Abstract
Evidence for a novel route of adult hematopoietic stem-cell lineage commitment through Lin−Sca-1+Kit+Flt3hi (LSKFlt3hi) lymphoid-primed multipotent progenitors (LMPPs) with granulocyte/monocyte (GM) and lymphoid but little or no megakaryocyte/erythroid (MkE) potential was recently challenged, as LSKFlt3hi cells were reported to possess MkE potential. Herein, residual (1%-2%) MkE potential segregated almost entirely with LSKFlt3hi cells expressing the thrombopoietin receptor (Mpl), whereas LSKFlt3hiMpl− LMPPs lacked significant MkE potential in vitro and in vivo, but sustained combined GM and lymphoid potentials, and coexpressed GM and lymphoid but not MkE transcriptional lineage programs. Gradually increased transcriptional lymphoid priming in single LMPPs from Rag1GFP mice was shown to occur in the presence of maintained GM lineage priming, but gradually reduced GM lineage potential. These functional and molecular findings reinforce the existence of GM/lymphoid-restricted progenitors with dramatically down-regulated probability for committing toward MkE fates, and support that lineage restriction occurs through gradual rather than abrupt changes in specific lineage potentials.
Introduction
Whereas considerable knowledge has been gained with regard to the identity and roles of extrinsic and intrinsic regulators of blood lineage development, much less is known about the molecular mechanisms regulating lineage commitment of hematopoietic stem cells (HSCs).1,2 Unraveling the involved molecular determinants and mechanisms of lineage restriction will be facilitated by, and most likely depend on, a more complete understanding of the cellular pathways of the lineage restriction process from pluripotent HSCs to lineage-restricted progenitor cells.
It remains unclear and debated2-4 exactly how the lineage commitment process from pluripotent HSCs to lineage-restricted progenitors occurs in adult bone marrow (BM), and even unequivocal evidence for one such pathway would not exclude the existence of alternative routes for HSC lineage commitment. In the prevailing model of HSC lineage commitment,1-3 HSCs (long-term and short-term) and multipotent progenitors (MPPs) distinguish themselves from each other, only through gradual loss of self-renewal potential while sustaining the same degree of pluripotentiality, with the first lineage commitment event resulting in a strict separation of myelopoiesis and lymphopoiesis. This model for HSC lineage commitment was supported by the identification of common myeloid and common lymphoid progenitors (CMPs and CLPs, respectively).5,6 However, the degree to which the identified CMPs and CLPs represent obligatory or even main intermediates for myeloid and lymphoid development in adult hematopoiesis remains to be established.
Although conclusively established for long-term HSCs (LT-HSCs),7,8 the existence of short-term HSCs (ST-HSCs) and MPPs in the BM Lin−Sca-1+Kit+ (LSK) stem and primitive progenitor cell compartment, with sustained pluripotentiality has yet to be demonstrated at the single cell level.3 Rather, more recent studies have uncovered considerable heterogeneity in the LSK MPP compartment. Through the use of different but overlapping markers such as FMS-like tyrosine kinase 3 (Flt3),9-11 vascular cell adhesion molecule-1 (Vcam-1),12,13 and an Ikaros-reporter,14 the existence of lymphoid-primed MPPs (LMPPs) with combined granulocyte/monocyte (GM) and lymphoid potentials, but little or no megakaryocyte (Mk)/erythroid (E) potentials has been proposed.3,10,12,14,15 Further, molecular analysis of putative LMPPs show down-regulated transcriptional priming of genes specific for the MkE lineage, and up-regulation of lymphoid-specific genes, not yet expressed in HSCs.15,16
The identification of a putative LMPP, representing the earliest lineage-restricted lympho-myeloid progenitor identified in adult hematopoiesis, has provided a potential avenue toward uncovering alternative HSC lineage commitment pathways.2,3,17 However, the existence of the LMPP remains contentious,2,3,18-20 largely reflecting functional heterogeneity of phenotypically defined candidate LMPPs.9,10,12-14 While the evidence for a large fraction of LSKFlt3hi BM cells sustaining (at the single cell level) combined and robust GM and lymphoid potentials are compelling,9,10,14,15 the experimental evidence for LMPPs having lost MkE potential has been questioned.18 In the original studies,10,11 1% to 3%, at most, of FACS-purified LSKFlt3hi BM cells were found to produce Mk and E progeny in various in vitro and in vivo clonal assays. While it was argued that one possible reason for the low MkE potential observed could be impurities of HSCs and/or MPPs in the sorted LSKFlt3hi cells,10 Forsberg et al18 demonstrated through double and triple FACS sorting that highly purified cells with a LSKFlt3hi phenotype do possess in vivo Mk and E potential. However, as most of the in vivo studies required transplantation of large numbers of LSKFlt3hi cells in nonclonal assays, the frequency of LSKFlt3hi cells with MkE potential was not established, and could therefore, in agreement with previous in vitro clonal studies,10,12,14 be compatible with only a small fraction of LSKFlt3hi cells having sustained MkE potential. If so, a large fraction of LSKFlt3hi cells would in fact represent LMPPs with combined GM and lymphoid but no MkE potential. However, an alternative and fundamentally different interpretation, even if only a small fraction of LSKFlt3hi cells would read out with MkE potential in various assays, would be that most if not all LSKFlt3hi cells possess reduced but significant MkE potential but that the assays used are inefficient at uncovering it. In this scenario, LSKFlt3hi cells, although being transcriptionally lymphoid-GM and not MkE primed,15 would in fact as conventional MPPs remain truly multipotent, and consequently as reinforced by Forsberg et al,18 the strict separation of myelopoiesis and lymphopoiesis through generation of CMPs and CLPs might still represent the first lineage commitment or restriction point of HSCs and MPPs. The only feasible approach to distinguish between these 2 fundamentally different explanations for the reproducible readout of MkE potential from a small fraction of LSKFlt3hi cells would be to either develop more efficient clonal assays, which would promote MkE potential of most LSKFlt3hi cells (to demonstrate that LSKFlt3hi cells are in fact truly multipotent), or to identify one or several additional markers, which would allow further separation of a fraction of LSKFlt3hi cells without significant in vitro or in vivo clonal MkE potential (to prove beyond reasonable doubt the existence of GM-lymphoid–restricted LMPPs).
Another fundamental and related question is whether HSC lineage restriction occurs in an abrupt manner as perhaps interpreted by strict branching points in most schemes for hematopoiesis,1 or alternatively through a gradual loss of specific lineage potentials. Previous studies showing changes in lineage potentials correlated, for instance, to changes in recombination activating gene 1 (Rag1) and Vcam-1 expression, providing findings compatible with the last scenario,12,13,21 but as clonogenic studies were not performed, it remains possible that the observed changes in lineage potentials reflected altered ratios of a mixture of progenitors of multiple lineages, rather than gradual changes in potentials for multiple lineages within multipotent progenitor cells.
Herein we describe an approach that allowed us to purify a fraction of LSKFlt3hi LMPPs without significant in vitro or in vivo clonal MkE activity. The strategy developed was based on 2 key findings common to our study and the Forsberg study.10,11,18 First, that a small fraction (0.4%-1.2%) of LSKFlt3hi cells are able to generate large MkE-enriched colonies in the spleen (spleen colony-forming unit [CFU-S]22 ) 11 to 12 days after transplantation, and second that a fraction of LSKFlt3hi cells express the thrombopoietin receptor Mpl (Mpl), strongly implicated as a key regulator of the MkE lineages.23 We found that the in vivo CFU-S as well as in vitro clonal Mk and E activity segregate almost entirely with LSKFlt3hiMplhi BM cells, although also most LSKFlt3hiMplhi cells (> 98%) lacked detectable MkE potential and therefore fulfill the criteria of GM-lymphoid–restricted LMPPs. LSKFlt3hiMpl− LMPPs also sustain combined GM-lymphoid potential but without any significant MkE potential. Furthermore, using reporter mice expressing green fluorescent protein (GFP) under control of the promoter for the lymphoid Rag1 gene,24 we demonstrate that gradually increased transcriptional lymphoid priming in LMPPs occurs in the presence of sustained transcriptional GM priming, but gradually reduced GM lineage potential.
Methods
FACS purification of LSK subpopulations
BM cells were harvested from 8- to 12-week-old C57BL/6 or heterozygous Rag1/GFP (Rag1GFP) knock-in reporter mice.24 Fetal liver (FL) cells were obtained from time-matched pregnancies.15 Kit-enriched cells were stained with lineage antibodies followed by antibodies against Sca-1, Kit, Flt3, and Mpl. LSKFlt3hiMpl− and LSKFlt3hiMplhi populations as well as LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cells were FACS purified on a BD FACSAria (BD Biosciences, San Jose, CA), resulting in high purities for all populations. LSKFlt3− or LSKCD34+Flt3− cells were sorted as previously described10,11 and used as positive controls for in vitro assays and CFU-S assay, respectively. For detailed information, see Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
CFU-S assay
All mouse experiments were approved by the ethical committee at Lund University. Fifty LSKCD34+Flt3− (highly enriched in CFU-S11 ) or 250 LSKFlt3hiMpl− or LSKFlt3hiMplhi cells were transplanted into lethally irradiated (900 cGy) recipients. Eleven days after transplantation, spleens were harvested and cells from macroscopic colonies picked for cytospin preparations and morphologic analysis. The number and the size of macroscopic colonies from each spleen were scored after fixation in Tellesniczky fixative, as previously described.11,22 See Document S1 for further details.
In vitro evaluation of MkE, GM, and lymphoid potentials
For evaluation of Mk potential, cells were sorted into X-vivo 15 (BioWhittaker) supplemented with cytokines (detailed information about cytokines can be found in Document S1). Cells were subsequently seeded at 50 cells/well in 96-well (flat-bottomed) plates and evaluated morphologically by May-Grünwald Giemsa (MGG)–stained cytospin preparations (multiple cytospins from each well to evaluate all cells generated; Document S1), after 4, 6, or 10 days of culture. The frequency of cells having Mk potential was calculated as follows: 1 − 10((log(frequency of negative wells))/50).
For evaluation of erythroid potential, 30 to 50 LSKCD34+Flt3−, LSKFlt3hiMplhi or LSKFlt3hiMpl− cells were seeded in 1 mL methylcellulose (GF M3434; StemCell Technologies, Vancouver, BC) containing cytokines (Document S1). To establish an optimal time-point for read-out of each cell population investigated, cells were cultured for 4, 6, 8, 10, or 12 days, at which time erythroid potential was evaluated using 2,7-diaminofluorene staining (DAF; Sigma-Aldrich, St Louis, MO).15,25 DAF-positive cells were identified as cells with intracellular blue granules and LSKFlt3hiMpl−-derived pure GM clones (generated in methylcellulose [M3134; StemCell Technologies] supplemented with cytokines, but not hEPO or hTHPO) were used as a negative control to confirm the specificity of DAF staining.
To evaluate the individual GM, T, and B lineage potentials of the different LSKFlt3hi subpopulations, single cells were seeded by an automated cell deposition unit (ACDU) coupled to a BD FACSAria (providing single cells in > 99% of the wells, and no wells with more than 1 cell) or occasionally manually at 1 cell per well (Document S1).
For evaluation of GM potential, single cells were sorted into 60-well Terasaki plates, with each well containing 20 μL X-vivo 15 with cytokines (Document S1). Wells were scored, with an inverted microscope, for clonal growth after 8 or 10 days of culture. Frequencies of clones containing G and/or M cells were scored by morphologic evaluation of MGG-stained cytospins (Document S1).
T- and B-cell potential was evaluated by sorting single cells onto approximately 80% confluent monolayers of OP9-DL1 and OP9 stromal cells respectively, as previously described.15,26 Clones were identified and picked at 3 to 4 weeks (depending on clonal size), and subsequently analyzed by FACS for the presence of T cell– and B cell–committed progeny.
In vivo multilineage reconstitution assay
Competitive reconstitution assays using congenic CD45.1 and CD45.2 mice were performed as previously described.11,15 See Document S1 for details about antibodies used for analyses. Briefly, 2000 LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, and LSKFlt3hiRag1GFPint/hi cells were sorted and transplanted together with 200 000 unfractionated support BM cells, into lethally irradiated (900 cGy) recipients, after which peripheral blood was withdrawn and analyzed at 3, 6, 9, and 12 weeks posttransplantation to study the total donor and donor-derived lineage reconstitution levels.
Gene-expression analysis of single cells by multiplex RT-PCR
Multiplex single-cell reverse transcription–polymerase chain reaction (RT-PCR) analysis was performed as previously described.10,15,27 See Document S1 for details, including primers.
Results
The CFU-S activity within LSKFlt3hi BM cells resides within the Mplhi and not Mpl− subfraction
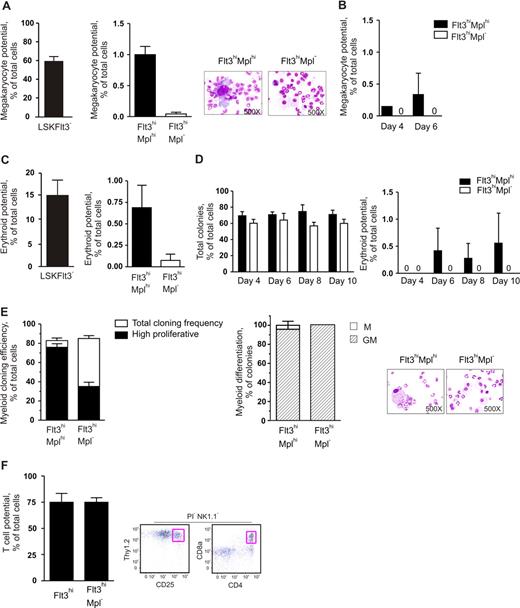
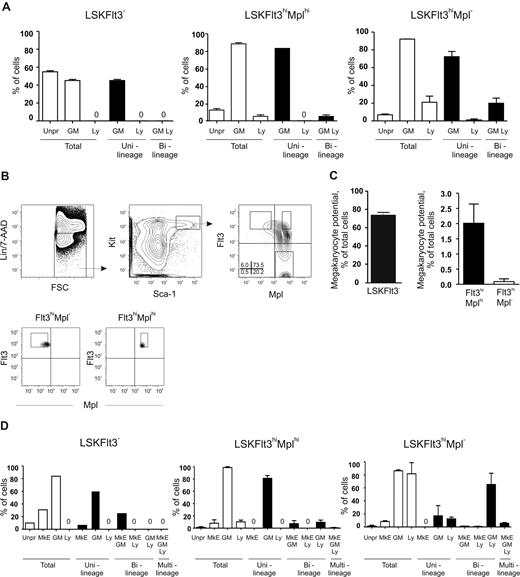
Although, in general, a low Mk and E potential was revealed in different clonal assays, both we and others noted in previous studies that a small fraction (0.4%-1.2%) of LSKFlt3hi BM cells possessed potent CFU-S activity,11,18 comparable to the low frequency of LSKFlt3hi cells also producing cells of the Mk and E lineages in vitro.10,12 To investigate whether LSKFlt3hi cells with CFU-S potential could be separated from LSKFlt3hi LMPPs without such activity, we analyzed previous Affymetrix array results.15 Whereas we had shown that most investigated genes affiliated with the Mk and E lineages are no longer expressed in LSKFlt3hi cells, we did find that Mpl was still expressed in a fraction of LSKFlt3hi cells, although at reduced levels and frequencies when compared with LSKCD34−Flt3− and LSKCD34+Flt3− cells.10,15 Of further note, the fraction of BM LSKFlt3hi cells expressing Mpl coexpressed genes of the GM lineage but predominantly not of the lymphoid lineage, in contrast to LSKFlt3hi cells negative for Mpl, which frequently coexpressed lymphoid and GM genes.15 Thus, we hypothesized that at least a fraction of LSKFlt3hiMpl+ cells might include an intermediate progenitor with sustained but restricted MkE potential in the pathway from pluripotent MPPs to LSKFlt3hiMpl− LMPPs, with little or no CFU-S or MkE potential. Thus, we investigated by FACS the coexpression of Flt3 and Mpl on LSK cells (Figure 1A). In agreement with previous gene-expression analyses,15,28 all LSKFlt3− HSCs expressed Mpl at uniform high levels, whereas a large fraction of LSKFlt3+ cells expressed Mpl with a gradual reduction in mean intensity with increasing Flt3 expression. Most notably, 37% ± 10% (mean ± standard error of the mean [SEM] of 8 experiments) of cells within the LSKFlt3hi LMPP population (25% highest Flt3-expressing LSK cells),10,15 lacked detectable Mpl expression (Figure 1A). We next purified LSKFlt3hiMpl− and LSKFlt3hiMplhi (approximately 12% of LSKFlt3hi LMPPs) BM cells (Figure 1A).
CFU-S activity within LSKFlt3hi cells resides in the Mplhi, not in the Mpl−, subpopulation. (A) Coexpression pattern of Mpl and Flt3 on Kit-enriched, lineage-negative (Lin−), Sca-1+ and Kit+ (LSK) BM cells. Gates denote the sorting strategies used to purify LSKFlt3hiMpl− (Flt3hiMpl−), LSKFlt3hiMplhi (Flt3hiMplhi), and LSKFlt3− cells. Percentages indicate mean quadrant frequencies within LSK cells from 8 experiments. Right panels show typical purity analysis for LSKFlt3hiMpl− and LSKFlt3hiMplhi cells. (B) Number and size distribution of day 11 CFU-S in mice that underwent transplantation with 50 LSKCD34+Flt3− (CD34+Flt3−) cells (n = 20), 250 LSKFlt3hiMplhi cells (n = 37), or 250 LSKFlt3hiMpl− cells (n = 34), respectively. Mean plus or minus the standard error of the mean (SEM) values from 3 experiments. (C) Left and middle panels show photographs and cell morphology (original magnification, ×500) of typical CFU-S colonies in spleens of mice that underwent transplantation with LSKCD34+Flt3− and LSKFlt3hiMplhi cells, respectively. Right panels show spleens and morphology of cells picked from CFU-S from mice that underwent transplantation with LSKFlt3hiMpl− cells. To the left, a typical spleen transplanted with 250 LSKFlt3hiMpl− cells and no CFU-S (31 of 34 mice), and to the right, one of the few cases (3 of 34 mice) in which a small CFU-S was observed in mice that underwent transplantation with LSKFlt3hiMpl− cells. Below, typical cell morphology of cells picked from a spleen without CFU-S (left), and from a small colony (right) derived from LSKFlt3hiMpl− cells.
CFU-S activity within LSKFlt3hi cells resides in the Mplhi, not in the Mpl−, subpopulation. (A) Coexpression pattern of Mpl and Flt3 on Kit-enriched, lineage-negative (Lin−), Sca-1+ and Kit+ (LSK) BM cells. Gates denote the sorting strategies used to purify LSKFlt3hiMpl− (Flt3hiMpl−), LSKFlt3hiMplhi (Flt3hiMplhi), and LSKFlt3− cells. Percentages indicate mean quadrant frequencies within LSK cells from 8 experiments. Right panels show typical purity analysis for LSKFlt3hiMpl− and LSKFlt3hiMplhi cells. (B) Number and size distribution of day 11 CFU-S in mice that underwent transplantation with 50 LSKCD34+Flt3− (CD34+Flt3−) cells (n = 20), 250 LSKFlt3hiMplhi cells (n = 37), or 250 LSKFlt3hiMpl− cells (n = 34), respectively. Mean plus or minus the standard error of the mean (SEM) values from 3 experiments. (C) Left and middle panels show photographs and cell morphology (original magnification, ×500) of typical CFU-S colonies in spleens of mice that underwent transplantation with LSKCD34+Flt3− and LSKFlt3hiMplhi cells, respectively. Right panels show spleens and morphology of cells picked from CFU-S from mice that underwent transplantation with LSKFlt3hiMpl− cells. To the left, a typical spleen transplanted with 250 LSKFlt3hiMpl− cells and no CFU-S (31 of 34 mice), and to the right, one of the few cases (3 of 34 mice) in which a small CFU-S was observed in mice that underwent transplantation with LSKFlt3hiMpl− cells. Below, typical cell morphology of cells picked from a spleen without CFU-S (left), and from a small colony (right) derived from LSKFlt3hiMpl− cells.
As we had previously found that the total LSKFlt3hi population contains some CFU-S day 11 (CFU-S d11) but no CFU-S day 8 (CFU-S d8) activity,11 we next compared the CFU-S d11 activity of purified LSKFlt3hiMpl− and LSKFlt3hiMplhi BM cells, using LSKCD34+Flt3− cells enriched for ST-HSC and CFU-S activity,11 as a positive control (Figure 1B,C). As previously described,11 LSKCD34+Flt3− cells were highly enriched in CFU-S d11 (1/21 cells). LSKFlt3hiMplhi cells contained less although significant CFU-S d11 activity (1/93 cells), whereas LSKFlt3hiMpl− cells contained virtually no CFU-S d11 (1/2833 cells), as illustrated by only a total of 3 spleen colonies being generated in as much as 34 mice transplanted with 250 cells each. Furthermore, while the majority of CFU-S d11 colonies derived from LSKCD34+Flt3− and LSKFlt3hiMplhi cells were larger than 1 mm (Figure 1B,C), the few LSKFlt3hiMpl−-derived colonies were small (≤ 1 mm). While morphologic evaluation of spleen colony cells derived from LSKFlt3hiMplhi cells (and LSKCD34+Flt3− cells), as expected, contained largely immature cells of the erythroid (nucleated) and megakaryocyte lineages (Figure 1C), cells from the small LSKFlt3hiMpl− colonies did not, and were indistinguishable from cells obtained from the spleens of nontransplanted mice (Figure 1C). These experiments conclusively demonstrate that the limited CFU-S activity previously ascribed to LSKFlt3hi cells11,18 is a property restricted to LSKFlt3hi cells expressing Mpl, although most LSKFlt3hiMplhi cells (98%-99%) lack such activity.
The limited in vitro erythroid and megakaryocyte potential of LSKFlt3hi BM cells resides within the Mplhi and not the Mpl− subfraction
In previous studies, we and others10,12 demonstrated through clonal in vitro assays that a very small fraction (1%-2%) of LMPPs or LSKFlt3hi cells read out with Mk and/or E progeny. Thus, we next tested whether the expression of Mpl would allow separation of the limited in vitro Mk and E potentials of LSKFlt3hi cells, as demonstrated for CFU-S (Figure 1).
In agreement with previous studies,10 LSKFlt3− cells, known to have potent MkE potential,10 generated megakaryocytes at high frequencies (Figure 2A, left panel). As expected, both LSKFlt3hiMplhi and LSKFlt3hiMpl− cells generated megakaryocytes at much reduced frequencies, but whereas LSKFlt3hiMplhi cells had a significant Mk potential (1/100 cells), virtually no (1/2350 cells) LSKFlt3hiMpl− cells possessed such potential in vitro (Figure 2A), although the cells were carefully investigated at multiple time points (Figure 2B).
The low megakaryocyte and erythroid potentials of LMPPs are highly enriched in LMPPs coexpressing cell-surface Mpl. (A) In vitro megakaryocyte (Mk) potential of BM LSKFlt3−, LSKFlt3hiMplhi (Flt3hiMplhi), and LSKFlt3hiMpl− (Flt3hiMpl−) cells, as described in “Methods” after 10 days of culture. Mean plus or minus SEM values from 7 experiments. Cell morphology pictures from typical cultures of LSKFlt3hiMplhi cells and LSKFlt3hiMpl− cells, respectively. (B) Mk potential of LSKFlt3hiMplhi and LSKFlt3hiMpl− BM cells, after 4 and 6 days of culture. Mean plus or minus SEM values from 2 experiments. (C) In vitro erythroid potential of BM LSKFlt3−, LSKFlt3hiMplhi, and LSKFlt3hiMpl− cells, as established by DAF staining of methylcellulose cultures after 12 days of culture, as described in “Methods.” Mean plus or minus SEM values from 4 experiments. (D) Total cloning frequencies (left) and erythroid potential (right) of LSKFlt3hiMplhi and LSKFlt3hiMpl− cells evaluated after 4, 6, 8, and 10 days of methylcellulose culture. Mean plus or minus SEM values from 3 experiments. (E) Left panel shows results from clonal assays of single-cell deposited LSKFlt3hiMplhi and LSKFlt3hiMpl− cells cultured in cytokines promoting GM development (“Methods”). Open bars show cloning frequencies as established after 10 days of culture, and black bars show frequency of high proliferative clones (covering > 50% of the well). Mean plus or minus SEM values from 3 experiments. Middle panel shows relative distribution between clones with monocyte (M) or combined granulocyte-monocyte (GM) contents, derived from single LSKFlt3hiMplhi and LSKFlt3hiMpl− cells as established by morphologic evaluation of MGG-stained cytospin preparations (right panels). Mean plus or minus SEM values from 2 experiments. (F) T-cell potential of single-cell deposited LSKFlt3hi (Flt3hi) and LSKFlt3hiMpl− BM cells grown for 3 to 4 weeks on OP9-DL1, as evaluated by FACS, and defined as NK1.1−Thy1.2hiCD25hi and/or NK1.1−CD4+CD8+ and negative for the viability dye propidium iodide (PI) as previously described.10,15 Mean plus or minus SEM values from 2 experiments (n = 24 per group and experiment). Right panel shows representative FACS profiles of analyzed clone derived from a single cell. The T-cell identity of NK1.1−Thy1.2hiCD25hi clones was as previously shown15 and also confirmed by nested PCR analysis demonstrating expression of CD3 antigen, epsilon polypeptide (Cd3e), and pre–T-cell antigen receptor alpha (Ptcra) (data not shown).
The low megakaryocyte and erythroid potentials of LMPPs are highly enriched in LMPPs coexpressing cell-surface Mpl. (A) In vitro megakaryocyte (Mk) potential of BM LSKFlt3−, LSKFlt3hiMplhi (Flt3hiMplhi), and LSKFlt3hiMpl− (Flt3hiMpl−) cells, as described in “Methods” after 10 days of culture. Mean plus or minus SEM values from 7 experiments. Cell morphology pictures from typical cultures of LSKFlt3hiMplhi cells and LSKFlt3hiMpl− cells, respectively. (B) Mk potential of LSKFlt3hiMplhi and LSKFlt3hiMpl− BM cells, after 4 and 6 days of culture. Mean plus or minus SEM values from 2 experiments. (C) In vitro erythroid potential of BM LSKFlt3−, LSKFlt3hiMplhi, and LSKFlt3hiMpl− cells, as established by DAF staining of methylcellulose cultures after 12 days of culture, as described in “Methods.” Mean plus or minus SEM values from 4 experiments. (D) Total cloning frequencies (left) and erythroid potential (right) of LSKFlt3hiMplhi and LSKFlt3hiMpl− cells evaluated after 4, 6, 8, and 10 days of methylcellulose culture. Mean plus or minus SEM values from 3 experiments. (E) Left panel shows results from clonal assays of single-cell deposited LSKFlt3hiMplhi and LSKFlt3hiMpl− cells cultured in cytokines promoting GM development (“Methods”). Open bars show cloning frequencies as established after 10 days of culture, and black bars show frequency of high proliferative clones (covering > 50% of the well). Mean plus or minus SEM values from 3 experiments. Middle panel shows relative distribution between clones with monocyte (M) or combined granulocyte-monocyte (GM) contents, derived from single LSKFlt3hiMplhi and LSKFlt3hiMpl− cells as established by morphologic evaluation of MGG-stained cytospin preparations (right panels). Mean plus or minus SEM values from 2 experiments. (F) T-cell potential of single-cell deposited LSKFlt3hi (Flt3hi) and LSKFlt3hiMpl− BM cells grown for 3 to 4 weeks on OP9-DL1, as evaluated by FACS, and defined as NK1.1−Thy1.2hiCD25hi and/or NK1.1−CD4+CD8+ and negative for the viability dye propidium iodide (PI) as previously described.10,15 Mean plus or minus SEM values from 2 experiments (n = 24 per group and experiment). Right panel shows representative FACS profiles of analyzed clone derived from a single cell. The T-cell identity of NK1.1−Thy1.2hiCD25hi clones was as previously shown15 and also confirmed by nested PCR analysis demonstrating expression of CD3 antigen, epsilon polypeptide (Cd3e), and pre–T-cell antigen receptor alpha (Ptcra) (data not shown).
Our previous studies using liquid-based, single-cell cultures to evaluate E potential of LSKFlt3hi cells revealed minimal potential for this lineage.10,15 Thus, we here compared the E potential of LSKFlt3hiMplhi and LSKFlt3hiMpl− cells using a conventional methylcellulose clonal assay, efficiently promoting growth and differentiation of erythroid progenitors (“Methods”). Whereas 1 of 135 plated LSKFlt3hiMplhi cells produced E progeny, only 1 of 1400 LSKFlt3hiMpl− cells did so after 12 days of culture (Figure 2C right panel). Furthermore, in a more extensive kinetic analysis, LSKFlt3hiMpl− cells, although showing a very high cloning efficiency, failed to produce erythroid progeny regardless of being assessed at 4, 6, 8, or 10 days of culture (Figure 2D). Thus, as for the in vivo clonal CFU-S activity, and with similar frequencies, expression of Mpl allows separation of LSKFlt3hi BM cells into a Mplhi subpopulation with a very restricted but reproducible in vitro Mk and E potential, and a Mpl− fraction virtually devoid of such activities.
LSKFlt3hi LMPPs have previously been demonstrated to have combined GM and lymphoid potential.10 Having demonstrated that the Mpl− fraction of LSKFlt3hi cells virtually lack in vivo CFU-S and in vitro Mk and E potentials, it was important to verify that LSKFlt3hiMpl− cells still possessed combined GM and lymphoid potential. As previously demonstrated for the total LMPP population,9,10 both LSKFlt3hiMplhi and LSKFlt3hiMpl− single cells generated large numbers of granulocytes as well as monocytes/macrophages (combined G and M potential) at high frequencies in vitro (83% and 85% respectively; Figure 2E). However, under these GM conditions, LSKFlt3hiMplhi cells were shown to have a significantly higher proliferative potential (Figure 2E), compatible with LSKFlt3hiMpl− cells being downstream progeny of LSKFlt3hiMplhi cells.
To establish that LSKFlt3hiMpl− BM cells also sustained lymphoid potential, we compared the T-cell potential of single LSKFlt3hiMpl− cells with that of total LSKFlt3hi cells, using the OP9-DL1 stromal cell line,10,26 resulting in as much as 75% of single LSKFlt3hiMpl− cells generating committed T-cell progeny under these conditions (Figure 2F). Thus, adult LSKFlt3hiMpl− BM cells represent multipotent progenitors with sustained GM and lymphoid but not in vivo CFU-S or in vitro Mk and E potentials.
Combined GM and lymphoid but not MkE transcriptional priming is a property of LSKFlt3hiMpl− rather than LSKFlt3hiMplhi cells
Previous studies have also presented molecular evidence for the existence of LMPPs, through identification of LSKFlt3hi cells10,15 (or closely related populations12,14 ) with combined GM and lymphoid but down-regulated or lost MkE transcriptional priming at the single-cell level. As only a fraction of LSKFlt3hi cells were lymphoid primed,15 one would predict, based on the functional data presented (Figure 2), that LSKFlt3hiMpl− cells would show enhanced lymphoid but sustained GM priming when compared with LSKFlt3hiMplhi cells. We explored this prediction through multiplex PCR analysis15,27 of single LSKFlt3hiMplhi and LSKFlt3hiMpl− adult BM cells, using LSKFlt3−-enriched HSCs as a control population shown to be not yet lymphoid primed.15 As previously described,15 primers for 2 to 3 genes specific for GM (colony stimulating factor 3 receptor [Csf3r, gene for G-CSF receptor] and myeloperoxidase [Mpo]) and lymphoid (interleukin 7 receptor [Il7r], sterile IgH transcript and Rag1) lineages were used. As predicted, both LSKFlt3hiMplhi and LSKFlt3hiMpl− cells were highly GM primed (88% and 92% of single cells, respectively) (Figure 3A). Whereas only a small fraction of LSKFlt3hiMplhi single cells expressed lymphoid genes (5%), as much as 21% of LSKFlt3hiMpl− BM cells were lymphoid primed, and almost all of these also coexpressed GM genes (Figure 3A).
Combined GM and lymphoid transcriptional priming in LSKFlt3hiMpl− BM cells. (A) Coexpression patterns of transcriptional lineage programs in single cells from BM LSK subpopulations. Cells were scored as expressing GM and/or lymphoid (Ly) programs based on the expression of one or more lineage-associated genes: GM: Csf3r and Mpo; lymphoid: Rag1, sterile IgH transcript, and Il7r. Mean plus or minus SEM values from 2 experiments with 88 cells investigated in each experiment. (B) Coexpression pattern of Mpl and Flt3 on Kit-enriched, lineage-negative (Lin−), Sca-1+ and Kit+ (LSK) fetal liver (day E14.5-E15.5) cells. Gates denote the sorting strategies used to purify LSKFlt3hiMpl− (Flt3hiMpl−), LSKFlt3hiMplhi (Flt3hiMplhi), and LSKFlt3− cells. Percentages indicate mean quadrant frequencies within LSK cells from 4 experiments. Panels below show typical purity analysis for LSKFlt3hiMpl− and LSKFlt3hiMplhi cells. (C) In vitro Mk potential of fetal liver LSKFlt3−, LSKFlt3hiMplhi, and LSKFlt3hiMpl− cells, investigated as described in “Methods” after 8 days of culture. Mean plus or minus SEM values from 4 experiments. (D) Coexpression patterns of lineage programs in single cells from fetal liver LSK subpopulations. Cells were scored as expressing MkE, GM, and/or lymphoid (Ly) programs based on the expression of one or more lineage associated genes: MkE: Gata1, VWF, and Epor; GM: Csf3r and Mpo; lymphoid: Rag1, sterile IgH transcript, and Il7r. Mean plus or minus SEM values from 2 experiments, with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells in which 88 cells were evaluated in total.
Combined GM and lymphoid transcriptional priming in LSKFlt3hiMpl− BM cells. (A) Coexpression patterns of transcriptional lineage programs in single cells from BM LSK subpopulations. Cells were scored as expressing GM and/or lymphoid (Ly) programs based on the expression of one or more lineage-associated genes: GM: Csf3r and Mpo; lymphoid: Rag1, sterile IgH transcript, and Il7r. Mean plus or minus SEM values from 2 experiments with 88 cells investigated in each experiment. (B) Coexpression pattern of Mpl and Flt3 on Kit-enriched, lineage-negative (Lin−), Sca-1+ and Kit+ (LSK) fetal liver (day E14.5-E15.5) cells. Gates denote the sorting strategies used to purify LSKFlt3hiMpl− (Flt3hiMpl−), LSKFlt3hiMplhi (Flt3hiMplhi), and LSKFlt3− cells. Percentages indicate mean quadrant frequencies within LSK cells from 4 experiments. Panels below show typical purity analysis for LSKFlt3hiMpl− and LSKFlt3hiMplhi cells. (C) In vitro Mk potential of fetal liver LSKFlt3−, LSKFlt3hiMplhi, and LSKFlt3hiMpl− cells, investigated as described in “Methods” after 8 days of culture. Mean plus or minus SEM values from 4 experiments. (D) Coexpression patterns of lineage programs in single cells from fetal liver LSK subpopulations. Cells were scored as expressing MkE, GM, and/or lymphoid (Ly) programs based on the expression of one or more lineage associated genes: MkE: Gata1, VWF, and Epor; GM: Csf3r and Mpo; lymphoid: Rag1, sterile IgH transcript, and Il7r. Mean plus or minus SEM values from 2 experiments, with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells in which 88 cells were evaluated in total.
We recently demonstrated that LSKFlt3hi cells in the fetal liver, as in adult BM, have combined GM and lymphoid potentials but down-regulated MkE potential.15 Furthermore, a similar pattern of multilineage transcriptional priming could be observed in fetal LSKFlt3hi cells with the distinction that a larger fraction of LSKFlt3hi cells from the fetal liver were transcriptionally primed compared with LSKFlt3hi adult BM cells.15 Thus, we also compared the multilineage transcriptional priming of LSKFlt3hiMpl− and LSKFlt3hiMplhi fetal liver (E14.5-E15.5) cells. The expression pattern of Mpl in relationship to Flt3 was comparable to that in adult BM, allowing purification of equivalent LSKFlt3hiMplhi and LSKFlt3hiMpl− populations (Figure 3B). As with adult BM, the Mk potential was limited in both populations, but whereas 2% of LSKFlt3hiMplhi cells generated Mk progeny, only 0.09% of LSKFlt3hiMpl− fetal liver cells produced Mk (Figure 3C). Multiplex single-cell RT-PCR analysis demonstrated, in line with the findings in adult BM, that only a small fraction (10%) of LSKFlt3hiMplhi cells were lymphoid primed, in contrast to as much as 82% of LSKFlt3hiMpl− cells, of which the great majority were also GM primed (Figure 3D). This finding further supports that lymphoid transcriptional priming is a property primarily of LSKFlt3hiMpl− cells. Of further interest, although the MkE priming (GATA binding protein 1 [Gata1], erythropoietin receptor [Epor], and Von Willebrand factor homolog [VWF]) was, as expected, low in both populations, a small fraction of LSKFlt3hiMplhi but virtually no LSKFlt3hiMpl− fetal liver cells had combined GM and MkE priming without lymphoid priming. This outcome is similar to what is observed more frequently in LSKFlt3− HSC-enriched cells with sustained MkE potential, whereas a small fraction of LSKFlt3hiMpl− cells were uniquely primed for MkE, GM, and lymphoid genes (Figure 3D). Collectively, these experiments provide compelling evidence for LSKFlt3hiMpl− cells representing pure LMPPs with sustained GM and lymphoid but down-regulated MkE potentials, with a corresponding down-regulated MkE transcriptional lineage priming and up-regulated lymphoid priming.
Gradual down-regulation of GM potential with increasing lymphoid transcriptional priming in LSKFlt3hi LMPPs
Although LSKFlt3hi cells appear to sustain GM potential after down-regulation of MkE potential,9,10,12,14 it remains unclear whether and how this GM potential is altered with increasing levels of lymphoid transcriptional priming. To investigate this as well as how GM transcriptional priming changes with increasing lymphoid transcriptional priming, we made use of a reporter mouse expressing enhanced green fluorescent protein (GFP) under control of the Rag1 promoter.24 We first investigated the coexpression pattern of Rag1GFP and Flt3 within the BM LSK compartment, and observed that Rag1GFP expression increased with increasing Flt3 expression and all cells with bright GFP levels were observed within the Flt3hi compartment (Figure 4A). To be able to investigate populations with different levels of Rag1GFP expression, LSKFlt3hi cells were separated by FACS into LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi populations (Figure 4A). Single-cell RT-PCR analysis of Rag1 expression in the different subpopulations confirmed the gradual increase in Rag1 expression from LSKFlt3− through LSKFlt3hiRag1GFPhi cells (Figure 4B). Other investigated early lymphoid genes (Il7r and sterile IgH transcript) also increased gradually from LSKFlt3− to LSKFlt3hiRag1GFPhi cells (Figure 4C).
Increasing lymphoid transcriptional priming in LMPPs expressing Rag1GFP. (A) Coexpression pattern of Rag1GFP and Flt3 on Kit-enriched LSK BM cells. Gates denote the sorting strategies used to purify LSKFlt3hiRag1GFP− (Rag1GFP−), LSKFlt3hiRag1GFPlo (Rag1GFPlo), LSKFlt3hiRag1GFPint (Rag1GFPint), LSKFlt3hiRag1GFPhi (Rag1GFPhi), and LSKFlt3− (Flt3−) cells. Percentages indicate mean frequencies of the sorted populations of total LSK cells from 4 experiments. Right panels show typical purity analysis. (B) Expression pattern of Rag1 transcripts in single cells from sorted BM LSKFlt3−, LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint and LSKFlt3hiRag1GFPhi cells. Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total. (C) Expression pattern of individual lymphoid genes (Il7r, sterile IgH transcript [IgH], and Rag1) in single cells from BM LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cells. To the right is shown the frequency of cells in each population coexpressing 2 or 3 of the investigated lymphoid genes (Multi-Ly). Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total.
Increasing lymphoid transcriptional priming in LMPPs expressing Rag1GFP. (A) Coexpression pattern of Rag1GFP and Flt3 on Kit-enriched LSK BM cells. Gates denote the sorting strategies used to purify LSKFlt3hiRag1GFP− (Rag1GFP−), LSKFlt3hiRag1GFPlo (Rag1GFPlo), LSKFlt3hiRag1GFPint (Rag1GFPint), LSKFlt3hiRag1GFPhi (Rag1GFPhi), and LSKFlt3− (Flt3−) cells. Percentages indicate mean frequencies of the sorted populations of total LSK cells from 4 experiments. Right panels show typical purity analysis. (B) Expression pattern of Rag1 transcripts in single cells from sorted BM LSKFlt3−, LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint and LSKFlt3hiRag1GFPhi cells. Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total. (C) Expression pattern of individual lymphoid genes (Il7r, sterile IgH transcript [IgH], and Rag1) in single cells from BM LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cells. To the right is shown the frequency of cells in each population coexpressing 2 or 3 of the investigated lymphoid genes (Multi-Ly). Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total.
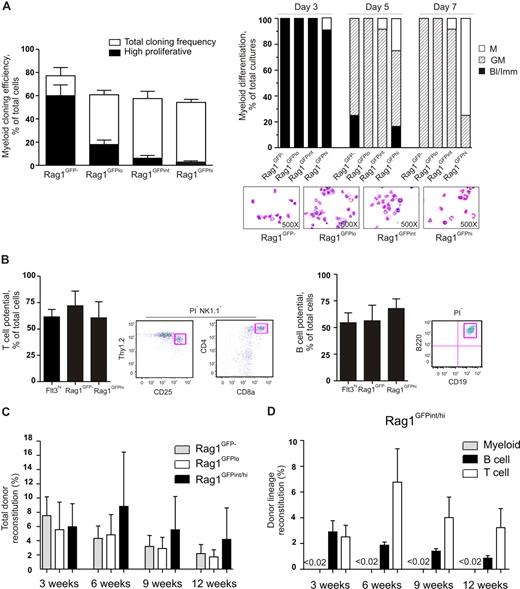
We next investigated whether and how the GM potential of LSKFlt3hi cells would change with gradually increasing lymphoid priming. LSKFlt3hiRag1GFP− BM cells presented the highest myeloid cloning frequency (77%), and most of the GM clones generated were very large (Figure 5A). Notably, not so much of the GM cloning frequency but rather the GM clonal size was reduced gradually when going from LSKFlt3hiRag1GFP− through LSKFlt3hiRag1GFPlo and LSKFlt3hiRag1GFPint to LSKFlt3hiRag1GFPhi cells (Figure 5A). A careful kinetic analysis did, however, reveal that all populations (including LSKFlt3hiRag1GFPhi cells) produced cells of the G and M lineages (Figure 5A). Furthermore, single-cell analysis on OP9-DL1 and OP9 stromal cells showed that LSKFlt3hiRag1GFP− and LSKFlt3hiRag1GFPhi BM cells possessed comparable T and B potentials (Figure 5B).
Gradual down-regulation of GM potential in LMPPs but sustained lymphoid potential in vitro and in vivo. (A) Left panel shows clonality of single-cell deposited BM LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cells cultured under GM conditions (“Methods”) for 8 days. Open bars show cloning frequencies and closed bars show the frequency of high proliferative clones (covering > 50% of well). Mean plus or minus SEM values from 3 experiments. Right panel shows results from morphologic evaluation of MGG-stained cytospin preparations of cells derived from LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cultures (20 cells each) after 3, 5, and 7 days in culture (“Methods”). Each bar shows relative distribution between wells containing only blast and immature myeloid cells (Bl/Imm), cells of both the granulocyte and monocyte lineages (GM), and cells of only the monocytic (M) lineage, as determined by morphologic evaluation of MGG-stained cytospin slides. Wells with only granulocytes were not detected. Note that although the peak for GM generation varies between the populations, all produce G and M cells. Mean plus or minus SEM values from 2 experiments, each performed with 6 replicate determinations (wells) per population. (B) T- and B-cell potential of single-cell deposited LSKFlt3hi, LSKFlt3hiRag1GFP−, and LSKFlt3hiRag1GFPhi BM cells grown for 3 to 4 weeks on OP9-DL1 and OP9 respectively, as evaluated by FACS. T cells were defined as NK1.1−Thy1.2hiCD25hi and/or NK1.1−CD4+CD8+ and B cells as B220+CD19+, both negative for the viability stain PI as previously described10,15 or DAPI. Mean plus or minus SEM values from 3 experiments (n = 16-48 per group and experiment). Right panels show representative FACS profiles of analyzed clone derived from a single cell. To verify the T-cell potential by molecular methods, 7 NK1.1−Thy1hiCD25hi clones were analyzed for expression of CD3 antigen, epsilon polypeptide (Cd3e), and pre–T-cell antigen receptor alpha (Ptcra). All clones were found to be positive for both T-cell genes, whereas clones grown on OP9 were negative for the T-cell genes but, as expected, positive for hypoxanthine guanine phosphoribosyl transferase (Hprt) and protein tyrosine phosphatase, receptor type C (Ptprc, gene for Cd45) (data not shown). (C) Reconstitution of lethally irradiated recipients that underwent transplantation with 2000 sorted cells of the indicated cell populations sorted from Rag1GFP mice, in competition with 200 000 BM cells. Results show mean plus or minus the standard deviation (SD) of reconstitution levels (total blood cells) at 3, 6, 9, and 12 weeks after transplantation from 2 experiments (n = 6–7). (D) Lineage analysis of mice that underwent transplantation with LSKFlt3hiRag1GFPint/hi cells. Myeloid cell reconstitution was below detection level (less than 0.02%) of total donor-derived cells at all time points after transplantation.
Gradual down-regulation of GM potential in LMPPs but sustained lymphoid potential in vitro and in vivo. (A) Left panel shows clonality of single-cell deposited BM LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cells cultured under GM conditions (“Methods”) for 8 days. Open bars show cloning frequencies and closed bars show the frequency of high proliferative clones (covering > 50% of well). Mean plus or minus SEM values from 3 experiments. Right panel shows results from morphologic evaluation of MGG-stained cytospin preparations of cells derived from LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cultures (20 cells each) after 3, 5, and 7 days in culture (“Methods”). Each bar shows relative distribution between wells containing only blast and immature myeloid cells (Bl/Imm), cells of both the granulocyte and monocyte lineages (GM), and cells of only the monocytic (M) lineage, as determined by morphologic evaluation of MGG-stained cytospin slides. Wells with only granulocytes were not detected. Note that although the peak for GM generation varies between the populations, all produce G and M cells. Mean plus or minus SEM values from 2 experiments, each performed with 6 replicate determinations (wells) per population. (B) T- and B-cell potential of single-cell deposited LSKFlt3hi, LSKFlt3hiRag1GFP−, and LSKFlt3hiRag1GFPhi BM cells grown for 3 to 4 weeks on OP9-DL1 and OP9 respectively, as evaluated by FACS. T cells were defined as NK1.1−Thy1.2hiCD25hi and/or NK1.1−CD4+CD8+ and B cells as B220+CD19+, both negative for the viability stain PI as previously described10,15 or DAPI. Mean plus or minus SEM values from 3 experiments (n = 16-48 per group and experiment). Right panels show representative FACS profiles of analyzed clone derived from a single cell. To verify the T-cell potential by molecular methods, 7 NK1.1−Thy1hiCD25hi clones were analyzed for expression of CD3 antigen, epsilon polypeptide (Cd3e), and pre–T-cell antigen receptor alpha (Ptcra). All clones were found to be positive for both T-cell genes, whereas clones grown on OP9 were negative for the T-cell genes but, as expected, positive for hypoxanthine guanine phosphoribosyl transferase (Hprt) and protein tyrosine phosphatase, receptor type C (Ptprc, gene for Cd45) (data not shown). (C) Reconstitution of lethally irradiated recipients that underwent transplantation with 2000 sorted cells of the indicated cell populations sorted from Rag1GFP mice, in competition with 200 000 BM cells. Results show mean plus or minus the standard deviation (SD) of reconstitution levels (total blood cells) at 3, 6, 9, and 12 weeks after transplantation from 2 experiments (n = 6–7). (D) Lineage analysis of mice that underwent transplantation with LSKFlt3hiRag1GFPint/hi cells. Myeloid cell reconstitution was below detection level (less than 0.02%) of total donor-derived cells at all time points after transplantation.
To evaluate whether the reduced GM clonal size reflected a general loss of proliferative capacity with increasing Rag1 expression or only a reduced propensity to produce cells of the GM lineage, 2000 LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, or LSKFlt3hiRag1GFPint/hi cells were transplanted into lethally irradiated recipients in a competitive setting. Peripheral blood analysis at 3, 6, 9, and 12 weeks after transplantation demonstrated robust and comparable reconstitution by all 3 populations (Figure 5C), and almost exclusively contribution to the B- and T-lymphoid lineages (Figure 5D).
The in vitro GM lineage potential assays as well as the in vivo transplantation assay suggested that the GM potential of LSKFlt3hi cells is gradually reduced with increasing levels of lymphoid transcriptional priming. Strikingly, whereas the frequency of transcriptionally lymphoid-primed cells increased gradually from LSKFlt3− (0%) to LSKFlt3hiRag1GFPhi (86%) cells, most cells from LSKFlt3hiRag1GFP− (95%) to the LSKFlt3hiRag1GFPhi (93%) populations remained GM primed, and in all populations almost all cells that were lymphoid primed were simultaneously GM-primed (Figure 6). Collectively, these data provide evidence, at the single-cell level, that with increasing lymphoid transcriptional priming, LMPPs remain GM transcriptionally primed (but not MkE) while displaying a gradually reduced propensity to form cells of the GM lineage.
LSKFlt3hi cells with increasing levels of transcriptional lymphoid priming continue to coexpress GM genes. Coexpression patterns of transcriptional lineage programs in single cells from BM LSK subpopulations separated based on levels of Rag1GFP expression. Cells were scored as expressing GM and/or lymphoid (Ly) programs based on the expression of one or more lineage-associated genes: GM: Csf3r and Mpo; lymphoid: Rag1, sterile IgH transcript, and Il7r. Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total.
LSKFlt3hi cells with increasing levels of transcriptional lymphoid priming continue to coexpress GM genes. Coexpression patterns of transcriptional lineage programs in single cells from BM LSK subpopulations separated based on levels of Rag1GFP expression. Cells were scored as expressing GM and/or lymphoid (Ly) programs based on the expression of one or more lineage-associated genes: GM: Csf3r and Mpo; lymphoid: Rag1, sterile IgH transcript, and Il7r. Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total.
Discussion
Recent studies demonstrating that a fraction of LSKFlt3hi BM cells possesses MkE potential were interpreted as support for the prevailing CMP-CLP model with “segregation of myeloid and lymphoid lineage potentials from multipotent progenitors,” questioning the existence of LMPPs with sustained GM and lymphoid but down-regulated MkE potential as an alternative lineage commitment pathway from HSCs.18 However, in the only clonal MkE analysis performed, only 0.4% of LSKFlt3hi BM cells revealed CFU-S d12 activity, in agreement with the reported low frequency of LSKFlt3hi cells with in vitro and in vivo clonal MkE potential.10-12,14 In contrast, the evidence presented previously9,10,15 and herein establish that a large fraction of LSKFlt3hi cells have exclusively a combined GM and lymphoid lineage potential. Although Forsberg et al failed to demonstrate that a large fraction of LSKFlt3hi BM cells have MkE potential as required of truly pluripotent MPPs, the evidence for at least a small fraction of LSKFlt3hi BM cells sustaining MkE potential was convincing, justifying re-evaluation of the proposed existence of LMPPs with combined GM and lymphoid but not MkE potentials.2,3
In the present studies we hypothesized and provided evidence for the residual MkE potential observed in in vitro as well as in vivo clonal assays,10,11,18 originating from a subpopulation of LSKFlt3hi cells sustaining expression of the Mpl, a critical regulator of the MkE lineage.23 Specifically, whereas 1 of 93 LSKFlt3hiMplhi cells generated CFU-S d11, virtually no CFU-S d11 cells (1/2833) were derived from LSKFlt3hiMpl− cells. Also, the reportedly rare in vitro clonal Mk and E activities of LSKFlt3hi cells almost entirely segregated with LSKFlt3hiMplhi cells and not with LSKFlt3hiMpl− cells. At least 85% of single LSKFlt3hiMpl− cells sustained GM potential and 75% lymphoid potential, translating into at least 60% of LSKFlt3hiMpl− cells sustaining a combined GM and lymphoid potential. Multiplex single-cell PCR analysis supported this conclusion, demonstrating that LSKFlt3hiMpl− cells, when compared with LSKFlt3hiMplhi cells, are highly enriched in cells with combined GM and lymphoid transcriptional priming.
What is the explanation for the extremely rare readout of Mk and E potential of LSKFlt3hiMpl− BM cells in in vitro and in vivo assays (maximum 1/1400 cells at any time point investigated), and what, if any, implications does this observation have for the integrity of the evidence for the existence of LMPPs? It is important to note that the evidence for the existence of LMPPs, as for any lineage-restricted progenitor cell,4 relies primarily on the integrity of negative (here MkE) data. Reflecting that lineage commitment is a process of loss rather than gain of potential, the possibility that the very low frequency of LSKFlt3hiMpl− cells with detectable MkE potential might reflect rare contamination of purified LSKFlt3hiMpl− cells with phenotypically distinct cells with MkE potential cannot be excluded, although LSKFlt3hiMpl− cells were sorted to very high purity. However, the present and recent molecular analysis of multilineage transcriptional priming in single LSKFlt3hi cells15 provide some clues for alternative interpretations. Although based on limited data, we did uncover rare cells in LSKFlt3hiMpl− fetal liver cells (which read out Mk potential with a similarly low frequency as their adult BM counterparts) that were MkE transcriptionally primed. However, these rare LSKFlt3hiMpl− cells were virtually always also GM as well as lymphoid primed, in contrast to LSKFlt3− (HSC-enriched) cells, in which we have never observed a single cell with such a pattern of lineage priming.15 In fact, we did not see such a pattern in the rare MkE-primed LSKFlt3hiMplhi fetal liver cells, which, like LSKFlt3− cells, did not typically coexpress lymphoid genes. Thus, the rare MkE-GM-lymphoid–primed LSKFlt3hiMpl− cells and the very rare but still rather consistent read-out of MkE potential, might reflect that a very small fraction of lymphoid-primed LSKFlt3hiMpl− LMPPs might sustain a very low but significant probability of committing toward the MkE lineage. In direct support of this, we have found that even LSKFlt3hiRag1GFPint/hi cells generate MkE progeny at similarly low frequencies (S.L. and S.E.W.J., unpublished observations, 2007). Regardless, the biologic and molecular data presented here demonstrate that LSKFlt3hiMpl− cells are LMPPs with combined and robust GM and lymphoid transcriptional priming and lineage potentials, but with at least a dramatically down-regulated MkE lineage priming and potential to commit and develop toward the MkE lineages.
While the purification of LSKFlt3hiMpl− cells, representing 35% to 40% of total LMPPs, provided more definitive evidence for the existence of GM-lymphoid–restricted adult and fetal LMPPs, it is important to emphasize that the evidence remains compelling for most LSKFlt3hiMpl+ cells also being LMPPs. In fact, regardless of the assays used, we could at most reveal a MkE potential of 1% to 2% of LSKFlt3hiMplhi cells, and we also demonstrate that transcriptional priming for the MkE lineage is largely down-regulated and lymphoid priming already initiated at the LSKFlt3hiMplhi stage. Our findings are in complete agreement with recent studies in which PU.1 reporter mice were used to further separate LMPPs into PU.1+ and PU.1lo subsets.29 Similar to LSKFlt3hiMpl− cells, PU.1+ LMPPs (around 40% of all LMPPs) lacked detectable MkE potential, but PU.1lo LMPPs revealed minimal (around 2%) MkE potential, suggesting that LSKFlt3hi cells represent predominantly LMPPs in which a minimal MkE potential is lost from Mplhi to Mpl− and from PU.1lo to PU.1hi LMPPs. It is important to also emphasize that most LSKFlt3+ cells are not LMPPs, as these are restricted to LSKFlt3hi (25% of LSK cells expressing the highest Flt3 levels) cells.9,10,15 While we, based on our and other studies,29 would argue that the LSKFlt3hi phenotype defines the LMPP, it is obvious that additional markers such as Mpl and Vcam-130 help to further subfractionate LMPPs.
In further support of the loss of lineage potentials of MPPs occurring in a gradual rather than abrupt manner, as might be inferred by strict branching points in models for hematopoiesis, LMPPs sorted from Rag1GFP mice demonstrated that gradually increased lymphoid transcriptional priming within single LSKFlt3hi LMPPs occurs without significant effect on the GM lineage priming, but nevertheless resulting in gradually reduced (but not lost) GM potential.
In conclusion, LSKFlt3hi LMPPs represent an early lineage commitment stage in adult hematopoiesis with sustained GM and lymphoid but down-regulated or absent MkE potential and lineage priming. Molecular and biologic data support that the loss of MkE and subsequent GM potential of MPPs occurs in a gradual manner, possibly reflecting the interaction and opposing actions of graded doses of essential lineage-instructive transcription factors promoting different lineage potentials and fate.2,3,31
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs A. Cumano and J.C. Zúñiga-Pflücker for the OP9 and OP9-DL1 cell lines, Dr N. Sakaguchi for the RAG1/GFP knockin reporter mice, and Kirin Brewery, Japan, for generously providing the anti-Mpl antibody. The expert advice and assistance of L. Thorén in cell sorting, S. Walsh in cell isolation, and A. Hultquist and R. Månsson in single-cell RT-PCR analysis are highly appreciated.
These studies were generously supported by grants from the EU project LHSB-CT-2003-503005 (EuroStemCell), ALF (Government Public Health Grant), the Swedish Research Council, the Swedish Foundation for Strategic Research (SSF), and The Göran Gustafsson's Foundation. The Lund Stem Cell Center is supported by a Center of Excellence grant from the SSF. S. Kharazi is supported by the Iranian Ministry of Health.
Authorship
Contribution: S.E.W.J. designed and supervised the research, analyzed data, and wrote the manuscript; S.L., K.A., S.K., and N.B.-V. performed most of the work and analyses of data and wrote the manuscript; C.B. performed the initiating experiments to the current work; C.J. contributed with expertise in FACS-related work; Z.M. performed the FACS; L.W. contributed with technical expertise in the animal work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sten Eirik W. Jacobsen, Lund University, BMC B10, Klinikgatan 26, 221, 84 Lund, Sweden; e-mail: sten.jacobsen@med.lu.se or sten.jacobsen@imm.ox.ac.uk.
References
Author notes
S.L., K.A., S.K., and N.B.-V. contributed equally to this report.


![Figure 4. Increasing lymphoid transcriptional priming in LMPPs expressing Rag1GFP. (A) Coexpression pattern of Rag1GFP and Flt3 on Kit-enriched LSK BM cells. Gates denote the sorting strategies used to purify LSKFlt3hiRag1GFP− (Rag1GFP−), LSKFlt3hiRag1GFPlo (Rag1GFPlo), LSKFlt3hiRag1GFPint (Rag1GFPint), LSKFlt3hiRag1GFPhi (Rag1GFPhi), and LSKFlt3− (Flt3−) cells. Percentages indicate mean frequencies of the sorted populations of total LSK cells from 4 experiments. Right panels show typical purity analysis. (B) Expression pattern of Rag1 transcripts in single cells from sorted BM LSKFlt3−, LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint and LSKFlt3hiRag1GFPhi cells. Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total. (C) Expression pattern of individual lymphoid genes (Il7r, sterile IgH transcript [IgH], and Rag1) in single cells from BM LSKFlt3hiRag1GFP−, LSKFlt3hiRag1GFPlo, LSKFlt3hiRag1GFPint, and LSKFlt3hiRag1GFPhi cells. To the right is shown the frequency of cells in each population coexpressing 2 or 3 of the investigated lymphoid genes (Multi-Ly). Mean plus or minus SEM values from 2 experiments with 88 single cells of each cell population investigated in each experiment, except for LSKFlt3− cells, in which 88 cells were evaluated in total.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/7/10.1182_blood-2007-08-108324/5/m_zh80070817450004.jpeg?Expires=1767742475&Signature=ogb3DsV4JZvHZ2HiNsXEf~eKtXD8rwmGicgEWJMea19ay5UJ7fjlwhuhmCFutivUBGHCLA~raRa-wuq8ahgC5ugy75tpfJNL6SZ7B5ri-4oSeoqZZRIplw2IwSDLr9FCT8SBIs6Rlg3IUYPxjZr2n3g94Qb7WPPmaKDCSf131MjiwzjaLOYUKQLjKvA9feMxgqPie8W82w5FKAv6F5pthuTAVjEpCVo38dQbtWjqMgFXa6QKbRMUIQvqWy1RHEEfyExsFUK3M8Xib24-oK3YO01TKCXt0DNkb8kKZ0BJ3qK3R28L22iIHSH1gWM2Mr6vmoxVw66UbaDyVNa3i9S33w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal