Abstract
Multiple myeloma (MM) remains incurable, and new drugs with novel mechanisms of action are still needed. In this report, we have analyzed the action of Zalypsis, an alkaloid analogous to certain natural marine compounds, in MM. Zalypsis turned out to be the most potent antimyeloma agent we have tested so far, with IC50 values from picomolar to low nanomolar ranges. It also showed remarkable ex vivo potency in plasma cells from patients and in MM cells in vivo xenografted in mice. Besides the induction of apoptosis and cell cycle arrest, Zalypsis provoked DNA double-strand breaks (DSBs), evidenced by an increase in phospho-histone-H2AX and phospho-CHK2, followed by a striking overexpression of p53 in p53 wild-type cell lines. In addition, in those cell lines in which p53 was mutated, Zalypsis also provoked DSBs and induced cell death, although higher concentrations were required. Immunohistochemical studies in tumors also demonstrated histone-H2AX phosphorylation and p53 overexpression. Gene expression profile studies were concordant with these results, revealing an important deregulation of genes involved in DNA damage response. The potent in vitro and in vivo antimyeloma activity of Zalypsis uncovers the high sensitivity of tumor plasma cells to DSBs and strongly supports the use of this compound in MM patients.
Introduction
Multiple myeloma (MM) is the second most frequent hematologic malignancy, with an estimated incidence of 56 new cases per million and year,1 and is the 14th cause of death by cancer when considering all tumors.2
The investigation of novel treatments for this disease and the subsequent clinical approval of some of them with demonstrated antimyeloma activity, such as thalidomide,3,4 bortezomib,5,6 or lenalidomide,7,8 has changed the outcome of MM patients in the last years.9 Nevertheless, most patients relapse and MM is still considered an incurable disease. Therefore, new treatments are still needed to achieve longer-lasting remissions and reach the promising objective of transforming MM into a chronic disease, and eventually to cure it. In this regard, one of the sources of drugs that has gained interest in the last years is the marine environment. The antitumoral activity of many compounds obtained from tunicates and other sea organisms is being tested in preclinical and clinical studies, and some of them, such as Yondelis, have already been approved for the treatment of advanced soft tissue sarcomas.10,11
Zalypsis (PM00104; N-[[(6aS,7R,13S,14S,16R)-5-(acetyloxy)-6,6a,7,13,14,16-hexahydro-8,14-dihydroxy-9-methoxy-4,10,17-trimethyl-7,13-imino-12H-1,3-dioxolo[7,8]isoquino[3,2-b][3]benzazocin-16-yl]methyl]-3-[3-(trifluoromethyl)phenyl]-2E-propenamide) (Figure 1A) is a new synthetic alkaloid related to Jorumycin12 (a compound originally isolated from Jorunna funebris), Renieramycins13,14 (isolated from sponges and tunicates), and Safracins and Saframycins (isolated from bacterial sources and marine sponges). Zalypsis has demonstrated significant in vitro activity against human solid and nonsolid tumor cell lines, as well as significant in vivo activity in several xenografted murine models using human cell types, such as breast, prostate, liver, bladder, and gastric.15-18 Zalypsis is currently under late phase 1 development in solid tumors, with preliminary evidence of antitumoral activity.
Zalypsis inhibits the viability of MM cells while preserving normal hematopoietic progenitor cells. (A) Chemical structure of Zalypsis. (B) Nine MM cell lines were incubated with different concentrations of Zalypsis for 24, 48, and 72 hours, and cell viability was analyzed by MTT uptake. The average proliferation values of control untreated samples were taken as 100%. Data are mean plus or minus SD of quadruplicates of an experiment that was repeated at least twice. (C) Freshly isolated BM cells obtained from 6 MM patients were plated in 6-well plates and treated ex vivo with Zalypsis (1-50 nM) for 18 hours. After the incubation period, cells were stained with the combination of annexin V–FITC and 3 monoclonal antibodies against plasma cell surface antigens (CD38, CD56, and CD45), which allows the analysis of the induction of apoptosis in the myelomatous population. Results are given as the percentage of annexin V–positive cells related to the percentage of viable cells in the untreated sample. (D) Freshly isolated BM cells obtained from an MM patient were treated ex vivo with 10 nM Zalypsis for 18 hours. After the incubation period, cells were stained with the combination of annexin V and 2 monoclonal antibodies, CD38 and CD34, to separately analyze the plasma cell (CD38++, CD34−; in red) and the hematopoietic progenitor cell (CD34+, CD38+d; in blue) compartments. The first graphs allow the identification of both populations with the 2 monoclonal antibodies; and in the second plot, the induction of apoptosis (by annexin V staining) in each compartment is displayed.
Zalypsis inhibits the viability of MM cells while preserving normal hematopoietic progenitor cells. (A) Chemical structure of Zalypsis. (B) Nine MM cell lines were incubated with different concentrations of Zalypsis for 24, 48, and 72 hours, and cell viability was analyzed by MTT uptake. The average proliferation values of control untreated samples were taken as 100%. Data are mean plus or minus SD of quadruplicates of an experiment that was repeated at least twice. (C) Freshly isolated BM cells obtained from 6 MM patients were plated in 6-well plates and treated ex vivo with Zalypsis (1-50 nM) for 18 hours. After the incubation period, cells were stained with the combination of annexin V–FITC and 3 monoclonal antibodies against plasma cell surface antigens (CD38, CD56, and CD45), which allows the analysis of the induction of apoptosis in the myelomatous population. Results are given as the percentage of annexin V–positive cells related to the percentage of viable cells in the untreated sample. (D) Freshly isolated BM cells obtained from an MM patient were treated ex vivo with 10 nM Zalypsis for 18 hours. After the incubation period, cells were stained with the combination of annexin V and 2 monoclonal antibodies, CD38 and CD34, to separately analyze the plasma cell (CD38++, CD34−; in red) and the hematopoietic progenitor cell (CD34+, CD38+d; in blue) compartments. The first graphs allow the identification of both populations with the 2 monoclonal antibodies; and in the second plot, the induction of apoptosis (by annexin V staining) in each compartment is displayed.
In the present paper, we have investigated the action of Zalypsis on MM cells. We show that Zalypsis has a potent antimyeloma activity in vitro, ex vivo, and in vivo, and overcomes drug resistance. It is noteworthy that Zalypsis is the most active antimyeloma agent tested in our laboratory, with IC50 values in the picomolar or low nanomolar range. Moreover, Zalypsis synergized with other agents currently used in the myeloma clinic. Studies on its mechanism of action indicate that Zalypsis provokes double-strand DNA breaks (DSBs) that trigger a DNA damage response (DDR). The potent and widespread antimyeloma action of Zalypsis together with the powerful induction of DSBs and the acceptable toxicity profile in phase 1 strongly support the development of clinical trials to analyze the potential use of this drug in MM patients.
Methods
Reagents and immunochemicals
Cell-culture media, serum, and penicillin-streptomycin were purchased from Invitrogen (Carlsbad, CA). Zalypsis was provided by PharmaMar (Madrid, Spain), bortezomib by Millennium Pharmaceuticals (Cambridge, MA), and lenalidomide by Celgene (Summit, NJ). Other antimyeloma drugs (dexamethasone, melphalan, and doxorubicin) were obtained from Sigma-Aldrich (St Louis, MO). The Annexin V–FITC Kit was from Bender MedSystems (Vienna, Austria). Z-VAD-FMK, Z-IETD-FMK, and Z-LEHD-FMK were from Calbiochem (San Diego, CA). Interleukin-6 (IL-6) and insulin-like growth factor I (IGF-I) were purchased from Strathmann Biotec (Hamburg, Germany). Other generic chemicals were purchased from Sigma-Aldrich, Roche Biochemicals (Mannheim, Germany), or Merck (Darmstadt, Germany).
The origins of the different monoclonal antibodies used in the Western blotting analyses were as follows: the anti-PUMA, anti-NOXA, anti GAPDH, anti-Bax, anti GADD45, and anti–caspase-3 were from Santa Cruz Biotechnology (Santa Cruz, CA); anti–caspase-8, anti–caspase-9, anti-cytochrome C, anti-AIF, anti–Bcl-X, anti-PARP, anti-Bcl-2, antibodies from BD Biosciences (San Jose, CA); anti–caspase-7, anti-Mcl1, anti-pH2AX, anti-pCHK2, and anti-p53 from Cell Signaling Technology (Danvers, MA). The anti-Endo G antibody was from Serotec (Oxford, United Kingdom). The horseradish peroxidase–conjugated secondary antibodies were from GE Healthcare (Little Chalfont, United Kingdom).
Cell-culture, cell-proliferation, cell-cycle, and apoptosis analyses and Western blotting and subcellular fractionation procedures
The source and the culture conditions of the MM cell lines have been previously described.19 MM1S-Luc cells were kindly provided by Dr Constantine Mitsiades (Dana-Farber Cancer Institute, Harvard University, Boston, MA). The detailed methodology to analyze proliferation of MM cells using methyl-thiazol-tetrazolium (MTT), cell cycle profiles, DNA laddering, cytometric evaluation of apoptosis in MM cell lines using annexin V–fluorescein isothiocyanate (FITC), mitochondrial membrane potential (ΔΨm), subcellular fractionation, and Western blotting can be also found in Maiso et al.19
Ex vivo experiments in freshly isolated patient cells
For cytometric analyses of apoptosis in bone marrow (BM) cell subpopulations from patients, samples were lysed with ammonium chloride to remove red blood cells, and white cells were maintained in RPMI 1640 containing antibiotics (penicillin 100 U/mL, streptomycin 100 μg/mL) and 20% fetal bovine serum. Subsequently, BM cells were incubated with different concentrations of Zalypsis in 6-well plates for 18 hours at 37°C. To discriminate between myelomatous plasma cells (PCs) and other BM cells, a multiparametric technique was performed in which cells were incubated for 15 minutes at room temperature in the dark with 5 μL annexin V–FITC (Bender MedSystems) together with a combination of monoclonal antibodies against myeloma-associated antigens (anti-CD56–PE, anti-CD45–APC, and anti-CD38–perCP/Cy5 [BD Biosciences]). A total of 50 000 cells were acquired on a FACSCalibur flow cytometer (BD Biosciences) and analyzed with the “Paint-a-Gate” program. Apoptosis was analyzed based on the annexin V positivity in the different populations: tumor PCs as well as in normal residual lymphocytes and granulomonocytes. The percentage of annexin V–positive cells after treatment with Zalypsis was calculated over the annexin V–negative cells (viable cells) in the control samples (without treatment).
The toxicity on CD34+ hematopoietic progenitor cells was analyzed with the same protocol described but including the following monoclonal antibodies: annexin V–FITC, anti-CD64–PE, anti-CD34–APC, anti-CD19–PerCP, anti-CD45–AmCyan, and anti-CD38–Alexa Fluor 700 (BD Biosciences). A total of 50 000 cells of the global population were acquired in a FACSCanto II Flow Cytometer (BD Biosciences); and in a second step, an acquisition gate was performed in the CD34+ population and in the plasma cell compartment in the MM.
Effect of IL-6, IGF-I, and BMSC on Zalypsis-induced growth inhibition
MM1S cells were incubated for 48 hours with Zalypsis, in the presence or absence of IL-6 or IGF-I. Proliferation of MM cells was then assessed by bromodeoxyuridine (BrdU) uptake.20 To evaluate the effect of Zalypsis on MM cells adherent to bone marrow stromal cells (BMSCs), the latter were plated in 96-well culture dishes (8000/well) and allowed to reach confluence during 48 hours. Then, medium was removed and 20 000 luciferase-expressing myeloma cells (MM1S-Luc) in RPMI 1640 containing 10% fetal bovine serum were plated on top of the BMSCs and treated for 48 hours with different concentrations of Zalypsis. After the incubation period, luciferin substrate (Caliper Life Sciences, Hopkinton, MA) at a final concentration of 150 μg/mL was added for 10 minutes and bioluminescence (photons/sec) was analyzed in a Xenogen IVIS Imaging System 50 Series (Caliper Life Sciences).
Evaluation of the potential synergism of Zalypsis with other antimyeloma agents
MM1S cells were treated for 72 hours with combinations of suboptimal doses of Zalypsis and other antimyeloma agents, such as dexamethasone, melphalan, doxorubicin, bortezomib, and lenalidomide, in double and triple combinations. Cell viability was analyzed by MTT assays. The potency of the combination was quantitated with the Calcusyn Software (Biosoft, Ferguson, MO), which is based in the Chou Talalay method and calculates a combination index (CI) with the following interpretation: CI > 1: antagonistic effect, CI = 1: additive effect and CI < 1 synergistic effect.
Microarray RNA analyses
MM cells treated in vitro with Zalypsis (5 nM) were harvested at the beginning of induction of cell death (15%–20% cell death as assessed by annexin V–FITC staining). Total RNA was extracted using TRIzol reagent (Invitrogen) and purified with the RNAeasy Mini Kit (QIAGEN, Valencia, CA). RNA integrity was verified with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Double-stranded cDNA and biotinylated cRNA were synthesized with T7-polyT primer and the BioArray RNA labeling kit (Enzo Diagnostics, New York, NY), respectively. The labeled RNA was then fragmented and hybridized to HG-U133 Plus 2.0 oligonucleotide arrays (Affymetrix, Santa Clara, CA), which were scanned in a Gene Array Scanner and analyzed using the DNA-Chip Analyzer software (DChip). Changes in gene expression greater than 2-fold were considered significant. All microarray data have been deposited with Gene Expression Omnibus under accession number GSE13662.
MM xenografts and immunohistochemistry
CB17-SCID mice (The Jackson Laboratory, Bar Harbor, ME) were subcutaneously inoculated into the right flank with 3 × 106 MM1S or OPM1 cells in 100 μL RPMI 1640 medium and 100 μL of Matrigel (BD Biosciences). When tumors became palpable, mice received Zalypsis or vehicle alone. Treatment with Zalypsis was given intravenously at doses of 0.8 mg/kg and 1 mg/kg once weekly for 3 doses. The control group received the vehicle alone (sterile water for injection plus saline). Caliper measurements of the tumor diameters were performed every day, and the tumor volume was estimated as the volume of an ellipse using the following formula: V = 4/3 π × (a/2) × (b/2)2, where “a” and “b” correspond to the longest and shortest diameter, respectively. Animals were killed when their tumors reached 2 cm. Differences in tumor volumes between control and treated groups were evaluated using 1-way analysis of variance and Bonferroni post-hoc tests. Time to endpoint was defined as the time from the day of initiation of treatment to death as a result of toxicity, tumor growth, or any other cause. Statistical differences were assessed by Kaplan-Meier curves with the log rank test. Statistical analyses were performed with the SPSS-15.0 software (SPSS, Chicago, IL), and statistical significance was defined as P < .05. All animal experiments were performed according to the protocol previously approved by the ethical committee of the University of Salamanca.
Immunohistochemical studies were performed on selected tumors excised from treated and control mice. After fixation for 24 hours in paraformaldehyde 10%, a tissue microarray was performed with a manual tissue arrayer (Beecher Instruments, Sun Prairie, WI), including 2 representative cylinders of each sample of 1 mm of diameter. Tissue microarray was paraffin embedded, and sections of 3-μM thickness were obtained. After deparaffinization in xylene and rehydration in increasing concentrations of ethanol, antigen retrieval and antibody incubation were performed with a semi-automatic Dako Autostainer (Dako North America, Carpinteria, CA) system. Primary antibodies included anti-p53 (DakoCytomation, Cambridgeshire, United Kingdom), and anti-cleaved PARP, anti-pH2AX, and anti–cleaved caspase-3 (Cell Signaling Technology). Secondary antibody was horseradish peroxidase Envision system (Dako North America, reference K5007). Staining was performed with the IHC DAB MAP system (Ventana Medical Systems, Tucson, AZ). Sections were then counterstained with hematoxylin and analyzed by standard light microscopy.
Results
Potent antimyeloma action of Zalypsis against cell lines and freshly isolated plasma cells from patients
To test the antiproliferative/cytotoxic effect of Zalypsis against MM cells, 9 MM cell lines were treated with increasing concentrations of the compound (0.1-50 nM) for 24, 48, and 72 hours, and viability was analyzed by MTT assays. As shown in Figure 1B, all cell lines were very sensitive to the drug, with IC50 values at 48 hours ranging from picomolar concentrations in some cell lines (MM1S, MM1R, and MM144) to low nanomolar (1-5 nM) in the less sensitive cell lines (RPMI8226, RPMI-LR5, U266, U266-LR7, OPM-1, and OPM-2). The sensitivity to Zalypsis was independent of the pattern of resistance of the cell lines to conventional antimyeloma agents, such as dexamethasone (all cell lines are resistant except for MM1S and MM144) or melphalan (RPMI-LR5 and U266-LR7 are resistant). Moreover, Zalypsis was demonstrated to be at least 10 times more potent than any of the antimyeloma agents we have tested so far (data not shown).
The effect of Zalypsis was further investigated ex vivo in cells isolated from BM samples obtained from 6 patients with MM. BM aspirates containing tumor plasma cells were incubated with different concentrations of Zalypsis (1-50 nM) for 18 hours, and the induction of apoptosis was analyzed by flow cytometry. Zalypsis induced cell death in all cases, including 2 plasma cell leukemias (cases 4 and 6; Figure 1C).
In addition, the toxicity of Zalypsis on nontumoral CD34+ BM progenitor cells was analyzed in 3 samples (1 obtained from a MM patient, and 2 from donors). In all cases, this cytotoxicity was clearly lower than that observed in tumor plasma cells. Figure 1D represents an example of the sample obtained from a MM patient, in which treatment with Zalypsis induced apoptosis in the plasma cell population (CD38+) while partially preserving the CD34+ cells. To further analyze the potential toxicity on the CD34+ compartment, we studied the effect of Zalypsis on the different stages of maturation of the hematopoietic progenitor cell compartment. Interestingly, we observed that, whereas the viability of the CD34+ cells committed to myeloid (CD34+, CD64+) or B-lymphoid (CD34+, CD19+) lineages was slightly affected by Zalypsis, the uncommitted progenitor cells (CD34+, CD64−, CD19−) were well preserved (data not shown). These findings indicate that the potential cytopenias that could be induced by Zalypsis would be reversible because the most immature hematopoietic precursor cells are preserved.
Zalypsis abrogates the survival advantage and drug resistance induced by the BM microenvironment
The presence of the BM microenvironment confers protection to myeloma cells through their adhesion or through the production of several cytokines, such as IL-6 or IGF-I.21 To test whether Zalypsis was able to inhibit this protective effect of the BM microenvironment, MM1S cells were incubated with IL-6 (1 nM) or IGF-I (10 nM), or cocultured with BMSCs and treated with increasing concentrations of Zalypsis for 48 hours. Proliferation was analyzed by BrdU uptake (for IL-6 and IGF-I) or by bioluminescence (for coculture with BMSCs). Despite the proliferative advantage to MM cells conferred by all these models, Zalypsis completely abrogated the effect of the soluble cytokines IL-6 and IGF-I (Figure 2A,B) and largely inhibited the protective effect resulting from adhesion of plasma cells to BMSCs (Figure 2C). In contrast, BMSCs were very resistant to the cytotoxic effect of Zalypsis (Figure 2D).
Zalypsis overcomes the protective effects of IL-6, IGF-I, and adherence to patient BMSCs. (A,B) MM1S cells were treated for 48 hours with the indicated concentrations of Zalypsis in the presence or absence of IL-6 (A) or IGF-I (B), and DNA synthesis was determined by measuring BrdU incorporation during the last 8 hours of 48-hour cultures. (C) MM1S-luc cells were treated for 48 hours with the indicated concentrations of Zalypsis in the presence or absence of BMSCs derived from a MM patient, and proliferation was analyzed by bioluminescence (photons/sec). (D) BMSCs were cultured with different doses of Zalypsis for 48 hours, and the cytotoxicity was analyzed by MTT assay. Data are mean plus or minus SD of quadruplicates.
Zalypsis overcomes the protective effects of IL-6, IGF-I, and adherence to patient BMSCs. (A,B) MM1S cells were treated for 48 hours with the indicated concentrations of Zalypsis in the presence or absence of IL-6 (A) or IGF-I (B), and DNA synthesis was determined by measuring BrdU incorporation during the last 8 hours of 48-hour cultures. (C) MM1S-luc cells were treated for 48 hours with the indicated concentrations of Zalypsis in the presence or absence of BMSCs derived from a MM patient, and proliferation was analyzed by bioluminescence (photons/sec). (D) BMSCs were cultured with different doses of Zalypsis for 48 hours, and the cytotoxicity was analyzed by MTT assay. Data are mean plus or minus SD of quadruplicates.
Zalypsis potentiates the efficacy of antimyeloma agents
As treatment of most cancers, including myeloma, is based on combinations of drugs with different mechanisms of action, we studied the effect of Zalypsis in double combinations with drugs normally used in the myeloma clinic, such as dexamethasone, melphalan, doxorubicin, bortezomib, and lenalidomide in MM1S cells (Figure 3A). Analyses of these data using the Chou and Talalay method22 indicated that Zalypsis was synergistic with dexamethasone (CI = 0.78), doxorubicin (CI = 0.64), and particularly melphalan (CI = 0.48), and lenalidomide (CI = 0.55). The combination indexes for bortezomib were in the additive range.
Zalypsis potentiates the antimyeloma action of conventional and novel antimyeloma agents. MM1S cells were treated for 72 hours with suboptimal concentrations of Zalypsis and other antimyeloma agents, such as dexamethasone, melphalan, doxorubicin, bortezomib, and lenalidomide in double (A) and triple combinations (B). Cell viability was analyzed by MTT assay.
Zalypsis potentiates the antimyeloma action of conventional and novel antimyeloma agents. MM1S cells were treated for 72 hours with suboptimal concentrations of Zalypsis and other antimyeloma agents, such as dexamethasone, melphalan, doxorubicin, bortezomib, and lenalidomide in double (A) and triple combinations (B). Cell viability was analyzed by MTT assay.
These promising results with double combinations prompted the investigation of triple combinations of the most synergistic compounds in the MM1S cell line. As shown in Figure 3B, triple combinations of Zalypsis plus dexamethasone plus any of the following: melphalan, doxorubicin, or lenalidomide, notably improved the efficacy of the respective double combinations, being the combination of Zalypsis plus lenalidomide plus dexamethasone especially attractive.
Zalypsis provokes changes in the cell cycle and induces apoptosis
To assess whether the decrease of the MTT uptake induced by Zalypsis was the result of cell cycle blockade, an increase in cytotoxicity, or both, MM1S cells were treated with Zalypsis (5 nM) for different times and cell cycle profiles and apoptotic induction were analyzed. Zalypsis provoked an increase in G0/G1 and a decrease of the G2/M phases of the cell cycle (Figure 4A). The effect of Zalypsis on apoptosis was analyzed by flow cytometry, which demonstrated a time-dependent increase of annexin V-positive cells already detectable after 3 hours of treatment (Figure 4B). In addition, Zalypsis provoked internucleosomal DNA fragmentation (Figure 4C).
Zalypsis induces changes in the cell cycle profile and apoptosis in MM1S cells. (A) MM1S cells were incubated with Zalypsis (5 nM) for 6, 12, and 24 hours, and the cell cycle profile was examined by flow cytometry after propidium iodide staining. Bars represent the percentage of cells in each phase of the cell cycle from a representative example. (B) MM1S cells were treated with Zalypsis (5 nM) for different time points, and the induction of apoptosis was analyzed by flow cytometry after staining with annexin V. (C) MM1S cells were treated with Zalypsis (5 nM) for the indicated times. DNA of the treated cells was isolated, and its fragmentation was analyzed by agarose gel electrophoresis. The position of the molecular weight markers is shown at the right. (D) MM1S cells were treated with Zalypsis (5 nM) for 12 and 24 hours, and the disruption of the mitochondrial membrane potential (ΔΨm) was analyzed by flow cytometry after staining with [DiOC6(3)low]. (E) MM1S cells were treated for the indicated times with Zalypsis, and cytochrome C, AIF, and endonuclease G in the mitochondrial fraction were analyzed by Western blotting. (F) The status of some Bcl-2 family members was analyzed by Western blotting after treatment of MM1S cells with 5 nM Zalypsis for the indicated time points in the presence or absence of the caspase inhibitor Z-VAD-FMK (50 μM for 60 minutes). (G) MM1S cells were treated with Zalypsis (5 nM) for the indicated times, and expression of PARP, caspase-3, caspase-7, caspase-8, and caspase-9 proteins was analyzed by Western blotting. (H) MM1S cells were plated and pretreated with the different caspase inhibitors (50 μM) for 60 minutes. Zalypsis (5 nM) was added to the corresponding samples, and the experiment was continued for 12 and 24 hours. Induction of apoptosis was analyzed by means of annexin V–FITC staining.
Zalypsis induces changes in the cell cycle profile and apoptosis in MM1S cells. (A) MM1S cells were incubated with Zalypsis (5 nM) for 6, 12, and 24 hours, and the cell cycle profile was examined by flow cytometry after propidium iodide staining. Bars represent the percentage of cells in each phase of the cell cycle from a representative example. (B) MM1S cells were treated with Zalypsis (5 nM) for different time points, and the induction of apoptosis was analyzed by flow cytometry after staining with annexin V. (C) MM1S cells were treated with Zalypsis (5 nM) for the indicated times. DNA of the treated cells was isolated, and its fragmentation was analyzed by agarose gel electrophoresis. The position of the molecular weight markers is shown at the right. (D) MM1S cells were treated with Zalypsis (5 nM) for 12 and 24 hours, and the disruption of the mitochondrial membrane potential (ΔΨm) was analyzed by flow cytometry after staining with [DiOC6(3)low]. (E) MM1S cells were treated for the indicated times with Zalypsis, and cytochrome C, AIF, and endonuclease G in the mitochondrial fraction were analyzed by Western blotting. (F) The status of some Bcl-2 family members was analyzed by Western blotting after treatment of MM1S cells with 5 nM Zalypsis for the indicated time points in the presence or absence of the caspase inhibitor Z-VAD-FMK (50 μM for 60 minutes). (G) MM1S cells were treated with Zalypsis (5 nM) for the indicated times, and expression of PARP, caspase-3, caspase-7, caspase-8, and caspase-9 proteins was analyzed by Western blotting. (H) MM1S cells were plated and pretreated with the different caspase inhibitors (50 μM) for 60 minutes. Zalypsis (5 nM) was added to the corresponding samples, and the experiment was continued for 12 and 24 hours. Induction of apoptosis was analyzed by means of annexin V–FITC staining.
Zalypsis deregulates mitochondrial permeability
Because mitochondria have an important role in the regulation of apoptosis,23 we explored the effect of Zalypsis on these organelles by measuring their membrane potential (ΔΨm). Zalypsis caused a decrease in ΔΨm in MM1S cells (Figure 4D). Loss of ΔΨm reflects increased mitochondrial outer membrane permeability (MOMP), which is accompanied by leakage of proapoptotic proteins that reside at the intermembranous mitochondrial space.24 Indeed, subcellular fractionation indicated that Zalypsis provoked a time-dependent loss of cytochrome C, apoptosis-inducing factor (AIF), and endonuclease G from mitochondria (Figure 4E). As Bcl-2 family members regulate the MOMP, the influence of treatment with Zalypsis on the expression of some of the most important Bcl-2 family members was also studied. As shown in Figure 4F, Zalypsis provoked a caspase-dependent down-regulation of Bcl-X and a decrease of Mcl-1, which was, at least partially, independent of the action of caspases. By contrast, no substantial effect was observed in Bcl-2 levels.
Zalypsis induces apoptosis through caspase-dependent and -independent mechanisms
The release of apoptotic mediators from mitochondria may induce procaspase cleavage and the ensuing activation of the intrinsic caspase-mediated apoptotic pathway, or activate caspase-independent routes.24,25 To investigate whether caspases were activated by Zalypsis, MM1S cells were treated with the drug for different times, and PARP (a caspase substrate) and caspase-3, -7, -8, and -9 were analyzed by Western blot. Treatment with Zalypsis induced the cleavage of initiator caspases, such as caspase-8, -7, and -9, after 6 hours and also provoked the cleavage of the effector caspase-3. PARP processing was detected after 12 hours of treatment (Figure 4G). The role of caspases in Zalypsis-induced cell death was analyzed by studying the influence of the pharmacologic inhibition of the activity of caspase-8 (with Z-IETD-FMK), caspase-9 (with Z-LEHD-FMK), and all caspases (with the pan-caspase inhibitor Z-VAD-FMK). MM1S cells preincubated with each of these caspase inhibitors for one hour were then treated with Zalypsis for 48 additional hours. The individual blockade of caspase-8 or caspase-9 only slightly inhibited the Zalypsis-induced apoptosis; whereas when all caspases were inhibited with Z-VAD-FMK, the apoptotic effect of Zalypsis was partially abrogated (Figure 4H).
Zalypsis triggers a DNA damage response
To further investigate the mechanism of action of Zalypsis, we analyzed the changes induced by the compound in the gene expression profiles of 2 MM cell lines. A total of 1458 genes were significantly deregulated in MM1S cells after treatment with Zalypsis, whereas it induced deregulation in 5278 genes in OPM-1 cells. Among them, 913 genes were commonly deregulated in both cell lines. Most of these genes were down-regulated after treatment with the drug, being the 3 most significantly deregulated functional categories, cell cycle, apoptosis, and DNA damage repair (Table 1; Tables S1,S2, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Genes involved in the “DNA damage” functional category, deregulated by Zalypsis in MM1S and OPM-1 cells
| Probe set . | Gene symbol . | MM1S FC . | OPM1 FC . | Description . |
|---|---|---|---|---|
| 212986_s_at | TLK2 | −2.65 | Tousled-like kinase 2 | |
| 203725_at | GADD45A | 2.58 | Growth arrest and DNA-damage-inducible, alpha | |
| 222879_s_at | POLH | 3.34 | Polymerase (DNA directed), eta | |
| 203409_at | DDB2 | 3.81 | Damage-specific DNA binding protein 2, 48kDa | |
| 236535_at | SMC6 | −16.06 | SMC6 structural maintenance of chromosomes 6-like 1 (yeast) | |
| 209849_s_at | RAD51C | −5.26 | RAD51 homolog C (S. cerevisiae) | |
| 209902_at | ATR | −4.74 | Ataxia telangiectasia and Rad3 related | |
| 228535_at | RAD1 | −4.69 | RAD1 homolog (S. pombe) | |
| 204146_at | RAD51AP1 | −4.59 | RAD51 associated protein 1 | |
| 210416_s_at | CHEK2 | −3.99 | CHK2 checkpoint homolog (S. pombe) | |
| 201589_at | SMC1A | −3.41 | SMC1 structural maintenance of chromosomes 1-like 1 (yeast) | |
| 201223_s_at | RAD23B | −2.95 | RAD23 homolog B (S. cerevisiae) | |
| 218527_at | APTX | −2.9 | Aprataxin | |
| 214426_x_at | CHAF1A | −2.65 | Chromatin assembly factor 1, subunit A (p150) | |
| 205024_s_at | RAD51 | −2.61 | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | |
| 219510_at | POLQ | −2.59 | Polymerase (DNA directed), theta | |
| 223261_at | POLK | −2.48 | Polymerase (DNA directed) kappa | |
| 201746_at | TP53 | 2.42 | Tumor protein p53 (Li-Fraumeni syndrome) | |
| 210886_x_at | TP53AP1 | 3.54 | TP53-activated protein 1 | |
| 205072_s_at | XRCC4 | −9.32 | −19.94 | X-ray repair complementing defective repair in cells 4 |
| 208393_s_at | RAD50 | −3.93 | −7.95 | RAD50 homolog (S cerevisiae) |
| 208643_s_at | XRCC5 | −2.5 | −14.01 | X-ray repair complementing defective repair in cells 5 |
| 235609_at | BRIP1 | −2.42 | −7.32 | BRCA1-interacting protein C-terminal helicase 1 |
| 239635_at | TMEM137 | 2.52 | 3.81 | RNA-binding motif protein 14 |
| 205875_s_at | TREX1 | 2.72 | 4.21 | Three prime repair exonuclease 1 |
| 238736_at | REV3L | 4.18 | 3.5 | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) |
| Probe set . | Gene symbol . | MM1S FC . | OPM1 FC . | Description . |
|---|---|---|---|---|
| 212986_s_at | TLK2 | −2.65 | Tousled-like kinase 2 | |
| 203725_at | GADD45A | 2.58 | Growth arrest and DNA-damage-inducible, alpha | |
| 222879_s_at | POLH | 3.34 | Polymerase (DNA directed), eta | |
| 203409_at | DDB2 | 3.81 | Damage-specific DNA binding protein 2, 48kDa | |
| 236535_at | SMC6 | −16.06 | SMC6 structural maintenance of chromosomes 6-like 1 (yeast) | |
| 209849_s_at | RAD51C | −5.26 | RAD51 homolog C (S. cerevisiae) | |
| 209902_at | ATR | −4.74 | Ataxia telangiectasia and Rad3 related | |
| 228535_at | RAD1 | −4.69 | RAD1 homolog (S. pombe) | |
| 204146_at | RAD51AP1 | −4.59 | RAD51 associated protein 1 | |
| 210416_s_at | CHEK2 | −3.99 | CHK2 checkpoint homolog (S. pombe) | |
| 201589_at | SMC1A | −3.41 | SMC1 structural maintenance of chromosomes 1-like 1 (yeast) | |
| 201223_s_at | RAD23B | −2.95 | RAD23 homolog B (S. cerevisiae) | |
| 218527_at | APTX | −2.9 | Aprataxin | |
| 214426_x_at | CHAF1A | −2.65 | Chromatin assembly factor 1, subunit A (p150) | |
| 205024_s_at | RAD51 | −2.61 | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | |
| 219510_at | POLQ | −2.59 | Polymerase (DNA directed), theta | |
| 223261_at | POLK | −2.48 | Polymerase (DNA directed) kappa | |
| 201746_at | TP53 | 2.42 | Tumor protein p53 (Li-Fraumeni syndrome) | |
| 210886_x_at | TP53AP1 | 3.54 | TP53-activated protein 1 | |
| 205072_s_at | XRCC4 | −9.32 | −19.94 | X-ray repair complementing defective repair in cells 4 |
| 208393_s_at | RAD50 | −3.93 | −7.95 | RAD50 homolog (S cerevisiae) |
| 208643_s_at | XRCC5 | −2.5 | −14.01 | X-ray repair complementing defective repair in cells 5 |
| 235609_at | BRIP1 | −2.42 | −7.32 | BRCA1-interacting protein C-terminal helicase 1 |
| 239635_at | TMEM137 | 2.52 | 3.81 | RNA-binding motif protein 14 |
| 205875_s_at | TREX1 | 2.72 | 4.21 | Three prime repair exonuclease 1 |
| 238736_at | REV3L | 4.18 | 3.5 | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) |
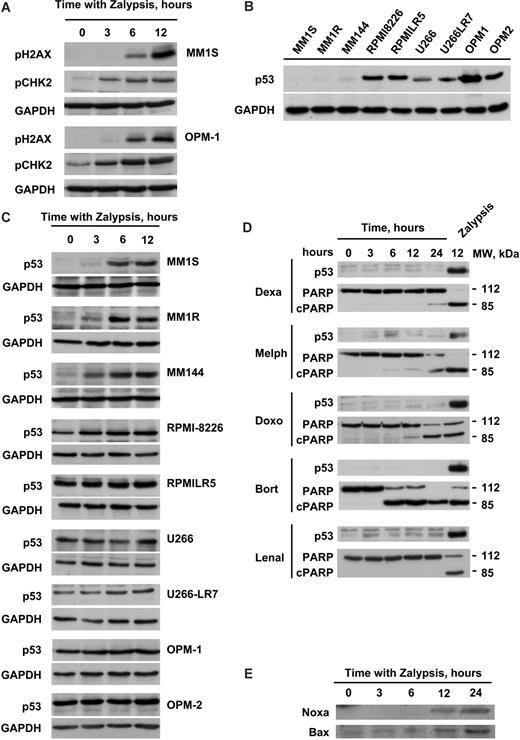
Although the deregulation of cell cycle and apoptotic mRNAs was not surprising, as anticipated by the biochemical and cell biologic studies, the important number of DDR genes deregulated by Zalypsis suggested that a DDR was indeed activated by this drug. In this sense, several genes implicated in the ATM repair pathway, such as TLK2, ATR, CHEK2, RAD5, and BRIP1, were all down-regulated after the treatment; in addition, there was also a deregulation of other mRNAs related to DNA repair, such as RAD23B, XPC, XRCC1, XRCC5, and GADD45A (Table 1). To further explore this mechanism, we initially evaluated the possibility that Zalypsis could provoke DNA DSBs, especially because all these ATM-related genes appeared affected by Zalypsis in the microarray gene studies. Generation of DSBs is recognized by the MRN protein complex that acts as a DSB sensor and recruits the ATM kinase to sites of broken DNA. Under these circumstances, ATM is activated and phosphorylates substrates, such as histone H2AX and CHK2.26 Treatment of MM1S and OPM-1 cells with Zalypsis induced histone H2AX and CHK2 phosphorylation (Figure 5A), suggestive of induction of DSBs by Zalypsis.
Zalypsis stimulates a p53-dependent DNA damage response through the induction of DSBs. (A) MM1S and OPM1 cells were treated with 5 nM Zalypsis for the indicated time points, cell protein extracts were obtained of each condition, and the phosphorylation of H2AX and CHK2 was analyzed by Western blotting. Equal loading was confirmed with an anti-GAPDH antibody. (B) Western blot showing the basal protein levels of p53 protein in the 9 MM cell lines tested. The 3 first cell lines are very sensitive to Zalypsis, whereas the 6 last ones are more resistant. Equal loading was confirmed with an anti-GAPDH antibody. (C) All the 9 MM cell lines were treated with 5 nM Zalypsis for 0, 3, 6, and 12 hours, and the induction of p53 protein levels was analyzed by Western blotting. Equal loading was confirmed with an anti-GAPDH antibody. (D) MM1S cells were treated with different antimyeloma agents at different doses (dexamethasone 1 μM, melphalan 10 μM, doxorubicin 100 nM, bortezomib 10 nM, and lenalidomide 10 μM) for 0, 3, 6, 12, and 24 hours, and changes in p53 protein levels compared with treatment with 5 nM Zalypsis for 12 hours were analyzed by Western blotting. (E) Western blot showing changes in the protein levels of some p53 targets (Bax and Noxa) after treatment of MM1S cells with 5 nM Zalypsis for different time points.
Zalypsis stimulates a p53-dependent DNA damage response through the induction of DSBs. (A) MM1S and OPM1 cells were treated with 5 nM Zalypsis for the indicated time points, cell protein extracts were obtained of each condition, and the phosphorylation of H2AX and CHK2 was analyzed by Western blotting. Equal loading was confirmed with an anti-GAPDH antibody. (B) Western blot showing the basal protein levels of p53 protein in the 9 MM cell lines tested. The 3 first cell lines are very sensitive to Zalypsis, whereas the 6 last ones are more resistant. Equal loading was confirmed with an anti-GAPDH antibody. (C) All the 9 MM cell lines were treated with 5 nM Zalypsis for 0, 3, 6, and 12 hours, and the induction of p53 protein levels was analyzed by Western blotting. Equal loading was confirmed with an anti-GAPDH antibody. (D) MM1S cells were treated with different antimyeloma agents at different doses (dexamethasone 1 μM, melphalan 10 μM, doxorubicin 100 nM, bortezomib 10 nM, and lenalidomide 10 μM) for 0, 3, 6, 12, and 24 hours, and changes in p53 protein levels compared with treatment with 5 nM Zalypsis for 12 hours were analyzed by Western blotting. (E) Western blot showing changes in the protein levels of some p53 targets (Bax and Noxa) after treatment of MM1S cells with 5 nM Zalypsis for different time points.
One of the main pathways activated by DSB-DDR is the p53 route. Western blotting analyses of p53 evidenced that sensitivity of the different MM cell lines to Zalypsis correlated with their basal expression of p53 (Figure 5B). Moreover, in those cell lines with low basal levels of p53 (MM1S, MM1R, and MM144), Zalypsis increased the levels of this protein (Figure 5C). By contrast, those cell lines with high basal levels of p53 (U266, RPMI-8226, OPM-1, OPM-2, U266-LR7, RPMI-LR5) displayed a more limited increase, if any, in the expression of the protein (Figure 5C). Of note, the increase in p53 induced by Zalypsis was higher than that provoked by other antimyeloma agents, including doxorubicin, another DNA-damaging agent currently used in MM treatment (Figure 5D). The mechanism of increase in p53 in MM1S levels is probably because of stabilization of the protein, as no evident changes in the mRNA levels were observed both in the microarray data analyses and by quantitative PCR studies (data not shown). In MM1S cells, p53 up-regulation was accompanied by an increase in the protein levels of target genes, such as Noxa or Bax (Figure 5E), whereas no significant changes were observed in the levels of other p53 controlled genes, such as Puma or GADD45α (data not shown).
In vivo antimyeloma efficacy of Zalypsis
The in vivo efficacy of Zalypsis was studied in a model of human plasmacytoma xenografted in CB17-SCID mice. These experiments were performed using the very sensitive cell line MM1S and the less sensitive one OPM-1. Two cohorts of 30 mice were subcutaneously injected with each cell line and mice were randomized into 3 groups receiving vehicle alone, Zalypsis 0.8 mg/kg, or Zalypsis 1 mg/kg intravenously once weekly for 3 doses. Both doses of Zalypsis decreased the growth of the plasmacytomas with statistically significant differences. Figure 6A shows the evolution of the MM1S plasmacytomas, after 15 days of treatment with Zalypsis: tumor volume was 1207 plus or minus 645, 420 plus or minus 242, and 176 plus or minus 78 mm3 (mean ± SD) for the cohorts receiving vehicle, 0.8 mg/kg, and 1 mg/kg, respectively (P < .001 for the global comparison and for each of the treated vs control comparisons). In the less sensitive and more rapidly growing OPM-1 cell line, results were similar with tumor volumes after 14 days of treatment of 4091 plus or minus 903, 1879 plus or minus 731, and 1042 plus or minus 596 for the same groups of mice (P < .001 for all comparisons; Figure 6B). This delay in tumor growth correlated with an increase in time to endpoint of treated mice compared with the controls (Figure 6C,D). In this sense, median plus or minus SE survivals in the MM1S-injected groups were 29 plus or minus 3.9, 52 plus or minus 4.8, and 59 plus or minus 5.5 days for vehicle control, Zalypsis 0.8 mg/kg, and Zalypsis 1 mg/kg, respectively (Figure 6C), whereas the times to endpoint for the same groups in the OPM-1 plasmacytomas were: 14 plus or minus 0, 21 plus or minus 0.8, and 23 plus or minus 1.1 (Figure 6D), with statistically significant differences for comparison between the vehicle control and both doses of Zalypsis in each xenograft model (P < .001).
In vivo antimyeloma effect of Zalypsis. (A) CB17-SCID mice subcutaneously inoculated with 3 × 106 MM1S or OPM1 cells in the right flank were assigned to the different treatment groups receiving Zalypsis at a dose of 0.8 mg/kg and 1 mg/kg intravenously once weekly for 3 doses (↑ indicates days of treatment) or to the control group receiving the vehicle alone. Tumor diameters were measured every other day, and tumor volume was estimated as the volume of an ellipse. (A,B) The evolution of the volume of MM1S and OPM-1 plasmacytomas for the indicated days. Statistical differences in each time point were analyzed for each dose compared with the vehicle control group with one-way analysis of variance and Bonferroni post hoc tests, and statistical significance was defined as P < .05. *First day in which differences were statistically significant for each dose. Bars represent SE. (C,D) For survival evaluation, mice were killed when their tumor diameters reached 2 cm or when they became moribund. Time to endpoint was defined as the time from the day of initiation of treatment to death as a result of toxicity, tumor growth, or any other cause. Statistical differences were analyzed in a Kaplan-Meier curve. *Significance was compared with the vehicle control group, defined as P < .05 in the log rank test. (E) Immunohistochemical analyses were performed with different apoptosis-related markers in selected control and treated tumors after the death of mice bearing MM1S and OPM-1 plasmacytomas. Panels (original magnification ×100) show that Zalypsis treatment induced PARP and caspase-3 expression (first 2 rows), an increase of H2AX phosphorylation (third row), and nuclear accumulation of p53 in tumors derived from both cell lines (fourth row), although basal p53 expression was higher in OPM-1 tumors.
In vivo antimyeloma effect of Zalypsis. (A) CB17-SCID mice subcutaneously inoculated with 3 × 106 MM1S or OPM1 cells in the right flank were assigned to the different treatment groups receiving Zalypsis at a dose of 0.8 mg/kg and 1 mg/kg intravenously once weekly for 3 doses (↑ indicates days of treatment) or to the control group receiving the vehicle alone. Tumor diameters were measured every other day, and tumor volume was estimated as the volume of an ellipse. (A,B) The evolution of the volume of MM1S and OPM-1 plasmacytomas for the indicated days. Statistical differences in each time point were analyzed for each dose compared with the vehicle control group with one-way analysis of variance and Bonferroni post hoc tests, and statistical significance was defined as P < .05. *First day in which differences were statistically significant for each dose. Bars represent SE. (C,D) For survival evaluation, mice were killed when their tumor diameters reached 2 cm or when they became moribund. Time to endpoint was defined as the time from the day of initiation of treatment to death as a result of toxicity, tumor growth, or any other cause. Statistical differences were analyzed in a Kaplan-Meier curve. *Significance was compared with the vehicle control group, defined as P < .05 in the log rank test. (E) Immunohistochemical analyses were performed with different apoptosis-related markers in selected control and treated tumors after the death of mice bearing MM1S and OPM-1 plasmacytomas. Panels (original magnification ×100) show that Zalypsis treatment induced PARP and caspase-3 expression (first 2 rows), an increase of H2AX phosphorylation (third row), and nuclear accumulation of p53 in tumors derived from both cell lines (fourth row), although basal p53 expression was higher in OPM-1 tumors.
Interestingly, no significant systemic toxicity was associated with Zalypsis treatment, and only a slight weight loss (∼ 10% of body weight compared with the controls) was observed with the highest of the doses. Nevertheless, an important local toxicity was caused at the site of injection of Zalypsis with ulcerations and necrosis of the tail in some of the mice.
All animal experiments were performed according to the protocol previously approved by the ethical committee of the University of Salamanca (Salamanca, Spain).
Immunohistochemical evidence of the DNA damage response in tumors excised from treated mice
To confirm the results obtained in the in vitro setting, tumors from selected mice bearing MM1S and OPM-1 xenografts were collected after death to perform some immunohistochemical studies. Zalypsis-induced PARP and caspase-3 cleavage were observed in the resected tumors from xenografts of both cell lines (Figure 6E). H2AX also showed an increase in phosphorylation in MM1S and OPM-1 xenografts, suggesting the induction of DSBs in the tumors on treatment with Zalypsis (Figure 6E). Concordant with the in vitro results in MM cell lines, p53 displayed a high basal expression in OPM-1 tumors compared with MM1S derived ones, and treatment with Zalypsis induced an increase of p53 in the nuclei of MM1S and OPM-1 cells (Figure 6E).
Discussion
In this work, we evaluated the action of Zalypsis in MM. This compound, which is undergoing phase 1 clinical studies, presented in our hands the strongest antimyeloma activity, both in vitro and in vivo, we have seen so far. The IC50 values were in the picomolar to low nanomolar range. Interestingly, Zalypsis was also equally efficient on cell lines, such as MM1R, RPMI-LR5, selected for their resistance to conventional antimyeloma treatments, indicating that this drug could be used to overcome drug resistance, a common situation found in treated MM patients. Furthermore, Zalypsis synergized with several antimyeloma treatments, supporting the possibility of using this drug in combination with other well-established antimyeloma agents, the combination of Zalypsis + lenalidomide + dexamethasone being particularly attractive. In addition to the in vitro results, in vivo animal studies confirmed the antimyeloma activity of Zalypsis. The drug appeared to be well tolerated and profoundly affected the growth of xenografted plasmacytomas of MM1S and OPM-1 in mice.
On analyzing the efficacy of a new agent, a mandatory experiment is to investigate its toxicity in normal cells. These experiments are usually conducted on normal peripheral blood samples. We have developed an ex vivo technique that allows to simultaneously analyze the cell death induced by a particular agent both on the malignant cells and the residual BM cells. Now, for the first time using multiparametric flow cytometry, we have analyzed the action of Zalypsis on the cell subset that mainly corresponds to the normal stem cells (CD34+, CD38−, CD33−) and have seen that Zalypsis does not affect this cell population, although it is toxic for the more mature myeloid population. Taken together, these studies support the clinical development of Zalypsis in MM, either alone or combined with treatments that have reached the myeloma clinic.
The mechanism of action of Zalypsis on MM cells is multifactorial and involves cell cycle blockade and stimulation of apoptotic cascades. An action on the cell cycle was revealed by the cell cycle profiling that showed that Zalypsis decreased the G2/M cellular population and increased the G0/G1 phase. In addition, Zalypsis down-regulated several genes involved in cell cycle progression, as indicated by the microarray studies. In addition to the action on cell cycle, Zalypsis also triggered a strong apoptotic response. This action was rapidly detected (within 6 hours, as indicated by annexin V staining) and was accompanied by the increase of MOMP and activation of a caspase-dependent signaling cascade that included PARP as well as procaspase-8, -7, -9, and -3 cleavages. The role of activation of caspases in Zalypsis-induced cell death was analyzed by studies using caspase inhibitors; however, their inhibitory effect was only partial, indicating that mechanisms other than caspase activation may also be responsible for the induction of cell death by Zalypsis. In this context, it is interesting to mention that the apoptotic mediators AIF and Endo G, which are released by Zalypsis from the mitochondria, may trigger cell death through caspase-independent routes.25
Microarray data interpretation led to the identification of DNA damage as a part of the mechanism of action of Zalypsis. The microarray gene expression studies identified several genes implicated in the ATM pathway, whose levels were down-regulated, in addition to other genes related to DNA repair. Western blotting studies indicated that Zalypsis up-regulated the levels of pH2AX, a surrogate marker of DSBs as well as pCHK2, both substrates of ATM. These studies confirmed that Zalypsis provoked a DDR response probably after inducing DSBs in the DNA. One of the principal routes involved in the DDR is the p53 pathway. Indeed, Zalypsis provoked a strong increase in p53 levels in the most sensitive MM cell lines, in which p53 is functional. This was evidenced by the up-regulation of classic p53 target genes, such as Bax or Noxa, that control cell cycle progression and apoptotic responses.27 Therefore, in the p53 wild-type MM cell lines, p53 could represent an important mediator of the action of Zalypsis, as these cells, particularly MM1S, are highly sensitive to the activation of the p53 route.28 It should be mentioned that the effect of Zalypsis on p53 levels in these cell types was higher than the effect of other MM treatments, including doxorubicin. This superiority in the induction of p53 represents an added value of Zalypsis, particularly in MM patients with p53 wild-type status in which in combination with lenalidomide, whose effect does not involve a p53 route, should be a highly efficient treatment.
In addition to the p53 route, Zalypsis may also induce cell death through a p53-independent pathway. In support of this is the fact that Zalypsis was still able to provoke cell death in cell lines with known p53 mutant status,28 although at slightly higher concentrations than in MM cells bearing wild-type p53, but still within the plasma levels reached in patients. In these p53 mutant cells, Zalypsis was still able to trigger DDR, as indicated by its action on pH2AX and pCHK2. Therefore, p53-independent routes can also be activated by Zalypsis in MM, and these routes probably act as salvage pathways that preserve an adequate DDR in the absence of functional p53.
The action of Zalypsis in MM therefore involves several interlaced pathways that move into an apoptotic response, probably initiated by direct DNA damage. These pathways may include p53-dependent and p53-independent routes that, by acting on caspases and other apoptotic signaling pathways, hamper MM cell proliferation and trigger apoptosis. The potent antimyeloma action of Zalypsis, together with the particular mechanism of action of this compound, strongly supports the initiation of clinical studies in MM patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Montserrat Martín for excellent technical assistance, Enrique Colado for his help with the flow cytometry experiments, and Diego Fernández-Lázaro for the luminescence assays.
This work was supported by a grant from the Ministry of Science and Innovation of Spain (BFU2006-01 813/BMC and RD06/0020/0041). The CIC receives support from the European Community through the regional development funding program (Federacion Espanola de Enfermedades Raras [FEDER]). P.M. was supported by the Fondo de Investigaciones Sanitarias (FIS)–FEDER through projects to J.F.S.-M. and a Spanish Myeloma Network Program (G03/136). E.M.O. was supported by the Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I + D + I) and by Instituto de Salud Carlos III-Fondo de Investigación Sanitaria (reference no. 400001). M.G. was supported by the Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I + D + I) and by Instituto de Salud Carlos III-Fondo de Investigación Sanitaria (expedient no. 05/0279). S.A.-F. was supported by the program FPU from the Ministry of Science and Innovation of Spain. J.C.M. was supported by the scientific foundation of the Spanish Association Against Cancer. J.F.S.-M. and A.P. received research funding from Pharmamar (Madrid, Spain).
Authorship
Contribution: E.M.O. and P.M. designed research, performed research, analyzed data, and wrote the paper; X.C., M.G., and S.A.-F. designed research, performed research, analyzed data, and contributed to the write-up of the paper; L.S.-S., D.V., L.L.-C., J.C.M., T.H.-I., and E.d.A. performed research and analyzed data; C.G., P.A., and C.C. contributed research tools, analyzed data and contributed to the write-up of the paper; and J.F.S.-M. and A.P. designed research, analyzed data, and contributed to the write-up of the paper.
Conflict-of-interest disclosure: C.G., P.A., and C.C. areemployees of PharmaMar (Madrid, Spain). The remaining authors declare no competing financial interests.
Correspondence: Enrique M. Ocio, University Hospital of Salamanca, Hematology department, 37007 Salamanca, Spain; e-mail: emocio@usal.es.
References
Author notes
*E.M.O. and P.M. contributed equally to this work.




![Figure 4. Zalypsis induces changes in the cell cycle profile and apoptosis in MM1S cells. (A) MM1S cells were incubated with Zalypsis (5 nM) for 6, 12, and 24 hours, and the cell cycle profile was examined by flow cytometry after propidium iodide staining. Bars represent the percentage of cells in each phase of the cell cycle from a representative example. (B) MM1S cells were treated with Zalypsis (5 nM) for different time points, and the induction of apoptosis was analyzed by flow cytometry after staining with annexin V. (C) MM1S cells were treated with Zalypsis (5 nM) for the indicated times. DNA of the treated cells was isolated, and its fragmentation was analyzed by agarose gel electrophoresis. The position of the molecular weight markers is shown at the right. (D) MM1S cells were treated with Zalypsis (5 nM) for 12 and 24 hours, and the disruption of the mitochondrial membrane potential (ΔΨm) was analyzed by flow cytometry after staining with [DiOC6(3)low]. (E) MM1S cells were treated for the indicated times with Zalypsis, and cytochrome C, AIF, and endonuclease G in the mitochondrial fraction were analyzed by Western blotting. (F) The status of some Bcl-2 family members was analyzed by Western blotting after treatment of MM1S cells with 5 nM Zalypsis for the indicated time points in the presence or absence of the caspase inhibitor Z-VAD-FMK (50 μM for 60 minutes). (G) MM1S cells were treated with Zalypsis (5 nM) for the indicated times, and expression of PARP, caspase-3, caspase-7, caspase-8, and caspase-9 proteins was analyzed by Western blotting. (H) MM1S cells were plated and pretreated with the different caspase inhibitors (50 μM) for 60 minutes. Zalypsis (5 nM) was added to the corresponding samples, and the experiment was continued for 12 and 24 hours. Induction of apoptosis was analyzed by means of annexin V–FITC staining.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-09-177774/7/m_zh80060930750004.jpeg?Expires=1763646785&Signature=TVYEmYXeUVZ~xv-g0FQnjPkfa7av1vqMvjnh7B5XoET1npOeFgqGUKxb1avbGvRYrIycoVlloxEPPZAYgH3FjLUl2b9AeZle2Swz3JO~bysDu8p6I6TNXd5Zn6D4QOAvZVIYcKMMcVZwyBemaDPrOaqhz1nOGJDZ1D23yZZ35prycGKWKHWJ8XP5frJArjNhHUaoPkYYtjvVt9t0Ng7l5f3HT~ReP5hGrve1-0Dk7lSZlMyRkpfqQO56A1JIYbq45uXVriwdA9Hr3UNK1Xze5ZXx9CqSJimKVcnNzoGT1I2kZdFUCLfvtDfMhuWBkB3fFHfu8hp3Z0vaBJBAxQzGDQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal