Abstract
Phagocytes such as macrophages and dendritic cells (DCs) engulf apoptotic cells to maintain peripheral immune tolerance. However, the mechanism for the recognition of dying cells by phagocytes is not fully understood. Here, we demonstrate that T-cell immunoglobulin mucin-3 (Tim-3) recognizes apoptotic cells through the FG loop in the IgV domain, and is crucial for clearance of apoptotic cells by phagocytes. Whereas Tim-4 is highly expressed on peritoneal resident macrophages, Tim-3 is expressed on peritoneal exudate macrophages, monocytes, and splenic DCs, indicating distinct Tim-mediated phagocytic pathways used by different phagocytes. Furthermore, phagocytosis of apoptotic cells by CD8+ DCs is inhibited by anti–Tim-3 mAb, resulting in a reduced cross-presentation of dying cell-associated antigens in vitro and in vivo. Administration of anti–Tim-3 as well as anti–Tim-4 mAb induces autoantibody production. These results indicate a crucial role for Tim-3 in phagocytosis of apoptotic cells and cross-presentation, which may be linked to peripheral tolerance.
Introduction
Apoptosis is a crucial process in the development and homeostasis of multicellular organisms.1,2 In the immune system, an enormous number of cells undergo apoptosis during development of lymphocytes and after interaction with antigens.3 Because apoptotic cells and secondary necrotic cells releasing intracellular contents could be autoantigens, phagocytes such as macrophages and dendritic cells (DCs) must engulf these dying cells rapidly and efficiently to prevent detrimental inflammatory responses and autoimmunity.1,4 To engulf apoptotic cells, macrophages use a variety of molecules, including Mer tyrosine kinase (MerTK),5 milk fat globule-EGF-factor 8 (MFG-E8),6 brain-specific angiogenesis inhibitor 1 (BAI1),7 and T-cell immunoglobulin and mucin domain-containing molecule 4 (Tim-4).8,9 However, their relative contributions to the phagocytosis remain to be elucidated. Multiple receptors may simultaneously recognize multiple “eat-me” signals on apoptotic cells. In addition, different subsets of macrophages may use different repertoires of receptors for the phagocytosis.
DCs are able to not only phagocytose apoptotic cells but also present dying cell-associated antigens with MHC class I molecules, which is termed as “cross-presentation.”1,10 It has been considered that, in steady state, cross-presentation of self-antigens by DCs stimulates CD8+ T cells to proliferate abortively, resulting in their deletion, which is crucial to maintain peripheral tolerance.10-14 Among mouse splenic DC subsets, CD8+ DCs are unique in their ability for efficient phagocytosis of apoptotic cells and cross-presentation.15,16 However, the mechanism for the recognition of apoptotic cells by CD8+ DCs is poorly understood. Scavenger receptor CD36 and mannose receptor (MR)/DEC205 are highly expressed on CD8+ DCs, but not CD8− DCs, however, these receptors are not required for cross-presentation of cell-associated antigens by this DC subset.16-18 Neither αvβ3 nor αvβ5 integrin that mediates phagocytosis of apoptotic cells by macrophages1 is essential for phagocytosis by CD8+ DCs.17 Thus, the phagocytic receptor for apoptotic cells linked to cross-presentation remains to be identified.
Tim-3 has been identified as a Th1-specific marker, and several in vivo studies have shown that Tim-3 regulates autoimmunity.19,20 We and others have reported that Tim-3 negatively regulates Th1-mediated inflammatory diseases such as experimental autoimmune encephalomyelitis (EAE), type I diabetes, and acute graft-versus-host diseases (aGVHD).21-23 Moreover, it has been reported that Tim-3 promotes tolerance induction.21,22 Recently, Zhu et al have identified galectin-9 as a Tim-3 ligand, and they have demonstrated that galectin-9 binds to the carbohydrate chains on Tim-3, and induces cell death of Th1 cells in vitro, which may explain the mechanism by which Tim-3 suppresses Th1 immunity.24 On the other hand, Anderson et al have reported that Tim-3 is expressed on DCs, and that galectin-9 activates the DCs through Tim-3, proposing that Tim-3 exacerbates EAE.25 Taken together, Tim-3 appears to have multiple roles for the immune regulation in vivo, however, it remains unknown whether these multiple functions of Tim-3 are mediated solely through galectin-9.
In this study, we demonstrate that Tim-3 recognizes apoptotic cells through the FG loop in the IgV domain. Although Tim-4 is reported to be crucial for the phagocytosis of apoptotic cells by peritoneal macrophages,8,9 we highlight here Tim-3 as the phagocytic receptor responsible for cross-presentation of dying cell-associated antigens by CD8+ DCs. We propose that this novel function of Tim-3 may be involved in autoimmune regulation and tolerance induction.
Methods
Mice and reagents
Five-week-old female C57BL/6 mice were obtained from Charles River Japan (Yokohama, Japan). OT-I mice expressing OVA257-264/H-2Kb–specific TCR were kindly provided by W. R. Heath (The Walter and Eliza Hall Institute, Melbourne, Australia) through H. Udono (RCAI, RIKEN, Yokohama, Japan). These mice were maintained under specific pathogen-free conditions, and used according to the guidelines of the institutional animal care and use committee established at Juntendo University and Tokyo Medical and Dental University. pcDNA3.1(−), pEF6/V5-TOPO, and pcDNA3.1-GFP-TOPO vectors were purchased from Invitrogen (Frederick, MD). PE-Mac1 (CD11b) mAb and control rat IgG2a were purchased from eBioscience (San Diego, CA). FITC-anti-CD11c mAb, PE-anti-Vα2 mAb, and allophycocyanin (APC)–anti-CD8α mAb were purchased from BD Biosciences (San Jose, CA). Alexa Fluor 647–anti-CD8α mAb was purchased from Biolegend (San Diego, CA). 5-(and-6)-Carboxyfluorescein diacetate succinimidyl ester (CFSE) and 5-(and-6)-carboxytetramethylrhodamine succinimidyl ester (TAMRA) were purchased from Invitrogen. TdT and biotin-16-dUTP were purchased from Roche (Indianapolis, IN).
Generation of Tim-Igs and mAbs
The expression vectors for Tim-1–Ig, Tim-2–Ig, Tim-3–Ig, and Tim-4–Ig were generated by linking the extracellular domains of Tim-1 (aa 1-236), Tim-2 (aa 1-230), Tim-3 (aa 1-191), or Tim-4 (aa 1-288) to the Fc portion of mouse IgG2a in the pcDNA3.1(−) vector. Tim-Ig proteins were produced by transfection of each vector into HEK293T cells. The anti–mouse Tim-1 mAb (RMT1-17, rat IgG2a, κ), Tim-2 mAb (RMT2-14, rat IgG2a, κ), and Tim-3 mAb (RMT3-23, rat IgG2a, κ) were generated immunizing SD rats with Tim-1–Ig, Tim-2–Ig, and Tim-3–Ig, respectively, as described before.23 Likewise, the anti–Tim-4 mAb (RMT4-54, rat IgG2a, κ) was generated by immunizing an SD rat with Tim-4–Ig, fusing lymph node cells with P3U1 myeloma cells, and screening the binding to CHO cells expressing Tim-4, but not parental CHO cells. For confocal microscopy, RMT3-23 was labeled with Alexa Fluor 594 using the mAb labeling kit (Invitrogen). Requests for mAbs should be addressed to H. Akiba (e-mail: hisaya@juntendo.ac.jp).
Cell lines
A normal rat kidney cell line (NRK-52E) was maintained in complete RPMI medium (RPMI 1640 supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine, 10 mM HEPES, and 50 μM 2-mercaptoethanol). HEK293T cells (ATCC, Manassas, VA) were maintained in complete DMEM medium (DMEM supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine). For construction of expression vectors, the entire coding region of mouse Tim-1, Tim-2, Tim-3, or Tim-4 was subcloned into pMKITneo. Tim-3 cDNA was also subcloned into pEF6/V5-TOPO vector. Several mutant forms were prepared by polymerase chain reaction (PCR)–based mutagenesis using Tim-3/pEF6V5-TOPO as a template. After confirmation of nucleotide sequences, Tim expression vectors were transduced into NRK cells by electroporation with a Gene Pulser (Bio-Rad Laboratories, Hercules, CA). After selection with 1 mg/mL G418, cell surface expression was estimated by respective anti-Tim mAbs. HEK293T cells were transiently transfected with these expression vectors using lipofectAMINE2000 (Invitrogen). Two days after the transfection, cell surface expression of Tim was estimated by flow cytometry.
Phagocytosis assay
For preparation of apoptotic cells, thymocytes from C57BL/6 mice were labeled with 1 μM CFSE or 10 μg/mL TAMRA, and then were UV irradiated (100 J/cm2). After UV irradiation, the cells were cultured in complete RPMI for 2 hours at 37°C, and then used for the phagocytosis assay. NRK cells (105 per well), NIH3T3 cells (5 × 104 cells per well), and HEK293T cells (105 cells per well) were plated onto 24-well plates, which were precoated with poly-l-lysine for HEK293T cells, a day before the phagocytosis assay. These cell lines were incubated with fluorescently labeled apoptotic cells (2 × 106 per well) at 37°C for the indicated periods. The recognition (binding and/or incorporation) of fluorescently labeled apoptotic cells by these cell lines was analyzed by flow cytometry using a FACSCalibur (BD Biosciences) and/or fluorescence microscopy using an Olympus FV1000 laser scanning confocal microscope (Melville, NY) equipped with 40× or 100× objective lens. For macrophages, peritoneal cells were harvested from C57BL/6 mice 3 days after intraperitoneal injection of 2 mL 3% wt/vol thioglycolate, or from untreated mice. These peritoneal cells (5 × 105 per well) were plated onto 48-well plate for 2 hours at 37°C, and then washed with PBS twice to remove floating cells. In some assay, macrophages were preincubated with the indicated mAb (30 μg/mL) for 60 minutes at 4°C, and then washed with PBS to remove unbound mAb. Macrophages were cultured with fluorescently labeled apoptotic cells (2.5 × 106 per well) for 30 minutes at 37°C, and then washed with PBS 3 times to remove unbound apoptotic cells. After trypsinization, cells were harvested and stained with PE-Mac1. CFSE fluorescence intensity in Mac1+ cells was analyzed on a FACSCalibur (BD Biosciences). As for an in vivo phagocytosis by PEM, sterile peritonitis was induced in C57BL/6 mice by intraperitoneal injection of 3% thioglycolate medium (2 mL per mouse). Three days later, these mice (n = 4-5 per group) were treated intraperitoneally with control rat IgG (rIgG), RMT3-23, or RMT4-54 (200 μg/mouse). Three hours later, mice were intraperitoneally injected with CFSE-labeled apoptotic cells (108 per mouse), and another 2 hours later, peritoneal cells were harvested. The recognition of apoptotic cells by Mac1+ cells was analyzed by flow cytometry. For statistic evaluation, the unpaired Student t test, 2-tailed was used. P values less than .05 were considered significant. For in vitro phagocytosis assay using splenic DCs, spleens were digested with 400 U/mL collagenase (Wako Biochemicals) in the presence of 5 mM EDTA and separated into low- and high-density fractions on Optiprep gradient (Axis-Shield, Oslo, Norway). Low-density cells were purified using anti-CD11c MACS beads (Miltenyi Biotec, Auburn, CA). After staining CD11c+ cells with APC-anti-CD8α mAb, these cells (5 × 105 per well) were cocultured with TAMRA-labeled apoptotic cells (2.5 × 106 per well) in 48-well plate for the indicated periods, and then the cells were stained with FITC–anti-CD11c mAb. TAMRA fluorescence intensity in CD8−CD11c+ and CD8+CD11c+ cells was analyzed on a FACSCalibur. As for in vivo phagocytosis by splenic DCs, mice (n = 3 per group) were treated intravenously with rIgG, or RMT3-23 and/or RMT4-54 (200 μg each/mouse), and then 2 hours later, with CFSE-labeled apoptotic splenocytes (2 × 107 per mouse). Mice were killed at the indicated time points, and the recognition of apoptotic cells by CD8+CD11c+ cells was analyzed by flow cytometry. For statistic evaluation, the unpaired Student t test, 2-tailed was used. P values less than .05 were considered significant.
In situ identification of nuclear DNA fragmentation
C57BL/mice (n = 4-6 per group) were treated intraperitoneally with rIgG, or RMT3-23 and/or RMT4-54 (200 μg each per mouse) twice a week for 4 weeks. Three days after the final injection, mice were killed. Serum was stocked for measurement of anti-dsDNA antibody levels. Brain, liver, spleen, and pancreas were immersion-fixed in 20% buffered formalin and embedded in paraffin. After being deparaffinized, tissue sections were stained using in situ TUNEL method with biotin-16-dUTP (Roche Diagnostics, Basel, Switzerland) and diaminobenzidine as a peroxidase substrate. Nuclei were counterstained with hematoxylin. The number of TUNEL-positive cells was quantified with KS400 Image Analysis System (KS400; Zeiss, Heidelberg, Germany).
Measurement of anti-dsDNA antibody levels in serum
Serum levels of anti-dsDNA IgG were determined as described previously.26 In brief, enzyme-linked immunosorbent assay (ELISA) plates were coated with 5 μg/mL dsDNA derived from calf thymus (Sigma-Aldrich, St Louis, MO). Anti-dsDNA antibody level was expressed in units, referring to standard curve obtained by serial dilution of a standard serum pool from (NZB × NZW) F1 mice older than 8 months, containing 1000 U activities/mL (kindly provided by S. Hirose, Juntendo University).
Flow cytometric analysis for Tim expression
Mouse macrophages or low-density splenocytes were pretreated with anti-FcR mAb (2.4G2), and then were incubated with 0.5 μg biotinylated mAb for 30 minutes at 4°C, followed by PE-streptavidin and FITC-anti-CD11b mAb (for macrophages) or PE-streptavidin, FITC–anti-CD11c mAb and APC–anti-CD8α mAb (for splenic DCs). After washing with PBS, Tim expression on CD11b+, CD8+CD11c+, or CD8−CD11c+ cells was analyzed on a FACSCalibur, and the data were analyzed using the CellQuest program (BD Biosciences).
In vitro cross-presentation assay
OVA-loaded dying cells were prepared by osmotic shock as described previously.12 For splenic DC subsets, CD11c+ cells purified using anti-CD11c MACS beads (> 92% CD11c+) were then stained with FITC–anti-CD11c and APC–anti-CD8α mAbs, followed by sorting into subsets (> 89% CD8+CD11c+; > 98% CD8−CD11c+) on FACSVantage (BD Biosciences). OVA-loaded (10 mg/mL) dying cells (2.5 × 106 per well) were cocultured with CD8+ DCs or CD8− DCs (5 × 105 per well) in presence of rIgG, RMT3-23, or RMT4-54 (30 μg/mL) on 48-well plate for 1.5 hours, and then DCs were sorted again using anti-CD11c MACS beads. CD8− or CD8+ DCs (both 104 or 2 × 103 per well) were cocultured with purified OT-I CD8+ T cells (105 per well) in 96-well flat-bottom plate. For the direct presentation, CD8− or CD8+ DCs (104 per well) were cocultured with 1 nM OVA257-264 SIINFEKL peptides (AnaSpec, San Jose, CA) and OT-I CD8+ T cells (105 per well) in presence of rIgG, RMT3-23, or RMT4-54 (30 μg/mL) in 96-well flat-bottom plate. Two days later, 50 μL supernatant was harvested and tested for IFN-γ production by sandwich ELISA (eBioscience). The cultures were then pulsed overnight with [3H]thymidine (0.5 μCi [0.0185 MBq]/well; GE Healthcare, Little Chalfont, United Kingdom) and the uptake was measured in a microbeta counter (Microbeta Plus; Wallac, Turku, Finland).
In vivo cross-presentation assay
In vivo cross-presentation assay was performed as described previously27 with minor modifications. CFSE-labeled OT-I cells (2 × 106 per mouse) were transferred intravenously into B6 mice. Next day, mice were injected intravenously with rIgG, or RMT3-23 and/or RMT4-54 (200 μg each per mouse), and then 2 hours later, with OVA-loaded (1 mg/mL) dying splenocytes (107 per mouse). Two days later, mice were killed, and splenocytes were stained with PE–anti-Vα2 and APC–anti-CD8. CFSE fluorescence intensity of CD8+Vα2+ cells was analyzed by flow cytometry.
Results
Tim-3 recognizes apoptotic cells and is recruited to phagosome
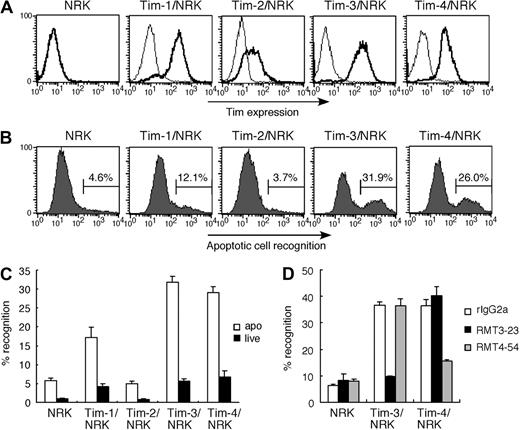
Because Tim-4 and Tim-1 are recently reported to recognize apoptotic cells,8,9 we first verified the binding activity of NRK cells stably expressing Tim family molecules (Figure 1A) to apoptotic cells. As for apoptotic cells, we used UV (100 J/cm2)–irradiated thymocytes because these cells show typical apoptotic phenotypes, which are annexin V+ and propidium iodide negative (PI−) (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article), and do not express Tim molecules (Figure S1B), excluding Tim-Tim interaction in this study. In addition to Tim-4/NRK and Tim-1/NRK, we found that Tim-3/NRK also efficiently bound apoptotic cells, but not live cells (Figure 1B,C), and that the binding of Tim-3/NRK and Tim-4/NRK cells to apoptotic cells was abrogated by anti–Tim-3 mAb RMT3-23 and anti–Tim-4 mAb RMT4-54, respectively (Figure 1D). These results suggest that Tim-3 as well as Tim-4 acts as a receptor for apoptotic cells.
Tim-3 recognizes apoptotic cells. (A) NRK cells stably expressing Tim-1, Tim-2, Tim-3, or Tim-4 were stained with biotinylated anti–Tim-1 mAb (RMT1-17), anti–Tim-2 mAb (RMT2-14), anti–Tim-3 mAb (RMT3-23), or anti–Tim-4 mAb (RMT4-54), respectively. Parental NRK cells were stained with a cocktail of all mAbs. Then cells were stained with PE-avidin, and analyzed by flow cytometry (thick histogram). Thin histograms indicate background staining with control rat IgG2a, followed by PE-avidin. (B) These NRK cells were cultured with CFSE-labeled apoptotic cells for 30 minutes at 37°C. Recognition of apoptotic cells by these NRK cells was quantified by flow cytometry. (C) These NRK cells were cultured with CFSE-labeled viable cells or apoptotic cells for 30 minutes at 37°C. Percentage of the recognition was quantified by flow cytometry. Data are represented as mean ± SD of triplicates. (D) These NRK cells were pretreated with 20 μg/mL control rIgG2a, RMT3-23, or RMT4-54 mAb, and then cultured with CFSE-labeled apoptotic cells for 30 minutes at 37°C. Percentage of the recognition was quantified by flow cytometry. Data are represented as mean ± SD of triplicates. Similar results were obtained in 3 (A-C) or 2 (D) independent experiments.
Tim-3 recognizes apoptotic cells. (A) NRK cells stably expressing Tim-1, Tim-2, Tim-3, or Tim-4 were stained with biotinylated anti–Tim-1 mAb (RMT1-17), anti–Tim-2 mAb (RMT2-14), anti–Tim-3 mAb (RMT3-23), or anti–Tim-4 mAb (RMT4-54), respectively. Parental NRK cells were stained with a cocktail of all mAbs. Then cells were stained with PE-avidin, and analyzed by flow cytometry (thick histogram). Thin histograms indicate background staining with control rat IgG2a, followed by PE-avidin. (B) These NRK cells were cultured with CFSE-labeled apoptotic cells for 30 minutes at 37°C. Recognition of apoptotic cells by these NRK cells was quantified by flow cytometry. (C) These NRK cells were cultured with CFSE-labeled viable cells or apoptotic cells for 30 minutes at 37°C. Percentage of the recognition was quantified by flow cytometry. Data are represented as mean ± SD of triplicates. (D) These NRK cells were pretreated with 20 μg/mL control rIgG2a, RMT3-23, or RMT4-54 mAb, and then cultured with CFSE-labeled apoptotic cells for 30 minutes at 37°C. Percentage of the recognition was quantified by flow cytometry. Data are represented as mean ± SD of triplicates. Similar results were obtained in 3 (A-C) or 2 (D) independent experiments.
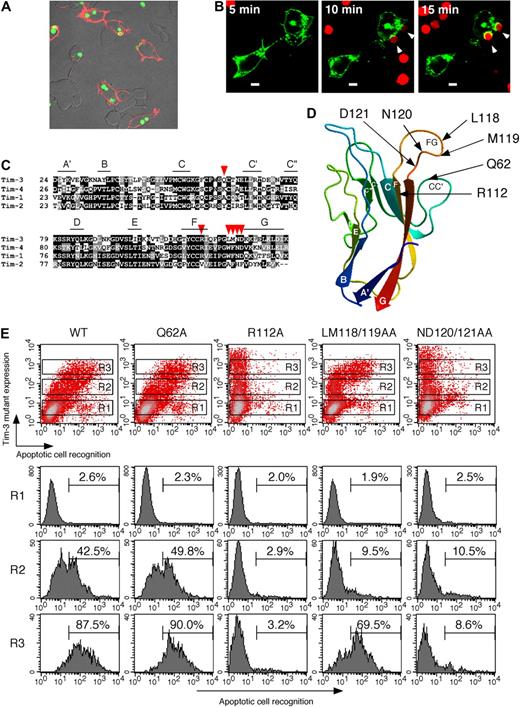
To further address whether expression of Tim-3 could confer the ability to internalize or just bind apoptotic cells, we next used HEK293T cell reconstitution system because although ectopic expression of an authentic phagocytic receptor FcγRIII with FcRγ chain enabled this cell line to internalize IgG-opsonized bacteria, the expression of scavenger receptor-A (SR-A)28 or paired Ig-like receptor-B (PIR-B),29 which is the nonphagocytic receptor for bacteria, conferred the binding without significant internalization (not shown). As shown in Figure 2A, Tim-3–positive cells efficiently internalized CFSE-labeled apoptotic cells, indicating that Tim-3 is a phagocytic receptor for apoptotic cells. Furthermore, we addressed Tim-3 localization upon initial contact with apoptotic cells. To visualize Tim-3, we generated an expression vector for Tim-3 fused at its C-terminus to green fluorescence protein (GFP), and then expressed this fusion receptor in HEK293T cells. Whereas Tim-3 was mostly expressed at the cell surface in resting conditions, upon recognition of TAMRA-labeled apoptotic cells, Tim-3 was recruited to the phagocytic cup (Figure 2B). This substantiates that Tim-3 mediates engulfment of apoptotic cells.
Tim-3 internalizes apoptotic cells through the FG loop in IgV domain. (A) HEK293T cells transiently expressing Tim-3 were cultured with CFSE-labeled apoptotic cells for 60 minutes at 37°C, and then cells were stained with biotinylated RMT3-23 and Alexa 594–avidin. (B) TAMRA-labeled apoptotic cells were added to HEK293T cells transiently expressing Tim-3-GFP under fluorescence microscope. Phagocytosis and Tim-3-GFP localization were analyzed at the indicated time points. White arrowheads indicate apoptotic cells internalized via Tim-3. White bars indicate 5 μm. (C) Alignment of IgV domain of Tim family molecules. The β-strands of Tim-3 were shown with lines. Mutated residues were indicated by red arrowheads. (D) Positions of mutated residues are indicated on 3-dimensional structure of Tim-3 IgV domain. The color of the protein main-chain is gradually changed along the sequence from blue (N-terminal) to red (C-terminal). (E) HEK293T cells transiently expressing wild-type or mutant Tim-3 were cultured with CFSE-labeled apoptotic cells for 60 minutes at 37°C, and then cells were stained with biotinylated RMT3-23 and PE-avidin. Recognition of apoptotic cells by gated HEK293T cells (R1; Tim-3−, R2; Tim-3low, R3; Tim-3high) was analyzed by flow cytometry. Similar results were obtained in 2 (A,B) or 3 (E) independent experiments.
Tim-3 internalizes apoptotic cells through the FG loop in IgV domain. (A) HEK293T cells transiently expressing Tim-3 were cultured with CFSE-labeled apoptotic cells for 60 minutes at 37°C, and then cells were stained with biotinylated RMT3-23 and Alexa 594–avidin. (B) TAMRA-labeled apoptotic cells were added to HEK293T cells transiently expressing Tim-3-GFP under fluorescence microscope. Phagocytosis and Tim-3-GFP localization were analyzed at the indicated time points. White arrowheads indicate apoptotic cells internalized via Tim-3. White bars indicate 5 μm. (C) Alignment of IgV domain of Tim family molecules. The β-strands of Tim-3 were shown with lines. Mutated residues were indicated by red arrowheads. (D) Positions of mutated residues are indicated on 3-dimensional structure of Tim-3 IgV domain. The color of the protein main-chain is gradually changed along the sequence from blue (N-terminal) to red (C-terminal). (E) HEK293T cells transiently expressing wild-type or mutant Tim-3 were cultured with CFSE-labeled apoptotic cells for 60 minutes at 37°C, and then cells were stained with biotinylated RMT3-23 and PE-avidin. Recognition of apoptotic cells by gated HEK293T cells (R1; Tim-3−, R2; Tim-3low, R3; Tim-3high) was analyzed by flow cytometry. Similar results were obtained in 2 (A,B) or 3 (E) independent experiments.
Tim-3 binds to PS
We next addressed whether Tim-3 also binds to phosphatidylserine (PS), a major “eat-me” signal,1,2 by solid-phase ELISA using soluble Ig fusion proteins with extracellular domains including both IgV and mucin domains of Tim-2, Tim-3, and Tim-4. In addition to the strong binding of Tim-4-Ig to PS, we observed that Tim-3–Ig weakly but substantially bound to PS, but not other phospholipids such as phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylcholine (PC) (Figure S2A). Tim-2–Ig did not bind any phospholipids even at a high dose. The Tim-3–Ig or Tim-4–Ig binding to PS was abrogated by RMT3-23 or RMT4-54, respectively, in a dose-dependent manner (Figure S2B). These results indicate that Tim-3 recognizes PS, although the affinity is lower than that of Tim-4.
Tim-3 recognizes apoptotic cells through the FG loop in IgV domain
We next explored the Tim-3 recognition site of apoptotic cells. Recently, Santiago et al have revealed a crystal structure of Tim-4 and demonstrated that Tim-4 binds PS through the metal ion–dependent ligand binding site (MILIBS) in the FG loop, which is conserved in all Tim family members except Tim-2.30 Thus, we specifically mutated some of amino acids locating around the FG loop of Tim-3 to alanine, and transiently transfected HEK293T cells with each mutant construct to address the relationship between expression level of each mutant and their phagocytic activities. We first replaced Gln62 or Arg112 to Ala because these amino acids, which locate in FG-CC′ cleft in the 3D structural model of Tim-3 IgV domain (Figure 2C,D), are critical for galectin-9–independent ligand binding.31 Substitution of Gln62 did not alter the phagocytic activity, although substitution of Arg112 completely abrogated the activity (Figure 2E). We next addressed the putative MILIBS, which locates on the FG surface loop (Figure 2D). The LM118/119AA mutant recognized apoptotic cells only at high expression level, suggesting that substitution of these residues to both Ala weakened the activity (Figure 2E). The ND120/121AA mutant completely lost the activity (Figure 2E). These results suggest that Tim-3 internalizes apoptotic cells through the FG loop in IgV domain, and that at least in part galectin-9–independent ligand binding31 might be linked to recognition of apoptotic cells.
Tim-3 and Tim-4 mediate phagocytosis of apoptotic cells by distinct macrophage subsets
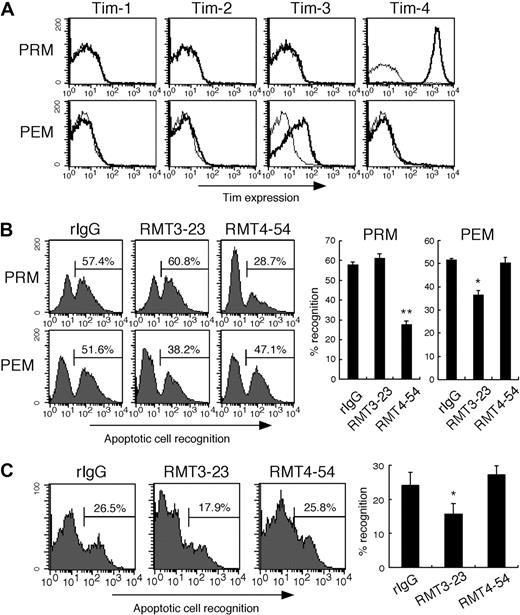
We next examined the cell surface expression of Tim molecules on mouse primary macrophages, and their contribution to the phagocytosis of apoptotic cells. We found Tim-3 expression on peritoneal exudate Mac1+ cells (PEMs), but not peritoneal resident Mac1+ cells (PRMs) from naive mice (Figure 3A). In contrast, Tim-4 was highly expressed on PRMs, but not PEMs, which is consistent with a previous report.8 Neither Tim-1 nor Tim-2 was expressed on both types of macrophages (Figure 3A). This result prompted us to investigate whether these different types of macrophages use different Tim molecules to recognize apoptotic cells. As shown in Figure 3B, PRMs efficiently phagocytosed apoptotic cells, and this was significantly inhibited by RMT4-54, but not by RMT3-23. In contrast, phagocytosis of apoptotic cells by PEMs was significantly inhibited by RMT3-23, but not RMT4-54. These results suggest that although these macrophage subsets use MerTK and MFG-E8 to recognize and internalize apoptotic cells,5,6,32 Tim-3 and Tim-4 also play a role in this process by PEMs and PRMs, respectively. Moreover, we used a mouse sterile peritonitis model to examine whether Tim-3 participates in the phagocytosis in vivo. As shown in Figure 3C, RMT3-23, but not RMT4-54, significantly inhibited the phagocytosis of apoptotic cells by peritoneal macrophages, suggesting that Tim-3 contributes to the phagocytosis of apoptotic cells in vivo.
Tim-3 mediates phagocytosis of apoptotic cells by peritoneal exudate macrophages. (A) Peritoneal resident Mac1+ cells (PRMs) and peritoneal exudate Mac1+ cells (PEMs) were stained with biotinylated RMT1-17, RMT2-14, RMT3-23, or RMT4-54, followed by FITC-anti-CD11b mAb and PE-avidin (thick histograms); then CD11b+ cells were analyzed by flow cytometry. Thin histograms indicate background staining with biotinylated control rat IgG2a. (B) Macrophages were pretreated with control rat IgG2a (rIgG), RMT3-23, or RMT4-54, and then cultured with CFSE-labeled apoptotic cells for 30 minutes at 37°C. Cells were stained with PE-Mac1, and percentage of recognition of CFSE-apoptotic cells by Mac1+ cells was quantified by flow cytometry. Columns represent mean ± SD of triplicates (*P < .05; **P < .01 compared with rIgG). (C) Peritonitis was elicited by intraperitoneal injection of thioglycolate. Three days later, the mice were intraperitoneally injected with rIgG, RMT3-23, or RMT4-54 (200 μg/head), and then with CFSE-labeled apoptotic cells. Two hours later, peritoneal cells were harvested, and recognition of CFSE-labeled apoptotic cells by Mac1+ cells was quantified by flow cytometry. The experiments (n = 4-5 per group) were performed 3 times independently with a similar result. Columns represent mean ± SD of 4 mice in a representative experiment (*P < .05 compared with rIgG). Similar results were obtained in 3 (A,C) or 2 (B) independent experiments.
Tim-3 mediates phagocytosis of apoptotic cells by peritoneal exudate macrophages. (A) Peritoneal resident Mac1+ cells (PRMs) and peritoneal exudate Mac1+ cells (PEMs) were stained with biotinylated RMT1-17, RMT2-14, RMT3-23, or RMT4-54, followed by FITC-anti-CD11b mAb and PE-avidin (thick histograms); then CD11b+ cells were analyzed by flow cytometry. Thin histograms indicate background staining with biotinylated control rat IgG2a. (B) Macrophages were pretreated with control rat IgG2a (rIgG), RMT3-23, or RMT4-54, and then cultured with CFSE-labeled apoptotic cells for 30 minutes at 37°C. Cells were stained with PE-Mac1, and percentage of recognition of CFSE-apoptotic cells by Mac1+ cells was quantified by flow cytometry. Columns represent mean ± SD of triplicates (*P < .05; **P < .01 compared with rIgG). (C) Peritonitis was elicited by intraperitoneal injection of thioglycolate. Three days later, the mice were intraperitoneally injected with rIgG, RMT3-23, or RMT4-54 (200 μg/head), and then with CFSE-labeled apoptotic cells. Two hours later, peritoneal cells were harvested, and recognition of CFSE-labeled apoptotic cells by Mac1+ cells was quantified by flow cytometry. The experiments (n = 4-5 per group) were performed 3 times independently with a similar result. Columns represent mean ± SD of 4 mice in a representative experiment (*P < .05 compared with rIgG). Similar results were obtained in 3 (A,C) or 2 (B) independent experiments.
Tim-3 is crucial for clearance of apoptotic cells in vivo
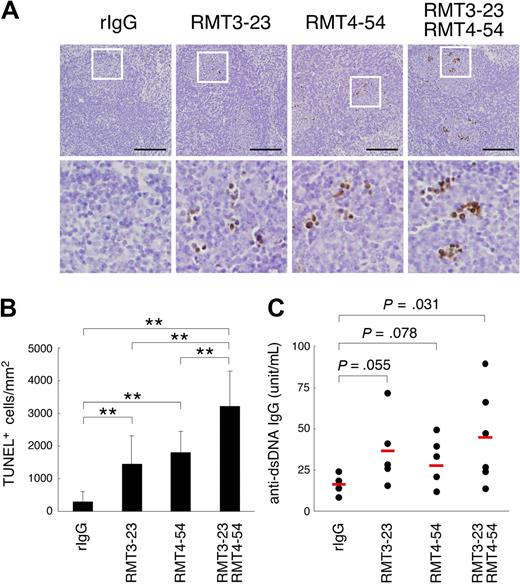
To evaluate the physiological role of Tim-3 and Tim-4 in vivo, we intraperitoneally injected RMT3-23 and/or RMT4-54 into C57BL/6 mice twice a week for 4 weeks, and stained apoptotic cells in various organs by terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) method. In spleen follicles, treatment with either RMT3-23 or RMT4-54 significantly increased TUNEL+ cells, and a remarkable increase was observed with the combination treatment (Figure 4A,B), whereas the administration of these mAbs did not increase the number of TUNEL+ cells in the liver, pancreas, or brain (not shown).
Involvement of Tim-3 in clearance of apoptotic cells in vivo. Mice (n = 4-6 per group) were treated with rIgG, RMT3-23, and/or RMT4-54 (200 μg each per mouse) twice a week for 4 weeks. (A) Apoptotic cells in paraffin-embedded spleen sections from these mice were detected by TdT-mediated dUTP nick-end labeling (TUNEL) method. Nuclei were counterstained with hematoxylin. Representative sections (top panels, ×20 magnification) were shown. Black bars indicate 100 μm. White squares mark the areas shown at a higher magnification (bottom panels, ×80). (B) The number of TUNEL-positive cells was counted in at least 10 randomly chosen follicles, represented as columns (**P < .01). (C) Anti-dsDNA antibody levels in serum were determined by ELISA. P values compared with rIgG are shown. Similar results were obtained in 2 independent experiments.
Involvement of Tim-3 in clearance of apoptotic cells in vivo. Mice (n = 4-6 per group) were treated with rIgG, RMT3-23, and/or RMT4-54 (200 μg each per mouse) twice a week for 4 weeks. (A) Apoptotic cells in paraffin-embedded spleen sections from these mice were detected by TdT-mediated dUTP nick-end labeling (TUNEL) method. Nuclei were counterstained with hematoxylin. Representative sections (top panels, ×20 magnification) were shown. Black bars indicate 100 μm. White squares mark the areas shown at a higher magnification (bottom panels, ×80). (B) The number of TUNEL-positive cells was counted in at least 10 randomly chosen follicles, represented as columns (**P < .01). (C) Anti-dsDNA antibody levels in serum were determined by ELISA. P values compared with rIgG are shown. Similar results were obtained in 2 independent experiments.
It has been reported that an impairment of clearance of apoptotic cells induces autoantibody production.5,33,34 Thus, we next measured serum level of autoantibodies in these mice. Consistent with a recent report,8 blocking of Tim-4 by RMT4-54 induced anti-dsDNA antibodies (Figure 4C). We found that serum level of anti-dsDNA antibodies was also increased by RMT3-23 (Figure 4C). These results suggest that Tim-3 as well as Tim-4 participates in the clearance of apoptotic cells in vivo, and that the disabling of this system leads to autoantibody production.
Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation by CD8+ DCs in vitro
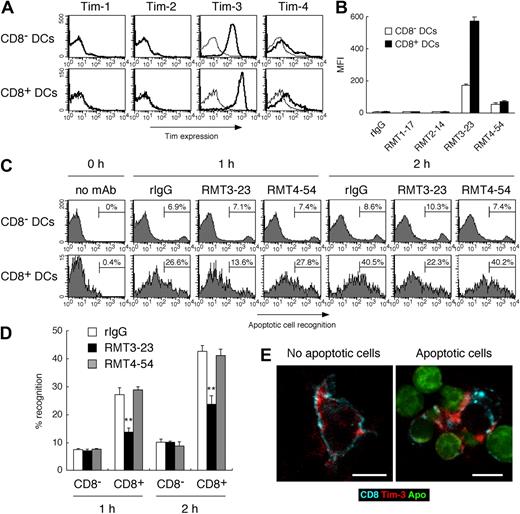
Because Tim-3 is involved in the clearance of apoptotic cells in not only inflammatory state (Figure 3C) but also steady state (Figure 4) in vivo, we further addressed the Tim-3 expression on several types of naive cells, and found that Tim-3 was highly expressed on peripheral blood monocytes and splenic DCs (Figure S3). Moreover, we noticed that the expression of Tim-3 on splenic CD8+ DCs was approximately 3-fold higher than that on CD8− DCs (Figure 5A,B). Several groups have demonstrated that, in mouse spleen, CD8+ DCs are uniquely able to recognize apoptotic cells, however, the receptor for apoptotic cells remains to be identified.16-18 These reports prompted us to address whether Tim-3 plays a role for the recognition of apoptotic cells by this DC subset. Consistent with previous reports,16,35 we observed that CD8+ DCs recognized apoptotic cells more efficiently than CD8− DCs (Figure 5C). Interestingly, masking of Tim-3 by RMT3-23 inhibited approximately 50% recognition of apoptotic cells by CD8+ DCs (Figure 5C,D), suggesting that CD8+ DCs use Tim-3 to efficiently recognize apoptotic cells. Confocal microscopy revealed that, although Tim-3 is largely expressed at cell surface of naive CD8+ DCs (Figure 5E left panel), upon recognition of apoptotic cells Tim-3 is recruited to the site of apoptotic cell apposition with the membrane (Figure 5E right panel), suggesting that Tim-3 mediates internalization of apoptotic cells by CD8+ DCs.
Tim-3 mediates phagocytosis of apoptotic cells by CD8+ DCs. (A) Low-density splenocytes were stained with biotinylated control rIgG2a (thin histograms), RMT1-17, RMT2-14, RMT3-23, or RMT4-54 (thick histograms), followed by PE-avidin, FITC–anti-CD11c mAb, and APC–anti-CD8α mAb; then Tim expression on CD8−CD11c+ or CD8+CD11c+ cells was analyzed by flow cytometry. The average of mean fluorescence intensity (MFI) ± SD of triplicates is represented in panel B. (C) Purified splenic CD11c+ cells prestained with APC–anti-CD8α mAb were treated with rIgG, RMT3-23, or RMT4-54, and then cultured with TAMRA-labeled apoptotic cells at 37°C. After the indicated time period, cells were stained with FITC–anti-CD11c mAb, and percentage recognition of TAMRA-labeled apoptotic cells by CD8−CD11c+ or CD8+CD11c+ cells was quantified by flow cytometry. Columns represent mean ± SD of triplicates in panel D (**P < .01 compared with rIgG). (E) (Left panel) Purified splenic CD11c+ cells were stained with Alexa 647–anti-CD8 mAb and Alexa 594–RMT3-23. (Right panel) Purified splenic CD11c+ cells prestained with Alexa 647–anti-CD8 mAb were cultured with CFSE-labeled apoptotic cells for 60 minutes at 37°C, and then cells were stained with Alexa 594–RMT3-23. Cells were analyzed by confocal microscopy. White bars indicate 5 μm. Similar results were obtained in 3 (A-D) or 2 (E) independent experiments.
Tim-3 mediates phagocytosis of apoptotic cells by CD8+ DCs. (A) Low-density splenocytes were stained with biotinylated control rIgG2a (thin histograms), RMT1-17, RMT2-14, RMT3-23, or RMT4-54 (thick histograms), followed by PE-avidin, FITC–anti-CD11c mAb, and APC–anti-CD8α mAb; then Tim expression on CD8−CD11c+ or CD8+CD11c+ cells was analyzed by flow cytometry. The average of mean fluorescence intensity (MFI) ± SD of triplicates is represented in panel B. (C) Purified splenic CD11c+ cells prestained with APC–anti-CD8α mAb were treated with rIgG, RMT3-23, or RMT4-54, and then cultured with TAMRA-labeled apoptotic cells at 37°C. After the indicated time period, cells were stained with FITC–anti-CD11c mAb, and percentage recognition of TAMRA-labeled apoptotic cells by CD8−CD11c+ or CD8+CD11c+ cells was quantified by flow cytometry. Columns represent mean ± SD of triplicates in panel D (**P < .01 compared with rIgG). (E) (Left panel) Purified splenic CD11c+ cells were stained with Alexa 647–anti-CD8 mAb and Alexa 594–RMT3-23. (Right panel) Purified splenic CD11c+ cells prestained with Alexa 647–anti-CD8 mAb were cultured with CFSE-labeled apoptotic cells for 60 minutes at 37°C, and then cells were stained with Alexa 594–RMT3-23. Cells were analyzed by confocal microscopy. White bars indicate 5 μm. Similar results were obtained in 3 (A-D) or 2 (E) independent experiments.
Because CD8+ DC is the splenic DC subset that plays a crucial role in cross-presentation,15 we next performed cross-presentation study using OT-I T cells specific for OVA257-264/H-2Kb. Consistent with previous reports,15,16 we confirmed that CD8+ DCs cultured with OVA-loaded, but not BSA-loaded, apoptotic cells could efficiently induce OT-I CD8+ T-cell proliferation, and that OT-I cells did not directly respond to OVA-loaded apoptotic cells (Figure S4). Then, we investigated the requirement of Tim-3 for this cross-presentation. As shown in Figure 6A, we observed that CD8+ DCs induced OT-I T-cell proliferation more vigorously than CD8− DCs, and that inhibition of the phagocytosis by RMT3-23 significantly abrogated the CD8+ DC-induced OT-I cell proliferation. Moreover, we observed a remarkable reduction in IFN-γ production by RMT3-23 (Figure 6B), whereas both DC subsets loaded with OVA257-264 peptides equally activated OT-I T cells, irrespective of masking Tim-3 by RMT3-23. Although splenic DCs express Tim-4 at low level, RMT4-54 did not inhibit the phagocytosis of apoptotic cells and the cross-presentation in vitro (Figures 5,6).
Tim-3–mediated phagocytosis of apoptotic cells is crucial for the cross-presentation by CD8+ DCs. (A) Purified CD8−CD11c+ or CD8+CD11c+ splenic DCs were pretreated with rIgG (white histograms), RMT3-23 (black histograms), or RMT4-54 (gray histograms) and then cultured with OVA-loaded apoptotic cells. After 2 hours, both DC subsets were purified again to remove dead cells, and then cocultured with OT-I CD8+ T cells at the indicated ratio. For the direct presentation, both DC subsets preincubated with OVA257-264 peptide (1 nM) in the presence of rIgG, RMT3-23, or RMT4-54 were cocultured with OT-I CD8+ T cells at a 1:10 (DC/T) ratio. [3H]thymidine (3H-TdR) uptake was measured at 48 to 60 hours. (B) Production of IFN-γ in the culture supernatant at 48 hours after addition of OT-I CD8+ T cells was measured by ELISA. Columns represent mean ± SD of triplicates (**P < .01 compared with rIgG). ND indicates not detectable. Similar results were obtained in 3 independent experiments (A,B).
Tim-3–mediated phagocytosis of apoptotic cells is crucial for the cross-presentation by CD8+ DCs. (A) Purified CD8−CD11c+ or CD8+CD11c+ splenic DCs were pretreated with rIgG (white histograms), RMT3-23 (black histograms), or RMT4-54 (gray histograms) and then cultured with OVA-loaded apoptotic cells. After 2 hours, both DC subsets were purified again to remove dead cells, and then cocultured with OT-I CD8+ T cells at the indicated ratio. For the direct presentation, both DC subsets preincubated with OVA257-264 peptide (1 nM) in the presence of rIgG, RMT3-23, or RMT4-54 were cocultured with OT-I CD8+ T cells at a 1:10 (DC/T) ratio. [3H]thymidine (3H-TdR) uptake was measured at 48 to 60 hours. (B) Production of IFN-γ in the culture supernatant at 48 hours after addition of OT-I CD8+ T cells was measured by ELISA. Columns represent mean ± SD of triplicates (**P < .01 compared with rIgG). ND indicates not detectable. Similar results were obtained in 3 independent experiments (A,B).
Tim-3 is crucial for phagocytosis of apoptotic cells and cross-presentation in vivo
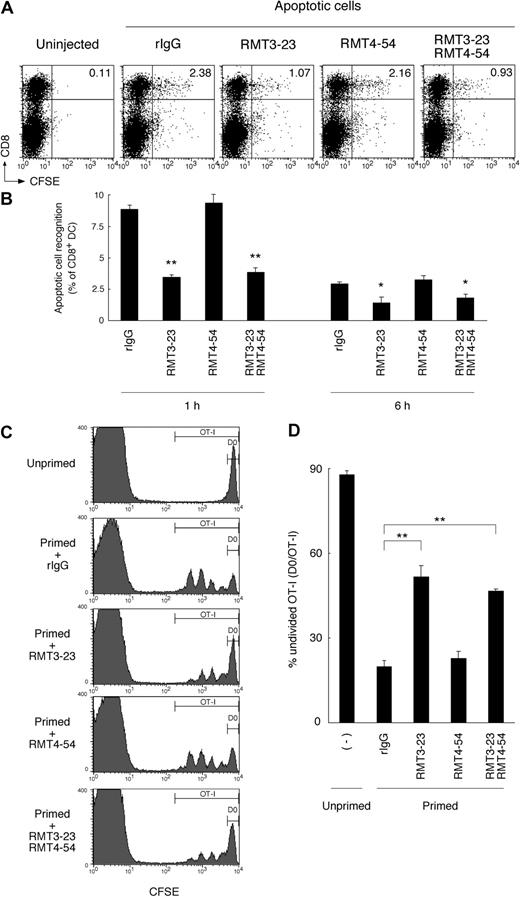
We further addressed the contribution of Tim-3 to the phagocytosis of apoptotic cells and cross-presentation by CD8+ DCs in vivo. It has been reported that intravenously injected apoptotic cells are taken up mainly by CD8+ DCs in mouse spleen.16,35 Consistently, we also observed that CD8+ DCs efficiently recognized CFSE-labeled apoptotic cells 1 hour after intravenous injection (Figure 7A,B). Interestingly, the recognition was significantly abrogated by RMT3-23. To rule out the possibility that binding of RMT3-23 to FcR on DCs might affect phagocytosis of apoptotic cells, we pretreated mice with anti-FcR (2.4G2). As shown in Figure S5C, 2.4G2 did not affect the blocking activity of RMT3-23. Moreover, we prepared F(ab′)2 fragments of RMT3-23, and observed that the F(ab′)2 fragments did not lose the blocking effect (Figure S5), indicating that the blocking effect of RMT3-23 is not mediated through FcRs. These results suggest that Tim-3 is crucial for the recognition of apoptotic cells by CD8+ DCs in vivo.
Tim-3 is crucial for the phagocytosis of apoptotic cells and cross-presentation in vivo. (A) Mice (n = 3 per group) were intravenously injected with the indicated mAb (200 μg each per head), and then 2 hours later with CFSE-labeled apoptotic splenocytes (2 × 107 per head). One hour later, collagenase-digested splenocytes were harvested, and recognition of CFSE-labeled apoptotic cells by splenic CD11c+ cells was analyzed by flow cytometry. Numbers indicate percentage of cells in top right quadrants. (B) Collagenase-digested splenocytes were harvested from mice treated as described in panel A at indicated time points, and recognition of apoptotic cells by splenic CD8+ DCs was analyzed by flow cytometry. Percentage recognition of CFSE-labeled apoptotic cells by CD8+ DCs (percentage CFSE+CD8+CD11c+ cells/percentage CD8+CD11c+ cells × 100) was calculated. Columns represent mean ± SD of triplicates (*P < .05; **P < .01 compared with rIgG). Similar results were obtained in 3 independent experiments. (C) CFSE-labeled OT-I CD8+ T cells (2 × 106 per head) were intravenously transferred into B6 mice (n = 3 per group). The next day, mice were intravenously injected with the indicated mAb (200 μg each per head), and then 2 hours later primed with OVA-loaded apoptotic cells (107 per head). Two days later, whole splenocytes were harvested, and CFSE intensity of CD8+Vα2+ OT-I cells was analyzed by flow cytometry. Percentage of undivided cells in total OT-I cells (D0 per OT-I in C) was calculated, and mean ± SD of triplicates was shown in panel D (**P < .01 compared with rIgG). Similar results were obtained in 3 independent experiments.
Tim-3 is crucial for the phagocytosis of apoptotic cells and cross-presentation in vivo. (A) Mice (n = 3 per group) were intravenously injected with the indicated mAb (200 μg each per head), and then 2 hours later with CFSE-labeled apoptotic splenocytes (2 × 107 per head). One hour later, collagenase-digested splenocytes were harvested, and recognition of CFSE-labeled apoptotic cells by splenic CD11c+ cells was analyzed by flow cytometry. Numbers indicate percentage of cells in top right quadrants. (B) Collagenase-digested splenocytes were harvested from mice treated as described in panel A at indicated time points, and recognition of apoptotic cells by splenic CD8+ DCs was analyzed by flow cytometry. Percentage recognition of CFSE-labeled apoptotic cells by CD8+ DCs (percentage CFSE+CD8+CD11c+ cells/percentage CD8+CD11c+ cells × 100) was calculated. Columns represent mean ± SD of triplicates (*P < .05; **P < .01 compared with rIgG). Similar results were obtained in 3 independent experiments. (C) CFSE-labeled OT-I CD8+ T cells (2 × 106 per head) were intravenously transferred into B6 mice (n = 3 per group). The next day, mice were intravenously injected with the indicated mAb (200 μg each per head), and then 2 hours later primed with OVA-loaded apoptotic cells (107 per head). Two days later, whole splenocytes were harvested, and CFSE intensity of CD8+Vα2+ OT-I cells was analyzed by flow cytometry. Percentage of undivided cells in total OT-I cells (D0 per OT-I in C) was calculated, and mean ± SD of triplicates was shown in panel D (**P < .01 compared with rIgG). Similar results were obtained in 3 independent experiments.
To study cross-presentation of apoptotic cell–associated antigens in vivo, we transferred CFSE-labeled OT-I T cells into C57BL/6 mice, and then 1 day later, we primed these mice with OVA-loaded dying splenocytes. Two days after the priming, we observed that OT-I cells proliferated vigorously in spleen (Figure 7C,D), although OT-I cells did not proliferate in unprimed mouse spleen, indicating that dying cell-associated OVA antigens were taken up by splenic CD8+ DCs and cross-presented to OT-I cells. Consistent with a crucial role of Tim-3 for the uptake of apoptotic cells by CD8+ DCs (Figure 7A,B), the proliferation of OT-I cells was also significantly reduced by RMT3-23, but not RMT4-54 (Figure 7C,D). These results suggest that Tim-3 plays a crucial role in phagocytosis of apoptotic cells and subsequent cross-presentation by CD8+ DCs in vivo.
Discussion
Phagocytes such as macrophages and DCs efficiently recognize and engulf apoptotic cells to maintain the peripheral tolerance. In this study, we demonstrate that Tim-3 mediates phagocytosis of apoptotic cells by PEMs and CD8+ DCs, and that disabling of the Tim-3 function in vivo induces autoantibody production. Likewise, several studies have shown that impairment of the clearance of apoptotic cells causes autoantibody production,5,8,34 however, the mechanism for the elicited immune response to dying cells remains to be elucidated. As the possible explanation, delay or impairment of clearance of apoptotic cells by macrophages causes secondary necrotic cells releasing intracellular contents, which could be endogenous “danger signal” activating immune system.1 However, chronic administration of apoptotic thymocytes to syngeneic mice induces more remarkable level of autoantibody production than that of the same dose of nonapoptotic cell lysate,36 suggesting that autoantibody production in response to dying cells could not be explained simply by the exposure of the intracellular contents. It has also been reported that, in mice, CD8+ DC subset is unique in its ability to recognize apoptotic cells and cross-present dying cell-associated antigens.15,16 It is considered that, in steady state, cross-presentation of dying cell-associated self-antigens by CD8+ DCs induces autoreactive CD8+ T-cell proliferation abortively, subsequently resulting in their deletion, which is important to maintain peripheral tolerance.10,13,14 Taken together, Tim-3–mediated phagocytosis of apoptotic cells by CD8+ DCs may be linked to peripheral tolerance, and a loss of Tim-3 function in DCs may exacerbate autoimmunity. Consistent with this hypothesis, we and others have reported that Tim-3 negatively regulates Th1-mediated inflammatory diseases such as EAE, type I diabetes, and aGVHD, and promotes tolerance induction.21-23
Alternatively, however, upon being activated by anti-CD40 mAb, endogenous danger signals such as heat shock proteins and uric acid, or pathogen-associated molecular patterns (PAMPs) such as LPS and CpG, DCs turn to cross-prime CD8+ T cells to generate effector cells.10,12 Given that Tim-3 is crucial for IFN-γ production in cross-presentation and that Tim-3 is expressed on sterile inflammatory macrophages, Tim-3 may promote induction of effector CD8+ T-cell proliferation and functional memory under pathological conditions. Indeed, Anderson et al have recently reported that Tim-3 expressed on DCs exacerbates a Th1-type autoimmune disease EAE.25 Although Tim-3 has been reported to have multiple functions,21-23 it would be important to further address whether this novel function of Tim-3 is linked to immune tolerance or activation under pathological conditions.
It remains unclear why CD8− DCs are not able to engulf apoptotic cells efficiently, although this DC subset expresses Tim-3. One possibility is that the phagocytic activity may be determined by relative expression level of receptors for “eat-me” signals and “don't eat-me” signals. Although we observed that expression level of Tim-3 on CD8+ DCs is approximately 3-fold higher than that on CD8− DCs, it has been reported that signal-regulatory protein (SIRP)α, a receptor for “don't eat-me” signal,37 is much more highly expressed on CD8− DCs than CD8+ DCs.38 Taken together, a high expression of Tim-3 may be required for the phagocytosis by DCs, and/or SIRPα may neutralize Tim-3 function on CD8− DCs. Moreover, phagocytic activity of each cell type may be determined not only by expression level of the phagocytic receptor, but also cell-intrinsic phagocytic machinery such as cytoskeletal architecture. Indeed, Tim-3 as well as Tim-1 is expressed on activated T cells,24,39 but these T cells are not able to recognize apoptotic cells.
In this study, ectopic expression of Tim-3 enabled HEK293T cells to engulf whole apoptotic cells. However, Miyanishi et al could not observe the phagocytic activity of Tim-3 expressed on NIH3T3 cells based on their engulfment assay, which quantified nuclear DNA degradation of apoptotic cells engulfed by NIH3T3 expressing Tim-3 and DNase II.8 To address this discrepancy, we also generated NIH3T3 cells expressing Tim-3, and did not observe the ability of Tim-3 to mediate engulfment of whole apoptotic cells, although we did observe the ability of Tim-3 to incorporate CFSE-labeled apoptotic cell debris (Figure S6). The reason why Tim-3 expressed on NIH3T3 cells is not able to efficiently phagocytose apoptotic cells remains to be elucidated. We cannot rule out the possibility that Tim-3 may recognize apoptotic cells in cooperation with some coreceptor, which may be expressed on HEK293T cells, macrophages, and CD8+ DCs, but not NIH3T3 cells or CD8− DCs. Likewise, it has been reported that although gene targeting of MerTK receptor results in remarkable defect of phagocytosis of apoptotic cells by macrophages,5 the receptor requires coexpression of αVβ5 integrin to enable NIH3T3 cells to recognize apoptotic cells.40 Although we did observe PS binding of Tim-3 by solid-phase ELISA, Miyanishi et al failed to observe this by PIP–strip binding assay.8 This discrepancy is probably due to a difference in assay sensitivity. Given that the affinity of Tim-3 to PS is much lower than that of Tim-4, Tim-3 might bind not only PS but also some other ligand to phagocytose apoptotic cells. Further studies are needed to elucidate the complexity of phagocytosis.
We show here a crucial role of Tim-3 for clearance of apoptotic cells in vivo and differential expression profile of phagocytic Tim molecules, suggesting that the pathophysiological roles of each Tim molecule appear to be different. Because Tim-1 is expressed on epithelial cells but not professional phagocytes, Tim-1 may contribute to remodeling of injured epithelia.9 Tim-4 is highly expressed on PRMs, and contributes to the phagocytosis of apoptotic cells during physiological tissue turnover.8 Our findings highlight Tim-3 as the phagocytic receptor responsible for cross-presentation by CD8+ DCs. This novel function of Tim-3 opens the door to new therapeutic approaches to combat infections, cancers, and autoimmune diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs William R. Heath for OT-I mice, Sachiko Hirose for aged (NZB × NZW) F1 mice serum, and Toshio Kitamura for pMXs-IRES-puro vector. We also thank Dr Tamami Sakanishi for cell sorting.
This work was supported by the Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (Tokyo, Japan), and by a grant from “High-Tech Research Center” Project for Private Universities: matching fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology (Tokyo, Japan).
Authorship
Contribution: M.N. designed and performed experiments; H.A. provided vital reagents; H.A., Y.K., and M.H. discussed experimental strategy and performed experiments; K.T. designed and discussed experimental strategy; M.A., H.Y., and K.O. supervised experiments and discussed the experimental strategy; M.N. wrote the paper; and K.T. and H.Y. edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masafumi Nakayama, Department of Immunology, Juntendo University School of Medicine, 2-1-1 Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; e-mail: nakayama@juntendo.ac.jp.

![Figure 6. Tim-3–mediated phagocytosis of apoptotic cells is crucial for the cross-presentation by CD8+ DCs. (A) Purified CD8−CD11c+ or CD8+CD11c+ splenic DCs were pretreated with rIgG (white histograms), RMT3-23 (black histograms), or RMT4-54 (gray histograms) and then cultured with OVA-loaded apoptotic cells. After 2 hours, both DC subsets were purified again to remove dead cells, and then cocultured with OT-I CD8+ T cells at the indicated ratio. For the direct presentation, both DC subsets preincubated with OVA257-264 peptide (1 nM) in the presence of rIgG, RMT3-23, or RMT4-54 were cocultured with OT-I CD8+ T cells at a 1:10 (DC/T) ratio. [3H]thymidine (3H-TdR) uptake was measured at 48 to 60 hours. (B) Production of IFN-γ in the culture supernatant at 48 hours after addition of OT-I CD8+ T cells was measured by ELISA. Columns represent mean ± SD of triplicates (**P < .01 compared with rIgG). ND indicates not detectable. Similar results were obtained in 3 independent experiments (A,B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/16/10.1182_blood-2008-10-185884/7/m_zh80190934740006.jpeg?Expires=1768524935&Signature=2UyBmwbKPtBTuIRutuLpGIaxlDCu1yzBPZ6ZsZvgtQpZyqc0BRgzhCFECZSrkEbfkHn7zC4DJhOKN4XP35hdQMx6lYlKVQcPe2k5ikyyVhAz~ncZD0ZU2dwnbzDCtbiTWwCJDeFKu4L~2rgZc6mF~fp2~tkUedD4qcY63c9Wigp~PmolUYxj3C1EVtjyH7W1iS1MrNVxdvwOEDa8bCqjnM5wrq0GKj-oz2ujE1jdnh6ZCbnOJmJ4~Y9-CZjioQN78eU0SJi~w783eGFbckCfNrsbVzxjmL2q7i7AqRvgOZTNJNbx9mX7pQQOanXjhXVYC-2j9Be06FfhvL3S1SmWQw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal