Abstract
A venous thromboembolism (VTE) with the subsequent risk of pulmonary embolism is a major concern in the treatment of patients with multiple myeloma with thalidomide. The susceptibility to developing a VTE in response to thalidomide therapy is likely to be influenced by both genetic and environmental factors. To test genetic variation associated with treatment related VTE in patient peripheral blood DNA, we used a custom-built molecular inversion probe (MIP)–based single nucleotide polymorphism (SNP) chip containing 3404 SNPs. SNPs on the chip were selected in “functional regions” within 964 genes spanning 67 molecular pathways thought to be involved in the pathogenesis, treatment response, and side effects associated with myeloma therapy. Patients and controls were taken from 3 large clinical trials: Medical Research Council (MRC) Myeloma IX, Hovon-50, and Eastern Cooperative Oncology Group (ECOG) EA100, which compared conventional treatments with thalidomide in patients with myeloma. Our analysis showed that the set of SNPs associated with thalidomide-related VTE were enriched in genes and pathways important in drug transport/metabolism, DNA repair, and cytokine balance. The effects of the SNPs associated with thalidomide-related VTE may be functional at the level of the tumor cell, the tumor-related microenvironment, and the endothelium. The clinical trials described in this paper have been registered as follows: MRC Myeloma IX: ISRCTN68454111; Hovon-50: NCT00028886; and ECOG EA100: NCT00033332.
Introduction
The introduction of thalidomide and other immunomodulatory drugs has revolutionized clinical management of patients with myeloma. Thalidomide treatment has achieved response rates of 30% at relapse and even higher rates at presentation.1 Investigation of the specific effects of thalidomide in myeloma remains an active area of research where up-regulation of ICAM-1,2 VCAM-1, IL-10, and 3,4 IL-12,5 and decreased levels of VEGF,6 βFGF,7-9 HGF,10 TNFα,11 IL-6,12 and soluble IL-6 receptor (sIL-6R)13 are thought to play a role in the mechanism of action, which suggests that thalidomide effects the myeloma cell directly as well as its microenvironment.14
The therapeutic use of thalidomide has focused attention on venous thrombotic events (VTEs). There appears to be a background rate of 5% to 10% VTE15,16 in myeloma possibly due to enhanced expression of tissue factor and VEGF,17 acquired cytokine-mediated activated protein C resistance,18 and down-regulation of thrombospondin.19 In intensively treated patients exposed to thalidomide, the rate of VTE increases to 10% to 15%16,20,21 ; the mechanisms leading to this are uncertain, but it is known that thalidomide regulates the level of COX-2,22-25 a well described prothrombotic factor. Thalidomide may also modulate the VTE risk by its effects on cytokine levels acting on the endothelial cell, a mechanism dependent on the differential apoptotic effects of thalidomide in myeloma plasma cells compared with endothelial cells, which are protected from apoptosis by decrease of VEGF by thalidomide.26-28 In this context, it is known that stressed human umbilical vein endothelial cells (HUVECs) up-regulate a number of procoagulant factors, including PAR-1, P-selectin, E-selectin, and tissue factor. Thalidomide protects these cells from apoptosis potentially enhancing these procoagulant effects, and there is some clinical evidence for this mechanism in non-myeloma settings.29-33
The risk of developing a VTE following thalidomide exposure depends upon a number of factors, including disease stage, the type of chemotherapy combination, and the supportive therapy used. Patient-specific variables also contribute to the excess risk of VTE, including immobility, poor performance status, and dehydration. An important clinical observation is that VTEs occur early after the initiation of thalidomide treatment. VTE rates are also increased in patients when used in conjunction with anthracycline and dexamethasone,34,35 and can decrease following exposure to bortezomib.36-40
The excess risk of thalidomide associated VTE in myeloma has been managed by a number of different strategies, ranging from the identification of high-risk patients suitable for prophylaxis to prophylactic anticoagulation for all patients.41 Aspirin has been suggested to be effective,42 but its use is controversial because of the lack of a readily applicable mechanism justifying its use. In this work, we have examined inherited genetic variation associated with VTE following thalidomide exposure in patients with myeloma using a custom array-based single nucleotide polymorphism (SNP) detection tool in an effort to elucidate the molecular mechanisms contributing to increased risk.
Methods
Clinical samples
Peripheral blood DNA samples were obtained from 544 patients with myeloma derived from 3 randomized clinical trials comparing standard induction treatment for presenting patients with thalidomide containing regimens derived from the Medical Research Council (MRC) Myeloma IX (1966 patients), the Eastern Cooperative Oncology Group (ECOG) EA100 (900 patients), and the Hovon-50 study (400 patients; Figure 1). The dose of thalidomide (100-200 mg daily) was comparable between the 3 studies, but the chemotherapy combinations used differed. The samples were used as the basis for 2 nested case-control comparisons examining the inherited genetic contribution to the risk of VTE as a consequence of thalidomide exposure. In a discovery set analysis, we compared the genotype results derived from 157 Myeloma IX patients with VTEs, of which 104 were related to thalidomide exposure and 53 were unrelated, to a control group of 315 age- and sex-matched patients with myeloma also in the trial, who did not develop a VTE (198 thalidomide-exposed patients and 117 non–thalidomide-exposed patients). To validate the frequency distributions, we carried out a second case-control comparison using 23 patients with VTE treated with thalidomide and 49 thalidomide-treated controls. To ensure homogeneity of allelic frequencies, only patients of European descent were included. This study has been approved by The United Kingdom Multicentre Ethics Committee.
Simplified treatment arms of ECOG EA100, Hovon-50, and Myeloma IX studies.
Clinical trials
The Myeloma IX study comprises 2 randomizations: an intensive pathway for younger, fitter patients comparing CVAD (cyclophosphamide 500 mg orally weekly, vincristine 0.4 mg intravenously on days [d] 1-4), doxorubicin 9.0 mg/m2 on d1-d4, dexamethasone 40 mg on d1-d4 and d12-d15), delivered by a central venous access device with oral CTD (cyclophosphamide, thalidomide, dexamethasone) using the same doses of cyclophosphamide and dexamethasone combined with 200 mg of thalidomide. The second randomization, for older, less-fit patients, compared an attenuated dose of CTD (thalidomide 100-200 mg) to melphalan (7.0 mg/m2 orally on d1-d4 every 28 days) and prednisolone (MP). All patients at high risk of VTE, defined by clinical criteria, were identified; prophylactic anticoagulation was considered by the treating physician, but it was not specified. The ECOG EA100 study randomized patients to either dexamethasone alone 40 mg daily from d1 to d4 and d12 to d15 or the same dose in combination with thalidomide 200 mg daily. In the study set, from which samples were available, no thromboprophylaxis was used on either arm. The Hovon-50 study randomized patients to either 3 cycles of VAD (vincristine 0.4 mg, intravenous rapid infusion on d1-d4; doxorubicin 9 mg/m2, intravenous rapid infusion on d1-d4; and dexamethasone 40 mg orally, d1-d4, d9-d12, and d17-d20) or the same regimen but with thalidomide replacing the vincristine (TAD). Thalidomide was given daily at a dose of 200 mg, but could be escalated to 400 mg. All patients in the TAD arm received thromboprophylaxis with low-molecular-weight heparin (LMWH). Incident cases of VTE were defined using clinical criteria, and no screening approach was used. The identification of VTE represents current clinical practice with initial clinical identification and subsequent confirmation and definition of the extent of thrombosis using a definitive radiologic investigation. Central venous thrombosis and line-related thrombosis were defined by clinical criteria and subsequently confirmed by ultrasound.
Genotyping, SNP selection, and chip design
DNA was extracted from frozen white blood cell pellets using the QIAGEN Flexigene kit (Valencia, CA) and quantified using a Nanodrop spectrophotometer (Wilmington, DE). Genotyping was performed using the Affymetrix targeted genotyping platform (Santa Clara, CA), which is based on a molecular inversion probe technology.43-45 Patient samples were assayed using a custom-built 3.0K panel comprising 3400 SNPs. SNPs were selected using a hypothesis-driven strategy. Pertinent candidate genes were nominated by myeloma groups in the International Myeloma Foundation–led “Bank On A Cure” (BOAC) consortium. An initial list was supplemented with referencing pathway databases, including BioCarta, Kyoto Encyclopedia of Genes and Genomes (KEGG),46,47 and Pathway Assist (Ariadne Genomics, Rockville, MD),48 generating a candidate gene list spanning some 67 molecular pathways important in the biology of myeloma, treatment response, and side effects to conventional and novel agents, which included important genes within the clotting and prothrombotic pathways. Taking the BOAC candidate genes, we completed a literature search49 to identify SNPs that had been previously reported as having a functional consequence or relevance in prior etiologic or treatment outcome studies. SNPs with a minor allele frequency (MAF) greater than 2% were then systematically selected from the candidate gene list using the following criteria: nonsynonymous SNPs present in dbSNP/SNP 50050 ; promoter variants present in homologous regions between human and mouse, in or adjacent to a transcription binding site using the Promolign database51 ; and promoter SNPs identified in the Functional Element SNPs Database (FESD).52 We then included Tag-SNPs in genes considered to be of particular relevance along with population discriminating admixture variants from the X chromosome.53 Finally, we included all nonsynonymous SNPs present in the dbSNP database in phosphatase, kinase, and transferase genes with a MAF greater than 2%. The genes and SNPs comprising this panel with allele frequencies are available online (see Johnson54 ).
Statistical analyses
We carried out a Fisher exact Hardy-Weinberg equilibrium (HWE) test at a P value less than or equal to .001 on all SNPs across the control samples and removed SNPs departing from HWE from the analysis to filter erroneously performing SNPs. We then carried out a “test of missing-ness” on patient and control status to control for any bias in missing data. We performed a basic Fisher (allelic) association test for disease trait based on a comparison of patients with controls. We then completed the analysis using 3 genetic models: additive (Cochran-Armitage trend test), dominant, and recessive. To account for multiple testing, we carried out label-swapping permutation procedures on each of the SNP assays, with their most significant models used to calculate an empirical P value for each SNP. The size of the dataset generated on the BOAC panel is much larger than a typical candidate-gene study; we therefore carried out this analysis in the program PLINK,55 an open-source whole genome association analysis toolset designed for large dataset analysis. The test for epistasis involved testing all pairwise combinations of SNPs. The output consists only of pairwise epistatic results above a P level less than .001 for each SNP. Combinations were restricted to SNPs more than 1 MB apart or on different chromosomes. This test is only an approximation of the extent of epistasis (SNP-SNP interaction), as it is a naive statistic that does not take linkage disequilibrium (LD) into account. We characterized the haplotypes using Haploview 4.0,56 and completed haplotype trend regression in Helix-tree (Bozeman, MT). Meta-analysis was performed in SPSS 14.0 (Chicago, IL) using a meta-analysis macro written by Garcia-Granero.57 Combined odds ratios were calculated using the Mantel Haenszel method for fixed events.
Biological relevance of the associated SNPs
To examine the possible functionality of the thalidomide-related VTE-associated SNPs, we used 2 complementary in silico algorithms for prediction of the putative impact of missense variants on protein function, PolyPhen58 (structural) and the SIFT59 (conservation), shown in Table S8 (available on the Blood website; see the Supplemental Materials link at the top of the online article). We then used a bioinformatics approach to define the pathways potentially deregulated by the associated and validated genes. We used the functional annotation tool on the DAVID Bioinformatics Resources/Database60 to characterize which pathways are most represented in associated gene groups from our single-point analysis. The gene coverage of the BOAC chip was used to form a template/background set, against which associated genes and validated associated SNPs with VTE were tested (Table S9).
Recursive partitioning
To develop a predictive model for the identification of patients at high risk of VTE, we first divided the combined dataset into a training and validation set. We then applied the method of recursive partitioning to the training set.61 In this approach, a regression tree is built by first finding the SNP which best splits the data into 2 groups (VTE, no VTE). This process is repeated over and over again for the individual subsets until the subgroups reach a minimum size or no improvement can be made. The second stage in recursive partitioning consists of cross-validation by trimming back (pruning) the typically complex full tree. The best pruned trees are examined to find which one has the largest classification rate while using the smallest number of SNPs. Sensitivity and specificity are determined for the training and validation set. A receiver operator characteristic (ROC) curve is used to determine the best sensitivity and specificity trade-off.
Results
Clinical results
The Myeloma IX analysis is based on 1966 randomized patients: 984 patients treated with CTD, 557 patients treated with CVAD, and 425 patients treated with MP. In the intensive pathway, the overall rate of VTE was identical in both arms (Table S1). However, there was a qualitative difference between the 2 arms, with deep vein thrombosis (DVT) predominant in the thalidomide-treated group and line-related thrombosis predominant in the CVAD group. In the nonintensive pathway, very few VTEs were seen in the MP group, whereas in the CTD group there was a 15.0% VTE rate. The median time to VTE in each of the groups was approximately 12 weeks from treatment initiation. The Hovon-50 study had VTE rates of 12.1% and 11.8% in the thalidomide-related and standard arms, respectively, with median time to first event of 8.9 weeks. In the ECOG EA100 study, the VTE rates were 17.0% and 3.0% in the thalidomide-related and standard arms, respectively.62
Panel, sample, and SNP assay validation
Affymetrix constructed and validated the SNP panel reagents. A total of 59 DNA samples from the extensively characterized and genotyped Coriell CEPH HAPMAP series were assayed to validate the call performance of the BOAC panel. A total of 58 Coriell CEPH HAPMAP samples were also used in a correlation analysis between the BOAC chip and HAPMAP study. We did not obtain Hapmap data for the remaining Coriell samples and did not perform a correlation analysis. A total of 2606 SNPs were present on both the chip and HAPMAP. There was a SNP call correlation of 96.1% at 95% confidence levels; SNPs falling below this were removed from the analysis (132 SNPs). The Coriell sample genotype validation was replicated in BOAC labs to ensure there was no differential bias in genotyping scoring between sites. Patients and controls were also genotyped together throughout the experiment to avoid any differential bias in genotype scoring. We observed complete agreement between the known sex and inferred SNP-based sex in all samples. We used Eigenstrat63 to highlight population stratification and removed 4 population outliers from the analysis. A number of admixture SNPs were included in the SNP panel.53 Genotype calls for these SNPs demonstrated that patients and controls reflected a sample set drawn from a European population.
Genotyping results and validation
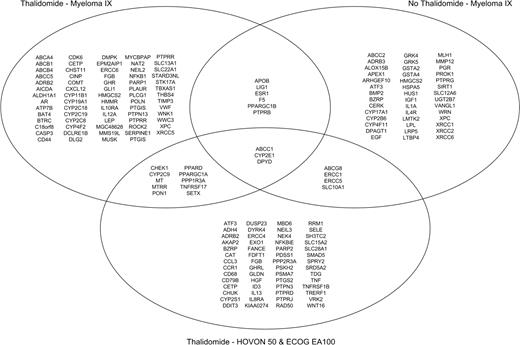
The MRC Myeloma IX study is the largest of the datasets in this study; to capitalize on this, we chose to focus our discovery set on this study and to validate the results on combined data sets from Hovon-50 and ECOG EA100 trials. A set of SNPs for validation was defined by separately determining the distribution of the most significant SNPs in the MRC myeloma IX, Hovon-50, and ECOG EA100 studies. Following testing allelic distributions using the Fisher test to an empirical P value less than .05 in the discovery set, 120 SNPs were found to be associated with thalidomide-associated VTE, involving 71 genes (Table S2). Further genetic model association analysis in Myeloma IX data are listed in Table S3; allelic and genetic model association for non–thalidomide-related VTEs are listed in Tables S4 and S5, with allelic and genetic model association for Hovon-50/EA100 trials shown in Tables S6 and S7. With the aim of identifying genes modulating the risk of thalidomide-related thrombosis, we compared the distribution of the SNPs identified in this analysis between 3 subgroups: thalidomide-associated VTEs from MRC Myeloma IX, non–thalidomide-associated VTEs from MRC Myeloma IX, and thalidomide-associated VTEs from the combined Hovon-50/ECOG EA100 studies. A Venn diagram depicting the overlapping associated genes in the 3 analyses is shown in Figure 2.
Venn diagram showing overlapping VTE-associated genes between thalidomide Myeloma IX, non-thalidomide Myeloma IX, and thalidomide Hovon-50/ECOG EA100 analyses.
Venn diagram showing overlapping VTE-associated genes between thalidomide Myeloma IX, non-thalidomide Myeloma IX, and thalidomide Hovon-50/ECOG EA100 analyses.
Validation of thalidomide-related VTE-associated SNPs
To validate the genotyping results in the discovery set, we discarded SNPs with conflicting frequency distributions between studies. This approach may have led to the removal of a number of true positives from the analysis because of the small sizes of the Hovon-50 and ECOG EA100 datasets. SNPs with small effect sizes were also removed from the analysis. As a result of this process we found 24 SNPs (Table 1) associated with VTE in the “discovery set” with consistent distributions in the 2 validation datasets. Haplotype analysis showed that 6 SNPs were in linkage with a stronger proxy SNP and as such were discarded, leaving 18 validated SNPs associated with thalidomide-related VTEs. A “forest plot” with odds ratios (ORs) and confidence intervals (CIs) for the combined and individual datasets of the validated SNPs was generated (Figure 3).
Allele distributions of cross-trial–validated associated thalidomide-related VTE SNPs
| SNP . | Chromosome . | Gene . | Functional class . | Minor allele . | MAF patients . | MAF controls . | Major allele . | P . | OR . | L95 . | U95 . | Empirical P . | Trial . | In Linkage . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2302387 | 7 | ABCB4 | Coding-synon | A | 0.085 | 0.1432 | G | .042 | 0.5557 | 0.3133 | 0.9857 | .041 | Myeloma IX | |
| A | 0.375 | 0.4 | G | .879 | 0.9 | 0.2331 | 3.475 | .857 | Hovon-50 | |||||
| A | 0 | 0.18 | G | .112 | NA | NA | NA | .149 | E1A100 | |||||
| rs1049216 | 4 | CASP3 | Untranslated | C | 0.19 | 0.2891 | T | .009 | 0.5769 | 0.3804 | 0.875 | .006 | Myeloma IX | |
| C | 0.2778 | 0.4545 | T | .251 | 0.4615 | 0.1221 | 1.745 | .345 | Hovon-50 | |||||
| C | 0.08333 | 0.42 | T | .029 | 0.1255 | 0.01503 | 1.049 | .087 | E1A100 | |||||
| rs506504 | 11 | CHEK1 | Coding-nonsynon | T | 0.06633 | 0.02865 | C | .031 | 2.409 | 1.059 | 5.481 | .044 | Myeloma IX | |
| T | 0.1111 | 0 | C | .109 | NA | NA | NA | .146 | Hovon-50 | |||||
| T | 0.25 | 0.06 | C | .046 | 5.222 | 0.9057 | 30.11 | .060 | E1A100 | |||||
| rs7011 | 14 | CINP | Coding-nonsynon | T | 0.3081 | 0.2266 | C | .032 | 1.52 | 1.034 | 2.233 | .036 | Myeloma IX | |
| T | 0.3889 | 0.25 | C | .358 | 1.909 | 0.4772 | 7.638 | .357 | Hovon-50 | |||||
| T | 0.5 | 0.22 | C | .051 | 3.545 | 0.9522 | 13.2 | .096 | E1A100 | |||||
| rs12022378 | 1 | DCLRE1B | Coding-nonsynon | T | 0.2323 | 0.1536 | C | .019 | 1.667 | 1.083 | 2.565 | .023 | Myeloma IX | |
| T | 0.2222 | 0.09091 | C | .247 | 2.857 | 0.4585 | 17.8 | .632 | Hovon-50 | |||||
| T | 0.25 | 0.22 | C | .823 | 1.182 | 0.2723 | 5.13 | .999 | E1A100 | |||||
| rs4253211 | 10 | ERCC6 | Coding-nonsynon | C | 0.1616 | 0.09896 | G | .028 | 1.755 | 1.059 | 2.909 | .035 | Myeloma IX | |
| C | 0.1667 | 0.04545 | G | .204 | 4.2 | 0.3973 | 44.4 | .261 | Hovon-50 | |||||
| C | 0.1667 | 0.1 | G | .512 | 1.8 | 0.3044 | 10.64 | .560 | E1A100 | |||||
| rs299295 | 5 | HMMR | Coding-nonsynon | T | 0.28 | 0.1979 | C | .024 | 1.576 | 1.059 | 2.346 | .041 | Myeloma IX | |
| T | 0.3889 | 0.2727 | C | .435 | 1.697 | 0.4472 | 6.439 | .999 | Hovon-50 | |||||
| T | 0.3333 | 0.32 | C | .929 | 1.063 | 0.2784 | 4.055 | .999 | E1A100 | |||||
| rs582537 | 3 | IL12A | Intron | A | 0.515 | 0.3995 | C | .007 | 1.596 | 1.132 | 2.251 | .005 | Myeloma IX | rs2227314 |
| A | 0.5 | 0.5 | C | .999 | 1 | 0.2877 | 3.476 | .999 | Hovon-50 | rs2227314 | ||||
| A | 0.5833 | 0.44 | C | .372 | 1.782 | 0.4973 | 6.385 | .591 | E1A100 | rs2227314 | ||||
| rs10249476 | 7 | LEP | Promoter | T | 0.405 | 0.3115 | G | .024 | 1.504 | 1.054 | 2.147 | .026 | Myeloma IX | |
| T | 0.5556 | 0.4091 | G | .356 | 1.806 | 0.5123 | 6.363 | .450 | Hovon-50 | |||||
| T | 0.3333 | 0.2083 | G | .360 | 1.9 | 0.4743 | 7.611 | .517 | E1A100 | |||||
| rs20579 | 19 | LIG1 | Untranslated | T | 0.065 | 0.1263 | C | .022 | 0.481 | 0.2544 | 0.9094 | .017 | Myeloma IX | |
| T | 0.05556 | 0.2727 | C | .072 | 0.1569 | 0.01696 | 1.451 | .113 | Hovon-50 | |||||
| T | 0 | 0.04 | C | .481 | NA | NA | NA | .999 | E1A100 | |||||
| rs13815 | 22 | MT | Coding-nonsynon | C | 0.2576 | 0.3679 | G | .007 | 0.5961 | 0.4078 | 0.8716 | .010 | Myeloma IX | |
| C | 0.2222 | 0.4545 | G | .125 | 0.3429 | 0.08519 | 1.38 | .126 | Hovon-50 | |||||
| C | 0.25 | 0.36 | G | .470 | 0.5926 | 0.142 | 2.473 | .857 | E1A100 | |||||
| rs2410558 | 8 | NAT2 | Locus | T | 0.215 | 0.3032 | C | .024 | 0.6295 | 0.4208 | 0.9416 | .024 | Myeloma IX | |
| T | 0.2222 | 0.5 | C | .071 | 0.2857 | 0.07114 | 1.148 | .226 | Hovon-50 | |||||
| T | 0.25 | 0.28 | C | .834 | 0.8571 | 0.202 | 3.636 | .999 | E1A100 | |||||
| rs3774968 | 4 | NFKB1 | Intron | A | 0.355 | 0.4581 | G | .017 | 0.651 | 0.4575 | 0.9263 | .017 | Myeloma IX | |
| A | 0.3333 | 0.3182 | G | .919 | 1.071 | 0.2838 | 4.046 | .999 | Hovon-50 | |||||
| A | 0.4 | 0.46 | G | .728 | 0.7826 | 0.1965 | 3.117 | .800 | E1A100 | |||||
| rs1805414 | 1 | PARP1 | Coding-synon | C | 0.25 | 0.3376 | T | .029 | 0.6539 | 0.4459 | 0.9592 | .033 | Myeloma IX | rs1002153, rs2048426, rs2267669 |
| C | 0.2222 | 0.4545 | T | .125 | 0.3429 | 0.08519 | 1.38 | .328 | Hovon-50 | rs1002153, rs2048426, rs2267669 | ||||
| C | 0.3333 | 0.36 | T | .862 | 0.8889 | 0.2346 | 3.367 | .999 | E1A100 | rs1002153, rs2048426, rs2267669 | ||||
| rs2267669 | 6 | PPARD | Intron | G | 0.1198 | 0.2139 | A | .006 | 0.5001 | 0.3031 | 0.8253 | .007 | Myeloma IX | |
| G | 0 | 0.2273 | A | .031 | NA | NA | NA | .037 | Hovon-50 | |||||
| G | 0.1667 | 0.1818 | A | .903 | 0.9 | 0.1643 | 4.929 | .999 | E1A100 | |||||
| rs2070682 | 7 | SERPINE1 | Intron | C | 0.365 | 0.4562 | T | .034 | 0.6852 | 0.4827 | 0.9728 | .029 | Myeloma IX | |
| C | 0.3333 | 0.3636 | T | .842 | 0.875 | 0.2362 | 3.241 | .999 | Hovon-50 | |||||
| C | 0.4167 | 0.42 | T | .983 | 0.9864 | 0.2749 | 3.539 | .999 | E1A100 | |||||
| rs12922317 | 16 | TNFRSF17 | Intron,Tag-SNP | G | 0.29 | 0.3958 | A | .011 | 0.6234 | 0.4317 | 0.9004 | .010 | Myeloma IX | rs11862958 |
| G | 0.2778 | 0.4 | A | .428 | 0.5769 | 0.1473 | 2.26 | .380 | Hovon-50 | rs11862958 | ||||
| G | 0.1667 | 0.36 | A | .198 | 0.3556 | 0.07006 | 1.804 | .800 | E1A100 | rs11862958 | ||||
| rs2440 | 2 | XRCC5 | Untranslated | T | 0.4286 | 0.3486 | C | .062 | 1.401 | 0.9828 | 1.998 | .047 | Myeloma IX | rs207932 |
| T | 0.25 | 0.1818 | C | .611 | 1.5 | 0.3131 | 7.186 | .857 | Hovon-50 | rs207932 | ||||
| T | 0.4167 | 0.2292 | C | .189 | 2.403 | 0.6351 | 9.088 | .226 | E1A100 | rs207932 |
| SNP . | Chromosome . | Gene . | Functional class . | Minor allele . | MAF patients . | MAF controls . | Major allele . | P . | OR . | L95 . | U95 . | Empirical P . | Trial . | In Linkage . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2302387 | 7 | ABCB4 | Coding-synon | A | 0.085 | 0.1432 | G | .042 | 0.5557 | 0.3133 | 0.9857 | .041 | Myeloma IX | |
| A | 0.375 | 0.4 | G | .879 | 0.9 | 0.2331 | 3.475 | .857 | Hovon-50 | |||||
| A | 0 | 0.18 | G | .112 | NA | NA | NA | .149 | E1A100 | |||||
| rs1049216 | 4 | CASP3 | Untranslated | C | 0.19 | 0.2891 | T | .009 | 0.5769 | 0.3804 | 0.875 | .006 | Myeloma IX | |
| C | 0.2778 | 0.4545 | T | .251 | 0.4615 | 0.1221 | 1.745 | .345 | Hovon-50 | |||||
| C | 0.08333 | 0.42 | T | .029 | 0.1255 | 0.01503 | 1.049 | .087 | E1A100 | |||||
| rs506504 | 11 | CHEK1 | Coding-nonsynon | T | 0.06633 | 0.02865 | C | .031 | 2.409 | 1.059 | 5.481 | .044 | Myeloma IX | |
| T | 0.1111 | 0 | C | .109 | NA | NA | NA | .146 | Hovon-50 | |||||
| T | 0.25 | 0.06 | C | .046 | 5.222 | 0.9057 | 30.11 | .060 | E1A100 | |||||
| rs7011 | 14 | CINP | Coding-nonsynon | T | 0.3081 | 0.2266 | C | .032 | 1.52 | 1.034 | 2.233 | .036 | Myeloma IX | |
| T | 0.3889 | 0.25 | C | .358 | 1.909 | 0.4772 | 7.638 | .357 | Hovon-50 | |||||
| T | 0.5 | 0.22 | C | .051 | 3.545 | 0.9522 | 13.2 | .096 | E1A100 | |||||
| rs12022378 | 1 | DCLRE1B | Coding-nonsynon | T | 0.2323 | 0.1536 | C | .019 | 1.667 | 1.083 | 2.565 | .023 | Myeloma IX | |
| T | 0.2222 | 0.09091 | C | .247 | 2.857 | 0.4585 | 17.8 | .632 | Hovon-50 | |||||
| T | 0.25 | 0.22 | C | .823 | 1.182 | 0.2723 | 5.13 | .999 | E1A100 | |||||
| rs4253211 | 10 | ERCC6 | Coding-nonsynon | C | 0.1616 | 0.09896 | G | .028 | 1.755 | 1.059 | 2.909 | .035 | Myeloma IX | |
| C | 0.1667 | 0.04545 | G | .204 | 4.2 | 0.3973 | 44.4 | .261 | Hovon-50 | |||||
| C | 0.1667 | 0.1 | G | .512 | 1.8 | 0.3044 | 10.64 | .560 | E1A100 | |||||
| rs299295 | 5 | HMMR | Coding-nonsynon | T | 0.28 | 0.1979 | C | .024 | 1.576 | 1.059 | 2.346 | .041 | Myeloma IX | |
| T | 0.3889 | 0.2727 | C | .435 | 1.697 | 0.4472 | 6.439 | .999 | Hovon-50 | |||||
| T | 0.3333 | 0.32 | C | .929 | 1.063 | 0.2784 | 4.055 | .999 | E1A100 | |||||
| rs582537 | 3 | IL12A | Intron | A | 0.515 | 0.3995 | C | .007 | 1.596 | 1.132 | 2.251 | .005 | Myeloma IX | rs2227314 |
| A | 0.5 | 0.5 | C | .999 | 1 | 0.2877 | 3.476 | .999 | Hovon-50 | rs2227314 | ||||
| A | 0.5833 | 0.44 | C | .372 | 1.782 | 0.4973 | 6.385 | .591 | E1A100 | rs2227314 | ||||
| rs10249476 | 7 | LEP | Promoter | T | 0.405 | 0.3115 | G | .024 | 1.504 | 1.054 | 2.147 | .026 | Myeloma IX | |
| T | 0.5556 | 0.4091 | G | .356 | 1.806 | 0.5123 | 6.363 | .450 | Hovon-50 | |||||
| T | 0.3333 | 0.2083 | G | .360 | 1.9 | 0.4743 | 7.611 | .517 | E1A100 | |||||
| rs20579 | 19 | LIG1 | Untranslated | T | 0.065 | 0.1263 | C | .022 | 0.481 | 0.2544 | 0.9094 | .017 | Myeloma IX | |
| T | 0.05556 | 0.2727 | C | .072 | 0.1569 | 0.01696 | 1.451 | .113 | Hovon-50 | |||||
| T | 0 | 0.04 | C | .481 | NA | NA | NA | .999 | E1A100 | |||||
| rs13815 | 22 | MT | Coding-nonsynon | C | 0.2576 | 0.3679 | G | .007 | 0.5961 | 0.4078 | 0.8716 | .010 | Myeloma IX | |
| C | 0.2222 | 0.4545 | G | .125 | 0.3429 | 0.08519 | 1.38 | .126 | Hovon-50 | |||||
| C | 0.25 | 0.36 | G | .470 | 0.5926 | 0.142 | 2.473 | .857 | E1A100 | |||||
| rs2410558 | 8 | NAT2 | Locus | T | 0.215 | 0.3032 | C | .024 | 0.6295 | 0.4208 | 0.9416 | .024 | Myeloma IX | |
| T | 0.2222 | 0.5 | C | .071 | 0.2857 | 0.07114 | 1.148 | .226 | Hovon-50 | |||||
| T | 0.25 | 0.28 | C | .834 | 0.8571 | 0.202 | 3.636 | .999 | E1A100 | |||||
| rs3774968 | 4 | NFKB1 | Intron | A | 0.355 | 0.4581 | G | .017 | 0.651 | 0.4575 | 0.9263 | .017 | Myeloma IX | |
| A | 0.3333 | 0.3182 | G | .919 | 1.071 | 0.2838 | 4.046 | .999 | Hovon-50 | |||||
| A | 0.4 | 0.46 | G | .728 | 0.7826 | 0.1965 | 3.117 | .800 | E1A100 | |||||
| rs1805414 | 1 | PARP1 | Coding-synon | C | 0.25 | 0.3376 | T | .029 | 0.6539 | 0.4459 | 0.9592 | .033 | Myeloma IX | rs1002153, rs2048426, rs2267669 |
| C | 0.2222 | 0.4545 | T | .125 | 0.3429 | 0.08519 | 1.38 | .328 | Hovon-50 | rs1002153, rs2048426, rs2267669 | ||||
| C | 0.3333 | 0.36 | T | .862 | 0.8889 | 0.2346 | 3.367 | .999 | E1A100 | rs1002153, rs2048426, rs2267669 | ||||
| rs2267669 | 6 | PPARD | Intron | G | 0.1198 | 0.2139 | A | .006 | 0.5001 | 0.3031 | 0.8253 | .007 | Myeloma IX | |
| G | 0 | 0.2273 | A | .031 | NA | NA | NA | .037 | Hovon-50 | |||||
| G | 0.1667 | 0.1818 | A | .903 | 0.9 | 0.1643 | 4.929 | .999 | E1A100 | |||||
| rs2070682 | 7 | SERPINE1 | Intron | C | 0.365 | 0.4562 | T | .034 | 0.6852 | 0.4827 | 0.9728 | .029 | Myeloma IX | |
| C | 0.3333 | 0.3636 | T | .842 | 0.875 | 0.2362 | 3.241 | .999 | Hovon-50 | |||||
| C | 0.4167 | 0.42 | T | .983 | 0.9864 | 0.2749 | 3.539 | .999 | E1A100 | |||||
| rs12922317 | 16 | TNFRSF17 | Intron,Tag-SNP | G | 0.29 | 0.3958 | A | .011 | 0.6234 | 0.4317 | 0.9004 | .010 | Myeloma IX | rs11862958 |
| G | 0.2778 | 0.4 | A | .428 | 0.5769 | 0.1473 | 2.26 | .380 | Hovon-50 | rs11862958 | ||||
| G | 0.1667 | 0.36 | A | .198 | 0.3556 | 0.07006 | 1.804 | .800 | E1A100 | rs11862958 | ||||
| rs2440 | 2 | XRCC5 | Untranslated | T | 0.4286 | 0.3486 | C | .062 | 1.401 | 0.9828 | 1.998 | .047 | Myeloma IX | rs207932 |
| T | 0.25 | 0.1818 | C | .611 | 1.5 | 0.3131 | 7.186 | .857 | Hovon-50 | rs207932 | ||||
| T | 0.4167 | 0.2292 | C | .189 | 2.403 | 0.6351 | 9.088 | .226 | E1A100 | rs207932 |
NA indicates not applicable; L95, lower 95% CI; and U95, upper 95% CI.
Forest plots showing distribution of validated SNPs associated with thalidomide-related VTEs across the Myeloma IX, Hovon-50, and ECOG EA100 trials. Error bars indicate upper and lower 95% CIs.
Forest plots showing distribution of validated SNPs associated with thalidomide-related VTEs across the Myeloma IX, Hovon-50, and ECOG EA100 trials. Error bars indicate upper and lower 95% CIs.
Gene-gene interactions
To examine gene-gene interactions, we looked for pairwise combinations mediating risk. The epistatic interactions with a P value less than .001 are shown in Table S10.
Recursive partioning analysis
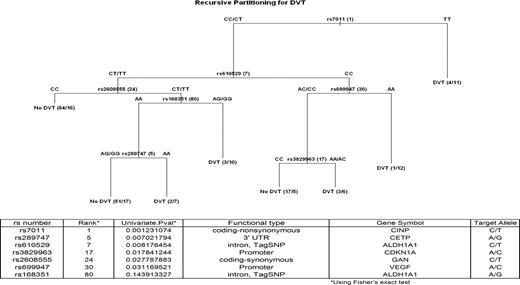
To maximize the size of the dataset and thus to maximize the ability to identify relevant SNPs, we combined all the datasets into one and randomly split it into a two-thirds training set and one-third validation set. The data were stratified by trial and VTE patients to ensure that the training and validation sets were comparable. These data included 165 subjects without VTE and 84 subjects with VTE in the training set and 82 subjects without VTE and 42 subjects with VTE in the validation set. The training set was used to identify the top associated genes and SNPs by association at the level of P less than .05 listed in Table S2. These SNPs were used in a recursive partitioning analysis carried out on the test set with the aim of finding the combination with the best sensitivity and specificity for the identification of VTE. We pruned the tree to find the tree with the highest classification and smallest number of SNPs. The results of this analysis (Figure 4) showed that using 7 SNPs (rs7011 in CINP, rs289747 in CETP, rs610529 in ALDH1A1, rs3829963 in CDKN1A, rs2608555 in GAN, rs699947 in VEGF, and andrs168351 in ALDH1A) it was possible to identify VTEs correctly in 70% of individuals with a specificity of 59% and sensitivity of 81%. This set of SNPs performed well in the validation set; the set was able to correctly classify VTEs in 61% of individuals with a specificity of 30% and sensitivity of 77% (Tables S12 and S13).
Predictive tree of thalidomide-related thrombosis in myeloma patients following recursive partitioning analysis.
Predictive tree of thalidomide-related thrombosis in myeloma patients following recursive partitioning analysis.
Discussion
This study has analyzed data from 3 large randomized clinical studies comprising 3100 patients, comparing induction treatment for newly presenting patients with myeloma with and without thalidomide. The results of this analysis show that the background rate of VTE in MP-treated patients is very low and significantly increases with the addition of thalidomide. In addition we provide further evidence that infusional regimes based on VAD increase VTE rates to around 15%, which is similar to the rates seen with oral thalidomide combinations. The nature of the thrombotic events is qualitatively different between regimes, with all events being either DVT or pulmonary embolism (PE) in the oral thalidomide-treated patients, whereas in the intravenous treatment, 50% of the events are central line related. There is a doubling of non–central line–related VTE rates in the thalidomide-treated patients compared with those receiving infusional induction regimens. The median time to VTE in each of the treatments is approximately 50 to 60 days after the initiation of treatment, a time reflecting the rapid dissolution of the myeloma clone. We have shown previously that response rate is enhanced in thalidomide-containing regimes compared with VAD-like regimes, and we postulate that this is important in determining the VTE risk.64 The mechanistic importance of increased response rates with VTE risk may explain the reduced numbers of VTEs seen in relapsed patients, who are frequently drug resistant and show lower response rates. It is also important not to discount increased VTE risk due to changes in the disease biology related procoagulant profiles of such relapsed patients.
Using a nested case-control design with readily defined exposure and clinical endpoint, this study has given useful information about inherited genetic variants with a moderate effect size affecting the thrombotic response to thalidomide exposure. We chose to use the MRC Myeloma IX study as our initial discovery set because it was the largest and had the most data available with it. Validation in the combined Hovon-50/ECOG study represents a pragmatic decision based on study size, study design, and our desire to identify penetrant variants that can be replicated with relevance to different studies and datasets.
Despite a comprehensive analysis of the genetic variation within the coagulation and prothrombotic pathways, we could not find evidence for a significant association of genetic variation within these pathways with VTE risk following thalidomide exposure. Although we found Factor 5 Leiden (rs6025) to be associated with an increased risk of VTE in this analysis, the thrombotic risk was not increased in patients treated with thalidomide; similar results were seen for polymorphisms in MTHFR and FGB. We saw no association with thalidomide-related VTE in commonly reported VTE risk alleles in F2-455G/A (rs3136430) splice variant 20210G/A (rs3136431). We did find weak associations with genes known to mediate the coagulation pathway, including MTRR, PLAUR, PPARD, PPARGC1A, PPARGC1B, THBS4, and WNK, but the associated risk was not high. We conclude that we can exclude a major contribution of genetic variation within the coagulation and prothrombotic pathways based on this targeted approach, although smaller contributions to the phenotype may be missed because of the study size and design. Our findings are consistent with previous clinical observations and work by some of the authors, who failed to identify relevant changes in functional assays investigating this pathway.65-67
The lack of a strong association with variation in the coagulation cascade suggests that VTE risk is mediated via alternative mechanisms. We identified the 18 SNPs, which validated across the 3 datasets (Figure 3). Using the whole BOAC panel as the background gene set in the DAVID Functional Annotation Clustering tool against the 18 validated genes, generated 3 major enriched annotation clusters. The annotation clusters consisted of 2 “response to stress” groups; a response to DNA damage group, including CHEK1, XRCC5, LIG1, ERCC6, DCLRE1B, and PARP1; a cytokine response group containing NFKB1, TNFRSF17, IL12B, and LEP; and a third related group of “apoptosis” with CASP3, PPARD, and NFKB1. These enrichment groups indicate that genetic variation in response to DNA damage and cytokine-mediated apoptosis modulates risk of developing a thalidomide-related thrombosis.
High-dose dexamethasone enhances hemostasis, increases platelet activation, and promotes von Willebrand factor (VWF) antigen–dependent thrombosis.68 Extremely high levels of factor VIII coagulant (FVIII:C) activity and VWF have been found in thalidomide-exposed patients.69 Patients that develop a subsequent VTE had higher VWF antigen (Ag) levels but not FVIII:C levels. High FVIII:C/VWF Ag levels are found in patients with active myeloma; this is probably a reflection of increased bone marrow angiogenesis in myeloma. These prothrombogenic circumstances would contribute to VTE during treatment with a thalidomide-dexamethasone combination.69 In line with VWF mediating the prothrombotic effects of dexamethasone in thalidomide-related VTE, we saw a protective effect of VWF nonsynonymous SNP (rs216321) and synonymous SNP (rs216902) in thalidomide-treated controls.
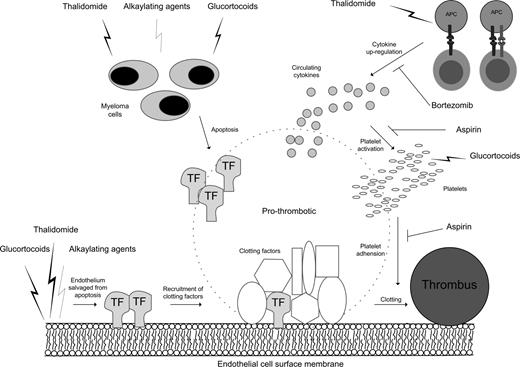
Although there is evidence to suggest thalidomide may damage DNA directly,70 it is important to note that most patients in this analysis were derived from the MRC and Hovon-50 studies, which included either cyclophosphamide or doxorubicin/adriamycin in the treatment regime, which may explain an association with DNA repair genes. Variation in DNA repair capacity could readily affect the response of the myeloma clone to treatment due to the direct relationship between the extent of DNA damage accumulation and the clinical response to alkylating agents.71 A rapid response and dissolution of myeloma clones with an impaired double-stranded DNA repair pathway would release greater prothrombotic factors that could be either microparticles with surface tissue factor or cytokines and tissue factor. The greater thrombogenesis due to increased dissolution of the myeloma clone may act additively with a dexamethasone-thalidomide interaction on plasma cells,72 giving rise to an increased number of VTEs in the MRC and Hovon-50 studies.73,74 An alternative mechanism to explain the increased risk of a VTE associated with DNA repair genes could be based on the observation that thalidomide can protect endothelial cells from doxorubicin-induced apoptosis by restoring PAR-1 expresssion,75 promoting subendothelial tissue factor exposure, endothelial dysfunction, and platelet activation, and consequently increasing the thrombosis risk.75-77 Under these conditions, decreased DNA repair capacity could promote clot formation at the endothelium (Figure 5).
Thalidomide treatment in combination with alkylating agents in myeloma promotes prothrombotic conditions at the endothelium surface via a combination of mechanisms. Mechanisms include rapid apoptosis of myeloma cells leading to circulating tissue factor (TF), exposed TF by endothelium cells salvaged from apoptosis, increased circulating cytokines (eg, TNFα) with T-cell activation by antigen-presenting cells (APCs), and activated platelets in response to increased circulating cytokines.
Thalidomide treatment in combination with alkylating agents in myeloma promotes prothrombotic conditions at the endothelium surface via a combination of mechanisms. Mechanisms include rapid apoptosis of myeloma cells leading to circulating tissue factor (TF), exposed TF by endothelium cells salvaged from apoptosis, increased circulating cytokines (eg, TNFα) with T-cell activation by antigen-presenting cells (APCs), and activated platelets in response to increased circulating cytokines.
The enrichment of cytokine-mediated apoptosis genes in SNPs associated with thalidomide-related thrombosis risk may also give clues to the role bortezomib and aspirin play in VTE management. Low rates of VTE are seen in patients with myeloma treated with bortezomib in thalidomide combinations,37,40,78 possibly through the prevention of the up-regulation of prothrombotic molecules such as thrombomodulin, cytokines, and E-selectin by bortezomib.79,80 A number of clinical studies have suggested that aspirin42,81-89 is effective at preventing the excess of VTEs seen in thalidomide-exposed individuals. Aspirin is classically thought to inhibit platelet COX-2, reducing platelet adhesiveness and modulating risk of arterial thrombosis. Aspirin can also lead to decreased levels of circulating TNF-α by inhibiting IKK and therefore NFκB. Higher levels of TNF-α and COX-2 lead to an increased risk of apoptosis in endothelial cells, which also become proadhesive to nonactivated platelets.90 In a thalidomide treatment setting, aspirin may be able to inhibit thalidomide VTE-mediated events by lowering circulating TNF-α.
Genetic analysis of the multifactorial phenotype that is thalidomide related venous thrombosis is challenging. To minimize experimental artifacts that can be found in many association studies,91 we have associated a discrete clinical outcome from a homogenous population of similarly treated patients with high-quality genotype data with stringent quality controls. We took a hypothesis-driven candidate gene approach rather than a whole genome scan (WGS)–based approach because it was clear that the number of events to be analyzed would be small, and we were aiming to identify pertinent functional loci variants with moderate to large effect size. We accept that future GWS and sequencing approaches may add relevant variants in unknown pathways. As part of the analysis, we took an exploratory approach to defining whether the SNPs identified could be used to identify patients at high risk of VTE and consequently guide clinical intervention. Guidelines have recently been established to govern clinical indicators for intervention, but these prognostic factors can be difficult to identify and use clinically.41 The US Food and Drug Administration (FDA) and European Medicines Evaluation Agency (EMEA) have published warnings suggesting the use of thromboprophylaxis with any immunomodulatory derivative of thalidomide (IMiD)–based regimen.92-94 The results of this recursive partitioning analysis have identified a limited number of SNPs that, when analyzed together, can predict the risk of VTE. Testing for these SNPs has the potential for being clinically useful for identifying high-risk patients for whom therapeutic intervention is required. For clinically defined high-risk patients intervention strategies may not change, but for patients at genetic high risk for whom aspirin was the chosen strategy, intervention with warfarin or LMWH would be more appropriate.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank the staff at the Haematological Malignancy Diagnostic Service, Leeds, and members of the MRC Myeloma IX Trial Management Group, as well as consultants and patients entered in the MRC Myeloma IX, ECOG, and Hovon-50 trials.
This study was supported by the International Myeloma Foundation (IMF) as part of the Bank on a Cure (BOAC) Consortium. We are also grateful for support received from Myeloma UK.
Authorship
Contribution: C.J. wrote the paper, performed research, and analyzed data; S.C. and C.R. performed research; A.H., K.H. N.J.D., J.H., J.A.C., S.E.B., G.H.J., and J.C. analyzed data; H.G., D.B., V.R., P.S., and B.V.N. designed the research; F.E.D. and G.J.M. designed the research and wrote the paper; and B.D. designed the research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: G. J. Morgan, Section of Haemato-Oncology, Institute of Cancer Research, Belmont, Sutton, Surrey, United Kingdom SM2 5NG; e-mail: gareth.morgan@icr.ac.uk.