Abstract
The Y chromosome encodes male-specific minor histocompatibility (H-Y) antigens that stimulate T- and B-lymphocyte responses after sex-mismatched allogeneic hematopoietic cell transplantation (HCT). A CD8+ cytotoxic T lymphocyte (CTL) clone that recognizes a novel HLA-B*2705–restricted H-Y antigen encoded by the DDX3Y gene was isolated from a male who had received a hematopoietic cell graft from his human leukocyte antigen (HLA)–identical sister. The antigenic peptide is a decamer that differs from the homologous DDX3X-encoded peptide at 4 positions. Expression of DDX3Y and of the H-Y epitope that it encodes was examined by quantitative polymerase chain reaction (PCR) and by CTL recognition assays. Expression of DDX3Y is detected in all myeloid and lymphoid leukemic cells that carry an intact Y chromosome. Moreover, the DDX3Y-encoded H-Y epitope is presented on the surface of both myeloid and lymphoid leukemic cells from male HLA-B*2705+ patients. DDX3Y-specific CTLs prevent engraftment of human acute leukemia in nonobese diabetic/severe combined immune deficient mice, demonstrating that the DDX3Y-encoded H-Y antigen is also expressed in leukemic stem cells. These results demonstrate that CD8+ T-cell responses against DDX3Y have the potential to contribute to graft-versus-leukemia (GVL) activity after female into male allogeneic HCT. This study is registered at http://clinicaltrials.gov as NCT00107354.
Introduction
Sex-mismatched allogeneic hematopoietic cell transplantation (HCT) represents a unique situation wherein T-cell responses against Y-chromosome–encoded minor histocompatibility (H-Y) antigens can potentially contribute to graft rejection,1,2 graft-versus-host disease (GVHD),3,4 and graft-versus-leukemia (GVL)5,6 activity. H-Y antigens in mice7-12 and humans1-3,13-19 are encoded by a specific class of Y-chromosome genes that have X-chromosome homologues and are expressed both in and variably outside the testis.20,21 In humans, there are 15 such genes, 6 of which—RPS4Y1,5,15 USP9Y,1,14 DDX3Y,3,4 UTY,2,18,19 TMSB4Y,16 and SMCY13,17 —have been shown to encode H-Y antigens. Although most of the human H-Y antigens identified to date are presented by major histocompatibility complex (MHC) class I molecules and recognized by CD8+ T lymphocytes, the DDX3Y3,4 and RPS4Y115 genes have been shown to encode H-Y antigens that are presented by MHC class II molecules and recognized by CD4+ T lymphocytes.
The clinical significance of H-Y–specific T-cell responses in female into male (F→M) HCT has not been completely defined. CD8+ SMCY peptide–specific T-cell clones mediated histologic changes of skin GVHD in an in vitro skin explant model.22 Studies using MHC class I HLA-A2 or HLA-B7 tetramers complexed with SMCY peptides also demonstrated that SMCY-specific CD8+ T cells were detectable in the peripheral blood of HLA-A2+ or HLA-B7+ F→M hematopoietic cell transplant recipients who developed acute GVHD.23 CD4+ T cells recognizing MHC class II–restricted peptides encoded by DDX3Y have been isolated from patients with severe acute3 and extensive chronic4 GVHD. In the latter case, the DDX3Y peptide–specific CD4+ T-cell response was temporally associated with a high-titer DDX3Y-specific IgG antibody response, demonstrating a coordinated B- and T-cell response to DDX3Y after sex-mismatched, F→M HCT. These observations have implicated SMCY- and DDX3Y-specific T-cell responses in GVHD that occurs in F→M transplant recipients. A role for H-Y–specific T-cell responses in GVHD is also supported by the observation that male recipients of female hematopoietic cell grafts exhibit the highest rates of both acute and chronic GVHD of any donor-recipient sex combination.24-26
Clinical observations also suggest, however, that H-Y–specific T-cell responses contribute to GVL responses. High frequencies in the peripheral blood of CD8+ SMCY peptide–specific cytotoxic T lymphocytes (CTLs) have been associated with regression of blast-phase chronic myelogenous leukemia (CML) after donor lymphocyte infusion.6 F→M hematopoietic cell transplant recipients also experience a lower risk of relapse than that observed in any other donor-recipient sex combination, which is apparent even after controlling for GVHD.24,26,27 The selective GVL effect seen in male recipients of female grafts is observed in patients who underwent transplantation for CML, acute myelogenous leukemia (AML), and acute lymphoblastic leukemia (ALL),26 and could potentially be explained by expression of some H-Y antigens in leukemic stem cells. This hypothesis is supported by previous studies in our laboratory, which demonstrated that CD8+ CTLs specific for an HLA-B8–restricted H-Y antigen encoded by UTY inhibited the engraftment of male AML in nonobese diabetic/severe combined immune deficient (NOD/SCID) mice.28
In this report, we describe a novel MHC class I–restricted H-Y antigen that is encoded by DDX3Y and recognized by CD8+ CTLs. In contrast to previous studies that reported the isolation of CD4+, DDX3Y-specific T cells from patients with severe GVHD,3,4 the CD8+ DDX3Y-specific CTL clone was derived from a male who had received a hematopoietic cell graft from his MHC-identical sister but did not develop histologic evidence of GVHD. We therefore investigated the expression of the DDX3Y-encoded H-Y antigen recognized by these CTLs to gain insight into the extent to which it might serve as a target for GVL and GVHD responses after F→M HCT.
Methods
Human subjects
After written informed consent was obtained in accordance with the Declaration of Helsinki, blood samples and skin biopsies were collected from 2 male patients—one (UPN 19492) with acute lymphoblastic leukemia (ALL) in first complete remission, and another (UPN 21234) with myelodysplasia/refractory anemia with excess blasts–2 (MDS/RAEB-2)—and from their MHC-identical sisters, all of whom were enrolled in a clinical trial approved by the Institutional Review Board at the Fred Hutchinson Cancer Research Center.
Cell culture
The CD8+, HLA-B*2705–restricted, donor-derived, recipient-specific CTL clone 68H7-819 was isolated from peripheral blood mononuclear cells (PBMCs) obtained on day +42 after a myeloablative, unmodified peripheral blood stem cell transplantation, and was maintained in vitro as previously described.29 Phytohemagglutinin (PHA)–stimulated T-cell blasts (PHA blasts),29 Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines (EBV-LCLs),30 and fibroblast lines31 were generated and maintained as described. COS-732 and WEHI 164 clone 13 cells33 were maintained in vitro as previously described.
Cytotoxicity assays
Cytotoxicity was measured in 51Cr release assays as previously described29 using target cells labeled overnight at 37°C with 100 μCi (3.7 MBq) 51Cr. EBV-LCL lines derived from males carrying constitutional deletions of the Y chromosome were infected at a multiplicity of infection of 10:1 with a recombinant vaccinia virus encoding HLA-B*2705 concurrent with 51Cr labeling. Specific lysis was calculated using the standard formula.31 H-Y peptides for epitope reconstitution studies were synthesized using standard Fmoc chemistry (SynPep, Dublin, CA) and pulsed onto 51Cr-labeled donor-derived EBV-LCLs for 30 minutes at 37°C before addition of CTLs.
Subgenic cloning and transfection studies
Subgenic fragments of the DDX3Y or USP9Y genes were amplified by polymerase chain reaction (PCR) and cloned into pcDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA). The HLA-B*2705 cDNA was amplified by PCR and cloned into pEAK10 (Edge Biosystems, Gaithersburg, MD). To identify the nucleotide sequence that encodes the H-Y epitope recognized by CTL 68H7-819, COS-7 cells previously seeded in 96-well flat-bottom plates at 1.5 × 104/well were transiently cotransfected in duplicate with plasmids encoding DDX3Y or USP9Y minigenes and the HLA-B*2705/pEAK10 plasmid. Transfection was carried out in Dulbecco modified Eagle medium containing 2 ng/μL each plasmid construct, 800 μg/mL diethylaminoethyl-dextran, and 200 μM chloroquine, at 37°C for 4 hours, followed by a 2-minute hyperosmotic shock with 10% dimethyl sulfoxide (Sigma-Aldrich, St Louis, MO) in phosphate-buffered saline (PBS; Invitrogen). The day following transfection, the medium was removed from the COS-7 transfectants, and 5 × 103 to 1 × 104 CTL 68H7-819 suspended in 100 μL COS-7 medium32 supplemented with 25 IU/mL interleukin-2 (IL-2; Chiron, Emeryville, CA) was added to each well. After 20 hours of coculture, the supernatants were evaluated for interferon-γ (IFN-γ) by enzyme-linked immunosorbent assay (ELISA) or for tumor necrosis factor by cytotoxicity.33
Quantitative real-time reverse transcription–PCR (RT-PCR)
Total RNA was purified from cultured cells, primary leukemic cells, and human tumor cell lines using the RNeasy Mini Kit (QIAGEN, Valencia, CA) or Trizol (Invitrogen). First-strand cDNA was synthesized from 1 to 4 μg total RNA using Oligo-dT and SuperScript II reverse transcriptase, initially treated with Dnase I (all from Invitrogen). First-strand cDNA from poly (A)+ RNA from selected normal human tissues and blood cell subsets, pooled from both males and females, was commercially obtained (human Multiple Tissue cDNA panels I and II, and human blood fractions; BD Biosciences/Clontech, Palo Alto, CA). Expression of DDX3Y was evaluated in triplicate and normalized to GAPDH expression. DDX3Y transcript level was expressed relative to the male AML cell line KG-1a (ATCC, Manassas, VA). Amplification was performed on 25 ng initial RNA for primary leukemic samples, or 2 ng commercially prepared cDNA, using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a total reaction volume of 25 μL. Primers were at a final concentration of 400 nM, and the sequences were as follows: DDX3Y: 5′-GACAGTTCAGGTTGGAGTTGC-3′ and 5′-TCACACCAACGACTATGTCCA-3′; GAPDH: 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. Amplification was performed on a Stratagene Mx3000P (La Jolla, CA), and relative expression was determined with the comparative Ct method,34 using the average Ct values for GAPDH and DDX3Y.
HLA typing
High-resolution HLA typing of UPN 19492 and UPN 21234 was performed by the Clinical Immunogenetics Laboratory of the Seattle Cancer Care Alliance. Identification of HLA-B*2705+ primary leukemia samples was performed using the Dynal RELI Sequence-Specific Oligonucleotide (SSO) HLA-A,B,C DRB1 and DQB1 Line Strip Typing System (Invitrogen) to determine HLA-B locus genotype from genomic DNA that was isolated using the Versagene Genomic DNA Purification Kit (Qiagen [formerly Gentra], Valencia, CA).
NOD/SCID leukemic engraftment assay
Primary leukemic cells were thawed, washed twice, and then cultured for 16 hours at 37°C in CTL medium29 supplemented with 25 IU/mL IL-2, in the absence or presence of DDX3Y-specific or irrelevant CTLs at a CTL/leukemia ratio of 5:1. Leukemia cells and CTL/leukemia cell mixtures were then washed once, resuspended in PBS, and injected via the tail vein into cohorts of 5 to 6 sublethally irradiated (350 cGy delivered by linear accelerator at 20 cGy/min) NOD/SCID mice. Control mice were injected with 200 μL PBS. For experiments using AML cells, NOD/SCID mice were injected with purified rat antimouse CD122 antibody (200 μg/mouse) into the peritoneal cavity immediately following irradiation to facilitate the engraftment of human leukemic cells.35 Mice were killed 6 weeks after injection, and bone marrow mononuclear cells (BMMCs), peripheral blood, spleen, liver, kidney, lung, and thymus were harvested and analyzed for engraftment by flow cytometry or by PCR using human Y-chromosome–specific primers.
Mouse BMMCs were washed and incubated with fluorescein isothiocyanate (FITC)–conjugated anti–HLA-B27 (One Lambda, Canoga Park, CA) and phycoerythrin (PE)–conjugated anti-CD33 or anti-CD45 (BD Pharmingen, San Diego, CA) antibodies for 30 minutes on ice, washed twice, and analyzed on a FACScan flow cytometer (BD Biosciences, Mountain View, CA). The data were analyzed with CellQuest software (BD Biosciences). PE- or FITC-conjugated isotype-matched antibodies were used as controls.
Genomic DNA was isolated using the QIAamp DNA Blood Mini Kit or QIAamp DNA Mini Kit (Qiagen) and analyzed by PCR with human Y-chromosome–specific sequence-tagged site sY14 primers. PCR was carried out using ABI Fast PCR mix (Applied Biosystems), 500 nM sY14 primers,36 deionized H20, and approximately 100 ng genomic DNA. Cycling conditions were 95°C for 10 seconds, 40 cycles of 95°C for 0 seconds, 61°C for 15 seconds, and 72°C for 30 seconds, followed by 72°C for 1 minute.
Quantitation of DDX3Y-specific CD8+ and CD4+ T-cell responses
Detection and enumeration of DDX3Y-specific T-cell responses were performed by IFN-γ enzyme-linked immunosorbent spot (ELISpot) assay. Assessment of subject UPN 19492 for T cells specific for the DDX3Y74-83 epitope presented by HLA-B*2705 was performed directly ex vivo, without in vitro stimulation, on PBMCs obtained on days +42, +62, +93, and +109 after transplantation. This patient was also evaluated for T-cell responses against the 2 previously identified MHC class II–restricted DDX3Y-encoded H-Y epitopes, DDX3Y30-48,4 and DDX3Y176-187,3 using PBMCs obtained on days +42, +62, +100, and +109 after transplantation that were analyzed both directly ex vivo and after 2 to 6 weekly in vitro stimulations with DDX3Y30-48- and DDX3Y176-187-pulsed (1 μM), γ-irradiated (35 Gy) recipient pretransplantation PBMCs. The in vitro stimulations were performed in CTL medium29 supplemented with 10 IU/mL IL-2. ELISpot assays were performed in 96-well Multi-Screen-IP plates (MAIPS4510; Millipore, Bedford, MA) coated overnight with antihuman IFN-γ antibody (M700A; Thermo Fisher Pierce Endogen, Rockford, IL) by culturing PBMC responders with peptide (2 μM)–pulsed donor PBMCs pulsed at a responder-stimulator ratio of 5:1 for 24 to 48 hours in 200 μL LCL medium.30 Spots were visualized by addition of an antihuman biotin-labeled antibody (M701B; Thermo Fisher Pierce Endogen), PolyHRP20-streptavidin (Fitzgerald Industries International, Concord, MA), and Vectastain AEC substrate (Vector Laboratories, Burlingame, CA), and counted on an automated ELISpot reader (Immuno Biosys, The Colony, TX).
Identification of novel DDX3Y-specific T-cell responses in subject UPN 21234 was performed by stimulating PBMCs obtained on day +127 after transplantation with γ-irradiated (80 Gy) donor-derived EBV-LCLs pulsed with a mixture of 36 overlapping pentadecapeptides37 that collectively encompassed the N-terminal 195 residues of DDX3Y (Table 1; NMI Peptides, Reutlingen, Germany). Stimulations were performed in 24-well plates at a responder/stimulator ratio of 4:1 in CTL medium29 supplemented with 12.5 IU/mL IL-2, 5 ng/mL IL-7, and 10 ng/mL IL-15, and with each peptide present at a final concentration of 2.5 μM. Responder cells were tested by ELISpot on days +7 to +9 after the second and third stimulations, against 12 pools of 6 pentadecapeptides each, with each pool derived from 1 of the 6 rows or 6 columns of a 6 × 6 array of the 36 pentadecapeptides (Table 2). The concentration of the constituent pentadecapeptides in each pool was 1.25 μM.
Overlapping DDX3Y pentadecapeptides for identification of novel epitopes
| Peptide . | Sequence . | Residues . |
|---|---|---|
| 1 | MSHVVVKNDPELDQQ | 1-15 |
| 2 | VVKNDPELDQQLANL | 5-19 |
| 3 | DPELDQQLANLDLNS | 9-23 |
| 4 | DQQLANLDLNSEKQS | 13-27 |
| 5 | ANLDLNSEKQSGGAS | 17-31 |
| 6 | LNSEKQSGGASTASK | 21-35 |
| 7 | KQSGGASTASKGRYI | 25-39 |
| 8 | GASTASKGRYIPPHL | 29-43 |
| 9 | IPPHLRNREASKGFH | 39-53 |
| 10 | LRNREASKGFHDKDS | 43-57 |
| 11 | EASKGFHDKDSSGWS | 47-61 |
| 12 | GFHDKDSSGWSCSKD | 51-65 |
| 13 | KDSSGWSCSKDKDAY | 55-69 |
| 14 | GWSCSKDKDAYSSFG | 59-73 |
| 15 | SKDKDAYSSFGSRDS | 63-77 |
| 16 | DAYSSFGSRDSRGKP | 67-81 |
| 17 | SFGSRDSRGKPGYFS | 71-85 |
| 18 | RDSRGKPGYFSERGS | 75-89 |
| 19 | GKPGYFSERGSGSRG | 79-93 |
| 20 | YFSERGSGSRGRFDD | 83-97 |
| 21 | RFDDRGRSDYDGIGN | 94-108 |
| 22 | RGRSDYDGIGNRERP | 98-112 |
| 23 | DYDGIGNRERPGFGR | 102-116 |
| 24 | IGNRERPGFGRFERS | 106-120 |
| 25 | ERPGFGRFERSGHSR | 110-124 |
| 26 | FGRFERSGHSRWCDK | 114-128 |
| 27 | ERSGHSRWCDKSVED | 118-132 |
| 28 | HSRWCDKSVEDDWSK | 122-136 |
| 29 | CDKSVEDDWSKPLPP | 126-140 |
| 30 | VEDDWSKPLPPSERL | 130-144 |
| 31 | YDDIPVEATGSNCPP | 161-175 |
| 32 | PVEATGSNCPPHIEN | 165-179 |
| 33 | TGSNCPPHIENFSDI | 169-183 |
| 34 | CPPHIENFSDIDMGE | 173-187 |
| 35 | IENFSDIDMGEIIMG | 177-191 |
| 36 | SDIDMGEIIMGNIEL | 181-195 |
| Peptide . | Sequence . | Residues . |
|---|---|---|
| 1 | MSHVVVKNDPELDQQ | 1-15 |
| 2 | VVKNDPELDQQLANL | 5-19 |
| 3 | DPELDQQLANLDLNS | 9-23 |
| 4 | DQQLANLDLNSEKQS | 13-27 |
| 5 | ANLDLNSEKQSGGAS | 17-31 |
| 6 | LNSEKQSGGASTASK | 21-35 |
| 7 | KQSGGASTASKGRYI | 25-39 |
| 8 | GASTASKGRYIPPHL | 29-43 |
| 9 | IPPHLRNREASKGFH | 39-53 |
| 10 | LRNREASKGFHDKDS | 43-57 |
| 11 | EASKGFHDKDSSGWS | 47-61 |
| 12 | GFHDKDSSGWSCSKD | 51-65 |
| 13 | KDSSGWSCSKDKDAY | 55-69 |
| 14 | GWSCSKDKDAYSSFG | 59-73 |
| 15 | SKDKDAYSSFGSRDS | 63-77 |
| 16 | DAYSSFGSRDSRGKP | 67-81 |
| 17 | SFGSRDSRGKPGYFS | 71-85 |
| 18 | RDSRGKPGYFSERGS | 75-89 |
| 19 | GKPGYFSERGSGSRG | 79-93 |
| 20 | YFSERGSGSRGRFDD | 83-97 |
| 21 | RFDDRGRSDYDGIGN | 94-108 |
| 22 | RGRSDYDGIGNRERP | 98-112 |
| 23 | DYDGIGNRERPGFGR | 102-116 |
| 24 | IGNRERPGFGRFERS | 106-120 |
| 25 | ERPGFGRFERSGHSR | 110-124 |
| 26 | FGRFERSGHSRWCDK | 114-128 |
| 27 | ERSGHSRWCDKSVED | 118-132 |
| 28 | HSRWCDKSVEDDWSK | 122-136 |
| 29 | CDKSVEDDWSKPLPP | 126-140 |
| 30 | VEDDWSKPLPPSERL | 130-144 |
| 31 | YDDIPVEATGSNCPP | 161-175 |
| 32 | PVEATGSNCPPHIEN | 165-179 |
| 33 | TGSNCPPHIENFSDI | 169-183 |
| 34 | CPPHIENFSDIDMGE | 173-187 |
| 35 | IENFSDIDMGEIIMG | 177-191 |
| 36 | SDIDMGEIIMGNIEL | 181-195 |
Underlined amino acids indicate the positions at which there is nonidentity between the DDX3Y-encoded pentadecapeptide and the homologous peptide encoded by DDX3X.
DDX3Y pentadecapeptide grid
| Peptide pool # . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . |
|---|---|---|---|---|---|---|
| 1 | 1 | 2 | 3 | 4 | 5 | 6 |
| 2 | 7 | 8 | 9 | 10 | 11 | 12 |
| 3 | 13 | 14 | 15 | 16 | 17 | 18 |
| 4 | 19 | 20 | 21 | 22 | 23 | 24 |
| 5 | 25 | 26 | 27 | 28 | 29 | 30 |
| 6 | 31 | 32 | 33 | 34 | 35 | 36 |
| Peptide pool # . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . |
|---|---|---|---|---|---|---|
| 1 | 1 | 2 | 3 | 4 | 5 | 6 |
| 2 | 7 | 8 | 9 | 10 | 11 | 12 |
| 3 | 13 | 14 | 15 | 16 | 17 | 18 |
| 4 | 19 | 20 | 21 | 22 | 23 | 24 |
| 5 | 25 | 26 | 27 | 28 | 29 | 30 |
| 6 | 31 | 32 | 33 | 34 | 35 | 36 |
DDX3Y pentadecapeptide grid illustrating the 12 peptide pools, each comprising one row or one column of the grid, used in ELISpot analysis to determine the pentadecapeptide specificity of responding PBMC. The sequences of peptides 1 through 36 are indicated in Table 1.
Results
CD8+ CTL clone 68H7-819 recognizes a HLA-B*2705–restricted H-Y antigen encoded by DDX3Y
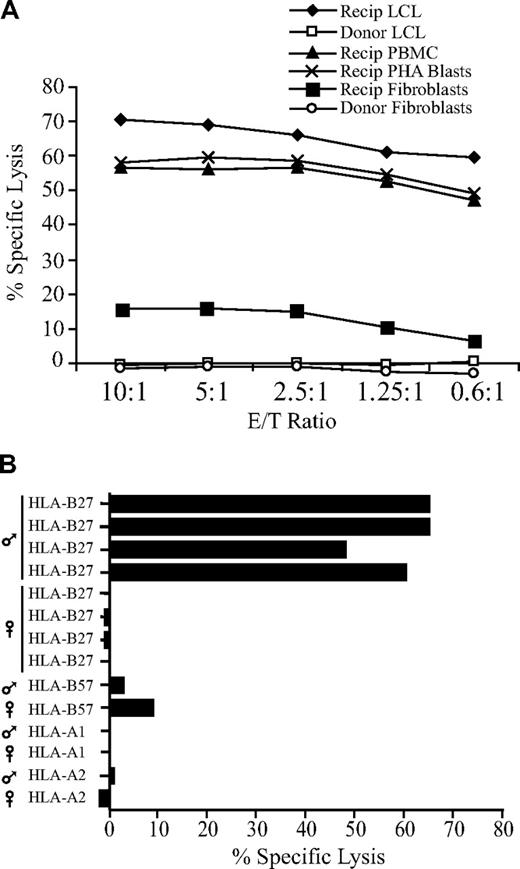
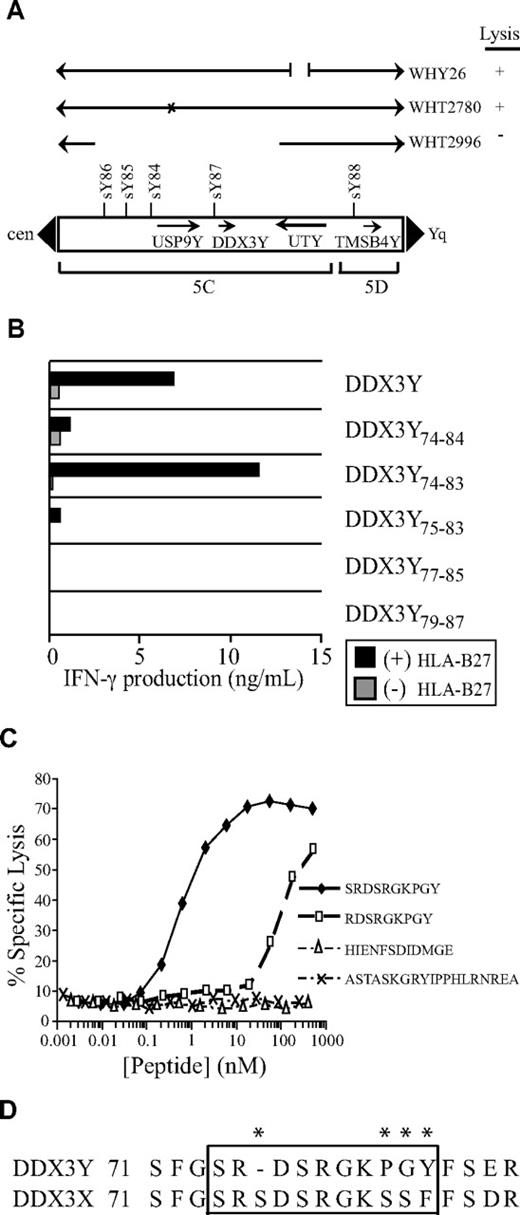
The CD8+ CTL clone 68H7-819 was isolated from UPN 19492, a male patient with ALL in first complete remission who had received an unmodified hematopoietic cell graft from his MHC-identical sister. The CTLs efficiently lysed EBV-LCLs, PBMCs, and PHA-stimulated T-cell blasts derived from the recipient, did not lyse EBV-LCLs derived from the donor, and weakly recognized recipient- but not donor-derived fibroblasts (Figure 1A). The CTLs also recognized EBV-LCLs derived from male but not female HLA-B27+ individuals (Figure 1B), suggesting that the clone was specific for a peptide encoded by a Y-chromosome gene and presented by HLA-B27. To identify the gene encoding this novel H-Y antigen, CTL 68H7-819 was tested for recognition of EBV-LCLs derived from individuals with constitutional Y-chromosome deletions18,36 after infection with a recombinant vaccinia virus encoding HLA-B*2705. CTL 68H7-819 showed no recognition of vac/HLA-B*2705–infected WHT2996, an EBV-LCL carrying a proximal Yq deletion encompassing USP9Y and DDX3Y in the AZFa locus, but did recognize vac/HLA-B*2705–infected WHT2780, which contains a point mutation in USP9Y that results in 90% truncation (Figure 2A).38 CTL 68H7–819 also recognized vac/HLA-B*2705–infected WHY26, which carries a Y-chromosomal break in UTY. These results suggested that the epitope recognized by CTL 68H7-819 is encoded by DDX3Y. This was confirmed by CTL 68H7-819 recognition of COS-7 cells cotransfected with HLA-B*2705 and DDX3Y cDNAs (Figure 2B).
CTL 68H7–819 recognizes a male-specific minor histocompatibility (H-Y) antigen presented by HLA-B*2705. (A) 51Cr release assay at the indicated effector-target (E/T) ratios showing cytolytic activity of CTL 68H7-819 against donor- and recipient-derived EBV-LCLs, recipient-derived unfractionated PBMCs and PHA-stimulated T-cell blasts, and donor- and recipient-derived dermal fibroblasts. (B) 51Cr release assay at E/T 5:1 showing cytolytic activity of CTL 68H7-819 against a panel of EBV-LCLs derived from unrelated male and female individuals who shared a single MHC class I allele with the donor-recipient pair from which CTL 68H7-819 was isolated. The shared MHC class I allele and sex of the individual is indicated.
CTL 68H7–819 recognizes a male-specific minor histocompatibility (H-Y) antigen presented by HLA-B*2705. (A) 51Cr release assay at the indicated effector-target (E/T) ratios showing cytolytic activity of CTL 68H7-819 against donor- and recipient-derived EBV-LCLs, recipient-derived unfractionated PBMCs and PHA-stimulated T-cell blasts, and donor- and recipient-derived dermal fibroblasts. (B) 51Cr release assay at E/T 5:1 showing cytolytic activity of CTL 68H7-819 against a panel of EBV-LCLs derived from unrelated male and female individuals who shared a single MHC class I allele with the donor-recipient pair from which CTL 68H7-819 was isolated. The shared MHC class I allele and sex of the individual is indicated.
DDX3Y encodes the H-Y antigen recognized by CTL clone 68H7-819. (A) Localization by Y-chromosome deletion mapping of the gene encoding the H-Y antigen recognized by CTL 68H7-819. The CTLs were tested for recognition of EBV-LCLs derived from males carrying Y chromosomes with constitutional deletions in a 51Cr release assay at E/T 10:1. EBV-LCLs were infected the day before the assay with a recombinant vaccinia virus carrying an HLA-B*2705 transgene. EBV-LCL WHT2996 is derived from an individual with a deletion encompassing genes DDX3Y and USP9Y.38 WHT2780 is derived from an individual with a splice site deletion that results in 90% truncation of the USP9Y gene, indicated by an X.38 WHY26 is from an individual with a chromosomal break in UTY. Arrows indicate the intact and aberrant segments of the Y chromosome. Y-chromosome landmarks include the boundaries of deletion intervals 5C and 5D, and selected sequence-tagged sites. + indicates lysis of 35% or more, and − indicates lysis of 4% or less. (B) Plasmids encoding DDX3Y minigenes were cotransfected into COS-7 cells with a plasmid encoding HLA-B*2705. On the following day, CTL 68H7-819 was added to the COS-7 transfectants, and IFN-γ release was measured in the supernatants by ELISA after 20 hours of coculture. (C) Epitope reconstitution assay to determine CTL 68H7-819 recognition of donor EBV-LCLs that had been pulsed for 30 minutes with the indicated synthetic peptides over the indicated range of concentrations; 4-hour 51Cr release assay, E/T 5:1. (D) Partial sequence alignment of the DDX3Y and DDX3X proteins spanning the region that includes the epitope recognized by CTL 68H7-819. Asterisks indicate disparate residues.
DDX3Y encodes the H-Y antigen recognized by CTL clone 68H7-819. (A) Localization by Y-chromosome deletion mapping of the gene encoding the H-Y antigen recognized by CTL 68H7-819. The CTLs were tested for recognition of EBV-LCLs derived from males carrying Y chromosomes with constitutional deletions in a 51Cr release assay at E/T 10:1. EBV-LCLs were infected the day before the assay with a recombinant vaccinia virus carrying an HLA-B*2705 transgene. EBV-LCL WHT2996 is derived from an individual with a deletion encompassing genes DDX3Y and USP9Y.38 WHT2780 is derived from an individual with a splice site deletion that results in 90% truncation of the USP9Y gene, indicated by an X.38 WHY26 is from an individual with a chromosomal break in UTY. Arrows indicate the intact and aberrant segments of the Y chromosome. Y-chromosome landmarks include the boundaries of deletion intervals 5C and 5D, and selected sequence-tagged sites. + indicates lysis of 35% or more, and − indicates lysis of 4% or less. (B) Plasmids encoding DDX3Y minigenes were cotransfected into COS-7 cells with a plasmid encoding HLA-B*2705. On the following day, CTL 68H7-819 was added to the COS-7 transfectants, and IFN-γ release was measured in the supernatants by ELISA after 20 hours of coculture. (C) Epitope reconstitution assay to determine CTL 68H7-819 recognition of donor EBV-LCLs that had been pulsed for 30 minutes with the indicated synthetic peptides over the indicated range of concentrations; 4-hour 51Cr release assay, E/T 5:1. (D) Partial sequence alignment of the DDX3Y and DDX3X proteins spanning the region that includes the epitope recognized by CTL 68H7-819. Asterisks indicate disparate residues.
Testing recognition of CTL 68H7-819 against COS-7 cells cotransfected with HLA-B*2705 cDNA and a panel of overlapping DDX3Y minigenes revealed recognition of a minigene encoding residues 1 through 93, but not a minigene encoding residues 1 through 72 (data not shown). An epitope prediction algorithm39 was used to identify nonameric or decameric peptides in this region expected to bind to HLA-B*2705 with high affinity. Minigenes encoding 3 such peptides—DDX3Y75-83 (RDSRGKPGY), DDX3Y74-83 (SRDSRGKPGY), and DDX3Y74-84 (SRDSRGKPGYF)—were recognized by CTL 68H7-819 in an HLA-B*2705–dependent manner (Figure 2B). Donor EBV-LCLs pulsed with serial dilutions of synthetic RDSRGKPGY and SRDSRGKPGY peptides were recognized by CTL 68H7-819 with half-maximal recognition seen at both 60 nM and 600 pM, respectively (Figure 2C). No recognition by CTL 68H7-819 of donor EBV-LCLs pulsed with either of the synthetic peptides corresponding to the 2 previously described DDX3Y-encoded, MHC class II–restricted H-Y epitopes3,4 was observed (Figure 2C). These data demonstrate that SRDSRGKPGY represents the HLA-B*2705–restricted H-Y antigen recognized by CTL 68H7-819. Comparison of the predicted DDX3Y protein sequence with that of the homologous X-chromosome–encoded DDX3X protein (Figure 2D) revealed that the SRDSRGKPGY epitope differs at 4 positions with the homologous DDX3X peptide.
DDX3Y expression in cultured cells, primary tissues, and primary leukemic cells
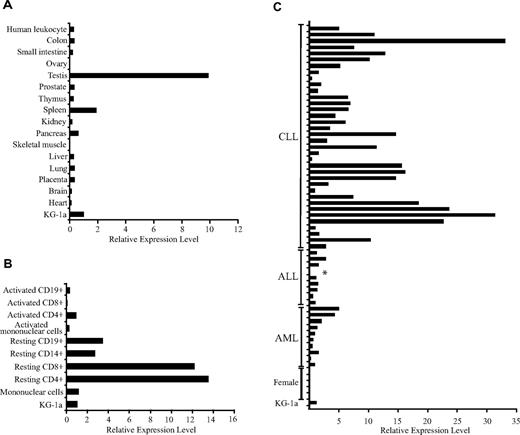
DDX3Y expression was evaluated by semiquantitative real-time PCR of cDNA from normal and malignant tissues of both hematopoietic and nonhematopoietic origin. Analysis of cDNA from a panel of normal human tissues, derived from a pool containing both males and females, revealed that DDX3Y transcript was detectable in all normal tissues tested except ovary and skeletal muscle, with the highest levels of expression seen in testis, followed by the spleen and pancreas (Figure 3A). DDX3Y transcript was also detectable in all subsets of resting and activated human blood cells, with the highest levels seen in resting blood fractions, and approximately 10-fold lower expression in activated fractions (Figure 3B). DDX3Y expression was detected in 54 of 55 primary male leukemic samples (Figure 3C). The relative DDX3Y expression levels were highest in male CLL samples (35 samples; mean: 9.1; median: 6.3; range: 0.4-33.5), followed by AML (11 samples; mean: 1.7; median: 1.1; range: 0.2-5.0) and ALL samples (9 samples; mean: 1.4; median: 1.3; range: 0.6-2.8). DDX3Y expression was undetectable in all 14 female malignant samples tested (Figure 3C and data not shown). DDX3Y expression in the recipient fibroblasts from which CTL 68H7-819 was derived was 0.3 compared with the reference sample KG-1a, and undetectable in the female donor fibroblast sample (data not shown). One leukemic sample obtained from a male patient with ALL did not express detectable DDX3Y transcript (indicated by asterisk in Figure 3C). Cytogenetic analysis of this sample revealed clonal loss of the Y chromosome (data not shown).
DDX3Y is transcribed outside the testis and is universally expressed in myeloid and lymphoid leukemia cells that carry a Y chromosome. Relative expression of DDX3Y in a panel of normal human tissues (A), normal blood cell fractions (B), and primary male ALL, CLL, and AML samples (C). Quantitative real-time PCR using SYBR green was carried out as described in “Methods.” Analysis of GAPDH expression was used to standardize samples for RNA quality and quantity, and the relative DDX3Y expression in the KG-1a AML cell line was arbitrarily defined as 1. The asterisk in panel C indicates a primary male ALL sample that had clonal loss of the Y chromosome by cytogenetic analysis and no detectable DDX3Y transcript. Five of 14 female samples are shown, all of which were negative.
DDX3Y is transcribed outside the testis and is universally expressed in myeloid and lymphoid leukemia cells that carry a Y chromosome. Relative expression of DDX3Y in a panel of normal human tissues (A), normal blood cell fractions (B), and primary male ALL, CLL, and AML samples (C). Quantitative real-time PCR using SYBR green was carried out as described in “Methods.” Analysis of GAPDH expression was used to standardize samples for RNA quality and quantity, and the relative DDX3Y expression in the KG-1a AML cell line was arbitrarily defined as 1. The asterisk in panel C indicates a primary male ALL sample that had clonal loss of the Y chromosome by cytogenetic analysis and no detectable DDX3Y transcript. Five of 14 female samples are shown, all of which were negative.
The DDX3Y-encoded H-Y epitope is presented on the surface of HLA-B*2705+ leukemias
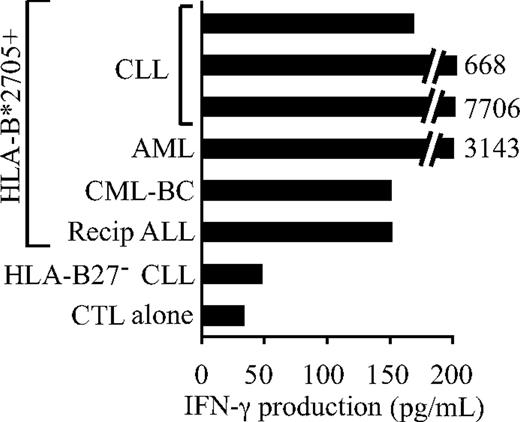
To investigate the expression of the DDX3Y-encoded H-Y epitope on the surface of primary leukemic cells, leukemic samples that natively expressed the restricting molecule HLA-B*2705 were identified by genotyping the HLA-B locus in each of the 54 primary male leukemic samples that carried a Y chromosome. Of these, 6 carried a HLA-B*2705 allele, consistent with the low frequency of this allele in the white population from which most of the samples were obtained.40 The 6 HLA-B*2705+ leukemic samples included BMMCs from the ALL patient from whom CTL clone 68H7-819 was derived, as well as PBMCs from 3 patients with CLL, 1 patient with CML in T-lymphoid blast crisis (CML-BC), and 1 patient with primary refractory AML. Each sample contained between 80% to 90% leukemic cells, and were reproducibly recognized by CTL 68H7-819 in an IFN-γ ELISA assay (representative data in Figure 4); no recognition of HLA-B*2705− or female leukemic samples was seen. These results demonstrate that the DDX3Y-encoded H-Y epitope is expressed on the surface of both acute and chronic leukemia cells of both myeloid and lymphoid lineage.
The DDX3Y-encoded H-Y antigen recognized by CTL 68H7-819 is expressed on the surface of HLA-B*2705+ leukemic cells. CTL 68H7-819 recognition of primary leukemic samples from males carrying the HLA-B*2705 allele was tested by IFN-γ ELISA after overnight coculture with CTL at a CTL/leukemia ratio of 1:5. The 6 samples consisted of BMMCs derived from the hematopoietic cell transplant recipient from whom CTL 68H7-819 was derived (Recip ALL), PBMCs from one patient each with primary refractory AML and with CML in T-lymphoid blast crisis (CML-BC), and 3 PBMC samples from CLL patients. Negative controls included a CLL sample that did not carry the HLA-B*2705 allele, and CTL 68H7-819 alone.
The DDX3Y-encoded H-Y antigen recognized by CTL 68H7-819 is expressed on the surface of HLA-B*2705+ leukemic cells. CTL 68H7-819 recognition of primary leukemic samples from males carrying the HLA-B*2705 allele was tested by IFN-γ ELISA after overnight coculture with CTL at a CTL/leukemia ratio of 1:5. The 6 samples consisted of BMMCs derived from the hematopoietic cell transplant recipient from whom CTL 68H7-819 was derived (Recip ALL), PBMCs from one patient each with primary refractory AML and with CML in T-lymphoid blast crisis (CML-BC), and 3 PBMC samples from CLL patients. Negative controls included a CLL sample that did not carry the HLA-B*2705 allele, and CTL 68H7-819 alone.
The DDX3Y-encoded H-Y epitope is expressed by leukemic stem cells
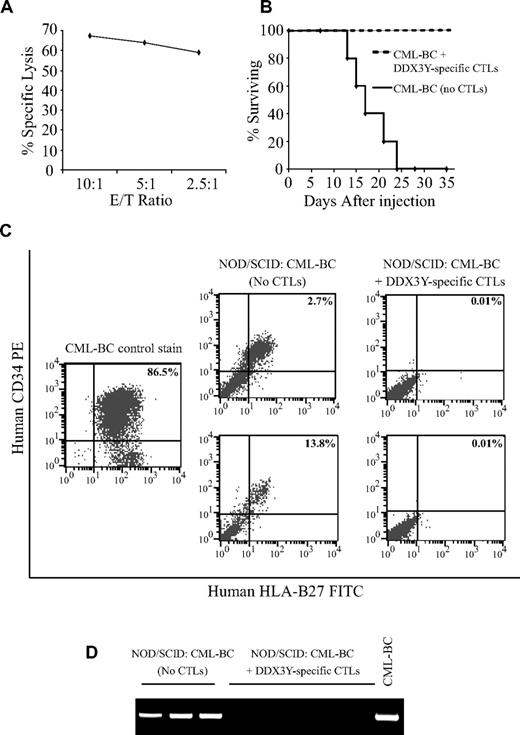
To determine whether the DDX3Y epitope is expressed on the putative leukemic stem cell that can establish human acute leukemia in NOD/SCID mice, CTL clone 68H7-819 was tested for its ability to inhibit the engraftment of HLA-B27+ primary leukemic cells. PBMCs from the 2 HLA-B*2705+ patients with T-lymphoid blast crisis of CML and primary refractory AML were used for these experiments. CTL 68H7-819 demonstrated in vitro recognition of both leukemic samples by both IFN-γ ELISA assay (Figure 4) as well as by chromium release assay (Figure 5A, and data not shown).
CTL 68H7-819 targets the leukemic stem cell in T-lymphoid blast phase CML (CML-BC). (A) Recognition of CML-BC cells by CTL 68H7-819 in a 51Cr release assay at the indicated E/T ratios. (B) Survival analysis of mice injected with CML-BC cells cultured overnight in medium alone or with CTL 68H7-819. Each group was composed of 5 mice. (C) Flow cytometric analysis using human PE-conjugated antihuman CD34 and FITC-conjugated anti–HLA-B27 antibodies of BMMCs from representative mice injected with CML-BC cells that had been cultured overnight in medium alone or with CTL 68H7-819. Uninjected CML-BC cells were used as a positive control. (D) Human Y-chromosome PCR analysis of genomic DNA extracted from mouse BMMCs to detect human male leukemic cells. Flow cytometric and PCR analysis was performed on mice that survived at least 15 days after injection.
CTL 68H7-819 targets the leukemic stem cell in T-lymphoid blast phase CML (CML-BC). (A) Recognition of CML-BC cells by CTL 68H7-819 in a 51Cr release assay at the indicated E/T ratios. (B) Survival analysis of mice injected with CML-BC cells cultured overnight in medium alone or with CTL 68H7-819. Each group was composed of 5 mice. (C) Flow cytometric analysis using human PE-conjugated antihuman CD34 and FITC-conjugated anti–HLA-B27 antibodies of BMMCs from representative mice injected with CML-BC cells that had been cultured overnight in medium alone or with CTL 68H7-819. Uninjected CML-BC cells were used as a positive control. (D) Human Y-chromosome PCR analysis of genomic DNA extracted from mouse BMMCs to detect human male leukemic cells. Flow cytometric and PCR analysis was performed on mice that survived at least 15 days after injection.
In the first NOD/SCID experiment, control mice injected with CML-BC cells alone did not survive beyond 24 days after injection, but mice that received equal doses of CML-BC cells that had been cultured overnight with CTL 68H7-819 uniformly survived to day 35, at which point they were killed (Figure 5B). Analysis of the 2 groups of mice for engraftment with human cells by flow cytometry and PCR with human Y-chromosome–specific primers confirmed the presence of human leukemic cells (Figure 5C) and human Y-chromosome DNA (Figure 5D) in the BMMCs of all mice that received CML-BC cells alone and survived at least 15 days after injection. In contrast, all mice that received CML-BC cells cocultured with CTL 68H7-819 had 0.01% or less human cells by flow cytometry (Figure 5C) and no detectable human Y-chromosome DNA by PCR (Figure 5D). CTL 68H7-819 had no inhibitory effect on the engraftment of leukemic cells obtained from a female AML patient (data not shown).
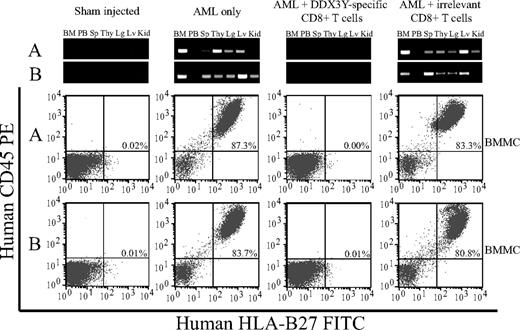
In the second NOD/SCID experiment, mice that received injections of AML cells from an HLA-B*2705+ male that were cultured overnight in medium alone exhibited high levels of bone marrow engraftment, ranging from 51% to 87% when assessed at the time of death at 6 weeks after injection (Figure 6). PCR analysis revealed human Y-chromosome DNA in bone marrow, peripheral blood, spleen, thymus, lung, liver, and kidney. In contrast, all mice injected with AML cells that had been cultured overnight with CTL 68H7-819 showed no detectable human cells in the bone marrow by flow cytometry and no detectable human Y-chromosome DNA by PCR (Figure 6), comparable with mice injected with PBS alone. Engraftment of AML cells was not inhibited by overnight culture with an irrelevant CD8+ CTL clone specific for a distinct minor histocompatibility antigen not expressed by the AML (Figure 6), ruling out nonspecific inhibition of engraftment by overnight coculture with CTLs. Moreover, CTL 68H7-819 once again did not inhibit the engraftment of a control AML sample derived from a female, demonstrating that the inhibitory effect was male specific (data not shown).
CTL 68H7-819 targets the leukemic stem cell in AML. Flow cytometric analysis using human PE-conjugated antihuman CD45 and FITC-conjugated anti–HLA-B27 antibodies of BMMCs from representative mice injected with PBS (sham), or with AML cells that had either been cultured overnight in medium alone, with CTL 68H7-819, or with an irrelevant CD8+ CTL clone that did not recognize the AML in vitro. Top panels show human Y-chromosome–specific PCR analysis of genomic DNA isolated from BMMCs (BM), peripheral blood (PB), spleen (Sp), thymus (Thy), lung (Lg), liver (Lv), and kidney (Kid). The flow cytometry and PCR data from 2 representative mice from each group are shown, and are designated as A or B.
CTL 68H7-819 targets the leukemic stem cell in AML. Flow cytometric analysis using human PE-conjugated antihuman CD45 and FITC-conjugated anti–HLA-B27 antibodies of BMMCs from representative mice injected with PBS (sham), or with AML cells that had either been cultured overnight in medium alone, with CTL 68H7-819, or with an irrelevant CD8+ CTL clone that did not recognize the AML in vitro. Top panels show human Y-chromosome–specific PCR analysis of genomic DNA isolated from BMMCs (BM), peripheral blood (PB), spleen (Sp), thymus (Thy), lung (Lg), liver (Lv), and kidney (Kid). The flow cytometry and PCR data from 2 representative mice from each group are shown, and are designated as A or B.
Quantitation of DDX3Y-specific CD8+ and CD4+ T-cell responses in UPN 19492
IFN-γ ELISpot analysis was performed to estimate the frequency of DDX3Y74-83-specific T cells in the peripheral blood of UPN 19492 at intervals during the first 100 days after transplantation, during which time he did not have histologic evidence of GVHD and also remained in complete remission. Analysis of CTL 68H7-819 serially diluted into polyclonal PBMCs revealed that the sensitivity of the ELISpot assay was approximately 1 in 3000 cells. Direct ex vivo analysis of PBMCs obtained on days +42, +62, +93, and +109 after transplantation did not detect DDX3Y74-83-specific responses, indicating that the frequency of DDX3Y74-83-specific T cells in the blood at these time points was less than 0.03%.
HLA typing of UPN 19492 revealed that he also expressed the MHC class II molecules, HLA-DQ5 and HLA-DRB1*1501, that have been shown in previous studies3,4 to present other DDX3Y-encoded H-Y peptides (DDX3Y176-187 and DDX3Y30-48, respectively) to CD4+ T cells. Since the CD8+ DDX3Y74-83-specific response was detected in PBMCs obtained on day +42 after transplantation, day +42 PBMCs were analyzed both directly ex vivo and after in vitro stimulation with the DDX3Y176-187 and DDX3Y30-48 peptides in ELISpot assays to determine whether CD4+ T-cell responses to the 2 MHC class II–restricted DDX3Y epitopes had developed concurrently. The DDX3Y-specific CD4+ T-cell responses were not detected in PBMCs obtained at days +42, +62, +100, and +109 after transplantation, when analyzed both directly ex vivo and after multiple (up to 6) cycles of in vitro stimulation with the DDX3Y176-187 and DDX3Y30-48 peptides. Therefore, T-cell responses to the MHC class II–restricted epitopes DDX3Y176-187 and DDX3Y30-48 were either not present in the recipient's peripheral blood at the indicated time points, or were present at a frequency below the detection limit of the ELISpot assay.
Identification of novel DDX3Y-specific T-cell responses in a F→M hematopoietic cell transplant recipient with active GVL
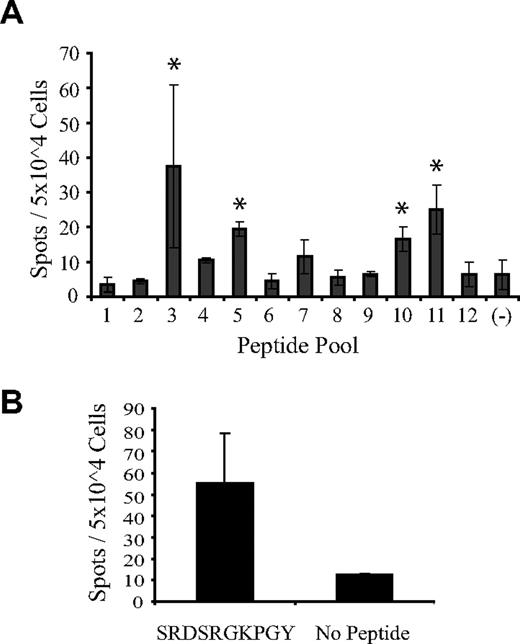
To determine whether DDX3Y-specific T cells are detectable in F→M hematopoietic cell transplant recipients who are actively experiencing GVL activity, we analyzed posttransplantation PBMCs from an HLA-B*2705–negative patient (UPN 21234) with MDS/RAEB-2 who relapsed on day +62 after he received a hematopoietic cell transplant from his MHC-identical sister, underwent withdrawal of immune suppression, and subsequently achieved a complete remission of his disease by day +160 after transplantation. Recipient PBMCs obtained on day +127, after relapse and withdrawal of immune suppression but before documentation of complete remission, were stimulated in vitro with a mixture of 36 DDX3Y-derived, overlapping pentadecapeptides that collectively spanned the amino-terminal 195 residues of the 660-residue protein (Table 1). Responder PBMCs were analyzed by ELISpot for reactivity against 12 peptide pools comprising the peptides in the 6 rows and 6 columns of a 6 × 6 array of the 36 pentadecapeptides (Table 2). Significant peptide-specific responses were detected against pools 3, 5, 10, and 11, with the largest responses against the orthogonal pools 3 and 11 (Figure 7A). These 2 pools share pentadecapeptide no. 17, with the sequence SFGSRDSRGKPGYFS in common (Tables 1, 2). An epitope prediction algorithm39 was then used to identify any peptides within this pentadecameric sequence that were predicted to bind to the recipient's MHC molecules, which included HLA-A*01, -A*02, -B*07, -B*15, -DRB1*0401/0404, and -DQB1*03 (data not shown). The peptide with the highest predicted binding affinity was the decamer SRDSRGKPGY (DDX3Y74-83), identical to the HLA-B*2705–restricted epitope recognized by CTL 68H7-819, which was predicted to bind to HLA-A*01. An aliquot of the T-cell line that had been stimulated with the pool of 36 DDX3Y-derived pentadecapeptides was then restimulated with the SFGSRDSRGKPGYFS pentadecamer, and subsequent ELISpot analysis demonstrated specific reactivity with the SRDSRGKPGY decamer (Figure 7B).
DDX3Y peptide–specific T-cell responses in a F→M hematopoietic cell transplant recipient experiencing GVL. PBMCs obtained from UPN 21234 (HLA-A*01, -A*02, -B*07, -B*15, -DRB1*0401/0404, and -DQB1*03) on day +127 after transplantation were stimulated in vitro with a pool of 36 overlapping DDX3Y-derived pentadecapeptides pulsed onto donor-derived EBV-LCLs and subsequently analyzed by IFN-γ ELISpot for DDX3Y peptide–specific responses (Tables 1, 2). (A) ELISpot analysis of day +127 PBMCs that had been stimulated in vitro with the entire pool of 36 DDX3Y-derived pentadecapeptides. Asterisks indicate the 4 peptide pools that stimulated spot formation above background. (B) An aliquot of the T-cell line analyzed in panel A was restimulated with pentadecapeptide no. 17, and subsequently analyzed by ELISpot for reactivity with the SRDSRGKPGY decamer, encompassed within the sequence of pentadecapeptide no. 17.
DDX3Y peptide–specific T-cell responses in a F→M hematopoietic cell transplant recipient experiencing GVL. PBMCs obtained from UPN 21234 (HLA-A*01, -A*02, -B*07, -B*15, -DRB1*0401/0404, and -DQB1*03) on day +127 after transplantation were stimulated in vitro with a pool of 36 overlapping DDX3Y-derived pentadecapeptides pulsed onto donor-derived EBV-LCLs and subsequently analyzed by IFN-γ ELISpot for DDX3Y peptide–specific responses (Tables 1, 2). (A) ELISpot analysis of day +127 PBMCs that had been stimulated in vitro with the entire pool of 36 DDX3Y-derived pentadecapeptides. Asterisks indicate the 4 peptide pools that stimulated spot formation above background. (B) An aliquot of the T-cell line analyzed in panel A was restimulated with pentadecapeptide no. 17, and subsequently analyzed by ELISpot for reactivity with the SRDSRGKPGY decamer, encompassed within the sequence of pentadecapeptide no. 17.
Discussion
The human DDX3Y gene is expressed at high levels in the testis and encodes a 660-residue protein that has only 92% sequence identity with the protein encoded by its X-chromosome homologue, DDX3X. Although the precise function of the DDX3Y protein is unknown, deletion of the AZFa locus on the Y chromosome that includes DDX3Y results in Sertoli-cell-only (SCO) syndrome and azoospermia.41 DDX3Y orthologues in C elegans,42 Drosophila,43 Xenopus,44 mice,45 and zebrafish46 are also implicated in the development or function of germ cells. DDX3Y, however, is also expressed outside the immunologic sanctuary of the testis, in both normal as well as malignant tissues, which enables presentation of DDX3Y to the afferent and efferent arms of the immune system. Consequently, in the setting of allogeneic F→M HCT, the female immune system encounters DDX3Y protein in extratesticular tissues, and this encounter can elicit CD4+ T-cell,3,4 B-cell,47,48 and, as shown for the first time in this study, CD8+ T-cell immunity.
Although the identification of a MHC class I–restricted H-Y antigen encoded by DDX3Y further illustrates the immunogenicity of its gene product in F→M HCT, the results of our study do not permit drawing any firm conclusions about the extent to which CD8+ DDX3Y-specific T-cell responses may contribute to GVHD. The hematopoietic cell transplant recipient from whom CTL clone 68H7–819 was isolated did not develop histologic evidence of either acute or chronic GVHD during the first 100 days after transplantation, but did receive a short course of prednisone therapy for diarrhea that was initially attributed to GVHD but subsequently attributed to C difficile infection. However, ELISpot analysis demonstrated that the frequency in the peripheral blood of T cells reactive with the DDX3Y-encoded SRDSRGKPGY peptide remained quite low (< 0.03%) throughout this interval. Thus, it is quite possible that the magnitude of the CD8+ DDX3Y-specific T-cell response was insufficient to trigger clinically significant manifestations of GVHD. Previous studies have reported an association of CD4+ T-cell responses to MHC class II–restricted epitopes derived from DDX3Y with severe acute3 or extensive chronic4 GVHD in F→M hematopoietic cell transplant recipients. ELISpot analysis did not detect T-cell responses against these MHC class II–restricted DDX3Y epitopes in UPN 19492, suggesting that any CD4+ DDX3Y-specific T-cell response that may have developed in this patient may also have been of insufficient magnitude to trigger clinically significant GVHD. Definitive assessment of the extent to which CD8+ DDX3Y-specific T cells can contribute to GVHD will likely require adoptive transfer studies49 in which the GVHD potential of the DDX3Y-specific cells can be more confidently distinguished from the effects of simultaneous T-cell responses against other alloantigens.
Several lines of evidence suggest that CD8+, MHC class I–restricted T-cell responses to DDX3Y could potentially contribute to GVL activity after F→M HCT. First, DDX3Y transcripts are detected in all male leukemic samples that retain a Y chromosome. Second, the DDX3Y74-83 peptide recognized by CTL 68H7-819 is expressed on the surface of both myeloid and lymphoid leukemic cells from HLA-B*2705+ males. Third, the HLA-B*2705–restricted DDX3Y74-83 epitope is expressed on the surface of the putative leukemic stem cell(s) that can establish AML and T-lymphoid blast phase CML in NOD/SCID mice. It is unclear whether UPN 19492, the patient from whom CTL 68H7-819 was isolated, might have benefited from any antileukemic effect mediated by CD8+ DDX3Y-specific T cells, since he underwent transplantation in complete remission, and remained in remission throughout the period of observation. Therefore, to determine whether DDX3Y-specific T-cell responses could be detected in F→M hematopoietic cell transplant recipients experiencing active GVL responses, we looked for DDX3Y-specific T cells in an HLA-B*2705–negative patient, UPN 21234, who suffered relapse of MDS/RAEB-2 on day +62 after transplantation and subsequently achieved a complete remission after withdrawal of immune suppression. Although the scope of this analysis was limited to less than a third of the total DDX3Y protein sequence, it demonstrated the presence of T cells reactive with several pools of DDX3Y-derived peptides and indicates that additional DDX3Y-encoded T-cell epitopes remain to be identified. In addition, the identification of a T-cell response specific for DDX3Y74-83 in a HLA-B*2705–negative male hematopoietic cell transplant recipient indicates that this peptide can be presented by additional MHC allele(s). Identification of these alleles and characterization of other novel DDX3Y-encoded epitopes, and their expression in normal and malignant tissues, are the focus of active research in our laboratory.
All human H-Y antigens identified to date50 are encoded by a specific class of Y-chromosome genes that have X-chromosome homologues and are expressed both in and variably outside the testis.15,20,21 There are 15 such genes on the human Y chromosome, and sequence analysis of their predicted protein products reveals 990 residues at which there is sequence nonidentity between the Y- and the X-encoded isoforms. Thus, it is very likely that the Y chromosome encodes a large number of H-Y antigens that remain to be identified, all of which would be expected to be in perfect linkage disequilibrium. The predicted 660-residue DDX3Y protein sequence, for example, contains more than 50 contiguous peptide sequences that (1) differ by one or more residues with the homologous peptides in DDX3X, and (2) are predicted to bind with high affinity to common class I MHC alleles, including HLA-A*0101, -A*0201, -A*0301, -B*0702, -B*0801, -B*4402, and -B*4403 (K.V.R. and E.H.W., unpublished observations, March 2007). The demonstration that a DDX3Y-specific CD8+ T-cell response can contribute to GVL activity provides an experimental basis for the clinical observation of a selective GVL effect associated with F→M HCT,24,26,27 and raises the prospect that further studies of H-Y immunity could eventually lead to strategies for selectively enhancing the GVL effect in recipients of sex-mismatched transplants.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank David Page and Laura Brown (Whitehead Institute and Massachusetts Institute of Technology, Cambridge, MA) and Elizabeth Simpson (MRC Clinical Sciences Center, London, United Kingdom) for generously providing EBV-LCLs derived from individuals carrying constitutional deletions of the Y chromosome. Assays in NOD/SCID mice were provided as a core service by Cynthia Nourigat.
This work was supported by the J. Orin Edson Foundation, a Lilly Clinical Investigator Award from the Damon Runyon Cancer Research Foundation (E.H.W.), National Institutes of Health grants CA106512 (E.H.W.), CA18029 (S.R.R.), and DK56465, a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Science, Sports, and Technology (Y.A.), the Poncin Scholarship Fund, and the University of Washington Medical Scientist Training Program.
National Institutes of Health
Authorship
Contribution: K.V.R., N.F., J.K.M., K.K.W.K., S.M.X., and E.H.W. performed experiments; O.S.-T., J.S.G., J.P.R., Y.A., and B.J.V.E. contributed vital new reagents; E.H.W., K.V.R., N.F., and S.R.R. analyzed the data; E.H.W. and K.V.R. designed the research, wrote the paper, and made the figures; all authors edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edus H. Warren, Program in Immunology, Fred Hutchinson Cancer Research Center, Seattle, WA 98109-1024; e-mail: ehwarren@u.washington.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal