Abstract
To investigate the cell of origin linking follicular (FL) and transformed (t-FL) lymphomas, we analyzed the somatic hypermutation (SHM) pattern of the variable region of the immunoglobulin heavy gene (IgH-VH) in 18 sequential FL/t-FL samples and a father (donor) and son (recipient), who developed FL and t-FL, after transplantation. Genealogic trees showed a pattern compatible with a common progenitor cell (CPC) origin in 13 cases. The identification of the t-FL clonotype in the previous FL sample and of the putative CPC sequence in both the FL/t-FL biopsies showed that the intraclonal diversity of FL and t-FL germinal centers (GCs) is more intricate than previously described, and all 3 clonotypes (CPC, FL, t-FL) may occur simultaneously within the same lymph node. On the basis of the father/son model, this CPC must be long-lived, providing a possible explanation for the incurable nature of this disease.
Introduction
Follicular lymphoma (FL) is a low-grade disorder characterized by multiple relapses1 and frequently by transformation (t-FL) to a more aggressive lymphoma.2 Although it has long been assumed that transformation reflects the emergence of an aggressive subclone of cells from the FL population, this explanation could be an oversimplification because FL and t-FL might also arise by divergent evolution from a more immature common progenitor cell (CPC).3 This is supported by recent cytogenetic, comparative genomic hybridization, and single nucleotide polymorphism data,4-6 suggesting that relapse and transformation may originate in more undifferentiated cells
FL originates from germinal center (GC) B cells and is characterized by somatic hypermutations (SHMs) responsible for the high intraclonal diversity characteristic of this malignancy. Novel N-glycosylation sites are introduced by SHMs in 85% of cases7 and positively selected during B-cell maturation.8 Analysis of the variable region of the immunoglobulin heavy gene (IgH-VH) has been used extensively to trace the stage of differentiation of B lymphocytes in lymphomas.9-12 Previous studies in paired FL/t-FL showed that these tumors are clonally related13 ; however, whether t-FL evolves from a FL subclone or both tumors originate from a more immature cell remains controversial. Now the existence of cancer-initiating cells is accepted in both hematologic and epithelial cancers; such cells share many biologic features, including the ability to self-renew and proliferate, and the resistance to chemical or physical insults.14-16
To elucidate the stage of B-cell ontogeny at which such a cell may arise, we investigated the IgH-VH SHMs in DNA from serial FL/t-FL and from a father (donor) and son (recipient), who developed FL and t-FL after bone marrow transplantation (BMT), respectively.17
Methods
Patient material
DNA from lymph nodes from 18 patients, collected before (FL) and after transformation (t-FL), was available. Clinical and molecular characteristics of patients are summarized in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). We also investigated DNA from a donor (father) with FL and a recipient (son) with t-FL, with the same t(14;18) translocation, who developed lymphomas 3 and 10 years after transplantation, respectively.17 This study received ethical approval from our Local Research Ethics Committee (REC no. 06/Q0605/69), and informed consent was obtained in accordance with the Declaration of Helsinki.
Identification of the IgH-VH clonal rearrangement
The IgH-VH region was investigated by polymerase chain reaction (PCR) with family-specific leader and a reverse JH consensus primers.18,19 The major tumor clone was isolated by heteroduplex analysis (HH). Homoduplex bands were purified19 and directly sequenced. In 8 cases the clonal IgH family PCR product was cloned, and inserts were sequenced using the M13 reverse primer. Sequences were compared with the IgH-VH germline sequence using IMGT/V-QUEST (IMGT; http://imgt.cines.fr).
Identification of the t-FL clonotype and of the CPC
For 8 patients (10 samples), t-FL clonotype-specific primers/probes were used to investigate the preceding FL by real-time quantitative PCR (RQ-PCR; Document S1). In 6 cases (based on the putative CPC sequences inferred from their genealogic trees) primers specific for the CPC SHMs were designed. A nested PCR of both the FL/t-FL samples was performed, and products cloned and sequenced.
Results and discussion
The mechanisms responsible for transformation of FL into t-FL remain a matter of debate. To shed light on this process and to trace the evolutionary pattern of FL/t-FL clones, genealogic trees were generated by HH and cloning. In 7 (87.5%) of 8 cases studied with both methods we observed a concordance between the 2 approaches (Table S2). The sequential and the father/son biopsies were all clonally related,9,20,21 and the IgH-VH family usage and frequency of SHMs were similar to other FL studies (data not shown). All cloned t-FL tumors showed a similar mutation rate compared with their FL counterparts, albeit a lower degree of oligoclonality (P = .04; Table S2).
The comparison of FL/t-FL homoduplex sequences confirmed the existence of a putative CPC in 12 (67%) of 18 pairs (Figure 1A; Figures S1,S2) and in the father/son cases (Figure S3). On the basis of the number of common SHMs we could infer that this hypothetical CPC derives from a GC B cell with homology to germline sequence ranging from 83.5% (patient 6) to 94.5% (patient 12). The remaining 6 patients (33%) had a transformation pattern compatible with direct evolution (Figure 1B), with 4 showing identical SHMs (data not shown). In these cases it seems likely that a FL cell attained additional abnormalities leading to a more aggressive phenotype.
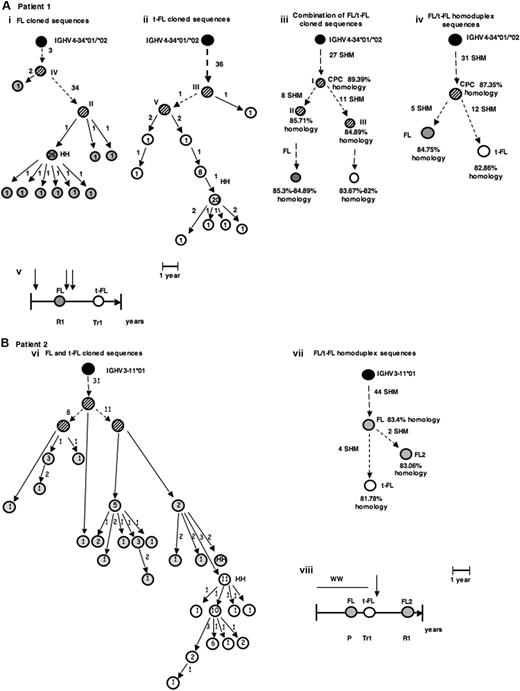
Patterns of evolution of FL and t-FL. (A,B) Two examples of genealogic trees generated by comparison of cloned IgH-VH PCR products and by heteroduplex (HH) analysis, from FL and t-FL samples. The putative CPC sequences were generated by comparison of the common pattern of SHMs observed in the paired FL and t-FL samples. The FL sequences are depicted as  , the t-FL clones as ○, and the germline sequence as ·. The putative common progenitor cell and the hypothetical intermediate sequences, which were not detected experimentally, are depicted as
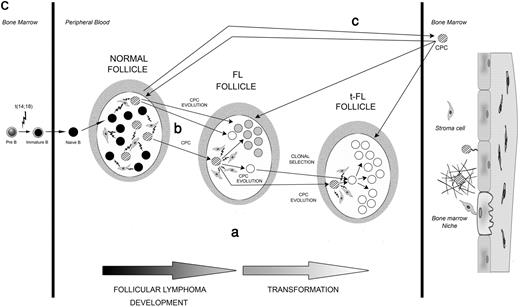
, the t-FL clones as ○, and the germline sequence as ·. The putative common progenitor cell and the hypothetical intermediate sequences, which were not detected experimentally, are depicted as  . HH corresponds to the sequence identified by HH analysis. Numbers inside the circles correspond to the number of clones with the same sequence, and the numbers beside the arrows represent the additional mutations in the subsequent clone. The roman numbers I, II, III, and so on, on the trees, refer to the intermediate clones. (A) Genealogic trees of patient 1, a case compatible with the existence of a CPC. Genealogic trees generated after comparison of sequences obtained after screening of the FL and t-FL cloned PCR products (i-iii) and after comparison of the homoduplex sequences (iv); (v) graphic timeline representation of the FL/t-FL biopsies investigated and of clinical evolution of disease: vertical arrow represents the line of therapy (either chemotherapy or radiotherapy); left-hand vertical bar, the time of diagnosis; the right-hand vertical bar, time of death; R1, the first relapse; Tr1, the first transformation. (B) Genealogic trees of patient 2, a case compatible with a direct evolution. In this case an additional FL sample (FL2) was investigated. Genealogic tree generated after comparison of the cloned sequences (vi) and of the 2 major clones identified by HH analysis (vii); graphic timeline representation of the sequential biopsies investigated and the clinical evolution of disease: each vertical arrow represents a line of therapy; the bar at the end of the horizontal line, time the death; ww, watch and wait; P, progression; Tr1, first transformation; and R1, first relapse. (C) Models for development and evolution of the CPC cell. The CPC, FL, and t-FL clones are depicted as previously described. Letters a, b, and c represent the 3 patterns of evolution. Pre-B cells acquire the t(14;18) translocation in the bone marrow and then migrate to the GC of the lymph node as a naive B cell. There, because of the interaction with cells of the microenvironment, they undergo the SHM process to give rise to mature B cells. The first hypothesis (i) is that CPC survives the different lines of therapy and coexist with FL and t-FL clones in the GC. The second hypothesis (ii) suggests that the CPC is present only in the GC of the prelymphoma (normal follicle) lymph node and not in the GC of FL or t-FL. The third hypothesis (iii) is that the CPC migrates to the BM (before or after FL) and is maintained in the BM niche before migrating to a secondary lymphoid organ where it evolves into FL or t-FL. The second (ii) and third (iii) evolution pathways both relate to the existence of “premalignant” niches (the first one in the GC and the second one in the BM) where CPC can independently give rise to FL or t-FL.
. HH corresponds to the sequence identified by HH analysis. Numbers inside the circles correspond to the number of clones with the same sequence, and the numbers beside the arrows represent the additional mutations in the subsequent clone. The roman numbers I, II, III, and so on, on the trees, refer to the intermediate clones. (A) Genealogic trees of patient 1, a case compatible with the existence of a CPC. Genealogic trees generated after comparison of sequences obtained after screening of the FL and t-FL cloned PCR products (i-iii) and after comparison of the homoduplex sequences (iv); (v) graphic timeline representation of the FL/t-FL biopsies investigated and of clinical evolution of disease: vertical arrow represents the line of therapy (either chemotherapy or radiotherapy); left-hand vertical bar, the time of diagnosis; the right-hand vertical bar, time of death; R1, the first relapse; Tr1, the first transformation. (B) Genealogic trees of patient 2, a case compatible with a direct evolution. In this case an additional FL sample (FL2) was investigated. Genealogic tree generated after comparison of the cloned sequences (vi) and of the 2 major clones identified by HH analysis (vii); graphic timeline representation of the sequential biopsies investigated and the clinical evolution of disease: each vertical arrow represents a line of therapy; the bar at the end of the horizontal line, time the death; ww, watch and wait; P, progression; Tr1, first transformation; and R1, first relapse. (C) Models for development and evolution of the CPC cell. The CPC, FL, and t-FL clones are depicted as previously described. Letters a, b, and c represent the 3 patterns of evolution. Pre-B cells acquire the t(14;18) translocation in the bone marrow and then migrate to the GC of the lymph node as a naive B cell. There, because of the interaction with cells of the microenvironment, they undergo the SHM process to give rise to mature B cells. The first hypothesis (i) is that CPC survives the different lines of therapy and coexist with FL and t-FL clones in the GC. The second hypothesis (ii) suggests that the CPC is present only in the GC of the prelymphoma (normal follicle) lymph node and not in the GC of FL or t-FL. The third hypothesis (iii) is that the CPC migrates to the BM (before or after FL) and is maintained in the BM niche before migrating to a secondary lymphoid organ where it evolves into FL or t-FL. The second (ii) and third (iii) evolution pathways both relate to the existence of “premalignant” niches (the first one in the GC and the second one in the BM) where CPC can independently give rise to FL or t-FL.
Patterns of evolution of FL and t-FL. (A,B) Two examples of genealogic trees generated by comparison of cloned IgH-VH PCR products and by heteroduplex (HH) analysis, from FL and t-FL samples. The putative CPC sequences were generated by comparison of the common pattern of SHMs observed in the paired FL and t-FL samples. The FL sequences are depicted as  , the t-FL clones as ○, and the germline sequence as ·. The putative common progenitor cell and the hypothetical intermediate sequences, which were not detected experimentally, are depicted as
, the t-FL clones as ○, and the germline sequence as ·. The putative common progenitor cell and the hypothetical intermediate sequences, which were not detected experimentally, are depicted as  . HH corresponds to the sequence identified by HH analysis. Numbers inside the circles correspond to the number of clones with the same sequence, and the numbers beside the arrows represent the additional mutations in the subsequent clone. The roman numbers I, II, III, and so on, on the trees, refer to the intermediate clones. (A) Genealogic trees of patient 1, a case compatible with the existence of a CPC. Genealogic trees generated after comparison of sequences obtained after screening of the FL and t-FL cloned PCR products (i-iii) and after comparison of the homoduplex sequences (iv); (v) graphic timeline representation of the FL/t-FL biopsies investigated and of clinical evolution of disease: vertical arrow represents the line of therapy (either chemotherapy or radiotherapy); left-hand vertical bar, the time of diagnosis; the right-hand vertical bar, time of death; R1, the first relapse; Tr1, the first transformation. (B) Genealogic trees of patient 2, a case compatible with a direct evolution. In this case an additional FL sample (FL2) was investigated. Genealogic tree generated after comparison of the cloned sequences (vi) and of the 2 major clones identified by HH analysis (vii); graphic timeline representation of the sequential biopsies investigated and the clinical evolution of disease: each vertical arrow represents a line of therapy; the bar at the end of the horizontal line, time the death; ww, watch and wait; P, progression; Tr1, first transformation; and R1, first relapse. (C) Models for development and evolution of the CPC cell. The CPC, FL, and t-FL clones are depicted as previously described. Letters a, b, and c represent the 3 patterns of evolution. Pre-B cells acquire the t(14;18) translocation in the bone marrow and then migrate to the GC of the lymph node as a naive B cell. There, because of the interaction with cells of the microenvironment, they undergo the SHM process to give rise to mature B cells. The first hypothesis (i) is that CPC survives the different lines of therapy and coexist with FL and t-FL clones in the GC. The second hypothesis (ii) suggests that the CPC is present only in the GC of the prelymphoma (normal follicle) lymph node and not in the GC of FL or t-FL. The third hypothesis (iii) is that the CPC migrates to the BM (before or after FL) and is maintained in the BM niche before migrating to a secondary lymphoid organ where it evolves into FL or t-FL. The second (ii) and third (iii) evolution pathways both relate to the existence of “premalignant” niches (the first one in the GC and the second one in the BM) where CPC can independently give rise to FL or t-FL.
. HH corresponds to the sequence identified by HH analysis. Numbers inside the circles correspond to the number of clones with the same sequence, and the numbers beside the arrows represent the additional mutations in the subsequent clone. The roman numbers I, II, III, and so on, on the trees, refer to the intermediate clones. (A) Genealogic trees of patient 1, a case compatible with the existence of a CPC. Genealogic trees generated after comparison of sequences obtained after screening of the FL and t-FL cloned PCR products (i-iii) and after comparison of the homoduplex sequences (iv); (v) graphic timeline representation of the FL/t-FL biopsies investigated and of clinical evolution of disease: vertical arrow represents the line of therapy (either chemotherapy or radiotherapy); left-hand vertical bar, the time of diagnosis; the right-hand vertical bar, time of death; R1, the first relapse; Tr1, the first transformation. (B) Genealogic trees of patient 2, a case compatible with a direct evolution. In this case an additional FL sample (FL2) was investigated. Genealogic tree generated after comparison of the cloned sequences (vi) and of the 2 major clones identified by HH analysis (vii); graphic timeline representation of the sequential biopsies investigated and the clinical evolution of disease: each vertical arrow represents a line of therapy; the bar at the end of the horizontal line, time the death; ww, watch and wait; P, progression; Tr1, first transformation; and R1, first relapse. (C) Models for development and evolution of the CPC cell. The CPC, FL, and t-FL clones are depicted as previously described. Letters a, b, and c represent the 3 patterns of evolution. Pre-B cells acquire the t(14;18) translocation in the bone marrow and then migrate to the GC of the lymph node as a naive B cell. There, because of the interaction with cells of the microenvironment, they undergo the SHM process to give rise to mature B cells. The first hypothesis (i) is that CPC survives the different lines of therapy and coexist with FL and t-FL clones in the GC. The second hypothesis (ii) suggests that the CPC is present only in the GC of the prelymphoma (normal follicle) lymph node and not in the GC of FL or t-FL. The third hypothesis (iii) is that the CPC migrates to the BM (before or after FL) and is maintained in the BM niche before migrating to a secondary lymphoid organ where it evolves into FL or t-FL. The second (ii) and third (iii) evolution pathways both relate to the existence of “premalignant” niches (the first one in the GC and the second one in the BM) where CPC can independently give rise to FL or t-FL.
The number of SHMs acquired by the putative CPCs after progression to FL and t-FL was high in the BMT model: 28 and 47 new mutations observed in the major clone of father and son samples, respectively (Figure S3). This difference was less striking in the paired biopsies, (median SHMs, 4 [FL] and 3 [t-FL], respectively) and could be a consequence of the different origin of these putative CPCs: in the familial case the CPC was transmitted at BMT, whereas in the sequential biopsies it may represent a cell that never left the GC.
On the basis of the pattern of SHMs as inferred from the genealogic trees, we investigated by qualitative PCR whether the putative CPC could be detected in the corresponding FL/t-FL samples and by RQ-PCR the presence of the t-FL clonotype in the preceding FL. Our data clearly showed an intricate pattern in the FL/t-FL's GCs, not compatible with a direct evolution, and suggested the existence of 3 different models (Figure 1C). First, the identification of the t-FL clonotype in the preceding FL sample in 5 of 8 cases (Table 1), together with the amplification of the putative CPC (patient 12 and patient 26) and of more immature clones (patient 1) in both the FL/t-FL (Table S3) shows that the CPC survives different lines of therapy and coexists with FL and t-FL cells within the same lymph node. The RQ-PCR data also suggest that a clone in the preceding FL, having the same SHMs as the t-FL cell, could be selected during transformation. The identification of this t-FL clonotype in FL samples from patients treated with different regimens (chemotherapy [patients 1, 6, and 27], radiotherapy [patient 26], and watch and wait [patient 2]) suggests that drug resistance is not the only mechanism responsible for clonal selection. Second, the CPC may be present only in the prelymphoma GC with the FL and the t-FL clones arising independently from this precursor. Finally, and in accordance with the father/son's data and the recent report describing the Sμ-region mutation pattern in sequential FL samples,22 the CPC could spread from the original lymph node and be maintained in the BM before migrating to secondary lymphoid organs. This mechanism, first suggested by studying the t(14;18)–positive cells in healthy persons, supposes the existence of BM niches in which the CPC could survive before acquiring new genomic lesions.23 Indeed, on the basis of the father/son model, this CPC must be long-lived; moreover, the identification of new glycosylation motifs in 10 hypothetical CPC sequences, together with the observation that in 13 of 14 cases these sites are conserved after transformation (Table S4), support an involvement of microenvironment during the evolution of the CPC.
Identification and quantification of the t-FL clonotype in the previous FL samples by RQ-PCR
| Patient, n = 8 . | Sample, n = 10 . | Total no. of primers . | Region* . | Sensitivity primers† . | Amplification level . |
|---|---|---|---|---|---|
| 1 | FL | 4 | CDR1-FR2/CDR2-FR3 | 10-4 | 10-2/10-3 |
| FL | CDR1/FR3 | 10-4 | 10-2/10-3 | ||
| 2 | FL | 3 | FR2-CDR2/FR3 | 10-4 | 10-3 |
| FL | FR2-CDR2/FR3 | 10-4 | 10-3 | ||
| 6 | FL | 4 | FR1-CDR1/FR3 | 10-4 | 10-4 |
| FL | FR1-CDR1/FR3 | 10-4 | 10-4 | ||
| 12 | FL | 4 | FR2-CDR2/FR3-CDR3 | 10-4 | Negative |
| FL | FR2-CDR2/FR3-CDR3 | 10-4 | Negative | ||
| 15 | FL | 3 | CDR1-FR2/FR3 | 10-4 | Negative |
| FL | CDR1-FR2/FR3 | 10-4 | Negative | ||
| 25 | FL | 3 | FR2-CDR2/FR3 | 10-4 | Negative |
| FL | 3 | FR2-CDR2/CDR3 | 10-4 | Negative | |
| FL2‡ | FR2-CDR2/FR3 | 10-4 | Negative | ||
| FL2‡ | FR2-CDR2/FR3 | 10-4 | Negative | ||
| 26 | FL | 3 | FR1-CDR1/FR3 | 10-4 | Negative |
| FL | 3 | FR1-CDR1/FR3 | 10-4 | Negative | |
| FL2‡ | FR1-CDR1/FR3 | 10-4 | 10-1/10-2 | ||
| FL2‡ | FR1-CDR1/FR3 | 10-4 | 10-1/10-2 | ||
| 27 | FL | 4 | FR2-CDR2/CDR3 | 10-4 | 10-1/10-2 |
| FL | FR2-CDR2/CDR3 | 10-4 | 10-1/10-2 |
| Patient, n = 8 . | Sample, n = 10 . | Total no. of primers . | Region* . | Sensitivity primers† . | Amplification level . |
|---|---|---|---|---|---|
| 1 | FL | 4 | CDR1-FR2/CDR2-FR3 | 10-4 | 10-2/10-3 |
| FL | CDR1/FR3 | 10-4 | 10-2/10-3 | ||
| 2 | FL | 3 | FR2-CDR2/FR3 | 10-4 | 10-3 |
| FL | FR2-CDR2/FR3 | 10-4 | 10-3 | ||
| 6 | FL | 4 | FR1-CDR1/FR3 | 10-4 | 10-4 |
| FL | FR1-CDR1/FR3 | 10-4 | 10-4 | ||
| 12 | FL | 4 | FR2-CDR2/FR3-CDR3 | 10-4 | Negative |
| FL | FR2-CDR2/FR3-CDR3 | 10-4 | Negative | ||
| 15 | FL | 3 | CDR1-FR2/FR3 | 10-4 | Negative |
| FL | CDR1-FR2/FR3 | 10-4 | Negative | ||
| 25 | FL | 3 | FR2-CDR2/FR3 | 10-4 | Negative |
| FL | 3 | FR2-CDR2/CDR3 | 10-4 | Negative | |
| FL2‡ | FR2-CDR2/FR3 | 10-4 | Negative | ||
| FL2‡ | FR2-CDR2/FR3 | 10-4 | Negative | ||
| 26 | FL | 3 | FR1-CDR1/FR3 | 10-4 | Negative |
| FL | 3 | FR1-CDR1/FR3 | 10-4 | Negative | |
| FL2‡ | FR1-CDR1/FR3 | 10-4 | 10-1/10-2 | ||
| FL2‡ | FR1-CDR1/FR3 | 10-4 | 10-1/10-2 | ||
| 27 | FL | 4 | FR2-CDR2/CDR3 | 10-4 | 10-1/10-2 |
| FL | FR2-CDR2/CDR3 | 10-4 | 10-1/10-2 |
Each sample is reported twice because the experiments were performed using 2 different combinations of primers and for patient 15 also 2 different probes.
This is the region of the IgH-VH gene where the clonotype primers were designed.
Maximal sensitivity was reported.
In 2 cases (patient 25 and patient 26), 2 sequential FL (FL2) samples were investigated. Patient 25 was negative for the t-FL clone in both the FL samples analyzed, whereas patient 26 was negative in the first FL biopsy and positive, at a level between 10−1 and 10−2, in the second.
In conclusion, the pattern of SHMs observed in our familial and sporadic models supports the existence of a CPC linking FL and t-FL, provides an alternative mechanism for transformation other than direct evolution, and helps to explain the discordant patterns of SHMs previously reported.24 These 2 mechanisms are not mutually exclusive and as seen for patient 26 (Figure S2) could be both independently active. Although the exact mechanism of CPC evolution, the stage of B-cell differentiation, where it occurs, or its role during lymphomagenesis is uncertain, the existence of these cells offers a new model to explain the clinical-molecular features of this disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rachel Waters for statistical analysis and M. Alikian for her contribution in editing the pictures.
This work was supported by grants from Cancer Research UK (London, United Kingdom) and Barts and The London Charitable Foundation (RAB05/F2).
Authorship
Contribution: E.C. planned the project, performed the molecular analysis, and wrote the manuscript; D.W. performed the molecular analysis; J.M. provided the clinical information; S.I. provided the previous molecular data; A.D. and S.M. followed the patients in the clinic; A.N. performed the histopathologic analysis; J.H. and R.L. provided the samples and the relative data of the familial case; J.G.G. and T.A.L. helped to plan the work and critically revised the manuscript; and J.F. planned the project, supervised the work, and helped to write the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Emanuela Carlotti, Centre for Medical Oncology, Institute of Cancer, Barts and The London School of Medicine and Dentistry, Charterhouse Sq, London EC1M 6BQ, United Kingdom; e-mail: emanuela.carlotti@cancer.org.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal