 120 kb, consists of 24 exons and undergoes complex alternative splicing. Two major transcripts were identified: type I encodes a protein of 647 amino acids and type II encodes a protein of 1365 amino acids.1 Both proteins share a common amino terminus. A third transcript initiated within a middle intron of MMSET, encoding a mRNA comprising the 3′ half of the MMSET gene was identified8 and encodes a protein named RE-IIBP.
The 1365 amino acid MMSET protein contains a SET domain that is found in many histone methyltransferases (HMTs) and determines their enzymatic activity. Histone methylation of chromatin yields docking sites for modules found on transcriptional regulators, attracting these proteins to chromatin. Depending on the histone site modified and genetic context, methylation may be associated with activation or repression of genes.9 Other potential functional motifs in the MMSET proteins include nuclear localization signals (NLSs), an HMG box (high mobility group) often representing a DNA-binding domain, 2 PWWP domains2,10 (proline-tryptophan-tryptophan-proline) found in other nuclear proteins and 4 PHD (plant homeodomain) zinc fingers recently defined as binding modules for methylated lysines.11,12 We found that the MMSET protein is strikingly up-regulated in myeloma cell lines harboring t(4;14). MMSET is concentrated in the nucleus, has specific HMT activity against core histones H3 and H4, and coimmunoprecipitates and interacts functionally with corepressors and histone deacetylases. Together, these data suggest that MMSET is a transcriptional cofactor whose aberrant expression could deregulate gene expression and play a role in the development of MM.
Methods
Plasmid construction
The cDNA used to derive pCEFL-MMSET-I and pCEFL-MMSET-II was described previously.1 Individual MMSET domains were amplified by polymerase chain reaction (PCR) using primers (sequences available on request) flanked with 5′ BamHI and 3′ XhoI restriction sites, digested and ligated into pBXG113 creating in-frame fusions to the Gal4 DNA-binding domain, or to the pGEX-5X-1 vector (GE Healthcare, Little Chalfont, United Kingdom) creating GST fusions. GST-G9a14 and GST-NSD1(1700-1987)15 were previously described. Flag-HDAC1 and Flag-HDAC2 were gifts of Ed Seto (H. Lee Moffitt Cancer Center, Tampa, FL). ShRNA against MMSET was constructed by cloning oligonucelotides 5′-GAGAGCAA-GACAGATGTTTTCAAGAGAAACATCTGTCTTGCTCTCTTTTTTT-3′ and 3′-CTCTCGTTCTGTCTACAAAAGTTCTCTTTGTAGACAGA-ACGAGAGAAAAAAA-5′ (ShRNA2) and 5′-CTCGAAACTGCCATT-GTGATTCAAGAGATCACAATGGCAGTTTCGAGTTTTTTT-3′, 3′-GAGCTTTGACGGTAACACTAAGTTCTCTAGTGTTACCGTCAAAGCTCAAAAAAA-5′ (ShRNA8; targeted sequence in bold) into pSIREN-RetroQ-ZsGreen (Clontech, Mountain View, CA).
Expression and preparation of GST fusion proteins
A GST expression plasmid linking amino acids 1000 to 1365 of MMSET was expressed in BL21-Gold pLysS competent Escherichia coli (Stratagene, La Jolla, CA). After addition of isopropyl-b-D-thiogalactopyranoside (0.5 mM), the bacterial cultures were incubated for 10 hours at 18°C. The bacterial were sonicated on ice in buffer supplemented with Complete protease inhibitors (Roche Applied Science, Indianapolis, IN). The supernatant was incubated with glutathione Sepharose beads for 2 hours and washed with lysis buffer containing 0.6% NP40, and the purity of the fusion protein was determined by SDS-PAGE.
Preparation of anti-MMSET antibodies
GST-MMSET-I fusion protein was used to immunize rabbits (Covance Research Products, Princeton, NJ) and mice (Hybridoma Core Facility, Mount Sinai). The rabbit polyclonal anti-MMSET serum was purified using protein A-Agarose beads. The mouse monoclonal antibody was obtained by purifying hybridoma supernatant through a protein G column.
In vitro histone methyltransferase assay
Purified GST-MMSET enzyme (5 μg) was added to a 30-μL reaction containing 0.2 μCi of S-adenosyl- [methyl-14C]-L-methionine, native histones from calf thymus (Roche Applied Science) or recombinant histones (Upstate, Charlottesville, VA) in methylation buffer (50 mM Tris pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 β-mercaptoethanol, 200 mM sucrose), and incubated for 2 hours at 37°C. The reactions were stopped by boiling in SDS buffer, and their contents separated by 15% SDS-PAGE. Proteins were stained with Coomassie blue and methylation was visualized by autoradiography for native histones or immunoblotting with anti-H4K20Me2, H4K20Me3, H3K36Me2, H3K4Me2, and -Me3 antibodies (Upstate) for recombinant histones.
Cell culture and transfection
Myeloma cell lines KMS11, H929, KMS28PE, SKMM2, KMS12BM, and MM.M116 were maintained in RPMI 1640 medium, 10% fetal bovine serum. 293T cells harboring the Gal4-tk-Luc reporter gene were maintained in Dulbecco modified Eagle medium, and transfected in 24-well plates using FuGENE6 (Roche Applied Science) in triplicate with various amounts of expression plasmids encoding the Gal4 fusion proteins. The cells were harvested 48 hours later and assayed for luciferase activity using a Dual Reporter Luciferase Assay Kit (Promega, Madison, WI). The results obtained were normalized for protein concentration from each well of transfected cells. Myeloma cell lines were transduced using an Amaxa nucleoporator (Gaithersburg, MD); 5 × 106 cells were incubated in solution T and 5 μg shRNA plasmid and transduced using program T20. At 3 and 5 days after transfection, transduced cells were stained with Texas red–conjugated annexin V (Clontech) and propidium iodide and analyzed by flow cytometery (LSR-II, BD Biosciences, San Jose, CA). To determine the extent of shRNA knockdown, transduced cells were sorted (FACSort, BD Biosciences) and total cell lysates were immunoblotted for MMSET.
Nuclear extract and chromatin preparation
Cells were washed with ice-cold phosphate-buffered saline (PBS) and resuspended in buffer A1 [10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), and Complete protease inhibitor cocktail (Roche Applied Science)] and incubated on ice for 10 minutes. Nuclei were pelleted by centrifugation and incubated in buffer B1 [20 mM Hepes-KOH (pH 7.9), 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM ethylenediaminetetraacetic acid, 0.5 mM DTT, and Complete protease inhibitor] on ice for 20 minutes, and nuclear extracts were cleared by centrifugation.17 For nuclear fractionation,18 cells were washed with PBS, resuspended in buffer A2 [10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 10% glycerol, 0.1% Triton X-100, and protease inhibitors] and incubated on ice for 7 minutes. The pelleted nuclei were resuspended in buffer B2 [0.2 mM EGTA (pH 8), 3 mM ethylenediaminetetraacetic acid (pH 8), 1 mM DTT, and protease inhibitors] and incubated on ice for 30 minutes with occasional vortexing. The insoluble chromatin fraction and the soluble nuclear fraction were separated by centrifugation in a microfuge at 4000g for 5 minutes.
Immunoprecipitation and immunoblotting
Cells were washed with PBS and lysed on ice for 15 minutes in buffer consisting of 1% NP40, 20 mM Tris-HCl (pH. 7.6), 150 mM NaCl, and protease inhibitors. Cell lysates were clarified by centrifugation at 13 000g in a microcentrifuge for 15 minutes, and supernatants were incubated with anti-MMSET, anti-mSin3A (K-20) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-HDAC (Upstate), anti-LSD1 (Upstate), or rabbit IgG (Invitrogen, Carlsbad, CA) at 4°C for 1 hour; 50 μL protein A-agarose was added to the immunoprecipitated samples and incubated overnight at 4°C. The beads were washed in cold lysis buffer 3 times, protein released by boiling in SDS buffer, and separated by 8% SDS/PAGE. The gel was transferred to Immobilon P membrane (Millipore, Billerica, MA) and immunoblotted with antibodies as indicated, followed by incubation with a 1:7000 dilution of secondary antibody (Chemicon, Temecula, CA) and detection using enhanced chemoluminescence.
Generation of GAL4 reporter cell line
A cassette containing the SV40 promoter, puromycin gene, and polyadenylation signals was amplified by PCR from pSEC-puro (Promega) between nucleotides 2166 and 3561 with the primers 5′-GGTTGGATCCCACCTATTGGTGTGGAAAGTC-3′ and 5′-GGTTGGATCCAAAGGGCCTCGTGATACGCCT-3′. The 1.4-kb PCR product was cleaved with BamH1 and introduced into pGL2-Gal4-E1b-LUC,19 using BamH1 adapters. The resulting plasmid was transfected into 293T cells; puromycin populations were selected and evaluated by Southern blot hydrization and by genomic DNA PCR for the presence of the reporter gene. Controls using human G9a and mouse Cbx7 fused to the GAL4 DNA-binding domain indicated the GAL4-directed occupation of the reporter as determined by chromatin immunoprecipitation.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays (ChIP) assays were performed according to the manufacturer's protocol (Upstate). Chromatin, sonicated to an average DNA size of 500 bp, from 2 × 107 transfected Gal4-reporter/293T cells was used for each immunoprecipitation with preblocked M2 agarose, or as indicated antibodies directed to GAL4, H4, H4K20Me1, Me2, Me3, H3K2Me2, Me3, H3K36Me2, H3K27Me3, H3, acetyl lysine, or rabbit IgG (Upstate). Immunoprecipitated DNA was analyzed by quantitative PCR using primers (5′GTCCAAACTCATCAATGTATC3′ and 5′CAGGCGATCTGACGGTTCAC3′) specific to the integrated GAL4 reporter.20
Immunofluorescence imaging
Myeloma cell were spun onto glass slides at 600g for 10 minutes in a Cytospin 2 centrifuge, fixed in ice-cold methanol for 5 minutes, permeabilized in 0.2% Triton X-100 in PBS for 10 minutes, and blocked with 5% goat serum in PBS for 10 minutes. The cells were stained for 1 hour at room temperature with anti-MMSET antibody, followed by a 30-minute incubation at room temperature with antirabbit Alexa Fluor 549 goat IgG (Invitrogen). The slides were mounted with Vectashield Medium containing DAPI (Vector Laboratories, Burlingame, CA) and observed in a Zeiss Axioscop 2 fluorescence microscope (100×/0.75 numeric aperture oil objective) and camera and images processed with Adobe Photoshop 7 (Adobe Systems, Mountain View, CA). NIH 3T3 cells, grown on glass cover slips were transfected with MMSET expression vectors using Fugene 6. The cells were fixed and stained as above.
Theoretical structural modeling
Results
Several MMSET isoforms are overexpressed in t(4;14) myeloma cell lines
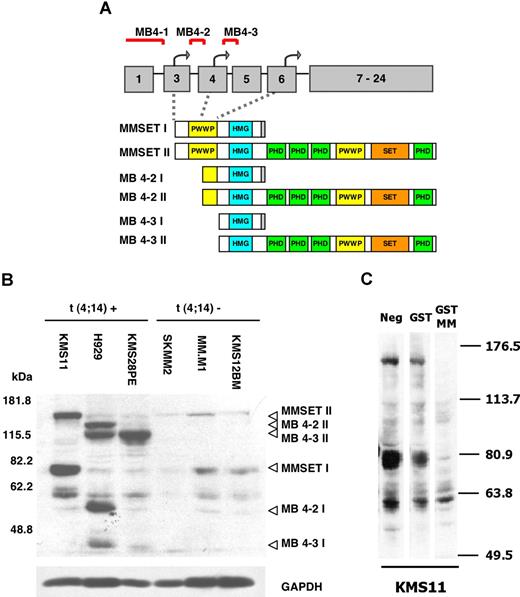
A rabbit polyclonal antibody, raised against the amino terminus shared by MMSET-I and II, demonstrated highly overexpressed levels of MMSET protein variants in cell lines carrying t(4;14), KMS11, H929, and KMS28PE compared with the cell lines SKMM2, MM.M1, and KMS12BM carrying t(11;14). The cell line KMS11 expresses both full-length MMSET-I and II, corresponding to transcripts originating from the MB 4-1 breakpoint (Figure 1A). This breakpoint is associated with translation from the true initiation site in exon 3 of the MMSET gene.23 The full-length MMSET-II protein has an apparent molecular weight of approximately 180 kDa, whereas the 645 amino acid long MMSET-I exhibits a mobility of approximately 80 kDa (Figure 1B lane 1). The specificity of the antibody was confirmed by preabsorption with the GST-MMSET immunogen, which abrogated the detection of MMSET isoforms (Figure 1C).
Myeloma cell lines carrying t(4;14) overexpress the MMSET protein. (A) The schematic map of the MMSET gene, its wild-type (MMSET-I and MMSET-II) and shorter transcripts resulting from variation in the position of the breakpoint in the t(4;14) translocation. Three transcription start sites (bent arrows) within the first 6 exons of the MMSET gene serve as initiation sites for different transcripts depending on the position of the breakpoint (MB4-1, MB4-2, MB4-3) (modified from Keats et al23 ). The different transcripts encode a different complement of chromatin-related MMSET domains (PWWP, HMG, PHD, and SET). (B) Immunoblot of nuclear extracts from t(4;14)-positive (lanes 1-3) and t(4;14)-negative (lanes 4-6) myeloma cell lines with a rabbit polyclonal anti-MMSET antibody. The KMS11 line overexpresses the full-length isoforms MMSET-I and MMSET-II (lane 1), whereas the H929 and KMS28PE lines overexpress shorter variants MB4-2 (I and II) and MB4-3 (I and II) originating from 2 alternative start sites (lanes 2 and 3). GAPDH is used as loading control. (C) An immunoblot of KMS11 cell extract with rabbit anti-MMSET antibody was coincubated with GST or the GST-MMSET immunogen to check antibody specificity.
Myeloma cell lines carrying t(4;14) overexpress the MMSET protein. (A) The schematic map of the MMSET gene, its wild-type (MMSET-I and MMSET-II) and shorter transcripts resulting from variation in the position of the breakpoint in the t(4;14) translocation. Three transcription start sites (bent arrows) within the first 6 exons of the MMSET gene serve as initiation sites for different transcripts depending on the position of the breakpoint (MB4-1, MB4-2, MB4-3) (modified from Keats et al23 ). The different transcripts encode a different complement of chromatin-related MMSET domains (PWWP, HMG, PHD, and SET). (B) Immunoblot of nuclear extracts from t(4;14)-positive (lanes 1-3) and t(4;14)-negative (lanes 4-6) myeloma cell lines with a rabbit polyclonal anti-MMSET antibody. The KMS11 line overexpresses the full-length isoforms MMSET-I and MMSET-II (lane 1), whereas the H929 and KMS28PE lines overexpress shorter variants MB4-2 (I and II) and MB4-3 (I and II) originating from 2 alternative start sites (lanes 2 and 3). GAPDH is used as loading control. (C) An immunoblot of KMS11 cell extract with rabbit anti-MMSET antibody was coincubated with GST or the GST-MMSET immunogen to check antibody specificity.
Two additional breakpoints, MB4-2 and MB4-3 (Figure 1A), result in the MMSET transcripts yielding proteins that initiate at 2 alternative start sites present in exons 4 and 6.23 Transcripts from either site undergo alternative splicing to generate 2 isoforms I and II, analogous to the full-length MMSET-I and II. Their corresponding translation products, MB4-2 (I and II), and MB4-3 (I and II), carry a truncated amino terminus, lacking either half or the entire first PWWP motif. All 4 variants were overexpressed H929 cells (Figure 1B lane 2). Only MB4-3 II but not MB4-3 I is overexpressed in the line KMS28PE (Figure 1B lane 3). In addition to the overexpressed MB4-2 and MB4-3 variants, the H929 and KMS28PE cell lines also express full-length MMSET-I and II at comparable levels to myeloma cells carrying t(11;14), most likely resulting from transcripts from the remaining untranslocated MMSET allele.
Overexpressed MMSET isoforms differ in their localization in myeloma cell lines
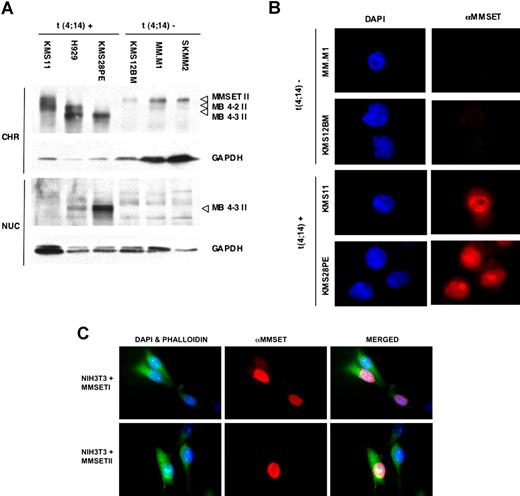
Nuclear extracts were prepared from myeloma cell lines and subfractionated. MMSET was found predominantly in the insoluble chromatin fraction within the nucleus with little in the soluble nuclear fraction. MMSET-II isoforms (MMSET-II, MB4-2 II, MB4-3 II) were found mainly in the chromatin pellet, whereas the overexpressed MB4-3 II protein, which lacks the amino terminus and the entire first PWWP motif, appears in both the soluble and insoluble nuclear fractions of H929 and KMS28PE cells (Figure 2A lanes 2 and 3). In t(4;14)-negative myeloma lines, endogenous MMSET-II is found mainly in the insoluble fraction (Figure 2A lanes 4-6).
MMSET isoforms exhibit different patterns of nuclear distribution. (A) Distribution of MMSET isoforms in the chromatin and nuclear fractions. The full-length MMSET-II protein of the line KMS11 and all t(11;14) lines as well as the MB4-2 II variant of line H929 are chromatin-bound (lanes 1-2 and lanes 4-6, respectively), whereas the MB4-3 II variant of lines H929 and KMS12PE is additionally found in the soluble nuclear fraction (lanes 2 and 3). GAPDH is used as a loading control. CHR indicates insoluble chromatin fraction; NUC, soluble nuclear fraction. (B) Overexpressed MMSET protein is localized to nuclei and shows a diffuse pattern of expression. The t(4;14)-positive myeloma cell lines KMS11 and KMS28PE stained with anti-MMSET antibody show higher levels of MMSET expression (red) in the cell nucleus (blue) than the t(11;14) lines MM.M1 and KMS12BM. The full-length MMSET protein present in KMS11 exhibits a diffuse speckled nuclear distribution and is excluded from the nucleolus, whereas the MB4-3 protein present in KMS28PE appears focalized mainly in nucleoli. (C) NIH3T3 cells were transfected with an expression vector for MMSET-I or MMSET-II, immunostained with anti-MMSET antibody, and counterstained with phalloidin and DAPI. Original magnification ×400 for all panels.
MMSET isoforms exhibit different patterns of nuclear distribution. (A) Distribution of MMSET isoforms in the chromatin and nuclear fractions. The full-length MMSET-II protein of the line KMS11 and all t(11;14) lines as well as the MB4-2 II variant of line H929 are chromatin-bound (lanes 1-2 and lanes 4-6, respectively), whereas the MB4-3 II variant of lines H929 and KMS12PE is additionally found in the soluble nuclear fraction (lanes 2 and 3). GAPDH is used as a loading control. CHR indicates insoluble chromatin fraction; NUC, soluble nuclear fraction. (B) Overexpressed MMSET protein is localized to nuclei and shows a diffuse pattern of expression. The t(4;14)-positive myeloma cell lines KMS11 and KMS28PE stained with anti-MMSET antibody show higher levels of MMSET expression (red) in the cell nucleus (blue) than the t(11;14) lines MM.M1 and KMS12BM. The full-length MMSET protein present in KMS11 exhibits a diffuse speckled nuclear distribution and is excluded from the nucleolus, whereas the MB4-3 protein present in KMS28PE appears focalized mainly in nucleoli. (C) NIH3T3 cells were transfected with an expression vector for MMSET-I or MMSET-II, immunostained with anti-MMSET antibody, and counterstained with phalloidin and DAPI. Original magnification ×400 for all panels.
Consistent with these results, immunostaining of myeloma cell lines with anti-MMSET antibody reveals an exclusive nuclear localization of the overexpressed MMSET protein (Figure 2B). The full-length MMSET protein in KMS11 myeloma cells is expressed in a diffuse pattern throughout the nucleus but is absent from the nucleoli. By contrast, in KMS28PE cells with MMSET deleted for the N-terminal PWWP domain, protein accumulates in the nucleolus as well as in the nucleus. These observations indicate that the N-terminal PWWP motif is important for chromatin affinity and nuclear distribution of the protein. Given that MMSET-I and MMSET-II are both expressed in myeloma and normal cells, we determined whether these 2 isoforms differ in nuclear pattern of expression. Immunostaining NIH3T3 cells transfected with MMSET-I or MMSET-II revealed a similar pattern of diffuse nuclear staining with some focal densities and a relative lack of staining of the nucleolus (Figure 2C).
The SET domain of MMSET is catalytically active
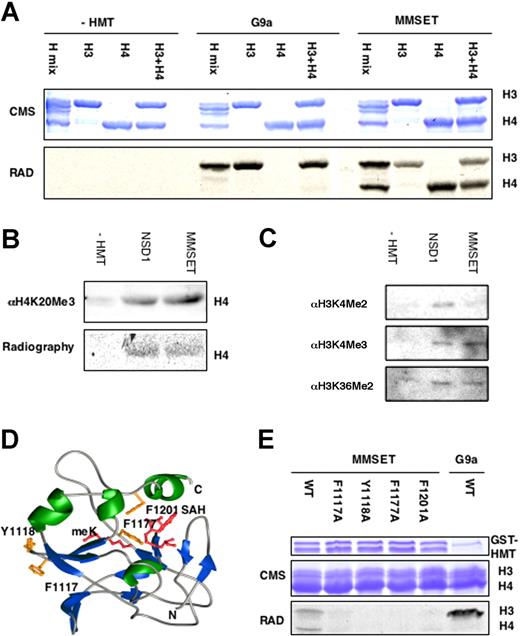
All t(4;14)-positive myeloma cell lines tested overexpress an MMSET isoform with an intact carboxyl terminus. The recombinant carboxyl terminus of the MMSET protein, consisting of the SET domain and the terminal PHD finger, was incubated with purified core histones as substrate and 14CH3-radiolabeled S-adenosyl methionine (SAM) as the methyl donor. MMSET exhibited methyltransferase activity for histones H3 and H4 (Figure 3A) among a purified histone mix containing H2A, H2B, H3, and H4. The enzyme methylated purified histone H3 or H4 alone; and purified H3 and H4 combined. By contrast, recombinant G9a14 led to methylation of histone H3 only. To determine the specificity of the SET domain, unmodified recombinant histone H4 was incubated with GST-NSD1 or GST-MMSET in the presence of labeled SAM. NSD1, a protein closely related to MMSET, was previously reported to methylate K20 in histone H415 as well as lysine 36 of histone 3. Like NSD1, MMSET yields modified histones reactive with an antibody specific for the trimethylated H4K20 (Figure 3B). Further immunoblot analysis (Figure 3C) showed that in vitro reaction of histone with MMSET was associated with trimethylation of H3K4. By contrast, incubation of histone with NSD1 led to dimethylation of H3K4, a novel target site for this enzyme. Incubation of NSD1 or MMSET also led to weak but reproducible dimethylation of H3K36.
The MMSET protein is a histone methyltransferase specific for histones H3 and H4 in vitro. (A) The SET domain of MMSET catalyzes the transfer of 14C-methyl groups to histones H3 and H4. The GST fusion of the SET domain and the terminal PHD finger of MMSET was incubated with the indicated histone substrates and 14C-SAM. The reaction mix was resolved by SDS-PAGE, stained with Coomassie blue, whereas radiolabel retention was visualized by autoradiography. (B) The SET domains of MMSET and NSD1 catalyze the trimethylation of K20 in histone H4 in vitro. The methylation state of the recombinant histone was assayed by immunoblotting with antitrimethyl K20 antibodies. (C) The SET domains of MMSET and NSD1 catalyze histone H3 methylation in vitro. The methylation state of the recombinant histone was assayed by immunoblotting with corresponding antibodies. (D) A model structure of the SET domain of MMSET in complex with a substrate lysine as well as methyl-donor cofactor. Four residues in the MMSET active were predicted to play an important role in catalysis by mediating binding to the substrate histone, substrate lysine, and SAM cofactor. (E) Mutations of the identified residues greatly reduce HMT activity. Recombinant wild-type or mutant in each of the 4 identified SET domain residues was assayed for in vitro HMT activity with 14C-SAM.
The MMSET protein is a histone methyltransferase specific for histones H3 and H4 in vitro. (A) The SET domain of MMSET catalyzes the transfer of 14C-methyl groups to histones H3 and H4. The GST fusion of the SET domain and the terminal PHD finger of MMSET was incubated with the indicated histone substrates and 14C-SAM. The reaction mix was resolved by SDS-PAGE, stained with Coomassie blue, whereas radiolabel retention was visualized by autoradiography. (B) The SET domains of MMSET and NSD1 catalyze the trimethylation of K20 in histone H4 in vitro. The methylation state of the recombinant histone was assayed by immunoblotting with antitrimethyl K20 antibodies. (C) The SET domains of MMSET and NSD1 catalyze histone H3 methylation in vitro. The methylation state of the recombinant histone was assayed by immunoblotting with corresponding antibodies. (D) A model structure of the SET domain of MMSET in complex with a substrate lysine as well as methyl-donor cofactor. Four residues in the MMSET active were predicted to play an important role in catalysis by mediating binding to the substrate histone, substrate lysine, and SAM cofactor. (E) Mutations of the identified residues greatly reduce HMT activity. Recombinant wild-type or mutant in each of the 4 identified SET domain residues was assayed for in vitro HMT activity with 14C-SAM.
We approximated the structure of the SET domain of MMSET complexed with the methyl-lysine product as well as SAM by arraying the primary amino acid sequence of the MMSET SET domain onto a template structure of SET7/9.22 This model predicts that MMSET residues F1117 and Y1118 mediate histone contacts, F1177 mediates substrate lysine binding, and F1201 mediates SAM binding (Figure 3D). Conversion of any of these amino acids in the recombinant SET domain to alanine either greatly impaired (F1117A, F1201A) or abolished (Y1118A, F1177A) enzymatic activity in vitro (Figure 3E). These findings suggest that the 4 residues play an important role in catalysis.
The endogenous MMSET protein is an active histone methyltransferase in vitro and in vivo
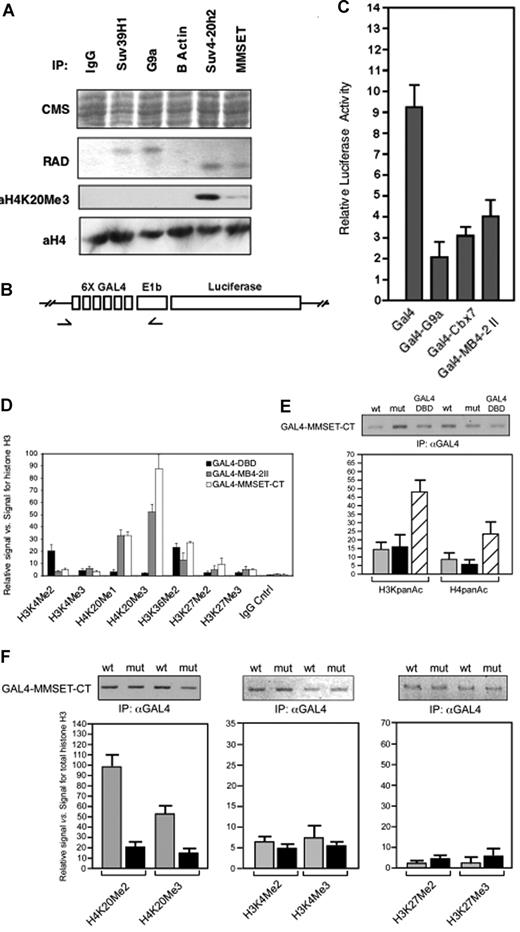
MMSET and various other endogenous histone methyltransferases were immunoprecipitated from nuclear extracts of the MM cell line KMS11 and tested for HMT activity in vitro. In accordance with previous findings,14,24 immunoprecipitated Suv39h1 and G9a led to the methylation of histone H3, whereas Suv4-20h225 methylated only histone H4 (Figure 4A). Immunoprecipitated MMSET also only led to methylation of histone H4, whereas control immunoprecipitations (Figure 4A IgG, β-actin) yielded no methyltransferase activity. Recombinant histone H4 incubated with either immunoprecipitated MMSET or Suv4-20h2 was reactive with an antibody specific for the trimethylated K20 form of histone H4.
MMSET mediates histone H4K20 trimethylation in vivo. (A) MMSET, Suv4-20h2, Suv39H1, and G9a were immunoprecipitated from KMS11 cells and incubated in vitro with histone substrates and 14C-SAM. Immunoprecipitated β-actin served as a negative control. Specific antibodies to trimethylated histone H4 indicate that both MMSET and Suv4-20h2 trimethylate H4K20. (B) Schematic diagram of the integrated Gal4 reporter gene, arrows show sites corresponding to PCR primers. (C) Luciferase activity from an integrated reporter gene was measured subsequent to transfection with Gal4 DNA binding domain, the C-terminal portion of MMSET (Gal4-MB4-2-II), and known repressors (GAL4-G9a, GAL4-Cbx7). (D) Chromatin configuration of an integrated Gal4 reporter gene in response to Gal4-MMSET. Chromatin from cells transfected with a Gal4-MMSET fusion or the Gal4 DNA binding domain was precipitated with the indicated antibody and a PCR fragment encompassing the Gal4 DNA binding sites and promoter region of the reporter gene was amplified. The fraction of input precipitated by each antibody was normalized to the fraction precipitated by antihistone H3 antibodies. Black bar, transfection with Gal4-DBD; gray bar, transfection with Gal4-MMSET-MB4-II; white bar, transfection with Gal4-MMSET (C terminal domain). The results shown are the average (± SD) of 4 independent experiments. (E) Relative levels of acetylation of H3 and H4 on the Gal4 target gene associated with transfection of Gal4-MMSET as shown by quantitative ChIP. Above the graphs, qualitative ChIP is displayed to show the level of recruitment of wild-type and mutant Gal4-MMSET to the target promoter. Gray bar, transfection with wild-type Gal4-MMSET; black bar, transfection with mutant Gal4-MMSET Y1118A; striped bar, transfection with Gal4. (F) Relative levels of methylated histone lysines on the Gal4 reporter associated with wild-type and mutant Gal4-MMSET as determined by quantitative ChIP was performed for each of the indicated modifications on histones H3 and H4. Promoter occupancy of both wild-type and mutant Gal4-MMSET as tested by immunoprecipitation with antibody to Gal4 is shown. Values represent relative signal versus total histone. Gray bar, transfection with wild-type Gal4-MMSET; black bar, transfection with mutant Gal4-MMSET Y1118A. Error bars represent SD.
MMSET mediates histone H4K20 trimethylation in vivo. (A) MMSET, Suv4-20h2, Suv39H1, and G9a were immunoprecipitated from KMS11 cells and incubated in vitro with histone substrates and 14C-SAM. Immunoprecipitated β-actin served as a negative control. Specific antibodies to trimethylated histone H4 indicate that both MMSET and Suv4-20h2 trimethylate H4K20. (B) Schematic diagram of the integrated Gal4 reporter gene, arrows show sites corresponding to PCR primers. (C) Luciferase activity from an integrated reporter gene was measured subsequent to transfection with Gal4 DNA binding domain, the C-terminal portion of MMSET (Gal4-MB4-2-II), and known repressors (GAL4-G9a, GAL4-Cbx7). (D) Chromatin configuration of an integrated Gal4 reporter gene in response to Gal4-MMSET. Chromatin from cells transfected with a Gal4-MMSET fusion or the Gal4 DNA binding domain was precipitated with the indicated antibody and a PCR fragment encompassing the Gal4 DNA binding sites and promoter region of the reporter gene was amplified. The fraction of input precipitated by each antibody was normalized to the fraction precipitated by antihistone H3 antibodies. Black bar, transfection with Gal4-DBD; gray bar, transfection with Gal4-MMSET-MB4-II; white bar, transfection with Gal4-MMSET (C terminal domain). The results shown are the average (± SD) of 4 independent experiments. (E) Relative levels of acetylation of H3 and H4 on the Gal4 target gene associated with transfection of Gal4-MMSET as shown by quantitative ChIP. Above the graphs, qualitative ChIP is displayed to show the level of recruitment of wild-type and mutant Gal4-MMSET to the target promoter. Gray bar, transfection with wild-type Gal4-MMSET; black bar, transfection with mutant Gal4-MMSET Y1118A; striped bar, transfection with Gal4. (F) Relative levels of methylated histone lysines on the Gal4 reporter associated with wild-type and mutant Gal4-MMSET as determined by quantitative ChIP was performed for each of the indicated modifications on histones H3 and H4. Promoter occupancy of both wild-type and mutant Gal4-MMSET as tested by immunoprecipitation with antibody to Gal4 is shown. Values represent relative signal versus total histone. Gray bar, transfection with wild-type Gal4-MMSET; black bar, transfection with mutant Gal4-MMSET Y1118A. Error bars represent SD.
To determine whether MMSET can modify chromatin in vivo, we generated a HEK293T cell line carrying a chromatin-embedded Gal4-UAS-thymidine kinase-luciferase reporter gene (Figure 4B). Gal4 fused to MMSET variant MB4-2 II (Figure 1A) or the C-terminal of MMSET (Figure 5A) carrying the SET domain, as well as control repressors G9a and Cbx7,26 repressed the reporter compared with the Gal4 DNA binding domain (Gal4DBD) alone (Figure 4C). To determine the chromatin changes correlated with MMSET-mediated repression, crosslinked chromatin fragments from transfected cells were immunoprecipitated with antihistone H3 or anti-Gal4 antibodies as normalization controls and anti–methyl H3K4, H3K27, H3K36, and H4K20 and amplified by quantitative PCR (Figure 4D). Recruitment of MMSET to the reporter led to a dramatic increase in H4K20 monomethylation and trimethylation, a modest increase in H3K27 monomethylation and dimethylation but no significant change in H3K36 dimethylation. Whereas levels of trimethylated H3K4 were not significantly affected, there was a significant decrease in the level of dimethylated H3K4. To confirm the enzymatic specificity of the SET domain on the target promoter, we tested the catalytically inactive Y1118A mutant. Although expression of the wild-type MMSET SET domain was associated with dimethylation and trimethylation at H4K20, the point mutant led to a loss of H4K20 methylation in vivo but no change in H3K4 or H3K27 methylation (Figure 4F). This suggested that the repression-associated changes in H3K4 and H3K27 methylation were caused indirectly through recruitment of other histone methylases and demethylases. This was consistent with the finding that the Y1118 mutant Gal4-MMSET construct was still able to repress the reporter (data not shown). Furthermore, the wild-type and SET domain mutant of MMSET both led to deacetylation of the reporter (Figure 4E).
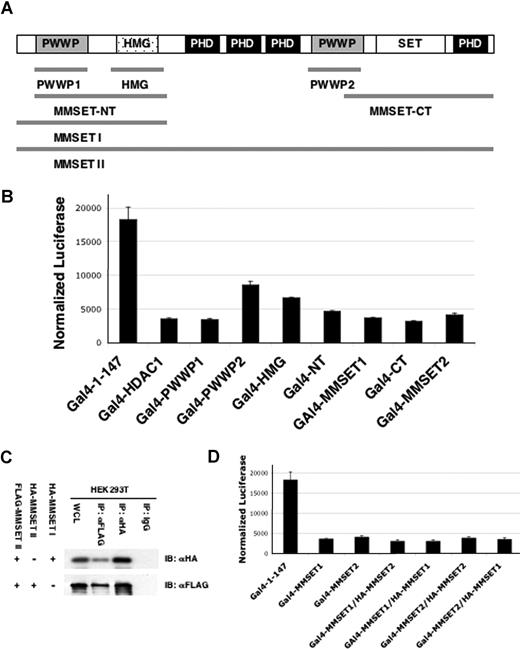
MMSET mediates repression of a model target gene. (A) Constructs used in the reporter assay are fusions of the respective MMSET fragments to the DNA-binding domain of Gal4 (Gal-DBD, residues 1-147). The MMSET domains present in each fusion fragment are shown on the top of the figure. (B) Plasmids (125 ng) expressing the Gal4 DNA binding domain or Gal4 fusions of the MMSET domains were transfected into the Gal4-tk-luciferase reporter cell line in 24-well dishes. A representative triplicate experiment showing luciferase activity normalized to protein concentration is shown. (C) MMSET-II coimmunoprecipitates with MMSET-I and II. FLAG-tagged MMSET-II was cotransfected in HEK293T cells either with HA-MMSET-I or HA-MMSET-II. Total cell lysates (WCL) were immunoprecipitated with anti-FLAG antibody and subjected to immunoblotting with anti-HA antibody. Immunoprecipitation with nonspecific IgG is shown as a control. (D) Gal4-MMSET-I or Gal4-MMSET-II plasmids (125 ng) were transfected into the Gal4 reporter cell line along with 250 ng of MMSET-I or MMSET-II expression plasmid as indicated. A representative triplicate experiment showing luciferase activity normalized to protein concentration is shown. Error bars represent SD.
MMSET mediates repression of a model target gene. (A) Constructs used in the reporter assay are fusions of the respective MMSET fragments to the DNA-binding domain of Gal4 (Gal-DBD, residues 1-147). The MMSET domains present in each fusion fragment are shown on the top of the figure. (B) Plasmids (125 ng) expressing the Gal4 DNA binding domain or Gal4 fusions of the MMSET domains were transfected into the Gal4-tk-luciferase reporter cell line in 24-well dishes. A representative triplicate experiment showing luciferase activity normalized to protein concentration is shown. (C) MMSET-II coimmunoprecipitates with MMSET-I and II. FLAG-tagged MMSET-II was cotransfected in HEK293T cells either with HA-MMSET-I or HA-MMSET-II. Total cell lysates (WCL) were immunoprecipitated with anti-FLAG antibody and subjected to immunoblotting with anti-HA antibody. Immunoprecipitation with nonspecific IgG is shown as a control. (D) Gal4-MMSET-I or Gal4-MMSET-II plasmids (125 ng) were transfected into the Gal4 reporter cell line along with 250 ng of MMSET-I or MMSET-II expression plasmid as indicated. A representative triplicate experiment showing luciferase activity normalized to protein concentration is shown. Error bars represent SD.
MMSET represses transcription and interacts with corepressors
Several putative functional domains of the MMSET protein, including the PWWP domain and HMG box as well as full-length MMSET-I and MMSET-II (Figure 5A), were transfected into 293T cells harboring a Gal4-tk-luciferase reporter.27 Gal4-MMSET-I and a full-length fusion of MMSET-II to Gal4 repressed the reporter gene. Multiple domains of MMSET, including the 2 PWWP domains and the HMG box, acted individually as repressors (Figure 5B). MMSET-I and MMSET-II are coexpressed in normal and myeloma cells. Coprecipitation experiments showed that the 2 major MMSET isoforms can interact (Figure 5C). However, repression mediated by the Gal4-tethered forms of MMSET was not affected by overexpression of untethered MMSET (Figure 5D), suggesting that the MMSET isoforms may act in concert and do not interfere with each other's function.
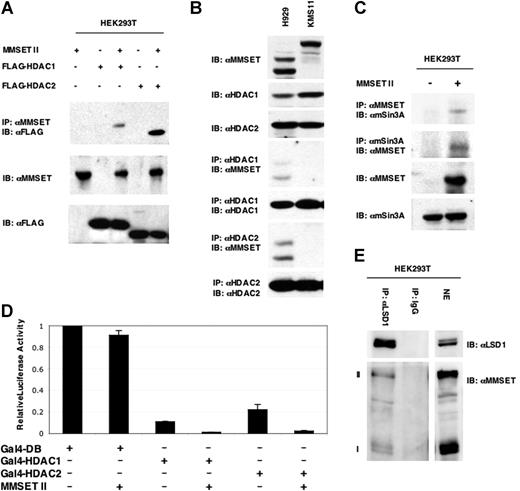
The fact that repression by MMSET occurred even when the SET domain was mutated and was associated with loss of acetylation of the reporter gene suggested that MMSET could interact with histone deacetylases (HDACs) and other cofactors.28 Therefore, MMSET-II was coexpressed in HEK293T cells along with epitope-tagged HDAC1 through HDAC8. Only HDAC1 and HDAC2 could be coimmunoprecipitated with MMSET-II (Figure 6A; data not shown). These interactions were also shown also for endogenous proteins in the t(4;14)-positive myeloma line H929 (Figure 6B). HDAC1 and 2 coimmunoprecipitated both MMSET variants MB4-2 II and MB4-3 II in H929 cells, but not the full-length MMSET-II in KMS11 cells under the extraction conditions used. This may reflect the tighter nuclear binding of MMSET in KMS11 cells. MMSET-II expressed in 293T cells could also interact with the endogenous sin3a corepressor29 (Figure 6C). Furthermore, repression mediated by Gal4-HDAC1 and Gal4-HDAC2 was augmented by cotransfection of MMSET-II (Figure 6D). Prompted by loss of H3K4 dimethylation noted on the reporter gene targeted by Gal4-MMSET, we assayed for interaction between MMSET and LSD1, a histone demethylase targeting monomethyl and dimethyl marks on histone H3K4.30 Precipitation of endogenous LSD1 in HEK293T cells (Figure 6E) led to the copurification of MMSET-I and MMSET-II, confirming the interaction between these 2 repressive enzymes and supporting the idea that MMSET isoforms I and II may act in concert.
MMSET interacts cofactors and histone deacetylases. (A) Total lysates of HEK293T cells transfected with HDAC1 or HDAC2 were immunoprecipitated with anti-MMSET antibody and probed with antibodies to epitope-tagged deacetylases. (B) Nuclear extracts from H929 and KMS11 cells were immunoprecipitated with anti-HDAC 1 and 2 antibodies and probed with anti-MMSET. (C) Total cell lysates from untransfected HEK293T cells and cells transfected with MMSET-II were immunoprecipitated with anti-mSin3A or anti-MMSET antibodies and analyzed by immunoblotting with the anti-MMSET and anti-Sin3A antibodies, respectively. (D) A Gal luciferase reporter gene was coexpressed in 293T cells along with Gal4-HDAC1 or Gal4-HDAC2 in the presence or absence of MMSET. Error bars represent SD. (E) MMSET-I and MMSET-II coimmunoprecipitate with the histone demethylase LSD1. Nuclear extracts (NE) from HEK293T cells were coimmunoprecipitated with either anti-LSD1 antibody or rabbit IgG, and immunoblotted with anti-MMSET antibody.
MMSET interacts cofactors and histone deacetylases. (A) Total lysates of HEK293T cells transfected with HDAC1 or HDAC2 were immunoprecipitated with anti-MMSET antibody and probed with antibodies to epitope-tagged deacetylases. (B) Nuclear extracts from H929 and KMS11 cells were immunoprecipitated with anti-HDAC 1 and 2 antibodies and probed with anti-MMSET. (C) Total cell lysates from untransfected HEK293T cells and cells transfected with MMSET-II were immunoprecipitated with anti-mSin3A or anti-MMSET antibodies and analyzed by immunoblotting with the anti-MMSET and anti-Sin3A antibodies, respectively. (D) A Gal luciferase reporter gene was coexpressed in 293T cells along with Gal4-HDAC1 or Gal4-HDAC2 in the presence or absence of MMSET. Error bars represent SD. (E) MMSET-I and MMSET-II coimmunoprecipitate with the histone demethylase LSD1. Nuclear extracts (NE) from HEK293T cells were coimmunoprecipitated with either anti-LSD1 antibody or rabbit IgG, and immunoblotted with anti-MMSET antibody.
MMSET plays a role in myeloma cell viability
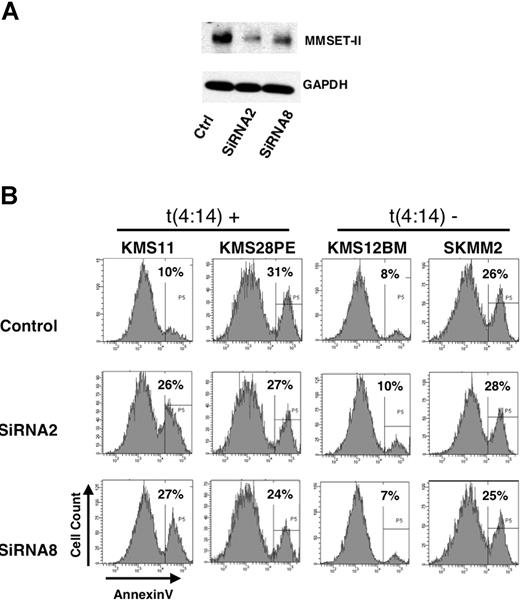
To determine the biologic role of MMSET in myleoma cell growth, t(4;14)-positive cell lines expressing high levels of MMSET as well as myeloma cells with low levels of MMSET were transduced with constructs harboring green fluorescent protein (GFP) and expressing shRNA directed against sequences encoding the second PHD domain of MMSET, only present in MMSET-II and to sequences in the 3′UTR of MMSET-II. These shRNA mediated a significant decrease in MMSET protein expression in KMS11 cells relative to a control shRNA directed against luciferase (Figure 7A). Five days after transfection, GFP+ cells harboring the shRNA were sorted by flow cytometry and assayed for viability. KMS11 cells transduced with both shRNA directed to MMSET showed a nearly 3-fold increase in apoptosis (Figure 7B, left). Knockdown of MMSET-II in KMS28PE cells did not change the rate of apoptosis in these cells, neither did shRNA lead to increased apoptosis of cells lines that did not harbor t(4;14). These data suggest that, at least in some myeloma cells with the t(4;14) rearrangement, high level MMSET expression may contribute to cell viability and growth.
Depletion of MMSET affects myeloma cell viability. (A) KMS11 cells transduced by nucleoporation with a control shRNA or 2 shRNA constructs directed against MMSET were grown in culture for 5 days, sorted for GFP expression, lysed, and immunoblotted with anti-MMSET antibody. (B) t(4;14)-positive and t(4;14)-negative myeloma cell lines were nucloeporated with the indicated shRNA, grown in culture for 5 days, and stained for annexinV expression. Histograms of annexin V staining of GFP+, propidium iodine-negative cells are displayed along with the percentage of cells staining positive for annexin V.
Depletion of MMSET affects myeloma cell viability. (A) KMS11 cells transduced by nucleoporation with a control shRNA or 2 shRNA constructs directed against MMSET were grown in culture for 5 days, sorted for GFP expression, lysed, and immunoblotted with anti-MMSET antibody. (B) t(4;14)-positive and t(4;14)-negative myeloma cell lines were nucloeporated with the indicated shRNA, grown in culture for 5 days, and stained for annexinV expression. Histograms of annexin V staining of GFP+, propidium iodine-negative cells are displayed along with the percentage of cells staining positive for annexin V.
Discussion
In this work, we describe properties of the MMSET protein encoded by the gene dysregulated in t(4;14)-positive cases of MM. By developing antibodies against MMSET, we show that known overabundance of MMSET transcripts in t(4;14)-positive MM cells23 leads to significantly elevated levels of the corresponding proteins. All the t(4;14) myeloma cell lines we tested overexpress either the full-length MMSET-II protein or the shorter variants MB4-2 II and MB4-3 II, which lack different portions of the amino terminus but carry an intact carboxyl terminus and SET domain. Prior work showed that GFP-tagged MMSET-I and II localized to the nucleus but not nucleoli, whereas the MB4-2 and MB4-3 proteins concentrate in nucleoli.23 Examining the pattern of expression of endogenous MMSET in t(4;14) myeloma cells by immunofluorescence and chromatin fractionation, we extended this finding, showing that tight and exclusive binding of MMSET to the chromatin fraction of the cell requires an intact N terminus, containing a PWWP motif. This motif was previously found to be necessary for targeting of DNA methyltransferases to chromatin.31 A significant fraction of N-terminal truncated MMSET remains in the insoluble chromatin fraction. This may be the result of the remaining PWWP domain or other chromatin-associated domains, such as the PHD and HMG domains within MMSET, and the ability of MMSET to self-associate. It is possible that truncated MMSET could form a complex with full-length MMSET expressed in myeloma cells from the unrearranged allele to guide the protein to the soluble chromatin compartment.
We demonstrated that MMSET is an HMT. Changes in the balance and activity of several HMTs are associated with malignancy. For example, EZH2 is overexpressed in some forms of MM, correlating with tumor burden during progression, and in metastatic, hormone-refractory prostate cancer.32 The MLL protein, fused to dozens of different partners in acute myeloid leukemia, targets its HMT activity to Hox genes.33 NSD1, a protein closely related to MMSET, is fused to Nup98 in rare cases of acute myeloid leukemia. Introduction of this fusion protein in murine marrow cells leads to immortalization-associated up-regulation of the Hoxa9 locus.34 Recombinant MMSET methylates both histones H3 and H4 in vitro, in contrast to the immunoprecipitated endogenous MMSET protein that methylates only histone H4. There are several possible explanations for this observation. The specificity of methylation may depend not only on the SET domain itself but may be modulated by other domains of the MMSET protein or by functional partners. Discordance between the in vitro and in vivo activity of SET domain containing HMTs was observed previously. For example, recombinant G9a protein methylates histone H3 at K9 and K27 in vitro,14 whereas immunoprecipitated protein methylates only H3K9.19 MMSET could also trimethylates histone H3K4 in vitro, a modification conventionally associated with gene activation.35 Our data do not show a role for this activity in vivo as Gal4-MMSET–mediated repression was associated with loss of H3K4 methylation. However, it remains possible that under other conditions this activity of MMSET might be observed such as when attracted to a promoter through association with a sequence specific transcription factor.
The SET domain of the MMSET protein trimethylates recombinant histone H4 at K20 in vitro. Methylation of K20 is generally associated with transcriptional repression and/or heterochromatin formation. Consistent with this notion, MMSET repressed a reporter gene, concurrent with trimethylation of H4K20 in vivo as shown by ChIP analysis. H4K20 methylation appeared to be performed directly by MMSET in vivo as mutation of the SET domain led to the loss of methylation of this residue on the Gal4 reporter gene. Targeting of MMSET to a reporter gene was also associated with loss of histone acetylation, an increase in H3K27 dimethylation and loss of H3K4 dimethylation, but not trimethylation. However, these latter activities did not depend on the catalytic activity of MMSET, suggesting that MMSET could recruit other effector enzymes to promoters. In accordance with this idea, MMSET interacted with sin3a, HDAC1, HDAC2, and LSD1, the latter able to demethylate dimethyl but not trimethyl H3K4.30,36 Hence, the histone modification signature of MMSET is complex and mediated by both direct action of MMSET and by interplay with other histone modifiers.
Our data corroborate and extend those of Todoerti et al37 who found that MMSET-I, the shorter form of the protein could immunopreciptate with HDAC1 but not HDAC2. By contrast, we show that both endogenous HDAC1 and HDAC2 coimmunoprecipitate with MMSET-II, the long isoform of MMSET in myeloma cells. However, we could only demonstrate this endogenous complex in H929 cells that express an N-terminal truncated form of the protein but not in KMS11 cells, which express the full-length MMSET protein. This might be explained by the fact that the full-length protein is very tightly bound to chromatin and not extracted from the cell under immunoprecipitation conditions, whereas the N-terminal truncated version is found in both the soluble and insoluble nuclear fractions and hence can be more readily detected. Although we have identified several MMSET partners, the exact nature of the native MMSET protein complex remains to be determined. In addition, MMSET can self-associate, making it possible that MMSET may exist as a higher order multimeric protein complex containing oligomers of MMSET. Of note, self-association was found to be critical for other oncogenic repressor proteins, including PML-RARα and RUNX1-MTG8.38
Methylation of H4K20 can be mediated by several other histone methyltransferases in vitro including NSD1,15 PR-Set7,39 Suv4-20h1, and Suv4-20h2.25 Nup98-NSD1, although related to MMSET, mediated activation of Hox gene transcription, stimulation of H3K36 dimethylation, a decrease in H3K27 methylation, and no increase in H4k20 methylation.34 By contrast in our system MMSET did not lead to stimulation of H3K36 dimethylation. Whether the differences between MMSET and NSD1 are due to amino acid differences in the SET domain that alters substrate specificity, differences in partner proteins or due to the Nup98 fusion remains to be determined.
Loss of global trimethylation of H4K20 is a hallmark of human cancer,40 perhaps posing a paradox for MMSET as H4K20-specific HMT overexpressed in a malignancy. However, we have found no significant differences in global levels of trimethylated H3K4 and H4K20 between t(4;14)-positive and -negative myeloma cell lines (data not shown). Furthermore, no studies have compared the state of H4K20 trimethylation in myeloma cells with that of normal plasma cells; hence, it is unknown whether global H4K20 methylation is altered in this malignancy. Dysregulated MMSET expression may affect the state of modification of specific regions of chromatin. MMSET overexpressing t(4;14) myeloma specimens have a distinct pattern of gene expression compared with other forms of the disease.41 Reevaluation of these array data (W. Berkofsky-Fessler, J.D.L., unpublished data, April 2, 2007) indicates that many of these alterations are independent of overexpression of FGFR3 in the tumor, suggesting that MMSET has a key role in the altered state of gene expression in t(4;14) myeloma.
Universal expression of MMSET in t(4;14) myeloma6 suggests that MMSET is critical for myeloma pathogenesis and or progression. In accordance with this notion, 2 different shRNA directed to MMSET led to a loss of viability of KMS11 t(4;14) cells but did not affect KMS28PE cells also overexpressing MMSET and did not affect the viability 2 myeloma cell lines without t(4;14). Why the MMSET-positive cell lines did not show equal sensitivity to depletion of MMSET is uncertain but could be related to the multiple genetic changes found in myeloma cell lines. For example, KMS11 cells have rearrangement of the c-myc and c-maf genes as well as MMSET, whereas KMS28 cells do not have the c-maf rearrangement.42 Further genomic analysis of myeloma specimens and cell lines shows diverse changes in pathways converging on the alternative NFκB pathway.43,44 For example, increased NFκB activity in KMS11 is associated with a deletion of the TRAF3 locus, leading to constitutive processing of NFκB2, whereas KMS28PE cells lack cIAP1/2 and also show increased NFκB activity. How these specific genetic anomalies might interact with MMSET to drive cancer cell viability and growth remains to be determined. Recently, Lauring et al45 showed that knockdown of MMSET in KMS11 cells led to cell-cycle arrest, decreased cell adhesion, decreased cell growth, colony formation in vitro, and tumorigenesis in immunodeficient mice. These workers found that depletion of MMSET also affected the growth of nonmyeloma cell lines as well as t(4;14)-negative cell lines that expressed appreciable levels of MMSET, suggesting that MMSET expression may support the growth of other malignant cells as well.
Collectively, our data indicate that MMSET possesses the properties of a transcriptional cofactor, including nuclear localization, histone methyltransferase activity, and the ability to bind other transcriptional effectors. The identification of the targets of MMSET and their role in cell growth in survival will be key to understanding how MMSET is associated with tumor development.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kristie Charoen for technical assistance.
This work was supported by a Burroughs Wellcome Clinical Scientist Award in Translational Research, a Multiple Myeloma Research Foundation Senior Award (J.D.L.), the Samuel Waxman Cancer Research Foundation, the Chemotherapy Foundation (J.D.L., S.W.), a Leukemia and Lymphoma Society Specialized Center of Research Grant (J.D.L.), and grants from the NHLBI and NCI and by the American Cancer Society (M.J.W.).
National Institutes of Health
Authorship
Contribution: J.M. designed and performed research and wrote manuscript; J.A.M. designed research, performed experiments, and analyzed data; M.S., H.N., and A.S. designed and performed research; Y.M.-M. performed research; M.C., P.L.B., and J.M. designed research and analyzed data; M.-M.Z. designed research; S.W. analyzed data; B.A.L. analyzed data and edited manuscript; M.J.W. designed research, analyzed data, and edited manuscript; J.D.L. designed research, analyzed data, and wrote and revised manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan D. Licht, Division of Hematology/Oncology, Northwestern University Feinberg School of Medicine, Robert H. Lurie Comprehensve Cancer Center, 303 East Superior Street, Lurie 5-123, Chicago, IL 60611; e-mail: j-licht@northwestern.edu.