Abstract
Vascular tissue engineering requires a ready source of endothelial cells and perivascular cells. Here, we evaluated human bone marrow–derived mesenchymal stem cells (hMSCs) for use as vascular progenitor cells in tissue engineering and regenerative medicine. hMSCs expressed a panel of smooth muscle markers in vitro including the cardiac/smooth muscle–specific transcription coactivator, myocardin. Cell-cell contact between endothelial cells and hMSCs up-regulated the transcription of myocardin. hMSCs efficiently stabilized nascent blood vessels in vivo by functioning as perivascular precursor cells. The engineered blood vessels derived from human umbilical cord vein endothelial cells and hMSCs remained stable and functional for more than 130 days in vivo. On the other hand, we could not detect differentiation of hMSCs to endothelial cells in vitro, and hMSCs by themselves could not form conduit for blood flow in vivo. Similar to normal perivascular cells, hMSC-derived perivascular cells contracted in response to endothelin-1 in vivo. In conclusion, hMSCs are perivascular cell precursors and may serve as an attractive source of cells for use in vascular tissue engineering and for the study of perivascular cell differentiation.
Introduction
Human bone marrow–derived mesenchymal stem cells (hMSCs) are intensively investigated for treating patients with ischemic heart disease and postmyocardial infarction. It has been hypothesized that MSCs, when injected into a failing heart, can differentiate into mature cardiomyocytes or secrete soluble factors that promote angiogenesis and enhance cardiomyocyte survival and functions.1 Based on promising results from preclinical animal models, a number of clinical trials have been initiated to test the clinical efficacy of hMSC injection. The early results of the clinical trial have been mixed, and the exact mechanism of the cardiac improvement remains unresolved.2 To maximize the therapeutic potential of hMSCs, a detailed analysis of the cellular functions of the injected hMSCs is needed
Data from animal studies have suggested that some of the injected hMSCs can differentiate into endothelial cells or smooth muscle cells. Some of the MSCs were found to express endothelial markers including CD31 and VWF, while others expressed smooth muscle markers including α-smooth muscle actin and desmin.3,4 These data suggest that hMSCs may function as vascular progenitors. These results raise several fundamental questions. First, can hMSCs differentiate into endothelial cells and smooth muscle cells in vitro? Second, if MSCs differentiate into endothelial cells and/or smooth muscle cells, can these cells be used to form functional blood vessels in vivo? Third, do the MSC-derived blood vessels respond appropriately to physiological stimulation? These are important outstanding questions that need to be addressed to maximize the therapeutic potential of MSCs. To address these questions, we used a tissue-engineered blood vessel model5-7 and monitored the cellular fate of implanted MSCs and their vasculogenic potential by intravital microscopy.
Methods
Cell culture
hMSCs were purchased from Cambrex and maintained in Mesenchymal Stem Cell Growth Medium (Cambrex, East Rutherford, NJ). MSCs were seeded at a density of 6000 cells/cm2 and the cells were passaged at 90% confluent. All experiments with wild-type hMSCs and EGFP-hMSCs were performed below 8 and 16 cell passages, respectively. Human umbilical cord vein endothelial cells (HUVECs) were obtained from Center of Excellence in Vascular Biology, Brigham and Women's Hospital (Boston, MA) and maintained in Endothelial Growth Medium (Cambrex). MS1, a mouse pancreatic endothelial cell line was obtained from American Type Culture Collection (ATCC; Rockville, MD). Human aortic vascular smooth muscle cells (CRL-1999) and human dermal fibroblasts (CRL-2575) were obtained from ATCC. 293ET packaging cells were a kind gift from Dr Brian Seed (Massachusetts General Hospital, Boston, MA). All cells were maintained at 37°C in a humidified 5% CO2 incubator.
Retrovirus packaging and transduction
The retrovirus vector for transducing enhanced green fluorescent protein (EGFP), PBMN-I-EGFP, was kindly provided by Dr Gary Nolan (Stanford, CA). For retrovirus packaging, the plasmids of PBMN-I-EGFP, Gag-pol, and VSVG (15 μg, 7 μg, and 5 μg, respectively) were mixed and cotransfected into 293ET cells with lipofectamine 2000 (Invitrogen, Carlsbad, CA) per the manufacturer's protocol. After overnight incubation, the 293ET cells were washed with PBS and replaced with fresh media. The next day, the supernatant-containing retrovirus was collected and fresh media were added; this step was repeated 3 more times. After the supernatant was collected, it was passaged through a 0.45-μm filter (Whatman, Middlesex, United Kingdom) and was either used immediately for infection or kept at −80°C.
Coculture of mouse endothelial cells and human mesenchymal stem cells
hMSCs were cultured either alone or cocultured with MS1 endothelial cells in Dulbecco modified Eagle medium (DMEM) with 2% fetal bovine serum (FBS) for 2 days. Briefly, 105 hMSCs in the hMSC-only group or 105 hMSCs and 105 MS1 endothelial cells in the coculture group were added to a 6-well plate. The plate was agitated to make sure the cells were well mixed and the plate was then placed in a 37°C incubator and cultured for 48 hours. For transwell culture, 105 hMSCs were placed in a 6-well plate and 105 MS1 endothelial cells were added onto a 0.4-μm pore membrane insert (Corning, Corning, NY) that was placed above the 6-well plate.
Reverse transcription–polymerase chain reaction (RT-PCR) and quantitative real-time RT-PCR
Total RNA was extracted with the Qiagen RNeasy mini kit (Qiagen, Valencia, CA), and cDNA was synthesized from RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems, University Park, IL); all procedures were performed according to the manufacturer's protocol. PCR for detection of endothelial and smooth muscle transcripts was performed with 2.5 × Mastermix (Eppendorf, Hamburg, Germany). PCR primer sequence and PCR condition can be found in the supplement. Quantitative RT-PCRs were performed on the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems). All experiments were performed in triplicate. Final quantification of each cDNA sample was relative to human GAPDH following the manufacturer's instructions (Applied Biosystems). Species-specific primers were designed using Primer Express or Primer3 (Whitehead Institute, Cambridge, MA).
Transwell migration assay
Cell migration was assessed using Falcon HTS FluoroBlok 24-well inserts (BD Biosciences, San Jose, CA) with 8-μm pores. EGFP-hMSC cells (2 × 104) suspended in 250 μL DMEM (Invitrogen) with 2% FBS were placed inside each insert, while 5 × 104 per well HUVECs suspended in 800 μL EGM (Cambrex) were plated on a 24-well plate. Eight hours later, all cell culture media were changed to DMEM with 2% FBS, and then the inserts were placed in the respective wells. To examine the effect of PDGFR inhibition on cell migration, 2, 5, or 10 μM imatinib mesylate (Gleevec; Novartis, Basel, Switzerland) was added to the medium. The number of migrated cells was quantified 24 hours later as previously described.8 Briefly, the image was threshold and the area covered by the migrated cells was quantified in ImageJ (version 1.38, http://rsb.info.nih.gov/ij/, National Institutes of Health).
Mesenchymal stem cell differentiation assay
Mesenchymal stem cells were cultured in chamber slides with adipogenic or osteogenic differentiating medium. Adipogenic medium consisted of DMEM, 10% FBS, 0.5 mM isobutylmethylxanthine, 10 μM insulin, 200 μM indomethacin, and 1% penicillin/streptomycin. Osteogenic medium consisted of DMEM, 10% FBS, 0.1 μM dexamethasone, 50 μM ascorbate-2-phosphate, 10 mM b-glycerophosphate, and 1% penicillin/streptomycin. After 30 to 40 days in culture, the cells were stained with oil red O and alizarin red or alkaline phosphatase for adipogenesis and osteogenesis, respectively.
Immunofluorescence
Mesenchymal stem cells were grown on chamber slides until confluent. The cells were washed with phosphate-buffered saline (PBS) and then fixed by incubating them in ice-cold methanol for 30 minutes. The chamber slides were then washed with PBS to remove all traces of methanol. To block nonspecific binding, the chamber slides were incubated in PBS with 3% bovine serum albumin (BSA) for 1 hour followed by overnight incubation with primary antibodies at 4°C. The following primary antibodies were used at 1:200 dilution: α-smooth muscle actin (Dako, Glostrup, Denmark), sm22α (Abcam, Cambridge, MA), desmin (Dako), and NG2 (Chemicon, Temecula, CA). The next day, the chamber slides were washed with PBS and incubated with appropriate Cy3-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA). The chamber slides were then washed and mounted in Vectashield containing DAPI (Vector Laboratories, Burlingame, CA). For whole-mount staining, collagen gels were extracted from cranial windows and incubated in 4% paraformaldehyde for 3 hours and then washed with PBS. The gels were initially blocked for endogenous mouse IgG with Vector Mouse on Mouse Kit (Vector Laboratories) for 2 hours following the manufacturer's protocol. The gels were then incubated overnight at 4°C in blocking solution containing 3% BSA, 5% normal rabbit serum, and 0.3% Triton X100 in PBS. The next day, primary antibodies (α-smooth muscle actin [Dako], Sm22α [Abcam], and desmin [Dako]) were diluted in blocking solution at a dilution of 1:400 and were added to the gel. After overnight incubation with primary antibodies at 4°C, the gels were washed extensively with PBS to remove unbound antibodies. The gels were then incubated with Cy5-conjugated secondary antibodies at 1:400 dilution for 3 hours at room temperature. After washing with PBS, the gels were mounted in Vectashield and imaged with a confocal microscope (model no. BX61WI; Olympus, Tokyo, Japan) using 20×/0.95 NA water objective. Image was acquired with Fluoview FV500 (Olympus).
Tissue-engineered blood vessel model
The engineered blood vessel model was prepared as previously described.5-7 Briefly, 106 HUVECs and 2 × 105 hMSCs; 106 HUVECs and 2 × 105 10T1/2 cells; or 106 HUVECs alone were suspended in 1 mL type 1 collagen-fibronectin solution. The next day, a skin puncher (4-mm diameter) was used to create a circular piece from the collagen gel. The circular piece of collagen gel was then implanted into a cranial window of a severe combined immunodeficient (SCID) mouse. The in vivo fate of the fluorescent protein-labeled endothelial cells and hMSCs was tracked by intravital imaging with multiphoton laser scanning microscopy at various time points9 (modified Axioskop 50 microscope [Carl Zeiss, Heidelberg, Germany] and MaiTai Ti:Sapphire laser [Spectra-Physics, Mountain View, CA]). Image was taken with 20×/0.50 NA water objective. Functional blood vessels were revealed by intravenous injection of 100 μL tetramethylrhodamine-conjugated dextran (2 000 000 MW at 10 mg/mL) via the tail vein. The same regions in each gel were tracked at different time points for consistency. The perfused vessel length density was quantified by manual tracing of perfused blood vessels lined by EGFP-HUVECs with a custom-written MATLAB macro (Mathworks, Natick, MA).
Arteriolar contractility assay
Arteriolar contractility was assessed by vasoactive response to endothelin-1 (ET-1). After careful removal of the cover glass, the cranial window was superfused with warm PBS. For vessel contrast enhancement, 100 μL 1% tetramethylrhodamine-dextran (MW 2 million) was injected intravenously. The engineered vessels were monitored by multiphoton laser scanning microscopy with a 20× water-immersion objective. After baseline measurements of vessel diameter, the cranial window was superfused with 100 nM ET-1 in PBS. Then, the same region of vessels was imaged every 5 minutes over a 20-minute period. The diameter of vessels was quantified manually with ImageJ.
Statistical analysis
All data were analyzed by ANOVA with the Fisher posthoc test. All data are reported as a mean with standard error. Statistical significance was set at P less than .05.
Results
Transcriptional profile for endothelial and smooth muscle markers
To investigate whether hMSCs exhibit endothelial phenotypes, we first checked by PCR for transcripts of endothelial markers, PECAM1 (CD31) and VE-cadherin. These endothelial-selective markers were not detectable in hMSCs, either at baseline or when cocultured with a mouse endothelial cell line (Figure 1A). Next, we assessed the transcription profile of a panel of smooth muscle markers in hMSCs and compared it with aortic smooth muscle cells and dermal fibroblasts (Figure 1B). All cell lines commonly transcribed mRNA of α-smooth muscle actin, sm22α, desmin, calponin, and smoothelin. Immunochemistry confirmed the expression of α-smooth muscle actin, SM22α, desmin, and NG2 in hMSCs (Figure 1C-F). Myocardin was recently discovered to be a cardiac/smooth muscle–specific coactivator for serum response factor in the regulation of a number of cardiac and smooth muscle genes.10 As expected, myocardin was transcribed in aortic smooth muscle cells. More interestingly, myocardin was transcribed by mesenchymal stem cells, but not by dermal fibroblasts. These data suggest that the expression of myocardin might potentially be a more specific marker for smooth muscle cell differentiation than the commonly used markers, such as α-smooth muscle actin and SM22α.
Transcriptional profile of endothelial and smooth muscle markers in hMSCs. (A) RT-PCR analysis of endothelial markers (VE-cadherin and PECAM1) in hMSCs at baseline and in hMSCs cocultured for 3 days with MS1 mouse endothelial cell line. (B) RT-PCR analysis of smooth muscle markers' mRNA in human mesenchymal stem cells, human aortic vascular smooth muscle cells (CRL-1999), and human dermal fibroblasts (CRL-2575). Water was used as negative control. (C-F) Protein expression of smooth muscle markers in hMSCs was confirmed by immunohistochemistry (α-smooth muscle actin [C], SM22α [D], desmin [E], and NG2 [F], yellow; DAPI, blue). Scale bar represents 50 μm.
Transcriptional profile of endothelial and smooth muscle markers in hMSCs. (A) RT-PCR analysis of endothelial markers (VE-cadherin and PECAM1) in hMSCs at baseline and in hMSCs cocultured for 3 days with MS1 mouse endothelial cell line. (B) RT-PCR analysis of smooth muscle markers' mRNA in human mesenchymal stem cells, human aortic vascular smooth muscle cells (CRL-1999), and human dermal fibroblasts (CRL-2575). Water was used as negative control. (C-F) Protein expression of smooth muscle markers in hMSCs was confirmed by immunohistochemistry (α-smooth muscle actin [C], SM22α [D], desmin [E], and NG2 [F], yellow; DAPI, blue). Scale bar represents 50 μm.
Endothelial cells up-regulate myocardin in hMSCs through cell-cell contact
Previous studies have suggested that endothelial cells can induce the differentiation of smooth muscle cells/pericytes.11,12 To determine whether endothelial cells could regulate the differentiation of hMSCs toward smooth muscle lineage, we cocultured them with a mouse endothelial cell line, MS1. Using human-specific primers, real-time RT-PCR was performed to examine the transcription of myocardin in hMSCs. Coculturing hMSCs with MS1 resulted in a statistically significant up-regulation of myocardin (Figure 2A). TGFβ1 has been suggested to be involved in the differentiation of smooth muscle cells/pericytes.11 We investigated whether addition of TGFβ1 could induce transcription of myocardin in hMSCs. TGFβ1 stimulation only modestly induced the transcription of myocardin, and the magnitude of the induction was much less compared with the coculture group (Figure 2A). To gain better understanding of the inductive signals, we checked whether the signals were mediated by soluble factors or cell-cell contact. To this end, we cultured hMSCs and endothelial cells in separate layers of a transwell culture. This allowed for the diffusion of soluble factors while preventing direct cell-cell contact between hMSCs and endothelial cells. The transwell culture did not induce myocardin transcription, suggesting that the inductive signal was provided by cell-cell contact (Figure 2A).
Induction of myocardin transcription and migration in hMSCs by endothelial cells. (A) Quantitative real-time PCR was performed to assess the induction of myocardin in hMSCs by various conditions after 2 days (hMSCs alone, hMSCs cultured with MS1 endothelial cells with contact, hMSCs cultured with MS1 without contact by transwell culture, and hMSCs stimulated with TGFβ1 [10 ng/mL]). Values expressed as fold increase above hMSC-alone levels and normalized by GAPDH. Representative data of 3 separate experiments. (B) Transwell migration assay was performed to assess endothelial cell–induced hMSC migration. Imatinib mesylate at 0.2 μM, 2 μM, and 10 μM was added to test for inhibition of hMSC migration. #P < .001 (medium vs HUVECs); *P < .001 (2 μM imatinib mesylate vs HUVECs); **P < .001 (10 μM imatinib mesylate vs HUVECs).
Induction of myocardin transcription and migration in hMSCs by endothelial cells. (A) Quantitative real-time PCR was performed to assess the induction of myocardin in hMSCs by various conditions after 2 days (hMSCs alone, hMSCs cultured with MS1 endothelial cells with contact, hMSCs cultured with MS1 without contact by transwell culture, and hMSCs stimulated with TGFβ1 [10 ng/mL]). Values expressed as fold increase above hMSC-alone levels and normalized by GAPDH. Representative data of 3 separate experiments. (B) Transwell migration assay was performed to assess endothelial cell–induced hMSC migration. Imatinib mesylate at 0.2 μM, 2 μM, and 10 μM was added to test for inhibition of hMSC migration. #P < .001 (medium vs HUVECs); *P < .001 (2 μM imatinib mesylate vs HUVECs); **P < .001 (10 μM imatinib mesylate vs HUVECs).
hMSCs migrate toward endothelial cells in response to PDGF
A necessary step in the process of vessel maturation is the recruitment of perivascular cells to stabilize an immature vessel. Therefore, we investigated whether HUVECs could induce hMSC migration in a transwell migration assay. In comparison with control (medium alone), HUVECs significantly increased the number of hMSCs that migrated across a transwell membrane (Figure 2B). Previous data suggested that PDGFR signaling mediated the process of perivascular cell migration and recruitment.11,13 To test whether PDGFR signaling is involved in hMSC migration, we added imatinib mesylate, a potent inhibitor of PDGFR, to the cell culture medium. Imatinib mesylate blocked hMSC migration and PDGF-Rβ phosphorylation in a dose-dependent manner (Figure 2B; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Mesenchymal stem cells functionally stabilize engineered blood vessels
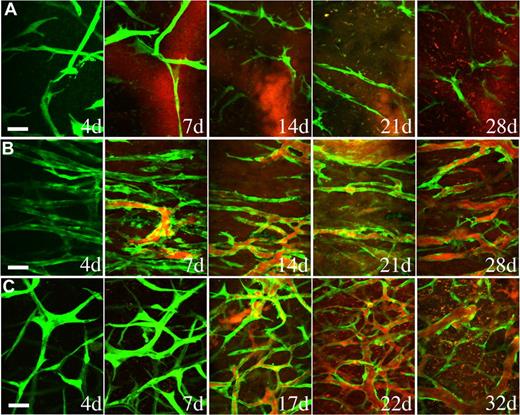
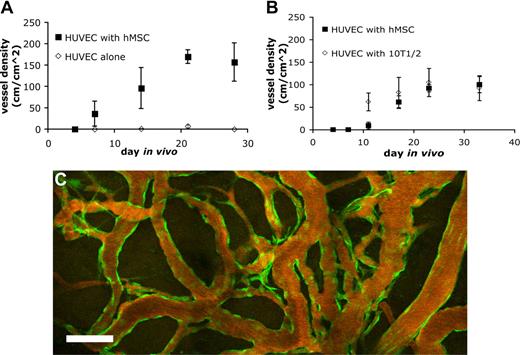
Endothelial cells implanted in vivo could form functional vessels, but such vessels quickly regressed unless the endothelial cells were ectopically transduced to express bcl2, an antiapoptotic gene.14 We previously demonstrated that coimplantation of endothelial cells with 10T1/2, a murine embryonic fibroblast cell line that differentiates into perivascular cells in vivo, could lead to the formation of a stable and functional microvascular network without the need for genetic modification of the endothelial cells.5 We investigated whether hMSCs could functionally replace 10T1/2 to stabilize engineered blood vessels. EGFP-labeled HUVECs were implanted alone or coimplanted with hMSCs or 10T1/2 cells in a collagen/fibronectin matrix in a mouse cranial window. The implanted endothelial cells were observed with multiphoton laser scanning microscopy to assess their ability to form functional blood vessels. When EGFP-labeled HUVECs were implanted alone, some vessels formed but they quickly regressed, similar to previous observations made by us and others (Figures 3A,4A).5,14 When EGFP-labeled HUVECs were coimplanted with hMSCs, the endothelial cells assembled into functional blood vessels that anastomosed to the host vascular network (Figures 3B,4A). The engineered blood vessel densities derived from HUVEC-hMSC and HUVEC-10T1/2 coimplantations were similar (Figures 3B,C,4B). Furthermore, the HUVEC-hMSC–engineered vessels were stable and remained functional for more than 130 days in vivo (Figure 4C).
hMSCs stabilized engineered blood vessels in vivo. EGFP-labeled HUVECs were either implanted alone (A) or coimplanted with hMSCs (B) or 10T1/2 cells (C) in a collagen gel onto cranial windows in SCID mice. Images were taken at periodic time points with multiphoton laser scanning microscope to monitor the in vivo dynamics of vascularization by the implanted endothelial cells. Green indicates HUVECs expressing EGFP; red, functional blood vessels contrast-enhanced by rhodamine-dextran. Scale bar represents 50 μm.
hMSCs stabilized engineered blood vessels in vivo. EGFP-labeled HUVECs were either implanted alone (A) or coimplanted with hMSCs (B) or 10T1/2 cells (C) in a collagen gel onto cranial windows in SCID mice. Images were taken at periodic time points with multiphoton laser scanning microscope to monitor the in vivo dynamics of vascularization by the implanted endothelial cells. Green indicates HUVECs expressing EGFP; red, functional blood vessels contrast-enhanced by rhodamine-dextran. Scale bar represents 50 μm.
Quantification of functional engineered blood vessel density. Quantification of functional vessel density in SCID mice implanted with HUVECs alone or HUVECs with hMSCs (A) and HUVECs with hMSCs or 10T1/2 (B) (results are mean ± SEM, n = 3 in panel A and n = 5 in panel B; different batches of HUVECs were used in panels A and B, resulting in variation in vessel density between the 2 experiments). (C) The hMSC-stabilized vascular network remained functional for more than 130 days in vivo. Scale bar represents 100 μm.
Quantification of functional engineered blood vessel density. Quantification of functional vessel density in SCID mice implanted with HUVECs alone or HUVECs with hMSCs (A) and HUVECs with hMSCs or 10T1/2 (B) (results are mean ± SEM, n = 3 in panel A and n = 5 in panel B; different batches of HUVECs were used in panels A and B, resulting in variation in vessel density between the 2 experiments). (C) The hMSC-stabilized vascular network remained functional for more than 130 days in vivo. Scale bar represents 100 μm.
Mesenchymal stem cells differentiate into perivascular cells in vivo and respond to endothelin-1 stimulation
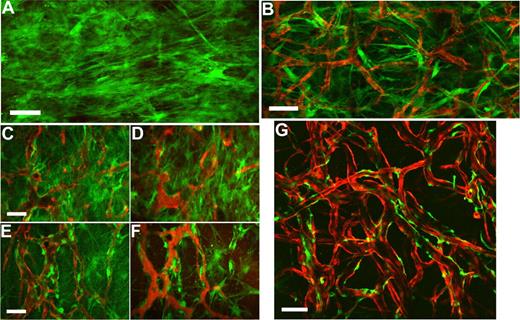
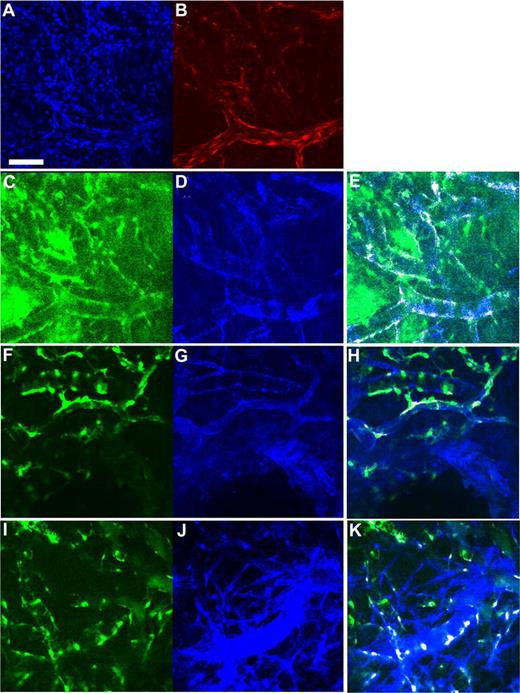
To examine the spatial orientation of hMSCs in relation to the engineered vessels, hMSCs and HUVECs were transduced with retrovirus to express EGFP and DsRed-Express (DsRed), respectively. Retroviral transduction of EGFP did not affect the multipotent nature of hMSCs as assessed by their ability to differentiate into adipocytes and osteocytes (Figures S2,S3). We then investigated whether EGFP-hMSCs implanted alone could self-assemble into functional vessels. We were not able to detect lumen formation or blood flow in EGFP-hMSCs derived cells if implanted alone even after 25 days in vivo (Figure 5A). In contrast, in mice coimplanted with DsRed-HUVECs and EGFP-hMSCs, EGFP-hMSCs were observed to elongate into thin slit structures and coalesced around the HUVEC-derived vessels (Figure 5B-F). The number of interstitial EGFP-hMSCs decreased with time, and most of them were associated with blood vessels by day 83 in vivo (Figure 5G). These EGFP-hMSC–derived cells behaved like perivascular cells with a single layer of cells wrapping around the endothelial tube. Furthermore, EGFP-hMSC–derived pericyte-like cells were positively stained for α-smooth muscle actin, sm22α, and desmin (Figure 6A-H).
Intravital monitoring of EGFP-hMSCs in a tissue-engineered vessel model. Fibronectin-collagen constructs with EGFP-hMSCs alone (A) or EGFP-hMSCs with DsRed-HUVECs (B-G) were implanted into cranial windows of SCID mice. Images were taken at different time points with multiphoton laser scanning microscopy (MPLSM) (B, day 7; C,D, day 19; E,F, day 31; G, day 83). Green indicates human mesenchymal stem cells expressing EGFP (A-G); red, HUVECs expressing DsRed-express fluorescent protein (B-E,G); and red, functional blood vessels contrast-enhanced by rhodamine-dextran (D,F). Scale bars represent (A,B,G) 100 μm; (C-F) 50 μm.
Intravital monitoring of EGFP-hMSCs in a tissue-engineered vessel model. Fibronectin-collagen constructs with EGFP-hMSCs alone (A) or EGFP-hMSCs with DsRed-HUVECs (B-G) were implanted into cranial windows of SCID mice. Images were taken at different time points with multiphoton laser scanning microscopy (MPLSM) (B, day 7; C,D, day 19; E,F, day 31; G, day 83). Green indicates human mesenchymal stem cells expressing EGFP (A-G); red, HUVECs expressing DsRed-express fluorescent protein (B-E,G); and red, functional blood vessels contrast-enhanced by rhodamine-dextran (D,F). Scale bars represent (A,B,G) 100 μm; (C-F) 50 μm.
Expression of smooth muscle cell markers in hMSC-derived cells incorporated into the tissue-engineered vessels. Whole-mount staining was performed for the extracted tissue-engineered vessel constructs. (A-H) Confocal microscopy images. (A) Blue indicates DAPI staining; (B) red, DsRed-HUVECs; (C,F,I) green, EGFP-hMSCs; (D) blue, α-smooth muscle actin staining; (G) blue, SM22α staining; and (J) blue, desmin staining. Region of colocalized staining of EGFP and smooth muscle markers is highlighted as white (E,H,K). Panels A-E, F-H, and I,J are the same location/construct. Scale bar represents 100 μm.
Expression of smooth muscle cell markers in hMSC-derived cells incorporated into the tissue-engineered vessels. Whole-mount staining was performed for the extracted tissue-engineered vessel constructs. (A-H) Confocal microscopy images. (A) Blue indicates DAPI staining; (B) red, DsRed-HUVECs; (C,F,I) green, EGFP-hMSCs; (D) blue, α-smooth muscle actin staining; (G) blue, SM22α staining; and (J) blue, desmin staining. Region of colocalized staining of EGFP and smooth muscle markers is highlighted as white (E,H,K). Panels A-E, F-H, and I,J are the same location/construct. Scale bar represents 100 μm.
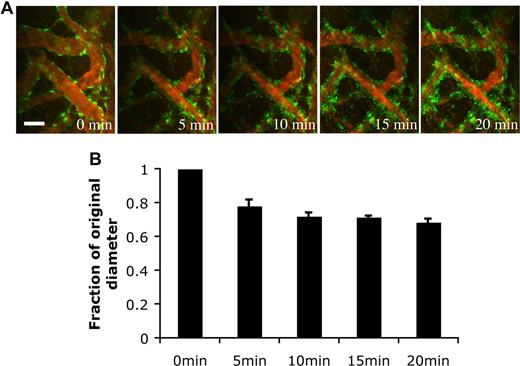
Pericytes have been shown to possess contractile abilities that directly regulate the blood-brain barrier and local tissue perfusion.15 To determine the function of the EGFP-hMSC–derived perivascular cells, we stimulated the blood vessels with endothelin-1, a potent contractile agent. The EGFP-hMSC–covered blood vessels constricted upon endothelin-1 stimulation (Figure 7A). The blood vessels narrowed within 5-minute exposure to endothelin-1 and remained constricted at 20 minutes (Figure 7A,B). These experiments support the notion that EGFP-hMSCs have the capacity to differentiate into perivascular cells that can properly respond to physiological stimulation.
Endothelin-1 stimulated vasoconstriction of the engineered blood vessels. HUVEC/hMSC-derived engineered blood vessels were superfused with 100 nM endothelin-1 to stimulate vasoconstriction in vivo. (A) Representative multiphoton microscopy images during endothelin-1 superfusion. Green indicates EGFP-hMSCs; red, functional blood vessels contrast-enhanced by rhodamine-dextran. Scale bar represents 50 μm. (B) The blood vessel diameter was quantified over a 20-minute period (n = 4). The value was expressed as a fraction of the original diameter. Scale bar represents 50 μm.
Endothelin-1 stimulated vasoconstriction of the engineered blood vessels. HUVEC/hMSC-derived engineered blood vessels were superfused with 100 nM endothelin-1 to stimulate vasoconstriction in vivo. (A) Representative multiphoton microscopy images during endothelin-1 superfusion. Green indicates EGFP-hMSCs; red, functional blood vessels contrast-enhanced by rhodamine-dextran. Scale bar represents 50 μm. (B) The blood vessel diameter was quantified over a 20-minute period (n = 4). The value was expressed as a fraction of the original diameter. Scale bar represents 50 μm.
Discussion
Cellular therapy with hMSCs has great potential for use in regenerative medicine and is currently in clinical development. MSCs are being investigated in treatment of bone and cartilage defects, and injured myocardium after acute infarction.16 The ability of MSCs to differentiate into several lineages of connective tissue, including osteocyte, chondrocyte, and adipocyte, is well documented.17 Recent evidence has suggested that MSCs may also differentiate into endothelial and vascular smooth muscle cells.18 In this report, we set out to clarify the cellular fate of implanted mesenchymal stem cells in participating in vasculogenesis in vivo. Our study showed that hMSCs did not spontaneously differentiate into endothelial tubes that were capable of carrying blood flow in vivo. On the other hand, hMSCs differentiated into and functioned as perivascular cells in vivo when coimplanted with endothelial cells. The hMSC-derived perivascular cells were able to stabilize nascent blood vessels, and the engineered blood vessels remained functional for more than 130 days in vivo. Furthermore, the hMSC-derived perivascular cells exhibited proper physiological function by constricting in response to stimulation by endothelin-1, an endogenous vasoconstrictive peptide.
It has long been known that endothelial cells and hematopoietic cells shared a common origin during development, but whether a common progenitor exists for endothelial cells and smooth muscle cells has been unclear.19 The first hint of the existence of vascular progenitor cells came from an in vitro differentiation study with mouse embryonic stem (ES) cells.20 Flk1-positive cells were generated and they were shown to have the potential to differentiate into either endothelial cells or smooth muscle cells depending on the growth factors in the culture medium. More recent evidence suggested such ES-derived cells might even be capable of differentiating into cardiomyocytes.21,22 Besides the work with ES cells, other data also hinted at the close link between endothelial cells and smooth muscle cells. Under certain in vitro culturing conditions, endothelial cells and smooth muscle cells could take on one another's phenotypic markers, but it remains unclear whether this was due to an artifact of in vitro culture or a process of transdifferentiation.23 Similarly, hMSCs have been induced to acquire endothelial and smooth muscle phenotype in vitro.18 An unresolved question is whether the hMSC-derived endothelial cells and smooth muscle cells can function properly with the acquired phenotype in vivo.
To systematically address these outstanding questions, we first began by evaluating the in vitro transcription of endothelial and smooth muscle markers. We were not able to detect endothelial markers in hMSCs at baseline and when cultured with a mouse endothelial cell line. The lack of endothelial marker expression suggested hMSCs had not differentiated into endothelial cells with the chosen culturing condition. We could not discount the possibility that we had not identified the optimal differentiation protocol for endothelial cells. However, one difference between our study and previous studies was that we used PCR to detect endothelial markers, a more sensitive and selective method compared with the immunofluorescence-based methods that were used in other studies.18
We found that hMSCs expressed a number of smooth muscle markers. When cocultured with endothelial cells, the cardiac- and smooth muscle–specific transcription coactivator myocardin was up-regulated more than 14-fold. The induction of myocardin was dependent on direct heterotypic cell-cell contact. Since TGF-β1 has been shown to be involved in smooth muscle differentiation and the activation of latent TGFβ1 requires cellular contact, we stimulated hMSCs with recombinant TGF-β1.11,24 The magnitude of myocardin induction induced by the given dose(s) of TGF-β1 is only a small fraction of the myocardin up-regulation by endothelial cells. Interestingly, the extent of myocardin induction by TGF-β1 in hMSCs was similar to the results obtained with multipotent adult progenitor cells.25 These data suggest other cell-contact mediators might be involved in smooth muscle cell differentiation.26
Our data suggest hMSCs efficiently differentiate into perivascular cells when coimplanted with endothelial cells in a tissue-engineered vessel model in vivo. The hMSC-engineered vessels may be an ideal model to study perivascular cell differentiation. While we had previously shown that 10T1/2 cells could function as perivascular cells in stabilizing engineered blood vessels, there are obvious drawbacks in using 10T1/2 cells in study of vessel maturation. 10T1/2 cells were shown to lack the expression of the transcription coactivator myocardin even when stimulated by TGFβ1.27 It was speculated that TGFβ1 stimulation of 10T1/2 cells might represent a myofibroblast response instead of true differentiation into vascular smooth muscle lineage.28 In contrast, hMSCs expressed a number of smooth muscle markers including myocardin. These cells could be easily manipulated in vitro, and unlike smooth muscle cells extracted from discarded tissue, hMSCs are in a relative primitive state of differentiation. Compared with embryonic stem cells, hMSCs do not carry the potential risks of teratoma formation and difficulty in obtaining a pure population of lineage-specific cells. hMSCs could potentially be genetically engineered for studying the vascular effects of embryonic lethal genes.
A previous study has investigated tissue engineered bone by coimplantation of endothelial cells and hMSCs.29 It was thought that secretion of BMP4 from endothelial cells stimulated the differentiation of hMSCs into osteocytes. In our study, we were not able to detect bone or cartilage formation in vivo. Most of the surviving hMSCs were associated with blood vessels, suggesting that heterotypic interaction between hMSCs and endothelial cells provided a survival advantage for both endothelial cells and hMSC-derived perivascular cells. One potential reason for the lack of bone or cartilage differentiation is that EGFP transduction affected the multipotential of hMSCs. The EGFP-labeled hMSCs were able to differentiate into both osteocytes and adipocytes in vitro, but it is unclear how EGFP transduction might affect cell differentiation in vivo. Another reason for the lack of bone formation could be attributed to the mechanical property of the tissue-engineered construct. Depending on the stiffness of the matrix, hMSCs could spontaneously differentiate into different cell lineages.30 In our study, we used a matrix (type I collagen/fibronectin) at a concentration with low stiffness, and this might have prevented osteocyte differentiation. This finding is especially important in the setting of revascularization therapy with hMSCs since ectopic bone formation has potentially untoward effects.
In conclusion, we demonstrated that hMSCs specifically differentiated into perivascular cells that could stabilize nascent blood vessels and responded appropriately to vasoactive stimuli. hMSCs provide an attractive cellular platform for vascular engineering and for studying perivascular cell differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially supported by an American Heart Association Predoctoral Fellowship (P.A.) and National Institutes of Health grants P01CA80124 (R.K.J., D.F.), R01CA96915 (D.F.), R01CA115767 (R.K.J.), R01CA85140 (R.K.J.), and R01CA126642 (R.K.J.).
National Institutes of Health
Authorship
Contribution: P.A. designed, performed, and analyzed research, and wrote the paper; J.T. performed and analyzed research; D.F. analyzed research and wrote the paper; R.K.J. designed and analyzed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rakesh K. Jain, Department of Radiation Oncology, Massachusetts General Hospital, 100 Blossom St, Cox-7, Boston, MA 02114; e-mail: jain@steele.mgh.harvard.edu.

![Figure 1. Transcriptional profile of endothelial and smooth muscle markers in hMSCs. (A) RT-PCR analysis of endothelial markers (VE-cadherin and PECAM1) in hMSCs at baseline and in hMSCs cocultured for 3 days with MS1 mouse endothelial cell line. (B) RT-PCR analysis of smooth muscle markers' mRNA in human mesenchymal stem cells, human aortic vascular smooth muscle cells (CRL-1999), and human dermal fibroblasts (CRL-2575). Water was used as negative control. (C-F) Protein expression of smooth muscle markers in hMSCs was confirmed by immunohistochemistry (α-smooth muscle actin [C], SM22α [D], desmin [E], and NG2 [F], yellow; DAPI, blue). Scale bar represents 50 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/9/10.1182_blood-2007-10-118273/4/m_zh80080818120001.jpeg?Expires=1767717151&Signature=jLTTzbvZyZJEKkX0x7K9ruOYpBxppgEGlHvio5uR2ixzeCRumxJeJqAV9AJT9isQAgJ6fyk91ORAyv80XhDntUg90kNaCUQyFb4qQbG25dv387bJngZFZU50581yxAtLPU3jKokmPSBQPCbD0ISnlu2hFR5cfCR0QnKtgkTcPit7xSKBI4nPkw32EuBO4qmjRKpRbvJoBur1K5~fCkTRtdFi5NAvAxzI4pbQ5PW0KQfBp3OrpNaUh8slXcXXTaUBNDF9mrOUC7YfvdYuHwynbi997TQB7R42qaLfJuK0lMNANSTtcwOdzJqyI8T~nnjab37xjR6Vc~xZbxXhcon7Dw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Induction of myocardin transcription and migration in hMSCs by endothelial cells. (A) Quantitative real-time PCR was performed to assess the induction of myocardin in hMSCs by various conditions after 2 days (hMSCs alone, hMSCs cultured with MS1 endothelial cells with contact, hMSCs cultured with MS1 without contact by transwell culture, and hMSCs stimulated with TGFβ1 [10 ng/mL]). Values expressed as fold increase above hMSC-alone levels and normalized by GAPDH. Representative data of 3 separate experiments. (B) Transwell migration assay was performed to assess endothelial cell–induced hMSC migration. Imatinib mesylate at 0.2 μM, 2 μM, and 10 μM was added to test for inhibition of hMSC migration. #P < .001 (medium vs HUVECs); *P < .001 (2 μM imatinib mesylate vs HUVECs); **P < .001 (10 μM imatinib mesylate vs HUVECs).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/9/10.1182_blood-2007-10-118273/4/m_zh80080818120002.jpeg?Expires=1767717151&Signature=O~qxdZywrrPAIizZcrqo5HDmoiYZUfXIIpYhmhpqCUcLZNTtH7shQDiW5Cypo2nJTFjApyoX4fPQ6n3~hN~12V3LTB1fCJp77Gvo4rdZfloPY53WIMfHSbiGVoYUaJe-liX4gxn2C3E0Q~qTTGjMMuHT24XeWOQ8v~PZtNgTi5QMrh3JtE8i9UJTglvB7cOCYZvBRuMhOgNaj8ISIBwGmazXSFKw8YaN5uinIi-QSpC1z0zwAbRnHJydPwgnv5tPXdQmXZQ0lEK7BtRDHwJqChyJPpcaesr6KaN53gfVzrrNENvwwYc2P52cC4LuUo9m1MC4u2zhU~cZBwMd1f3E8g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal