Follicular lymphoma (FL) is characterized by constitutive expression of Bcl-2 as a consequence of t(14;18). Evidence suggests factors in the lymph node microenvironment, related to intratumoral T cells, macrophages, and dendritic cells, play a role in the disease process. We generated proteomic cytokine profiles of FL (N = 50) and follicular hyperplasia (FH; N = 23). A total of 10 cytokines were assayed using ultrasensitive multiplex enzyme-linked immunosorbent assays: IL-1β, IL-2, IL-4, IL-5, IL-8, IL-10, IL-13, IL-12p70, tumor necrosis factor-α, and interferon-γ. Each cytokine showed overall lower protein concentrations in FL, with the exception of IL-4, which was nearly 5 times higher in FL than FH (P = .005). Using reverse-phase protein microarrays (RPMAs), we evaluated the activation state of several intracellular signaling proteins downstream of cytokine receptors. Basal Erk phosphorylation was approximately 4 times greater in FL than FH (P < .001), with similar findings for Mek; Stat-6 showed weak basal phosphorylation that was approximately twice as high in FL than in FH (P = .012). In conclusion, the FL microenvironment contains increased levels of IL-4, with prominent tumor basal phosphorylation of Erk. These findings suggest IL-4, Erk, and possibly Stat-6 may play a role in the biology of FL and may serve as targets for future therapies.
Introduction
Follicular lymphoma (FL) is a non-Hodgkin lymphoma of germinal center B-cell origin. FL is the second most common non-Hodgkin lymphoma and represents 20% of all lymphomas in the adult population.1 Although the course of disease is indolent, it is largely incurable, with a median survival of 7 to 10 years. With time, FL typically transforms into a high-grade lymphoma. FL is characterized by the t(14;18) (q32;q21) translocation,2,–4 which leads to constitutive expression of the antiapoptotic Bcl-2 protein, conferring survival advantage to FL cells.5
The BCL-2/JH translocation is required but not sufficient for lymphomagenesis, underscored by the fact that the BCL2/JH translocation has been identified in B cells from lymph nodes (LNs) and blood in healthy individuals.6,7 8 Hence, there are likely other genetic alterations within tumor cells that cooperate with Bcl-2 to drive the malignant phenotype in FL.
There is compelling evidence that FL tumor cells are dependent upon the LN microenvironment for continued survival and proliferation.9,–11 Two distinct gene expression signatures that predict patient survival independent of clinical prognostic variables have been identified by gene expression profiling.11 The expression profile designated immune response 1 (IR1) includes T cell– and macrophage-restricted genes and was predictive of a favorable clinical outcome. In contrast, IR2 is associated with genes primarily expressed by monocytes and dendritic cells and was predictive of a poorer outcome. Other studies have also implicated the importance of the tumor-host microenvironment in FL. Patients with spontaneous regression of FL reportedly demonstrated increased numbers of T-helper cells with no apparent differences in cytotoxic T cells or macrophages when compared with patients with FL exhibiting no regression.9 Increased numbers of CD57+ T cells have been associated with a higher frequency of B symptoms and bone marrow involvement.12 The presence of high numbers of CD68+ macrophages (> 15 per high-power field) was an independent negative predictor of survival in one study.13
Intratumoral T cells and macrophages presumably exert effects on the FL microenvironment via secretion of cytokines and/or by direct interaction with tumor cells. Studies have shown FL cells to proliferate in vitro in the presence of autologous CD4+ T cells,14 and in the presence of CD4+ T-cell clones that recognize alloantigens expressed by FL cells.10 Polymorphisms of several cytokines have been implicated in susceptibility to non-Hodgkin lymphoma, including tumor necrosis factor (TNF) and interleukin-10 (IL-10).15 Single-nucleotide polymorphisms (SNPs) of IL-8, IL-2, IL-12β, and the IL-1 receptor were shown in one study to be predictive of survival in patients diagnosed with FL.16
Cytokines play key roles in the regulation of hematopoietic differentiation, proliferation, and apoptosis. Cytokines commonly trigger signal transduction in target cells via the Janus kinase (Jak)–signal transducer and activator of transcription (Stat) pathway,17 with downstream nuclear translocation of specific Stat transcription factors. Cross-talk demonstrated between the Jak-Stat pathway and other signaling pathways such as the mitogen-activated protein (MAP) kinase pathway,18 the NFκB signaling pathway,19 and the TGFβ signaling pathway20 underscore the complexity of signaling networks activated by cytokine stimulation.
We hypothesized that the cytokine microenvironment might play a role in FL by influencing tumoral signaling networks that regulate survival, proliferation, and differentiation. To address this question, we performed cytokine proteomic profiling of FL from diagnostic LN biopsies obtained prior to therapy, and compared the results to cases of benign follicular hyperplasia (FH). We subsequently assayed the phosphorylated and total protein levels of several downstream intracellular signaling proteins. We detected overall low levels of cytokine protein expression in FL for 9 of the 10 cytokines that we studied. In contrast, IL-4 protein concentration was remarkably high in the FL microenvironment compared with FH. One of the downstream targets of IL-4, the MAP kinase Erk, showed prominent basal phosphorylation in FL, which was largely absent in FH. These findings shed light on potential cooperative mechanisms of disease in FL and have implications for treatment targets in FL.
Methods
Patient samples
Fresh frozen archival pretreatment FL lymph nodes were obtained with informed consent given in accordance with the Declaration of Helsinki between 1999 and 2005 from patients enrolled in Institutional Review Board–approved protocols at the National Institutes of Health (NIH) (Bethesda, MD). Formalin-fixed hematoxylin and eosin (H&E)–stained sections were histologically reviewed. Biopsies from patients with FL were classified into grades 1, 2, 3a, or 3b, based on World Health Organization (WHO) diagnostic criteria.21 Immunohistochemistry for Bcl-2, CD20, CD10, CD3, and MIB-1 was performed on FL samples. Tonsillar FH is commonly used as a nonmalignant normal control for FL studies,11,22,–24 as FH is rich in normal germinal center B cells. In this study, we used 23 samples of fresh frozen tonsils representing FH as controls, obtained with informed consent again in accordance with the Declaration of Helsinki from Children's National Medical Center (Washington, DC).
Lysate preparation
Frozen tissue blocks embedded in Tissue Tek OCT (optimal cutting temperature) compound (Sakura Finetek, Torrance, CA) were sectioned 20 μm thick with manual removal of OCT. Tissue sections were lysed in a Triton X-100–based lysis buffer (1% Triton, 20 nM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, protease inhibitors, and phosphatase inhibitors). Protein concentrations were determined using the bicinchoninic acid (BCA) Protein Assay (Pierce, Rockford, IL). Portions of lysate for use in cytokine detection were diluted in Triton X-100 buffer to 1 mg/mL. For reverse-phase protein microarray (RPMA), aliquots of the lysate were supplemented with 1% lithium dodecyl sulfate (LDS) and heated at 70°C for 10 minutes.
Cytokine detection
A total of 10 cytokines were chosen for analysis of FL samples based on their association with Th1 and Th2 immune responses and availability of validated antibodies in a multiplex enzyme-linked immunosorbent assay (ELISA) format. The Triton-based lysates were analyzed using a Th1/Th2-cell multiplex sandwich immunoassay and an electrochemiluminescent detection system according to the manufacturer's instructions (K11010C; Meso Scale Discovery [MSD], Gaithersburg, MD) for detection of the following cytokines: interferon (IFN)–γ, IL-1β, IL-2, IL-4, IL-5, IL-8, IL-10, IL-12p70, IL-13, and TNF-α. Quantification of cytokine concentrations was obtained using a SECTOR Imager 6000 (MSD) with subsequent analysis using GraphPad software (San Diego, CA).
Construction of RPMAs
Lysates in LDS-based buffer were printed onto Whatman FAST nitrocellulose-coated slides (Schleicher & Schuell, Sanford, ME) using a solid-pin 2470 Arrayer (Aushon Biosystems, Burlington, MA). Briefly, 20 μL of each sample lysate was loaded into a 384-well microtiter plate. Multiple depositions of each lysate per array spot were printed in series (16 depositions, 8, 4, 2, and 1). Cultured cells from the RAMOS cell line—untreated and treated with IL-4 for 30 minutes and 24 hours—were used as positive controls for STAT6 phosphorylation. HeLa cells treated and untreated with epidermal growth factor (EGF) for 10 minutes were used as controls for Erk and Mek phosphorylation.
Immunostaining and analysis of RPMAs
Microarrays were hydrated, treated for 10 minutes with Miser Antibody Extender Solution (Pierce), and washed in Tris-buffered saline with 0.1% Tween 20 and 0.1% BSA for a total of 15 minutes. Slides were blocked for 2 hours in 5% milk, washed, and incubated for 2 hours with the following primary antibodies and concentrations: rabbit anti–Stat-6 1:500 (H-4, sc-981, lot no. L3003; Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti–phosphoStat-6 Tyr641 1:250 (no. 9361S; Cell Signaling Technology, Danvers, MA); rabbit anti-Mek1/2 1:2000 (no. 9122; Cell Signaling Technology); rabbit anti-phosphoMek Ser217/221 1:1000 (no. 9121S; Cell Signaling Technology); rabbit anti-Erk, or p44/42 MAP kinase, 1:500 (no. 9102; Cell Signaling Technology); and rabbit anti-phosphoErk Thr202/Tyr204 1:2000 (no. 9101S; Cell Signaling Technology). Slides were washed for 15 minutes and incubated for 1 hour in anti-rabbit horseradish peroxidase polymer (Biocare Medical, Concord, CA). Following a final washing, ECL (enhanced chemiluminescence) Plus Detection compounds (GE Healthcare, Little Chalfont, United Kingdom) and coverslips were added to the slides. Within 1 hour of ECL addition, slides were imaged on a Kodak 4000MM Digital Imaging Station (Kodak, Rochester, NY); ECL was excited with a 365 nm UV light source. Relative signal intensities were calculated using Microvigene 2.9.9.7 software (VigeneTech, Carlisle, MA). Total protein was estimated using Sypro Ruby Protein Blot Stain (Invitrogen Molecular Probes, Eugene, OR), Kodak imager, and Microvigene software. Immunostain data were normalized to total protein content.
Statistical analysis
The assays for proteomic cytokine profiles and RPMA were repeated 3 times. The concentration levels from the 3 repeated assays for each cytokine or signaling protein per specimen were averaged and used for statistical analysis. The Mann-Whitney rank test was used to compare the concentration level of each cytokine/protein between the FL and FH samples. The Kruskal-Wallis rank test, analogous to analysis of variance (ANOVA), was used to compare the concentration levels between the 3 grades of FL samples. In order to adjust for multiple comparisons, the significance level for P values was set at .01. Heatmap depicts the fold change of the concentration level of each specimen relative to the median of each cytokine or signaling protein. The displayed fold changes were truncated at 3-fold above and below the median.
Results
Cytokine protein profiles were generated from LN biopsies from 50 patients diagnosed with FL (32 men and 18 women). The average age of patients was 52.6 years, ranging from 30 to 75 years. Of the 50 samples, 20 were diagnosed as grade 1 FL, 18 a grade 2 FL, and 12 as grade 3 FL (9 with grade 3A and 3 with grade 3B). A total of 19 of 20 samples with FL grade 1 were Bcl-2+ by immunohistochemistry (IHC); 18 out of 18 samples with grade 2 FL were Bcl-2+; and 8 of 12 samples with grade 3 FL were positive for Bcl-2.
Macrophage/monocyte-associated cytokines IL-1β, IL-8, IL-10, and IL-12p70 protein concentrations are decreased in the FL microenvironment
IL-1β.
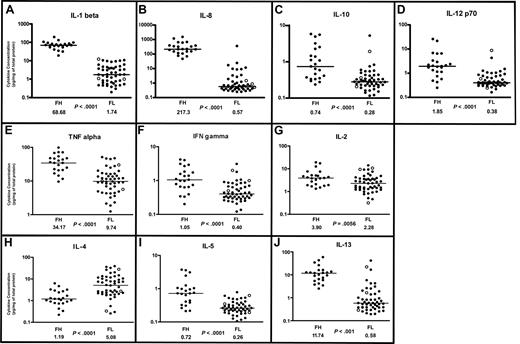
IL-1β is primarily secreted by macrophages and dendritic cells and can also be secreted by B cells. IL-1β promotes B-cell maturation, proliferation, and activation.25 SNPs in the IL-1 receptor gene were found predictive of survival in FL.16 We found average IL-1β protein concentrations to be more than 35 times higher in FH than in FL (P < .001), with a distinct separation between the FH and FL populations (Figure 1A, Table 1).
Cytokine profiles in follicular lymphoma and follicular hyperplasia. Scatterplots of designated cytokine protein concentrations in FL (N = 50) and FH (N = 23). Cytokine protein concentrations (picogram/mg of total protein for each tissue lysate) were determined by multiplex sandwich immunoassay system (“Methods”) and are depicted on a log10 scale. Median cytokine concentrations for FL and FH are designated with horizontal bars on the scatterplots and are listed beneath the plot for each group, with the P value for comparison of the 2 groups. FL samples representing grade 3B are indicated by ○. (A-D) Monocyte/macrophage-associated cytokine profiles in follicular lymphoma and follicular hyperplasia. (A) IL-1β. (B) IL-8. (C) IL-10. (D) IL-12 p70. (E-G) Th1-associated cytokine profiles in FL and FH. (E) TNF-α. (F) Interferon-γ. (G) IL-2. (H-J) Th2-associated cytokine profiles in FL and FH. (H) IL-4. (I) IL-5. (J) IL-13.
Cytokine profiles in follicular lymphoma and follicular hyperplasia. Scatterplots of designated cytokine protein concentrations in FL (N = 50) and FH (N = 23). Cytokine protein concentrations (picogram/mg of total protein for each tissue lysate) were determined by multiplex sandwich immunoassay system (“Methods”) and are depicted on a log10 scale. Median cytokine concentrations for FL and FH are designated with horizontal bars on the scatterplots and are listed beneath the plot for each group, with the P value for comparison of the 2 groups. FL samples representing grade 3B are indicated by ○. (A-D) Monocyte/macrophage-associated cytokine profiles in follicular lymphoma and follicular hyperplasia. (A) IL-1β. (B) IL-8. (C) IL-10. (D) IL-12 p70. (E-G) Th1-associated cytokine profiles in FL and FH. (E) TNF-α. (F) Interferon-γ. (G) IL-2. (H-J) Th2-associated cytokine profiles in FL and FH. (H) IL-4. (I) IL-5. (J) IL-13.
Cytokine concentrations in FL compared with FH
| Cytokine . | FH cytokine concentrations (N = 23)* . | FL cytokine concentrations, all grades (N = 50)* . | P† . | ||||
|---|---|---|---|---|---|---|---|
| Median . | SD . | Range . | Median . | SD . | Range . | ||
| IFN-γ | 1.05 | 1.15 | 0.2-4.4 | 0.40 | 0.51 | 0.12-3.07 | < .001 |
| IL-1β | 68.68 | 35.59 | 18.86-191.26 | 1.74 | 3.22 | 0.19-11.95 | < .001 |
| IL-2 | 3.90 | 4.75 | 1.31-18.93 | 2.28 | 2.9 | 0.31-13.89 | .006 |
| IL-4 | 1.19 | 1.48 | 0.22-6.21 | 5.08 | 9.39 | 0.25-38.6 | < .001 |
| IL-5 | 0.72 | 1.05 | 0.21-3.70 | 0.26 | 0.14 | 0.11-0.78 | < .001 |
| IL-8 | 217.33 | 358.99 | 46.36-1509.9 | 0.57 | 49.71 | 0.24-352.28 | < .001 |
| IL-10 | 0.74 | 1.74 | 0.23-5.81 | 0.28 | 0.74 | 0.12-5.25 | < .001 |
| IL-12 p70 | 1.85 | 6.15 | 0.23-24.51 | 0.38 | 1.23 | 0.15-8.26 | < .001 |
| IL-13 | 11.74 | 13.61 | 2.48-58.1 | 0.58 | 6.73 | 0.19-40.99 | < .001 |
| TNF-α | 34.17 | 23.49 | 0.33-97.39 | 9.74 | 12.06 | 1.23-50.8 | < .001 |
| Cytokine . | FH cytokine concentrations (N = 23)* . | FL cytokine concentrations, all grades (N = 50)* . | P† . | ||||
|---|---|---|---|---|---|---|---|
| Median . | SD . | Range . | Median . | SD . | Range . | ||
| IFN-γ | 1.05 | 1.15 | 0.2-4.4 | 0.40 | 0.51 | 0.12-3.07 | < .001 |
| IL-1β | 68.68 | 35.59 | 18.86-191.26 | 1.74 | 3.22 | 0.19-11.95 | < .001 |
| IL-2 | 3.90 | 4.75 | 1.31-18.93 | 2.28 | 2.9 | 0.31-13.89 | .006 |
| IL-4 | 1.19 | 1.48 | 0.22-6.21 | 5.08 | 9.39 | 0.25-38.6 | < .001 |
| IL-5 | 0.72 | 1.05 | 0.21-3.70 | 0.26 | 0.14 | 0.11-0.78 | < .001 |
| IL-8 | 217.33 | 358.99 | 46.36-1509.9 | 0.57 | 49.71 | 0.24-352.28 | < .001 |
| IL-10 | 0.74 | 1.74 | 0.23-5.81 | 0.28 | 0.74 | 0.12-5.25 | < .001 |
| IL-12 p70 | 1.85 | 6.15 | 0.23-24.51 | 0.38 | 1.23 | 0.15-8.26 | < .001 |
| IL-13 | 11.74 | 13.61 | 2.48-58.1 | 0.58 | 6.73 | 0.19-40.99 | < .001 |
| TNF-α | 34.17 | 23.49 | 0.33-97.39 | 9.74 | 12.06 | 1.23-50.8 | < .001 |
Median cytokine concentrations in follicular lymphoma (FL; N = 50) compared to follicular hyperplasia (FH; N = 20) determined using a multiplex sandwich immunoassay system (“Methods”), reported as pg/mg of total protein in LN tissue lysates. Standard deviations of the mean (SD) and ranges are reported in adjacent columns.
P values for comparisons between FL, all grades, and FH determined using the Mann-Whitney rank test with significance set at P = .01 due to multiple comparisons.
IL-8.
IL-8 is secreted by macrophages, is a chemoattractant for neutrophils, and has proangiogenic activity. SNPs in the IL-8 gene were shown to be predictive of survival in FL.16 In our study, we found average IL-8 concentrations were more than 300 times higher in FH than in FL (P < .001; Figure 1B, Table 1).
IL-10.
IL-10 is primarily secreted by macrophages and is associated with both inhibitory and stimulatory functions. IL-10 is also expressed by CD4+/CD25+ T cells of the regulatory subtype,26 and is postulated to enhance tumor immune evasion by down-regulating MHC class I molecules.27 When secreted by Th2 cells, IL-10 can promote B-cell activation. SNPs in the IL-10 gene were associated with an increased risk of non-Hodgkin lymphoma.15 We found average IL-10 concentrations to be more than twice as high in FH than FL (P < .001; Figure 1C, Table 1). There was a broad distribution of IL-10 concentrations in the FH samples. The differences in median IL-10 concentrations between FH and FL were not as great as those observed with IL-1β or with IL-8.
IL-12.
IL-12 is primarily secreted by macrophages but can also be secreted by B cells. IL-12 stimulates T-cell differentiation along the Th1 subtype28 and enhances the cytotoxicity of natural killer (NK) cells and CD8+ cytotoxic cells. SNPs in the IL-12 gene were found to be predictive of survival in FL.16 We found average IL-12 concentrations to be more than 4 times higher in FH than in FL (P < .001; Figure 1D, Table 1). There was also a broad distribution of IL-12 concentrations in the FH population, and the magnitude of difference was not as great as observed for IL-8 and IL-1β.
Th1-associated cytokines IL-2, TNF-α, and IFN-γ protein concentrations are decreased in the FL microenvironment
Th1 CD4+ helper T cells support a cellular immune response and secrete a number of cytokines, including TNF-α, IFN-γ, and IL-2.
TNF-α is a regulator of a host of immune functions. TNF-α is secreted by Th1 cells and can also be secreted by macrophages and dendritic cells. Gene polymorphisms in the TNF locus have been shown to be a genetic risk factor for FL29 and for diffuse large B-cell lymphoma.15 TNF-α receptors are reportedly up-regulated in FH and in high-grade non-Hodgkin lymphomas.30 In our study, we found average TNF-α cytokine protein concentrations to be more than 3 times higher in FH than FL (P < .001; Figure 1E, Table 1).
IFN-γ is secreted by Th1 cells and by NK cells. In addition to proinflammatory functions, there is evidence that IFN-γ plays an important role the inhibition of tumor development in the immunocompentent host.31 Sustained low levels of IFN-γ have been associated with tumor development, while sustained high levels of IFN-γ are associated with an antitumor effect.32 In our study, average IFN-γ protein concentrations were 50% lower in FL relative to FH (P < .001), with overlap in the distribution patterns of the 2 populations (Figure 1F, Table 1).
IL-2 is secreted by Th1 cells and stimulates the growth, survival, and differentiation of cytotoxic T cells. SNPs in the IL-2 gene were predictive of survival in FL.16 In our study, average IL-2 concentrations were more than 1.5 times higher in FH than FL (P = .006; Figure 1G, Table 1). There was overlap in the distribution patterns of IL-2 concentrations between FH and FL, and the magnitude of difference between FL and FH was not as striking as for other cytokines.
The Th2-associated cytokine IL-4 is the only cytokine profiled that showed significantly higher protein concentrations in FL than FH; other Th2-associated cytokines (IL-5 and IL-13) are decreased in FL
Th2 cells are CD4+ helper T-cells that promote humoral immunity and secrete several cytokines, including IL-4, IL-5, and IL-13. IL-4 regulates proliferation, differentiation, and apoptosis in B cells and mediates immunoglobulin class-switching to IgE and IgG4. IL-4 is reported to have antiproliferative effects on freshly isolated non-Hodgkin B-cell lymphomas,33,34 acute lymphoblastic leukemia,35 and human carcinoma cells.36 In contrast, IL-4 is reported to be a potent stimulator of FL cells in vitro.37 Components of the IL-4 signaling pathway have been reported to be up-regulated in the germinal center B-cell (GCB)–like subtype of diffuse large B-cell lymphoma (DLBCL)38 and in primary mediastinal large B-cell lymphoma.39 Gene expression profiles show up-regulation of the IL-4 receptor α gene in FL cells.40 In our study, IL-4 was the only cytokine in the panel that was consistently higher in FL than FH. Average IL-4 cytokine protein concentrations were more than 4 times higher in FL than in FH (P < .001; Figure 1H).
IL-5 is expressed predominantly by Th2 cells, mast cells, and eosinophils. IL-5 stimulates B-cell growth and immunoglobulin expression, and is associated with allergic responses. We found IL-5 levels to be on average more than twice as high in FH than FL (P < .001; Figure 1I).
IL-13 has several features in common with IL-4. Both receptors for IL-4 and IL-13 share a common chain and can activate signal transduction, resulting in the phosphorylation of Stat-6. There is overlap in the hematopoietic functions of IL-4 and IL-13; however, IL-13 is additionally associated with allergic responses. In this study, we found IL-13 protein concentrations to be more than 10 times greater in FH than in FL (P < .001; Figure 1J).
FL grades 1, 2, and 3 show similar cytokine profiles
According to current WHO guidelines, FL is typically subclassified into histologic grades 1 through 3 based on the number of centroblasts present per 400× microscopic field.21 We found no statistically significant difference in LN cytokine profiles between FL grades 1, 2, and 3 for 9 of 10 of the cytokines profiled (Table 2). IFN-γ was the only cytokine that showed average protein concentrations approximately twice as high in grade 3 FL compared with both grades 1 and 2 (P = .009). Grade 3 FL is further subclassified into grades 3A and 3B. Grade 3B FL is considered by many to represent the equivalent of DLBCL,21 and hence may represent a separate diagnostic entity. In this study, we had 3 samples with grade 3B FL that were used in the cytokine profiles and are indicated with open circles on the cytokine scatterplot figures (Figure 1). The 3 grade 3B cases were not significantly different from the remaining samples with FL for 9 of 10 of the cytokines tested. IFN-γ was the only cytokine in which the 3 samples with grade 3B FL were all above the median.
Cytokine concentrations of FL by grade
| Cytokine . | FL grade 1 cytokine concentrations (N = 20)* . | FL grade 2 cytokine concentrations (N = 18)* . | FL grade 3 cytokine concentrations (N = 12)* . | P† . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median . | SD . | Range . | Median . | SD . | Range . | Median . | SD . | Range . | ||
| IFN-γ | 0.33 | 0.27 | 0.14-1.05 | 0.38 | 0.21 | 0.12-1.00 | 0.75 | 0.83 | 0.29-3.07 | .009 |
| IL-1β | 0.90 | 2.48 | 0.37-10.59 | 2.95 | 3.21 | 0.29-9.88 | 2.63 | 4.07 | 0.19-11.95 | .084 |
| IL-2 | 1.57 | 1.66 | 0.46-7.38 | 2.27 | 2.16 | 0.97-9.02 | 3.29 | 4.34 | 0.31-13.89 | .053 |
| IL-4 | 6.30 | 6.70 | 1.18-26.20 | 6.43 | 7.40 | 0.29-32.08 | 2.56 | 14.83 | 0.25-38.60 | .8 |
| IL-5 | 0.27 | 0.16 | 0.12-0.78 | 0.26 | 0.11 | 0.11-0.52 | 0.21 | 0.17 | 0.12-0.62 | .83 |
| IL-8 | 0.54 | 2.96 | 0.3-11.39 | 0.53 | 82.87 | 0.24-352.28 | 1.74 | 7.77 | 0.25-24.15 | .11 |
| IL-10 | 0.29 | 0.19 | 0.12-0.83 | 0.28 | 0.12 | 0.12-0.57 | 0.26 | 1.48 | 0.14-5.25 | .94 |
| IL-12 p70 | 0.41 | 0.35 | 0.21-1.41 | 0.38 | 0.22 | 0.27-1.14 | 0.40 | 2.43 | 0.15-8.26 | .72 |
| IL-13 | 0.45 | 1.44 | 0.19-5.75 | 0.53 | 9.51 | 0.21-40.99 | 1.41 | 7.08 | 0.25-21.87 | .058 |
| TNF-α | 9.33 | 5.36 | 1.23-24.71 | 9.24 | 11.05 | 1.85-47.05 | 15.39 | 17.17 | 3.57-50.80 | .12 |
| Cytokine . | FL grade 1 cytokine concentrations (N = 20)* . | FL grade 2 cytokine concentrations (N = 18)* . | FL grade 3 cytokine concentrations (N = 12)* . | P† . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median . | SD . | Range . | Median . | SD . | Range . | Median . | SD . | Range . | ||
| IFN-γ | 0.33 | 0.27 | 0.14-1.05 | 0.38 | 0.21 | 0.12-1.00 | 0.75 | 0.83 | 0.29-3.07 | .009 |
| IL-1β | 0.90 | 2.48 | 0.37-10.59 | 2.95 | 3.21 | 0.29-9.88 | 2.63 | 4.07 | 0.19-11.95 | .084 |
| IL-2 | 1.57 | 1.66 | 0.46-7.38 | 2.27 | 2.16 | 0.97-9.02 | 3.29 | 4.34 | 0.31-13.89 | .053 |
| IL-4 | 6.30 | 6.70 | 1.18-26.20 | 6.43 | 7.40 | 0.29-32.08 | 2.56 | 14.83 | 0.25-38.60 | .8 |
| IL-5 | 0.27 | 0.16 | 0.12-0.78 | 0.26 | 0.11 | 0.11-0.52 | 0.21 | 0.17 | 0.12-0.62 | .83 |
| IL-8 | 0.54 | 2.96 | 0.3-11.39 | 0.53 | 82.87 | 0.24-352.28 | 1.74 | 7.77 | 0.25-24.15 | .11 |
| IL-10 | 0.29 | 0.19 | 0.12-0.83 | 0.28 | 0.12 | 0.12-0.57 | 0.26 | 1.48 | 0.14-5.25 | .94 |
| IL-12 p70 | 0.41 | 0.35 | 0.21-1.41 | 0.38 | 0.22 | 0.27-1.14 | 0.40 | 2.43 | 0.15-8.26 | .72 |
| IL-13 | 0.45 | 1.44 | 0.19-5.75 | 0.53 | 9.51 | 0.21-40.99 | 1.41 | 7.08 | 0.25-21.87 | .058 |
| TNF-α | 9.33 | 5.36 | 1.23-24.71 | 9.24 | 11.05 | 1.85-47.05 | 15.39 | 17.17 | 3.57-50.80 | .12 |
Median cytokine concentrations in follicular lymphoma (N = 50) by histologic grade determined using a multiplex sandwich immunoassay system (“Methods”), reported as pg/mg of total protein in LN tissue lysates. Standard deviations of the mean (SD) and ranges are reported in adjacent columns.
P values for comparisons among grades of FL determined using the Kruskal-Wallis rank test with significance at P = .01 due to multiple comparisons.
Erk is basally phosphorylated in FL in vivo
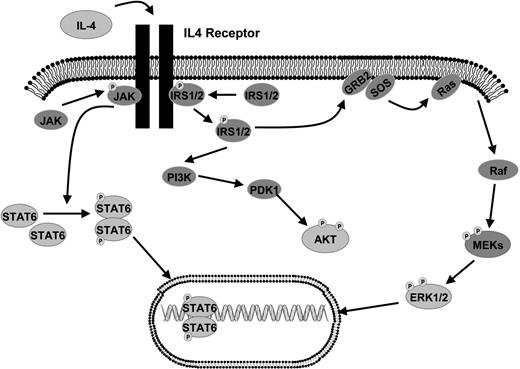
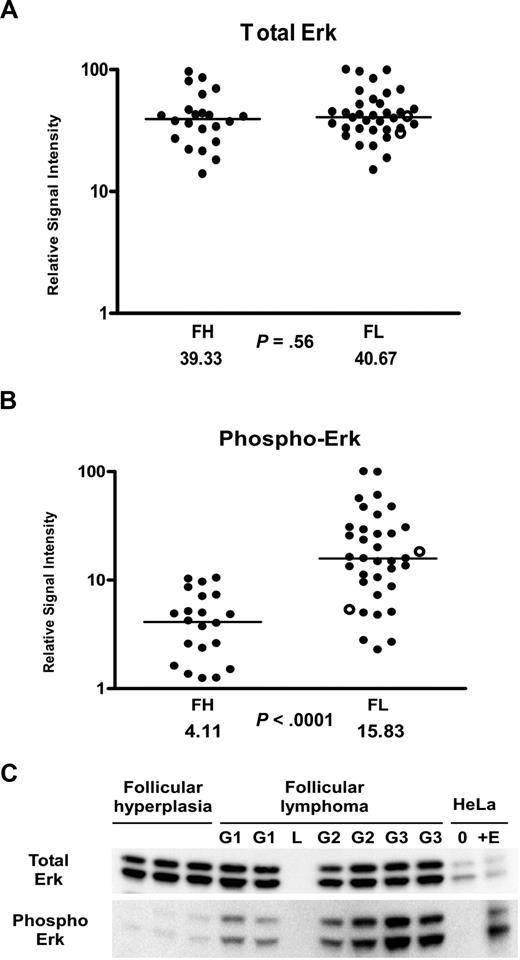
Given the overall decreased levels of cytokines and increased levels of IL-4 in the FL microenvironment, we hypothesized that activation of key intracellular signaling proteins would be different in FL in comparison to FH. We began by exploring the activation states of proteins within the IL-4 and downstream MAP kinase signaling pathway (Figure 2), which are activated by cytokines and are important in regulating proliferation. We used RPMA41 analysis of a large subset of our FH and FL tissue lysates that were previously analyzed in the cytokine profiling assays. Protein microarray tissue lysates included 36 FL samples (13 grade 1, 14 grade 2, and 9 grade 3 [7 grade 3A and 2 grade 3B]) and 22 FH samples. Measurement of total Erk protein levels was performed using antibodies that recognized both the phosphorylated and non-phosphorylated forms of Erk1/2 (“Methods”). The total Erk protein concentrations were similar in FL and FH (P = .56; Figure 3A). However, striking differences were observed using antibodies that recognized Erk1/2 phosphorylated on Thr202/Tyr204 residues. Phospho-Erk levels were on average approximately 4 times higher in FL versus FH (P < .001; Figure 3B). In order to cross-validate the RPMA findings, we performed Western blot analysis on a total of 27 of the FL samples and on 15 of the FH samples (Figure 3C). Similar to RPMA findings, FL and FH both showed strong bands for total Erk protein levels by Western blot. Erk showed prominent basal phosphorylation in 26 (95%) of 27 FL samples. In contrast, levels of phospho-Erk in the majority of FH samples were not sufficient for detection on Western blot; faint bands corresponding to relatively low levels of phospho-Erk were observed in only 4 (27%) of 15 samples with FH. Similar findings were obtained when analyzing the phosphorylation of Mek on Ser217/221, which is upstream of Erk in the MAP kinase signaling pathway (Table 3). By RPMA analysis, relative phospho-Mek levels were 1.7 times higher in FL versus FH (P = .002), while levels of total Mek were approximately the same in the 2 populations (Table 3).
Erk activation in follicular lymphoma. (A,B) Tissue lysates of FL (N = 36) and FH (N = 22) were printed on RPMAs and probed with antibodies for Erk (A) and phosphorylated Erk on Thr202/Tyr204 (B), as described in “Methods.” Scatterpoints represent relative signal intensity averaged over 3 runs and are depicted on a log10 scale. Median group values for FL and FH are designated with horizontal bars on the scatterplots and are listed beneath each plot, with the P value for comparison of the 2 groups. FL samples representing grade 3B are indicated by ○. (C) A total of 6 samples of FL (G1 indicates grade 1; G2, grade 2; G3, grade 3A) were compared with 3 samples of FH on Western blot, using antibodies for detection of total Erk and phospho-Erk Thr202/Tyr204. Serum-starved HeLa cells (0) and HeLa cells treated with epidermal growth factor (+E) served as negative and positive controls for Erk activation. L represents the lane used for the molecular weight ladder.
Erk activation in follicular lymphoma. (A,B) Tissue lysates of FL (N = 36) and FH (N = 22) were printed on RPMAs and probed with antibodies for Erk (A) and phosphorylated Erk on Thr202/Tyr204 (B), as described in “Methods.” Scatterpoints represent relative signal intensity averaged over 3 runs and are depicted on a log10 scale. Median group values for FL and FH are designated with horizontal bars on the scatterplots and are listed beneath each plot, with the P value for comparison of the 2 groups. FL samples representing grade 3B are indicated by ○. (C) A total of 6 samples of FL (G1 indicates grade 1; G2, grade 2; G3, grade 3A) were compared with 3 samples of FH on Western blot, using antibodies for detection of total Erk and phospho-Erk Thr202/Tyr204. Serum-starved HeLa cells (0) and HeLa cells treated with epidermal growth factor (+E) served as negative and positive controls for Erk activation. L represents the lane used for the molecular weight ladder.
Relative intensities of signaling proteins in FL compared with FH
| Signaling protein . | Relative signal intensity in FH (N = 22) . | Relative signal intensity in FL, all grades (N = 36) . | P* . | ||||
|---|---|---|---|---|---|---|---|
| Median . | SD . | Range . | Median . | SD . | Range . | ||
| Phospho Erk Thr202/Tyr204 | 4.11 | 3.09 | 0.96-10.46 | 15.83 | 23.19 | 2.28-100 | < .001 |
| Erk, total | 39.33 | 22.12 | 13.87-95.74 | 40.67 | 21.45 | 14.99-100 | .56 |
| Phospho-Mek 1/2 Ser217/221 | 14.79 | 10.77 | 4.68-44.44 | 25.37 | 23.75 | 7.5-100 | .003 |
| Mek 1/2, total | 39.29 | 13.68 | 21.61-69.88 | 41.99 | 23.86 | 18.24-100 | .58 |
| Phospho–Stat-6 Tyr641 | 3.44 | 3.34 | 0.58-12.06 | 6.24 | 18.28 | 1.25-100 | .012 |
| Stat-6, total | 17.54 | 19.38 | 4.95-100 | 19.53 | 18.94 | 3-95.11 | .88 |
| Signaling protein . | Relative signal intensity in FH (N = 22) . | Relative signal intensity in FL, all grades (N = 36) . | P* . | ||||
|---|---|---|---|---|---|---|---|
| Median . | SD . | Range . | Median . | SD . | Range . | ||
| Phospho Erk Thr202/Tyr204 | 4.11 | 3.09 | 0.96-10.46 | 15.83 | 23.19 | 2.28-100 | < .001 |
| Erk, total | 39.33 | 22.12 | 13.87-95.74 | 40.67 | 21.45 | 14.99-100 | .56 |
| Phospho-Mek 1/2 Ser217/221 | 14.79 | 10.77 | 4.68-44.44 | 25.37 | 23.75 | 7.5-100 | .003 |
| Mek 1/2, total | 39.29 | 13.68 | 21.61-69.88 | 41.99 | 23.86 | 18.24-100 | .58 |
| Phospho–Stat-6 Tyr641 | 3.44 | 3.34 | 0.58-12.06 | 6.24 | 18.28 | 1.25-100 | .012 |
| Stat-6, total | 17.54 | 19.38 | 4.95-100 | 19.53 | 18.94 | 3-95.11 | .88 |
RPMA of 36 samples of FL and 22 samples of FH, probed with antibodies for the proteins listed (“Methods”). Relative signal intensities representing protein content were generated using Microvigene imaging analysis softwear, normalized to total protein and scaled to the maximum value in the group. Standard deviations of the mean relative signal intensity and ranges are reported next to the median relative signal intensity for each protein.
The Mann-Whitney rank test was used to compare FL and FH with P values set at P = .01 for determination of significance, due to multiple comparisons.
Phosphorylation of Stat-6 in FL in vivo
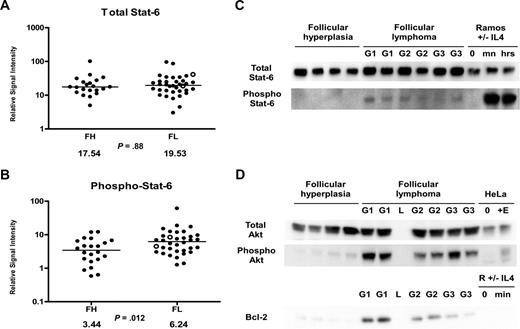
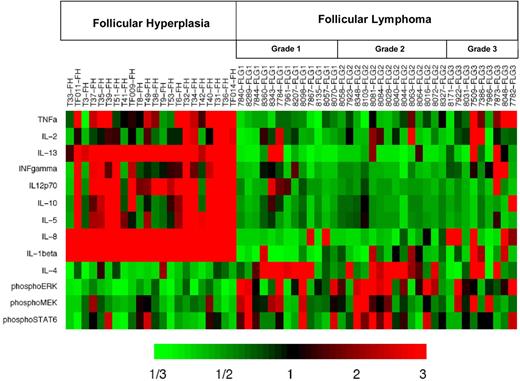
Given the significantly increased levels of IL-4 we observed in FL (Figure 1H), we hypothesized that Stat-6 may be activated in FL in vivo. RPMA analysis was used to compare total Stat-6 and phosphorylated Stat-6 levels between FL and FH. Levels of total Stat-6 protein were not significantly different between FL and FH (P = .88; Figure 4A). When we assayed the activated form of Stat-6 with antibodies that recognized only Stat-6 proteins phosphorylated on tyrosine 641, we detected average concentrations of phospho–Stat-6 that were nearly twice as high in FL as compared with FH (Table 3; Figure 4B) by RPMA analysis. The P value for this RPMA comparison was .012, which is borderline significant. Western blot analysis of a subset samples showed weak yet detectable phospho–Stat-6 in 14 (74%) of 19 of the FL samples and in 0 (0%) of 11 of the FH samples (Figure 4C). As noted previously, both IL-4 and IL-13 can activate Stat-6. However, cytokine protein profiles (Figure 1H,J) showed IL-4 concentrations to be much higher in FL than IL-13 concentrations. Hence, basal phosphorylation of Stat-6 observed in FL is more likely to result from IL-4 than from IL-13. A heatmap displays the combined cytokine and signaling protein results in Figure 5.
Stat-6 phosphorylation in FL and FH. (A,B) Tissue lysates of FL (N = 36) and FH (N = 22) were printed on RPMAs and probed with antibodies for Stat-6 (A) and phosphorylated Stat-6 on Tyr641 (B) as described in “Methods.” Scatterpoints represent relative signal intensity averaged over 3 runs and are depicted on a log10 scale. Median group values for FL and FH are designated with horizontal bars on the scatterplots and are listed beneath each plot, with the P value for comparison of the 2 groups. FL samples representing grade 3B are indicated by ○. (C) A total of 6 samples of FL (G1 indicates grade 1; G2, grade 2; G3, grade 3A) were compared with 4 samples of FH on Western blot, using antibodies for detection of total Stat-6 and phospho–Stat-6 Tyr641. Ramos cells untreated (0) and treated with IL-4 for 30 minutes (mn) and 24 hours (hrs) were used as negative and positive controls for Stat-6 activation. (D) A total of 6 samples of FL were compared with 4 samples of FH on Western blot, using antibodies for detection of total Akt, phospho-Akt Ser473, and Bcl-2. HeLa cell control lysates were used as described in Figure 3C.
Stat-6 phosphorylation in FL and FH. (A,B) Tissue lysates of FL (N = 36) and FH (N = 22) were printed on RPMAs and probed with antibodies for Stat-6 (A) and phosphorylated Stat-6 on Tyr641 (B) as described in “Methods.” Scatterpoints represent relative signal intensity averaged over 3 runs and are depicted on a log10 scale. Median group values for FL and FH are designated with horizontal bars on the scatterplots and are listed beneath each plot, with the P value for comparison of the 2 groups. FL samples representing grade 3B are indicated by ○. (C) A total of 6 samples of FL (G1 indicates grade 1; G2, grade 2; G3, grade 3A) were compared with 4 samples of FH on Western blot, using antibodies for detection of total Stat-6 and phospho–Stat-6 Tyr641. Ramos cells untreated (0) and treated with IL-4 for 30 minutes (mn) and 24 hours (hrs) were used as negative and positive controls for Stat-6 activation. (D) A total of 6 samples of FL were compared with 4 samples of FH on Western blot, using antibodies for detection of total Akt, phospho-Akt Ser473, and Bcl-2. HeLa cell control lysates were used as described in Figure 3C.
Heatmap display of cytokine protein profiling and signaling protein analysis highlights the difference in patterns underlying distinct biologic processes embodied in FL and FH. Results are displayed for FL (N = 36) and FH (N = 22) samples. Heatmap depicts the “fold change” of the designated protein concentration of each specimen relative to the median value of the designated cytokine or signaling protein across all specimens. The color red indicates relatively high protein concentration and the color green indicates relatively low protein concentration.
Heatmap display of cytokine protein profiling and signaling protein analysis highlights the difference in patterns underlying distinct biologic processes embodied in FL and FH. Results are displayed for FL (N = 36) and FH (N = 22) samples. Heatmap depicts the “fold change” of the designated protein concentration of each specimen relative to the median value of the designated cytokine or signaling protein across all specimens. The color red indicates relatively high protein concentration and the color green indicates relatively low protein concentration.
The results of signaling proteins obtained by RPMA and Western blot varied to some degree due to inherent differences in the 2 technologies. In RPMA, tissue lysates are analyzed without prior separation of proteins on the basis of size.41 For this reason, the background for a given validated antibody can be higher in RPMA than in Western blot, with potentially less resolution between 2 different groups, depending on the antibody, the target protein, and the difference in abundance of the target protein in the 2 groups being compared. The advantage of RPMA is that it is high-throughput and quantitative.
Akt is basally phosphorylated in FL in vivo
Several B-cell lymphoma cell lines demonstrate rapid phosphorylation of Stat-6 in response to IL-4 treatment. Cell lines representing the GCB subtype of DLBCL are reported to show Stat-6 phosphorylation in response to IL-4 without inducing phosphorylation of Akt.38 In contrast, cell lines representing the activated B-cell (ABC) subtype of DLBCL are reported to show preferential phosphorylation of Akt in response to IL-4 treatment without inducing phosphorylation of Stat-6.38 We previously reported that Akt was phosphorylated on serine 473 in FL patient samples using RPMA analysis.22,42 Given that FL and the GCB subtype of DLBCL share similar immunophenotypic features and are believed to arise from germinal center B cells, we wanted to evaluate the phosphorylation state of Akt in a subset of our samples by Western blot (Figure 4D). Western blot analysis confirmed basal phosphorylation of Akt in vivo on serine 473 in 6 (100%) of 6 of the FL samples tested, with presence of strong bands. In contrast, 4 FH samples showed little detectable phospho-Akt. Levels of total Akt were approximately equal in FH and FL. Bcl-2 protein was assayed in parallel; Western blot analysis of Bcl-2 protein was negative in FH and positive in FL grades 1 and 2, with weaker to negative expression in FL grade 3 lymphomas.
While normal admixed T cells in FH express Bcl-2, the overall level of Bcl-2 expression in FH is not sufficient for detection by Western blot. Bcl-2 expression in FL is significantly higher than in FH due to constitutive expression of Bcl-2 as a consequence of the t(14;18), and is detectable by Western blot. Similarly, some degree of activation of signaling proteins would be expected in activated cells within FH; however, the overall abundance of the phosphorylated proteins that we assayed was not sufficient for detection by Western blot, in contrast to FL for these same proteins.
Discussion
IL-4 was the only cytokine in our panel to show significantly higher levels of protein expression in FL versus FH. This finding has not been previously reported in FL. Prior cDNA array studies identified up-regulation of the IL4RA gene in FL cells in comparison to normal germinal center B cells.40 Together, these findings in suggest a role for IL-4 in FL. Based on other gene expression studies, the role of IL-4 in FL may or may not be relevant as a predictor of survival, as IL-4 gene expression was not identified in the gene expression profiling studies by Dave et al.11 It is also important to note that gene expression does not necessarily correlate with levels of protein expression, abundance, or presence of posttranslational modifications. Our study specifically focused on quantitative analysis of cytokine protein profiles and posttranslational modifications of selected intracellular signaling proteins, which may not be accurately reflected in gene expression studies.
We detected significant basal phosphorylation of Erk in FL samples. Previous cDNA microarray studies have implicated the involvement of multiple signaling pathways in the transformation of FL to DLBCL.43 Our study indicates that Erk and the MAP kinase pathway are already basally activated in the indolent form of FL. Erk is a downstream target for multiple receptor signaling complexes that signal via the MAP kinase cascade.44 Hence, many cytokine receptors, growth factor receptors, and the B-cell receptor are able to activate the MAP kinase Ras/Raf/Mek/Erk cascade when extracellular ligand is bound. We postulate that there may be a relationship between the relatively high IL-4 levels in FL and the sustained basal activation of Erk that we detected. FL cells are reported to up-regulate the IL-4 receptor α.40 In addition, IL-4 has been demonstrated to be a potent stimulator of FL cell proliferation in vitro,37 and Erk is an established regulator of proliferation within the MAP kinase pathway.45 Further studies are needed to elucidate the roles of IL-4 and Erk in FL. Unfortunately, in vitro functional studies are limited by the fact that there are no human cell lines that represent the nontransformed indolent form of FL.
It is also possible that other receptor signaling complexes play a role in the basal activation of Erk in FL. Previous studies showed increased sensitivity of FL cells to ex vivo stimulation through the B-cell receptor, with potentiation of signal intensity and increased duration of signal transduction, when compared with nonmalignant tumor infiltrating B cells.46 A recent study also reported increased expression of the IL-21 receptor on FL tumor cells, suggesting a potential role for IL-21 in FL.47
Constitutive phosphorylation of p70S6 kinase in FL LN tissue in comparison to FH23 has been reported. Basal p70S6 kinase activation was attributed to Syk-dependent mTOR activation based on additional in vitro studies in the DLBCL Karpas-422 cell line containing the t(14;18) translocation. Recent evidence suggests that PKC-ζ activates mTOR in FL cells via a MAP kinase–dependent mechanism involving Mek.48 Rituximab reportedly inhibits MAP kinase activation of mTOR by inhibition of PKC-ζ, resulting in an antitumor response. The results of these studies, together with our findings of basal in vivo activation of Erk and Mek in FL, suggest a role for Erk and Mek in maintenance of the malignant phenotype in FL, with implications for targeted therapeutic strategies. This may be particularly relevant given the development of new small-molecule Mek inhibitors, which are being introduced in clinical trials.49,–51
Stat-6 has been shown to be a primary downstream transcriptional activator of IL-4–dependent signaling.52,–54 IL-4 and Stat-6 protein expression have been associated with other B-cell malignancies.55 Primary mediastinal large B-cell lymphoma cell lines and patient tumors were found to demonstrate high levels of activated Stat-6 and Jak239 and transcriptional activation of IL-4/Stat-6 gene targets.56 Lymphocyte-predominant Hodgkin lymphoma was reported to show high expression levels of Jak2 with constitutive activation of Stat-6 in a portion of cases studied by one group; these findings were accompanied by somatic hypermutation of SOCS1.57 Constitutive activation of Stat-6 has also been reported in Hodgkin Reed Sternberg (HRS) cells in classical Hodgkin lymphoma and was attributed to IL-13 expression by HRS cells.58
We found that Stat-6 showed weak yet detectable basal phosphorylation in 74% of FL samples by Western blot in contrast to 0% of the FH samples tested. This finding is consistent with immunohistochemistry studies, showing that normal germinal center cells in FH are largely devoid of phopho–Stat-6.58 Given our findings of higher levels of IL-4 in the FL microenvironment and the reported up-regulation of IL-4Rα on FL cells,40 it is reasonable to postulate that the increased basal phospho–Stat-6 that we detected in FL involves FL tumor cells.
Previous studies with cell lines representing the GCB versus ABC subtype of DLBCL suggest that Stat-6 is preferentially activated in the GCB subtype in response to IL-4 without concurrent activation of Akt.38 In contrast, in the ABC subtype, Akt is activated in response to IL-4 without detectable activation of Stat-6, putatively due to nuclear T-cell protein tyrosine phosphatase (TCPTP) expressed in the ABC subtype, which targets Stat-6.59 Our analysis suggested that in vivo, both Akt and Stat-6 are frequently basally phosphorylated in FL. We did see some individual patient samples that contained relatively high levels of IL-4 with minimal detectable phospho–Stat-6 (Figure 5). In future analysis, it may be useful to determine whether TCPTP is expressed in these patient samples.
Overall, we did not detect significant differences between grades 1, 2, and 3 of FL. IFN-γ was the only cytokine that showed greater protein levels in grade 3 FL versus grades 1 and 2. Grade 3B FL in particular is thought by many to represent a different entity than the other grades of FL. Even so, we found the cytokine profiles of grade 3B to be similar to the profiles of grade 3A FL, which overall were similar to grades 1 and 2, with the exception of IFN-γ, as noted.
The overall decreased levels of cytokine protein expression observed in FL in comparison to FH may reflect the presence of residual normal inhibitory immune response mechanisms that help curb inappropriate tumor growth. Consistent with this possibility are findings that CD4+/CD25+/FOXP3+ T-regulatory cells isolated from FL tumors have an enhanced ability to suppress cytokine expression by autologous and allogeneic nodal CD4+ T cells when compared with CD4+/CD25+/FOXP3+ T-regulatory cells isolated from benign hyperplastic LNs.60 Thus, T-regulatory cells in FL, as part of an immune response, may contribute to the lower overall cytokine levels that we observed.
In summary, our study revealed the FL microenvironment to be characterized by high levels of IL-4 in a background of overall low cytokine concentrations in comparison to the microenvironment of the benign counterpart FH. We also detected prominent basal phosphorylation of Erk in vivo in FL, indicating dysregulation of signaling networks controlling proliferation in vivo in FL. These findings have implications for understanding the biology of FL and future development of effective molecularly targeted therapies in FL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Liang Cao for use of the SECTOR Imager 6000, Adnan Siddiqui for assistance in lysate preparation, and Dr David Levens and Dr Hye-Jung Chung for invaluable discussions.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute.
National Institutes of Health
Authorship
Contribution: K.R.C. designed research, analyzed and interpreted data, and wrote the paper; B.D. performed research and analyzed data; A.K. performed research, analyzed data, and helped write the paper; C.D. performed research and developed analytical tools; R.B. and A.B. performed experiments; J.S. performed statistical analysis; and E.S.J. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katherine R. Calvo, NIH Clinical Center, Department of Laboratory Medicine, 10 Center Drive, Bldg 10/2C306, Bethesda, MD 20892-1500; e-mail: calvok@mail.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal