Abstract
The human T-cell leukemia/lymphoma virus type 1 (HTLV-1) ORF-I encodes a 99–amino acid hydrophobic membrane protein, p12I, that affects receptors in different cellular compartments. We report here that proteolytic cleavage dictates different cellular localization and functions of p12I. The removal of a noncanonical endoplasmic reticulum (ER) retention/retrieval signal within the amino terminus of p12I is necessary for trafficking to the Golgi apparatus and generation of a completely cleaved 8-kDa protein. The 8-kDa protein in turn traffics to the cell surface, is recruited to the immunologic synapse following T-cell receptor (TCR) ligation, and down-regulates TCR proximal signaling. The uncleaved 12-kDa form of p12I resides in the ER and interacts with the β and γc chains of the interleukin-2 receptor (IL-2R), the heavy chain of the major histocompatibility complex (MHC) class I, as well as calreticulin and calnexin. Genetic analysis of ORF-I from ex vivo samples of HTLV-1–infected patients reveals predominant amino acid substitutions within ORF-I that affect proteolytic cleavage, suggesting that ER-associated functions of p12I may contribute to the survival and proliferation of the infected T cells in the host.
Introduction
Human T-cell leukemia/lymphoma virus type 1 (HTLV-1) is the etiologic agent of a rare but aggressive hematopoietic malignancy of T cells, designated adult T-cell leukemia/lymphoma (ATLL), as well as a progressive myelopathy defined as HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP). In addition to structural and enzymatic proteins, the viral RNA genome encodes other proteins through alternative splicing, such as p40Tax, p27Rex, p13II, p30II, p12I, and HBZ, from an antisense mRNA.1,2
Among those, the p12I protein encoded by open reading frame I (ORF-I) is a 99–amino acid highly hydrophobic protein containing 4 putative SH3 binding motifs, 1 leucine zipper domain, 1 leucine zipper-like domain, and 2 putative transmembrane domains (TM1, TM2).3 Evidence of the expression and importance of this protein in HTLV-1 pathogenesis is at present indirect. The singly spliced mRNA encoding p12I is found in infected cells in vitro4,5 and in ex vivo samples from HTLV-1–infected patients.5 In addition, experimentally infected animals produce antibodies that recognize recombinant p12I6 and HTLV-1–infected individuals mount a cytotoxic T lymphocyte (CTL) response to ORF-I.7
Two isoforms of p12I have previously been described, the p12IK88 contains a lysine at position 88 that is ubiquitinated and targeted for degradation by the proteasome,8 whereas the more stable p12IR88 protein encodes an arginine at position 88 and is therefore not ubiquitinated. However, no specific disease association of the 2 p12I isoforms has previously been established.8,9
Several functions have been ascribed to p12I. Ectopically expressed p12I resides in the endoplasmic reticulum (ER) and Golgi compartments10-12 and forms homodimers via its TMs.8 The p12I protein associates with the 16-kDa subunit of V-ATPase in vitro,13 and in the ER p12I physically binds to cellular receptors such as the β and γc chains of IL-2R, which increase T-cell activation and responsiveness to IL-2.14-16 The p12I protein binds the heavy chain of the MHC class I and down-regulates its surface expression11,17 and it also interacts with the ER-resident proteins calreticulin and calnexin.18 In conjunction with agonists such as phorbol myristate acetate (PMA), p12I increases nuclear factor of activated T cells (NFAT) activation in a linker for activation of T cells (LAT)–independent manner.18 In contrast, following TCR ligation, p12I down-regulates proximal TCR signaling and this effect is LAT dependent.19 In addition, p12I enhances the lymphocyte function–associated antigen-1 (LFA-1)–mediated T-cell adhesion and down-regulates ICAM-1 and ICAM-2, but not ICAM-3.17,20 Both TCR and LFA-1 are present in the lipid rafts that have been implicated in the regulation of the immunologic synapse by allowing local assembly of related molecules such as MHC class II.
In an attempt to reconcile the seemingly disparate functions of this viral protein, we found that the p12I protein undergoes complex posttranslational modifications that include proteolytic cleavage between amino acid positions 9 and 10 followed by a second cleavage between the amino acids 29 and 30. The first proteolytic cleavage removes a noncanonical ER retention/retrieval signal at the amino terminus of p12I and allows for further trafficking of this viral protein to the Golgi apparatus and the lipid rafts. Importantly, we found a high frequency of genetic mutations in the ORF-I of provirus from HTLV-1–infected individuals causing ER retention of p12I, suggesting an important role for p12I functions in the ER in vivo.
Methods
Expression plasmids and antibodies
The pME18S p12IΔSL expression plasmid has been described previously.19 p12I and its mutants were generated by polymerase chain reaction (PCR) or by the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using site-specific mutagenic oligonucleotides following the manufacturer's instructions. The pAB-GTG molecular clone mutant was generated by ligation of the Cla-I Sal-I DNA fragment from the pBST molecular clone21 into the pACH equivalent restriction sites22 and the initiation codon of p12 mutagenized as previously described.19
The following oligonucleotides were used and the sequence of plasmid clones was analyzed to confirm the mutations: L9A-F: 5′-GCCTTCTCAGCCCCGCGTCTCCACTTGCGCT-3′; L9A-R: 5′-AGCGCAAGTGGAGACGCGGGGCTGAGAAGGC-3′; S10A-F: 5′-CCTTCTCAGCCCCTTGGCTCCACTTGCGCTCA-3′; S10A-R: 5′-TGAGCGCAAGTGGAGCCAAGGGGCTGAGAAGG-3′; G29S-F: 5′-TGCTTTCTCCGGGCGACGTCAGCAGCCTTCTTCTC-3′; G29S-R: 5′-GCGGAGAAGAAGGCTGCTGACGTCGCC-3′; Δ24-F: 5′-AGCTCTCGAGATGGGCGACGTCAGCGGCCTTCT-3′; Δ29-F: 5′-GTGGCTCGAGACCATGCTTCTTCTCCGCCCGCCTC-3′; Δ24 and Δ29-R: 5′-TCGGTCTAGAAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCA-3′; S23P-F: 5′-GCTCCTGCTCTTCCTGCTTCCTCCGGGCGACGTCAGCG-3′; and S23P-R: 5′-CGCTGACGTCGCCCGGAGGAAGCAGGAAGAGCAGGAGC-3′.
The pAcGFP1-Mem vector (Clontech, Mountain View, CA) encodes the Aequorea coerulescens GFP containing 20 amino acids of neuromodulin (GAP-43) fused to its N-terminus. pAcGFP1-Mem is targeted to cellular membranes and especially to the plasma membrane. We use the term GFP-Mem for pAcGFP1-Mem. The p12I1-32-GFP-Mem and the p12IG29S1-32-GFP-Mem were generated by PCR amplification using wild-type p12I and p12IG29S as templates, respectively. The p12I1-15-GFP-Mem and the p12IS10A1-15-GFP-Mem were generated using wild-type p12I and p12IS10A, respectively. All primers were flanked 5′ by KpnI and 3′ by AgeI. The p12I1-32, p12IG29S1-32, p12I1-15, and p12IS10A1-15 PCR products were digested and ligated into the KpnI- and AgeI-digested pAcGFP1-Mem vector. The forward primer used was p12I1-32/1-15-F: 5′-AATTGGTACCACCATGCTGTTTCGCCTTCTC-3′ and reverse primers were p12I1-32-R: 5′-TTAAACCGGTGCGAGAAGAAGGCCGCTGAC-3′, p12IG29S1-32-R: 5′-TTAAACCGGTGCGAGAAGAAGGCTGCTGAC-3′, and p12I1-15-R: 5′-TTAAACCGGTGCCGTGAGCGCAAGTGG-3′. The GFP-Mem-KDEL construct was generated by introducing in the reverse primer AAG GAC GAA TTA (KDEL) followed by a stop codon. The cloning sites used were BamHI and XbaI and the digested PCR product was ligated into the BamHI- and XbaI-digested GFP-Mem vector. The forward primer used was 5′-GGATCCCCCGGGATCCACCGGTCGCCAC-3′ and the reverse primer was 5′-TTCTCTAGAGTCGCGGCCGCTCTATAATTCGTCCTTCTTGTACAGCTCATCCATGC-3′. The p12I1-5-GFP-Mem construct was generated by introducing ATGCTGTTTCGCCTT (MLFRL) in the forward primer. The cloning sites used were BamHI and XbaI and the PCR product was digested and ligated into the BamHI- and XbaI-digested GFP-Mem vector. The forward primer used was 5′-TTCCGGATCCACCATGCTGTTTCGCCTTCTGTGCTGTATGAGAAGAACAAA-3′ and the reverse primer was 5′-TTCTCTAGAGTCGCGGCCGCTTCACTTGTACAGC-3′. All constructs were verified by sequencing. The psdred2-ER (Clontech) construct expresses the red fluorescent protein from Discosoma sp, which has the ER targeting sequence of calreticulin fused to the 5′ and the ER retention signal KDEL fused to the 3′ of the pDsRed2.
The analysis of 9 previously unpublished clinical samples (1 TSP/HAM and 8 ATLL) was performed on genomic DNA obtained from peripheral blood mononuclear cells (PBMCs) following amplification by PCR using the following primers: p30II-ORF(+)-1: 5′-CAGTCGACATGGCACTATGCTGTTTCGCCTTCTCAGCC-3′, p30II-ORF(−)-1: 5′-ATGGATCCGAGGTTCTCTGGGTGGGGAAGGAGGGGAGT-3′. The DNA fragments were cloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA) and sequenced.
Because the forward primer overlaps with ORF-I, the first 7 amino acids of p12I do not reflect the actual amino acid sequence in those samples. The remaining 295 clinical samples were amplified and sequenced as described.3,23-28
The anti-HA1 antibody clones 12CA5 and 3F10-hrp were obtained from Roche Applied Science (Indianapolis, IN); the 6E2 clone with Alexa-488 conjugation was obtained from Cell Signaling Technology (Danvers, MA). Rabbit polyclonal mannosidase II antibody–Golgi marker, mouse monoclonal (RL90) to PDI–ER marker, and mouse monoclonal antibody (FMC 75) to calreticulin were all obtained from Abcam (Cambridge, MA).
Cell culture and DNA transfection
Jurkat T cells were grown in RPMI 1640 and 293T, COS-7, and HeLa cells were grown in Dulbecco modified Eagle medium (DMEM) and both media were supplemented with 10% FCS, 2 mM penicillin-streptomycin, and 5 mM l-glutamine (Invitrogen), respectively.
Transfections were performed using DMRIE-C (Invitrogen) or the Cell Line Nucleofector Kit V (Amaxa, Gaithersburg, MD) for Jurkat T cells, Effectene (QIAGEN, Valencia, CA) for 293T-cells, and FuGENE-6 or FuGENE HD (Roche, Indianapolis, IN) for COS-7 and HeLa cells following the manufacturers' protocols.
For the TCR stimulation, Jurkat T cells were centrifuged and treated with 1 μg/mL anti-CD3ϵ (UCH-T1) from EMD Biosciences (La Jolla, CA) in fresh media. The cells and culture media were harvested at the indicated times.
Immunofluorescence and confocal microscopy
For the formation of the immunologic synapse, Jurkat T and Raji cells were prepared separately at the concentration of 106 cells/mL in the fresh medium. Raji cells were prepulsed with or without 10 μg/mL SEE from Toxin Technology (Sarasota, FL) for 1 hour at 37°C. Cells were combined at a 1:1 ratio and fixed by adding 4% paraformaldehyde (PFA) at indicated time point. Cells (105) were analyzed on a glass slide coated with poly-l-lysine. Cells were washed in PBS and permeabilized in ice-cold methanol for 20 minutes at −20°C followed by staining with indicated antibodies.
HeLa cells and COS-7 cells were seeded on φ12-mm coverslips and transfected the following day by FuGENE HD or FuGENE-6. Cells on coverslips were fixed 48 hours after transfection with 4% PFA in PBS for 20 minutes, washed in PBS, and permeabilized in ice-cold methanol for 20 minutes at −20°C. For immunofluorescence the fixed cells were washed 3 times in PBS before being inverted onto a nescofilm containing a 25-μL droplet of 0.5% BSA in PBS blocking buffer for 15 minutes. The cells were further incubated for 1 hour with 25-μL droplets containing primary antibodies in appropriate dilutions, washed 3 × 5 minutes in PBS, and incubated in anti–mouse or anti–rabbit Alexa-conjugated secondary antibodies for 1 hour in the dark followed by 3 × 5 minutes washes in PBS. Finally, the cells were rinsed quickly in water before being mounted in 5 μL mounting media on a glass slide before being examined by Zeiss laser scanning confocal microscope LSM 510 (Carl Zeiss MicroImaging, Thornwood, NY) with a 63×/100×/1.3 NA Plan Apochromat oil objective.
Luciferase assay
The Dual-Luciferase Reporter Assay System was obtained from Promega (Madison, WI) and all the measurements were performed according to the manufacturer's protocol. Luciferase values were normalized to protein content and protein concentration was determined using the Bio-Rad (Hercules, CA) Protein Assay. All experiments were repeated at least 3 independent times and representative data are shown as average and standard deviation of triplicates.
Quantification of HTLV p19 Gag
The HTLV p19 Gag enzyme-linked immunosorbent assay (ELISA) kit from ZeptoMetrix (Buffalo, NY) was used to quantify p19 Gag in the supernatant. Direct or diluted cell-culture supernatants were used for the analyses to obtain concentrations within the standard range. Experiments were repeated at least 3 times and all the measurements performed according to the manufacturer's protocol are shown as average and standard deviation of duplicates.
Amino acid sequence analysis
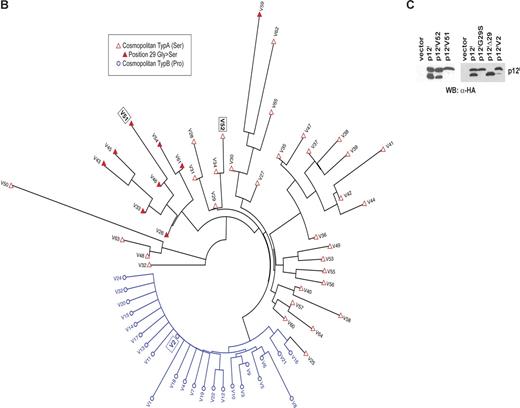
A total number of 304 p12I sequences for subtypes A and B (cosmopolitan A and B) were analyzed. These sequences were derived from ex vivo proviral DNA samples of HTLV-1–infected patients.3,23-28 The 203 subtype B samples could be grouped into 24 different variants and the remaining 101 subtype A samples were grouped into 41 different variants (Figure 2A). We used European Molecular Biology Open Software Suite (EMBOSS) for sequence handling and formatting.29 The Ape package for the R project for statistical computing (Foundation For Statistical Computing, http://www.R-project.org) was used to calculate genetic distance (Jukes-Cantor genetic distance model) and to generate the unrooted neighbor-joining tree (Figure 2B).
Results
Proteolytic cleavage dictates the cellular localization of the 12-kDa and 8-kDa forms of the p12I complex
Expression of the singly spliced ORF-I cDNA (Figure 1A) consistently results in the expression of 2 or more protein bands that are usually referred to as p12I (Figure 1C lane 2), whose nature and derivation are unknown. Computer program analysis (PSORT; http://psort.nibb.ac.jp; Kenta Nakai, Human Genome Center, Institute for Medical Science, University of Tokyo, Tokyo, Japan) and ELM30 (http://elm.eu.org) of the p12I amino acid sequence predicted 2 possible signal peptide cleavage sites at positions L9↓S10 and VSG29↓L30L (Figure 1A top). Indeed mutation of serine at position 10 to alanine resulted predominantly in the production of the 12-kDa protein (Figure 1B,C lane 4), whereas mutation of leucine at position 9 to alanine appeared to favor the generation of the 8-kDa protein (Figure 1C lane 3). In addition, the G29→S mutant yielded a single uncleaved protein product of 12 kDa (Figure 1B,C lane 7), whereas substitution of leucines 30 and 31 with alanine did not affect the generation of the 8-kDa protein band (data not shown). Additional substitution of leucine with alanine at position 9 in the p12IG29S mutant partially restored cleavage (Figure 1B,C lane 8), whereas a mutant lacking the first 29 amino acids (p12IΔ29) yielded a protein of 8 kDa (Figure 1B,C lane 10), and a mutant lacking the first 24 amino acids (p12IΔ24) (Figure 1B) resulted in similar migration (data not shown). Altogether these data are consistent with the finding that, at steady state, the p12I complex is constituted by an uncleaved 12-kDa precursor molecule and a completely cleaved 8-kDa protein.
Mutations of amino acids at positions 10 and 29 affect p12I cleavage and cellular localizations. (A) Single-letter amino acid code of p12I from HTLV-1. The arrows indicate the computer-predicted cleavage sites. (B) Schematic representation of the p12I mutants. Changes in amino acid positions are indicated on the top. L9 and S10 were changed to alanines. Glycine 29 was changed to serine. The Δ symbol signifies deletion of amino acids at the amino terminus for the truncated mutants Δ24 and Δ29. (C) Cell lysates from the 293T cells transfected with the wild-type p12I or the p12I mutants were analyzed by Western blot with anti-HA antibody. (D) Localization of p12I, p12IG29S, and p12IΔ29 in Jurkat T cells stained with anti-HA antibody. (E) HeLa cells expressing p12I and p12IG29S were costained with an anti-PDI antibody. (F,G) Localization of p12I, p12IG29S, and p12IΔ29 in HeLa cells compared with ER and Golgi markers. Red-ER is used as the ER marker, whereas an antibody to mannosidase II (Mann II) is used to label the Golgi apparatus.
Mutations of amino acids at positions 10 and 29 affect p12I cleavage and cellular localizations. (A) Single-letter amino acid code of p12I from HTLV-1. The arrows indicate the computer-predicted cleavage sites. (B) Schematic representation of the p12I mutants. Changes in amino acid positions are indicated on the top. L9 and S10 were changed to alanines. Glycine 29 was changed to serine. The Δ symbol signifies deletion of amino acids at the amino terminus for the truncated mutants Δ24 and Δ29. (C) Cell lysates from the 293T cells transfected with the wild-type p12I or the p12I mutants were analyzed by Western blot with anti-HA antibody. (D) Localization of p12I, p12IG29S, and p12IΔ29 in Jurkat T cells stained with anti-HA antibody. (E) HeLa cells expressing p12I and p12IG29S were costained with an anti-PDI antibody. (F,G) Localization of p12I, p12IG29S, and p12IΔ29 in HeLa cells compared with ER and Golgi markers. Red-ER is used as the ER marker, whereas an antibody to mannosidase II (Mann II) is used to label the Golgi apparatus.
The p12I protein complex has been reported to localize to the ER, Golgi, and the lipid rafts.11,18,19 To assess how the proteolytic cleavage of p12I affects its cellular localization, we transfected Jurkat T, HeLa, or COS-7 cells with the wild-type p12I, the p12IG29S mutant, as well as the deletion mutant p12IΔ29 (Figure 1B). In Jurkat T cells, the p12I wild-type protein and the p12IΔ29 mutant localized diffusely and in dots in the cytoplasm (Figure 1D). This localization was also observed for the p12IΔ24 mutant (data not shown). In contrast, when proteolytic cleavage was abrogated by mutation at amino acid position 29, p12I accumulated near the nucleus (Figure 1D). To further study the cellular localization of this mutant, we compared the distribution of p12I and the p12IG29S mutant to the localization of the ER-resident proteins, protein disulphide isomerase (PDI) and calreticulin. We noticed a much lower staining intensity of PDI (Figure 1E) and calreticulin (data not shown) in cells expressing p12I and the p12IG29S mutant (Figure 1E). Similar observations have been reported for the BPV E5 protein,31 a functional analog of p12I13 that alters the ER distribution. To circumvent this problem, HeLa cells were cotransfected with pDsRed2-ER (Red-ER). The Red-ER construct expresses the red fluorescent protein from Discosoma sp, which has the ER targeting sequence of calreticulin fused to the 5′ and the ER retention signal KDEL fused to the 3′ of the pDsRed2. To define the Golgi apparatus, we used an antibody to mannosidase II, which is a medial Golgi localized protein. The wild-type p12I and the p12IΔ29 mutant colocalized with Red-ER and partially colocalized with mannosidase II (Figure 1F,G). The expression of the p12IG29S mutant caused a redistribution of the Red-ER protein to a dotted pattern (Figure 1F). Furthermore, the p12IG29S mutant appeared to be excluded from the Golgi apparatus since no colocalizaton was observed with mannosidase II (Figure 1G).
The 12-kDa uncleaved form of p12I predominates in ex vivo samples from HTLV-1–infected individuals
The data presented here demonstrate that mutation at the cleavage sites of p12I interferes with its cellular localization. To investigate the relevance of these findings to HTLV-1 infection, we studied genetic polymorphism at the protein level of p12I obtained from 304 ex vivo proviral DNA samples from HTLV-1–infected individuals with either subtype A (nA = 101, 33.2%) or subtype B (nB = 203, 66.8%). The subtypes are characterized by the amino acid at position 23 (proline in subtype B and serine in subtype A). The 203 samples for subtype B could be grouped into 24 different variants, whereas the 101 samples for subtype A were more diverse and could be grouped into 41 different variants (Figure 2A). Seventy percent of all variants are represented by only one sequence sample, whereas V2 with 167 sequences is the most frequent variant in our dataset (Figure 2A). The average pairwise genetic distance was 4.38 for subtype A variants and 1.91 for subtype B variants using Jukes-Cantor correction method. The difference in the number of variants between the subtypes could be due to a sampling bias. Amino acid sequence near the first putative proteolytic cleavage site at position 9 shows a low level of genetic polymorphisms for both subtypes (Figure 2A), whereas the second cleavage site around position 29 is more variable for subtype A and more conserved for subtype B. In addition, position 29 itself is conserved within subtype B but not in subtype A. Twenty-two percent of the variants for subtype A have a serine instead of a glycine at this position. Although the low level of polymorphism among the 65 variants does not provide great statistical power for a detailed phylogenetic analysis, in a best tree analysis the different variants group according to their subtypes except for variant V25 (Figure 2B). It also shows that samples with a serine instead of the glycine at position 29 cluster together. These results suggest that the sequence differences might reflect emergence of functionally distinct variants.
Amino acid sequence analysis of HTLV ORF-I from ex vivo proviral DNA samples from HTLV-1–infected individuals. (A) Amino acid sequence alignment for p12I variants from ex vivo proviral DNA samples from HTLV-1–infected individuals. DNA provirus fragment corresponding to ORF-I was amplified by PCR from PBMCs of 9 new patients, and the putative amino acids sequence of p12I was compared with previously published p12I sequence from our and other laboratories.3,23-28 Consensus sequence is based on cosmopolitan type B samples (proline at position 23). Sequences V1-V24 are cosmopolitan type B variants and sequences V25-V65 are cosmopolitan type A variants (serine at position 23). The number of identical samples for each variant is given in the right column. Matches identical to the consensus sequences are represented by “.”; nonidentical matches are either in lowercase (if similar residues) or capital letters. There are 4 variants (V6, V9, V30, and V36) with premature termination represented by “−”. The shaded sequences indicate a cluster of changes proximal to the second proteolytic cleavage site. The 9 samples amplified with the primers described in “Methods” belong to variants V29, V46, V48-52 (1 sample each), and V34 (2 samples).
Amino acid sequence analysis of HTLV ORF-I from ex vivo proviral DNA samples from HTLV-1–infected individuals. (A) Amino acid sequence alignment for p12I variants from ex vivo proviral DNA samples from HTLV-1–infected individuals. DNA provirus fragment corresponding to ORF-I was amplified by PCR from PBMCs of 9 new patients, and the putative amino acids sequence of p12I was compared with previously published p12I sequence from our and other laboratories.3,23-28 Consensus sequence is based on cosmopolitan type B samples (proline at position 23). Sequences V1-V24 are cosmopolitan type B variants and sequences V25-V65 are cosmopolitan type A variants (serine at position 23). The number of identical samples for each variant is given in the right column. Matches identical to the consensus sequences are represented by “.”; nonidentical matches are either in lowercase (if similar residues) or capital letters. There are 4 variants (V6, V9, V30, and V36) with premature termination represented by “−”. The shaded sequences indicate a cluster of changes proximal to the second proteolytic cleavage site. The 9 samples amplified with the primers described in “Methods” belong to variants V29, V46, V48-52 (1 sample each), and V34 (2 samples).
(B) Unrooted neighbor-joining tree for 65 variants. Cosmopolitan type B variants are represented by open circle and highlighted in blue, and cosmopolitan type A variants are represented by red triangles. The closed red triangles indicate samples with a frequent glycine to serine mutation at position 29. (C) Lysates from 293T cells transfected with plasmids encoding the p12IV51, p12IV52, and p12IV2 variants were analyzed by Western blot with the anti-HA antibody.
(B) Unrooted neighbor-joining tree for 65 variants. Cosmopolitan type B variants are represented by open circle and highlighted in blue, and cosmopolitan type A variants are represented by red triangles. The closed red triangles indicate samples with a frequent glycine to serine mutation at position 29. (C) Lysates from 293T cells transfected with plasmids encoding the p12IV51, p12IV52, and p12IV2 variants were analyzed by Western blot with the anti-HA antibody.
Prototype expression plasmids were generated from variant 51 (V51) that carries the G29S mutation, variant 52 (V52) that has no mutations in this region, and variant 2 (V2) that carries the S23P subtype determining polymorphism. Expression in 293T cells of these prototype p12I variants demonstrated that the p12IV52 yielded the expression of the 12-kDa and 8-kDa form as observed for p12I, whereas p12IV51 expression resulted exclusively in the production of the uncleaved 12-kDa form (Figure 2C). The p12IV2 was not efficiently cleaved yielding a higher ratio of the 12-kDa protein versus the 8-kDa protein (Figure 2C). Whereas variant 2 and 51 account for 55.3% of the patients' samples studied, the total number of variants containing S23P or G29S changes account for 70.7% of the patients' samples, demonstrating that the uncleaved form of p12I predominates in the cohort of patients studied here.
A 2-step proteolytic cleavage is required to remove a noncanonical ER retention/retrieval signal within the first 5 amino acids of p12I
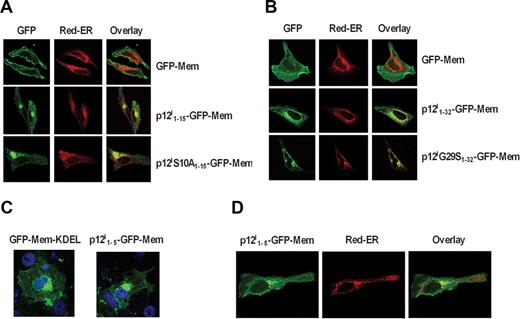
The data presented in Figure 1 are consistent with the hypothesis that a portion of the ORF-I protein product, the 12-kDa form, is retained/retrieved in the ER, whereas the other, the 8-kDa form, may be generated by a 2-step proteolytic cleavage and traffics to the cell surface. To provide further support to the notion that there are 2 cleavage sites within p12I, we generated constructs whereby different portions of the amino terminus of p12I were juxtaposed at the 5′ of the GFP-Mem construct. The GFP-Mem contains 20 amino acids of the plasma membrane targeting signal from neuromodulin fused to the 5′ of green fluorescent protein (GFP) and is targeted to the plasma membrane, as demonstrated in HeLa cells (Figure 3A,B).
p12I contains a noncanonical ER retention/retrieval signal at its amino terminus. (A) Localization of the GFP-Mem, p12I1-15-GFP-Mem, and p12IS10A1-15-GFP-Mem and Red-ER in HeLa cells. (B) Localization of the GFP-Mem, p12I1-32-GFP-Mem, and p12IG29S1-32-GFP-Mem compared with Red-ER in HeLa cells. (C) Localization of GFP-Mem-KDEL and p12I1-5-GFP-Mem in COS-7 cells. (D) Localization of the p12I1-5-GFP-Mem compared with Red-ER in HeLa cells.
p12I contains a noncanonical ER retention/retrieval signal at its amino terminus. (A) Localization of the GFP-Mem, p12I1-15-GFP-Mem, and p12IS10A1-15-GFP-Mem and Red-ER in HeLa cells. (B) Localization of the GFP-Mem, p12I1-32-GFP-Mem, and p12IG29S1-32-GFP-Mem compared with Red-ER in HeLa cells. (C) Localization of GFP-Mem-KDEL and p12I1-5-GFP-Mem in COS-7 cells. (D) Localization of the p12I1-5-GFP-Mem compared with Red-ER in HeLa cells.
We fused the GFP-Mem protein with either the first 15 amino acids of wild-type or of the S10A mutant that is not cleaved (Figure 1C). Expression of p12I1-15-GFP-Mem in HeLa cells demonstrated plasma membrane and intracellular distribution that colocalized with the Red-ER (Figure 3A). In contrast, the p12IS10A1-15-GFP-Mem was predominantly localized to the ER (Figure 3A), suggesting that the introduction of alanine at position 10 in the p12IS10A1-15-GFP-Mem construct caused ER retention/retrieval. Similarly, fusion of the first 32 amino acids of either the wild-type p12I or the p12IG29S mutant to the GFP-Mem protein resulted in retention/retrieval of the p121-32G29S-GFP-Mem protein in the ER of both HeLa cells (Figure 3B). These results were also confirmed in COS-7 cells (data not shown).
Thus, these data support the existence of 2 cleavage sites at the amino terminus of p12I and suggest the presence of an ER retention/retrieval signal within the first 9 amino acids of the protein. Indeed, computer analyses predicted a putative noncanonical ER retention/retrieval signal within the first 5 amino acids at the amino terminus: MLFRL. To investigate this, the first 5 amino acids of p12I were fused at the 5′of the GFP-Mem construct, generating p12I1-5-GFP-Mem. As a positive control, the canonical ER retention/retrieval signal KDEL was fused to the C-terminus of the GFP-Mem protein. Addition of KDEL at the carboxy terminus of GFP-Mem resulted in accumulation of GFP-Mem in the ER (Figure 3C); similarly addition of the first 5 amino acids of p12I to GFP-Mem indicated ER accumulation (Figure 3C), which was confirmed by comparison and colocalization with Red-ER (Figure 3D). Thus, the amino terminus of p12I contains a noncanonical ER retention/retrieval signal that is removed by a 2-step cleavage to allow trafficking of the 8-kDa form to the cell surface.
The 8-kDa form of p12I, but not the uncleaved 12-kDa form, is recruited to the immunologic synapse and mediates proximal TCR signaling down-regulation
Ligation of the TCR by peptides from MHC to antigen-presenting cells (APCs) leads to the formation of a complex structure that is termed the immunologic synapse (IS), which includes the TCR, protein tyrosine kinases, and LAT. Since we previously have shown that p12I is recruited to the IS upon TCR ligation, we hypothesized that the 8-kDa but not the 12-kDa form down-regulates TCR signaling. To test this, we used Jurkat T cells and Raji cells, as an in vitro model of CD4+ T-cell and APC interaction, whereby polarization of the TCR can be easily visualized with anti-CD3 following cocultivation of Jurkat T cells with Staphylococcus aureus enterotoxin E (SEE) prepulsed B cells. Within 15 minutes of coculture, p12I and the p12IΔ29 mutant were recruited to the IS formed between the T cells and the B cells (Figure 4A), whereas the p12IG29S mutant was not observed in the IS, suggesting that the 8-kDa protein is the isoform recruited to the IS.
The 8-kDa p12I protein down-regulates TCR signaling and is recruited to the immunologic synapse. (A) Jurkat T cells expressing the wild-type p12I, p12IG29S, or p12IΔ29 deletion mutants were cocultivated with SEE prepulsed B cells for 15 minutes. Cells were costained with anti-CD3 and anti-HA antibodies. The arrows indicate the IS found between the B cell and the T cell. (B-D) Jurkat T cells were cotransfected with the p12I knockout proviral clone, pAB-GTG, p12I, the p12I mutants, and the HTLV-LTR–driven luciferase reporter plasmid. Cells were collected 24 hours later, resuspended in fresh media, and stimulated with or without anti-CD3 antibody. Tax-driven luciferase activity (B), p19 Gag production in the supernatant (C), and p12I expression (D) at 48 hours are shown.
The 8-kDa p12I protein down-regulates TCR signaling and is recruited to the immunologic synapse. (A) Jurkat T cells expressing the wild-type p12I, p12IG29S, or p12IΔ29 deletion mutants were cocultivated with SEE prepulsed B cells for 15 minutes. Cells were costained with anti-CD3 and anti-HA antibodies. The arrows indicate the IS found between the B cell and the T cell. (B-D) Jurkat T cells were cotransfected with the p12I knockout proviral clone, pAB-GTG, p12I, the p12I mutants, and the HTLV-LTR–driven luciferase reporter plasmid. Cells were collected 24 hours later, resuspended in fresh media, and stimulated with or without anti-CD3 antibody. Tax-driven luciferase activity (B), p19 Gag production in the supernatant (C), and p12I expression (D) at 48 hours are shown.
To investigate how cellular localization affects p12I function, the abilities of the mutants p12IG29S, p12IS10A, p12IΔ24, p12IΔ29, and wild-type p12I to down-regulate proximal TCR signaling and viral replication were assessed in Jurkat T cells. The HTLV-1 LTR-driven luciferase construct was cotransfected with the HTLV-1 molecular clone pACH-GTG, whereby p12I expression was eliminated by a genetic mutation19 together with p12I and p12I mutants. In this experimental model, ligation of TCR with the anti-CD3 antibody increases Tax activity and viral expression. Because p12I dampens TCR signaling, a readout of p12I function is reduction of Tax activity and HTLV-1 replication. Tax activity was measured by the luciferase assay (Figure 4B) and viral replication by p19 Gag antigen capture assay of the culture supernatant (Figure 4C). The expression of p12I mutants was verified by Western blot (Figure 4D). Decreased Tax-mediated transactivation was observed after expression of p12I, p12IΔ24, and p12IΔ29. However, this was not observed after expressing the mutants p12IG29S and p12IS10A, demonstrating that proteolytic cleavage is essential for down-regulation of proximal TCR signaling and that this function is mediated by the 8-kDa form of p12I.
Discussion
Viruses that carry limited genetic information exploit complex posttranslational modification to generate protein products with diverse functions, and HTLV-1 is no exception. Here, we show that the HTLV-1 ORF-I 99–amino acid product undergoes complex posttranslational modification that yields 2 protein forms: one that mostly remains in the ER and the other that traffics to the cell surface. Since both mutations at serine 10 and glycine 29 result in loss of the 8-kDa form of p12I, our results suggest that the production of the 8-kDa protein occurs through a 2-step proteolytic cleavage, where the first cleavage occurs between amino acids 9 and 10 (PL↓SP) and the second between amino acids 29 and 30 (VSG↓LL). Our data support the hypothesis that the first cleavage is necessary to remove the amino terminus ER retention signal LFRL and allow trafficking of the partly cleaved protein to another cellular compartment, likely the Golgi, where the second cleavage can occur.
We provide 3 separate lines of evidence demonstrating that proteolytic cleavage is essential for plasma membrane localization of p12I. First, modification of serine at position 10 and/or glycine at position 29 results in a loss of the 8-kDa form of p12I and neither of these p12I mutants localize to the plasma membrane. Second, fusion of the GFP-Mem to the noncleavable first 15 or 32 amino acids from the p12IS10A or p12IG29S mutants, respectively, impaired trafficking of the GFP-Mem protein to the plasma membrane, demonstrating that cleavage at both positions is necessary for plasma membrane localization of these chimeric proteins. Third, we have identified a noncanonical ER retention/retrieval signal within the first 5 amino acids of p12I.
The data presented here suggest a model on how this small viral protein exerts its pleiotropic functions (Table 1; Figure 5). The p12I protein binds to and down-regulates surface expression of the β and γc chains of IL-2R by retaining both IL-2R chains in the ER, and increases signal transducer and activator of transcription 5 (STAT5) phosphorylation. By doing so, p12I decreases the threshold for T-cell activation and IL-2 requirement.16,32 The p12I protein also interacts with and down-regulates the cell surface expression of the heavy chain (Hc) of MHC class I. Indeed, the fraction of Hc-MHC class I and IL-2R chains that binds p12I is endoglycosidase H (EndoH) sensitive, demonstrating that this interaction occurs before the trafficking of p12I to the Golgi apparatus. Similarly, the interaction of p12I with the ER-resident proteins calreticulin and calnexin most likely occurs with the uncleaved form of p12I.
Comparison of functions and localization of the 12-kDa and 8-kDa isoforms of p12I
| p12I isoform . | ↓ TCR signaling . | ↓ Viral replication . | Recruitment to IS . | Localization . |
|---|---|---|---|---|
| 12 kDa | − | − | − | ER |
| 8 kDa | + | + | + | Plasma membrane |
| p12I isoform . | ↓ TCR signaling . | ↓ Viral replication . | Recruitment to IS . | Localization . |
|---|---|---|---|---|
| 12 kDa | − | − | − | ER |
| 8 kDa | + | + | + | Plasma membrane |
p12I function in different cellular compartments. (A) Summary of the 2-step proteolytic cleavage of p12I. (B) Interaction of p12I in the ER with the IL-2R chains, MHC class I, calnexin, and calreticulin and trafficking of the completely cleaved p12I to the Golgi and plasma membrane.
p12I function in different cellular compartments. (A) Summary of the 2-step proteolytic cleavage of p12I. (B) Interaction of p12I in the ER with the IL-2R chains, MHC class I, calnexin, and calreticulin and trafficking of the completely cleaved p12I to the Golgi and plasma membrane.
In contrast, as we demonstrate here, membrane localization of p12I is essential for TCR proximal signaling down-regulation. Indeed, it is the 8-kDa protein, but not the uncleavable p12I G29→S mutant, that is recruited to the immunologic synapse following TCR engagement and down-regulates proximal TCR signaling. The results reported here explain the contrasting effects of p12I on TCR signaling. It is likely that the uncleaved form of p12I, together with PMA, increases cytoplasmic calcium and NFAT activity, and this effect is LAT-independent.18 In contrast, after ligation of TCR, p12I decreases proximal signaling in a LAT-dependent manner.19 Here, we show that this effect is mediated exclusively by the 8-kDa protein.
Collectively our results suggest a model whereby the effects of p12I on IL-2R chains, calreticulin, and calnexin as well as MHC-I-Hc may be mediated by the uncleaved form of p12I that resides in the ER, whereas down-regulation of the TCR is mediated by the completely cleaved 8-kDa protein (Figure 5). The availability of the p12I mutants generated in the present work will enable us to dissect the role of the uncleaved and the cleaved form of p12I on other molecules affected by p12I, such as LFA-1, ICAM-1, ICAM-2, and the vacuolar ATPase.13,17,20
Importantly, we found that both the naturally occurring mutation S23→P and G29→S observed in HTLV-1–infected individuals inhibit cleavage and the generation of the 8-kDa protein. Interestingly, within the HTLV-1 provirus both of these mutations do not result in amino acid sequence changes in the overlapping ORF-II or in the antisense reading frame encoding HBZ. A predominance of the uncleaved form of p12I in vivo would cause increased STAT activation, calcium release, and T-cell proliferation with resulting increase in the number of infected T cells. In turn, the uncleaved form of p12I, by interacting and down-regulating the MHC class I-Hc in the ER would favor escape from immune recognition of virus infected cells. Lastly, whether the uncleaved form of p12I also mediates down-regulation of ICAM-1 and ICAM-217 remains to be investigated.
In conclusion, the findings reported here provide the rationale for the design of a prospective study that will attempt to correlate different functions of p12I with proviral load and disease development in HTLV-1–infected individuals.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Tom Misteli for helpful discussions; Robyn Washington Parks, Jonathan T. Magruder, Talisa A. Goss, Joanna Ostas, and Tad Guszcznski for assistance with some of the studies; Tatiana Karpova for helpful suggestions with the Zeiss 510 Meta microscope; and Steven Snodgrass and Ryan Kelly for editorial assistance.
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. R.F. was supported in part by the Japan Society for the Promotion of Science (Tokyo, Japan).
National Institutes of Health
Authorship
Contribution: R.F., V.A., I.B., and V.C. designed and performed research and analyzed data; J.M.N., C.N., and V.W.V. performed research; A.G. contributed samples; J.-C.W. analyzed the protein sequences; and G.F. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Genoveffa Franchini, Building 41, Rm D804, Bethesda, MD 20892-5065; e-mail: franchig@mail.nih.gov.
References
Author notes
*R.F. and V.A. contributed equally to this work.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal