Abstract
Proteolytic resistance of Notch prior to ligand binding depends on the structural integrity of a negative regulatory region (NRR) of the receptor that immediately precedes the transmembrane segment. The NRR includes the 3 Lin12/Notch repeats and the juxtamembrane heterodimerization domain, the region of Notch1 most frequently mutated in T-cell acute lymphoblastic leukemia lymphoma (T-ALL). Here, we report the x-ray structure of the Notch1 NRR in its autoinhibited conformation. A key feature of the Notch1 structure that maintains its closed conformation is a conserved hydrophobic plug that sterically occludes the metalloprotease cleavage site. Crystal packing interactions involving a highly conserved, exposed face on the third Lin12/Notch repeat suggest that this site may normally be engaged in intermolecular or intramolecular protein-protein interactions. The majority of known T-ALL–associated point mutations map to residues in the hydrophobic interior of the Notch1 NRR. A novel mutation (H1545P), which alters a residue at the crystal-packing interface, leads to ligand-independent increases in signaling in reporter gene assays despite only mild destabilization of the NRR, suggesting that it releases the autoinhibitory clamp on the heterodimerization domain imposed by the Lin12/Notch repeats. The Notch1 NRR structure should facilitate a search for antibodies or compounds that stabilize the autoinhibited conformation.
Introduction
Notch proteins are transmembrane receptors that transmit signals in response to transmembrane ligands expressed on adjacent cells (see Bray for a recent review1 ). Signals transduced by Notch receptors influence cell fate decisions during development and also contribute to tissue homeostasis in the mature organism.
Mammalian Notch receptors are processed by a furinlike protease at an external site (S1) while en route to the cell surface, yielding a mature heterodimer composed of 2 noncovalently associated subunits.2,3 The receptor is normally held in a resting, protease-resistant conformation by a negative regulatory region (NRR) that contains 3 Lin12/Notch repeats and a heterodimerization domain that flanks the S1 cleavage site4,5 (Figure 1). Canonical Notch signaling is normally initiated when a ligand of the Delta/Serrate/Lag-2 family binds to the receptor6 and induces several additional proteolytic cleavages. The first of these cleavages occurs within the C-terminal portion of the heterodimerization domain at site 2 (S2), and is catalyzed by ADAM-type metalloproteases such as TACE.7,8 This creates a short-lived transmembrane intermediate variously termed NEXT or NTM*, which is rapidly cleaved within the membrane by γ-secretase.9-13 γ-Secretase cleavage releases the intracellular portion of Notch (ICN) from the membrane, allowing it to be transported to the nucleus, where it enters into a nuclear complex that participates in the induction of target gene transcription.1,14-16
Evidence that aberrant Notch signaling is associated with T-cell acute lymphoblastic leukemia lymphoma (T-ALL) first emerged when the human Notch1 gene was cloned from the breakpoint of a t(7;9) chromosomal translocation found in a minor subset of T-ALLs.17 These rare translocations result in the production of ICN-like polypeptides that result in constitutive and unregulated Notch signaling. More recently, point mutations and small insertions or deletions in Notch1 were found in more than half of human T-ALLs by our group18 and others.19-24 Notch1 mutations also occur in many different murine T-ALL models, making Notch1 perhaps the most frequently mutated gene in this type of leukemia.25
Notch1 mutations associated with human T-ALL cluster in 2 general regions of the protein. One cluster lies at the C-terminal end of the receptor, and consists of nonsense or frameshift mutations that result in the deletion of a PEST domain that regulates ICN1 degradation.26 It appears that these mutations increase Notch activity by stabilizing ICN1.
The second cluster of mutations maps to the heterodimerization domain of the NRR and the region at the boundary between the extracellular and transmembrane regions of the protein. This group includes the most common Notch1 mutations found in human T-ALL.18-22,24 Mutations in this region cause ligand-independent Notch1 signaling, and fall into at least 2 mechanistic classes.27 Class I mutations are single amino acid substitutions or short insertions or deletions that cause increased sensitivity of Notch1 heterodimers to subunit dissociation under either native or mildly denaturing conditions. Class II mutations consist of insertions of at least 12 residues near the C-terminal end of the heterodimerization domain that duplicate the S2 cleavage site; these produce ligand-independent S2 cleavage and strong increases in signaling without any detectable destabilization of the heterodimeric receptor.27 A third group of mutations in this region, recently reported to be present in the Jurkat cell line and certain primary T-ALLs,23 create insertions at the boundary between the extracellular and transmembrane regions of the protein; these resemble class II mutations in terms of their structural consequences, but may activate signaling through a novel mechanism.
We recently reported the structure of the NRR of human Notch2, which provided insight into the mechanism by which Notch receptors are normally held in the “off-state” prior to ligand-mediated activation.28 To determine the molecular basis underlying the ligand-independent signaling accompanying T-ALL mutations in the NRR, examine whether the structural basis for Notch autoinhibition is shared by different mammalian Notch receptors, and provide a structural template for the development of agents designed to stabilize the autoinhibited conformation of the receptor, we solved the structure of the Notch1 NRR by X-ray crystallography. We report here the structure of the Notch1 NRR determined to 2.0-Å resolution, map the sites of tumor-associated mutations onto the structure, and analyze a recently identified novel point mutation found in a T-ALL patient that is located in the third LNR,21 a position distinct and distant from the typical NRR mutations found in the heterodimerization domain. Together, these studies give new insights into the mechanism of pathogenic Notch1 signaling in human T-ALLs.
Methods
Protein expression and purification
The NRR from human Notch1 (GenBank sequence ID AF308602,29 also used to assign residue numbers for the protein) was subcloned into a pET 15b vector containing a hexahistidine tag and a custom TEV site. The crystallized protein has 47 residues removed from the nonconserved, unstructured loop (residues 1623-1669) containing the furin cleavage site, and also contains an additional glycine at the N-terminus resulting from TEV cleavage to release the tag. The protein was produced recombinantly in Rosetta(DE3)pLysS bacteria and recovered from the insoluble fraction after centrifugation using 5 M urea. The protein was affinity purified on a nickel column, eluted with imidazole in 1 M urea, and treated with TEV protease to remove the His tag. The protein was refolded in vitro by dialysis in a redox buffer containing 5 mM cysteine and 1 mM cystine, and purified by anion exchange chromatography after folding was complete (as monitored by reversed-phase high-performance liquid chromatography [HPLC]) to obtain a homogenous species.
Crystallization
The Notch1 NRR crystallized in 100 mM sodium acetate (pH 4.5), 1 to 1.5 M NaCl, and 10% (vol/vol) glycerol. Crystals were cryoprotected in liquid nitrogen after rapid transfer into the same buffer containing 2 M NaCl and glycerol at a final concentration of 35% (vol/vol).
Data collection and scaling
Diffraction data were collected at beamline 24ID at the Advanced Photon Source of Argonne National Laboratory (Argonne, IL). Data were indexed and scaled using the program HKL2000 (HKL Research, Charlottesville, VA).30 Because 2 lattices were observed in the diffraction pattern, the pick peaks feature was used to select the peaks from a single lattice for processing and subsequent analysis.
Phasing
Initial phases were calculated using molecular replacement and the program Phaser (Read group, Univeristy of Cambridge, Cambridge, United Kingdom)31 using the coordinates of the Notch2 NRR as a search model. To construct the working model for refinement, we used the program Coot32 (Paul Emsley, York Structural Biology Laboratory, University of York, York, United Kingdom) to add the residues of Notch1 that were not present in the Notch2 model.
Refinement
CNS33 was used to carry out multiple rounds of simulated annealing. Subsequent refinement and model building were performed iteratively using noncrystallographic symmetry restraints in Refmac34 (Garib Murshudov, York Structural Biology Laboratory) and Coot.32 The NCS restraints were fully released by the end of refinement. The final model contains 289 molecules of solvent (282 water, 7 glycerol) with an overall Rcryst/Rfree of 21.3%/25.2% at 2.0-Å resolution. The percentage of protein residues in core, allowed, generous, and disallowed regions of the Ramachandran plot are 82.9, 17.1, 0.0, and 0.0, respectively.
Analysis
Structure superpositions were performed using Dali-lite35 (European BioInformatics Institute, Cambridge, United Kingdom), Coot,32 and SwissModel (Swiss Institute of Bioinformatics, Basel, Switzerland). Individual residue RMSD values were obtained from SwissModel and manually added in the B-factor column for Figure 2B. The program Pisa36 (European BioInformatics Institute) was used to evaluate crystallographic interfaces, as well as to calculate accessible surface areas. Buried surface area for individual T-ALL mutations was calculated by comparing the accessible surface area of a given residue to the known ASA of the relevant amino acid in the context of a Gly-X-Gly tripeptide.37
Western blots
Luciferase reporter/urea sensitivity assays
Reporter assays were performed essentially as described previously.27 Receptors used to evaluate ligand-independent signaling in Figure 6 (H1545P and other point mutations) and in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) were in the pcDNA3 vector. The reporter plasmid for these assays contained 4 iterated CSL binding sites, and luciferase activity was measured 44 to 48 hours after transfection. Full-length Notch1/Gal4 chimeras (containing the Gal4 DNA-binding domain in place of the RAM and ankyrin repeat domains) in pcDNA5 were used to investigate the effect of mutating the crystal contact interface on LNR-C. The luciferase reporter gene used for these studies contained 4 copies of the GAL4 binding site. In these experiments, U2OS cells transfected with Notch constructs and reporter plasmids were overlaid onto 3T3 cells alone or 3T3 cells stably expressing the Jagged2 ligand 24 hours after transfection, and luciferase activity was measured after an additional 24 hours. The urea sensitivity assays were conducted as described previously.27
Coordinates
Results
Initial crystallization trials using the uncleaved form of the natural Notch1 NRR gave rise to crystals that were fragile and diffracted poorly. To increase the stability of the crystals and improve their diffraction properties, we excised a 47-residue sequence from the poorly conserved loop that contains the S1 cleavage site to create a single-chain mimic of the normal S1-cleaved heterodimer (Figure 1). The resulting protein, hereafter referred to as the Notch1 NRR, yielded crystals in the C2 space group diffracting to 2.0 Å with 2 independent copies of the protein in the asymmetric unit (Table 1). The root-mean-square deviation (RMSD) of the alpha-carbon atoms of the 2 copies is 0.21 Å when they are overlaid, with the largest deviations occurring in the LNR-A module.
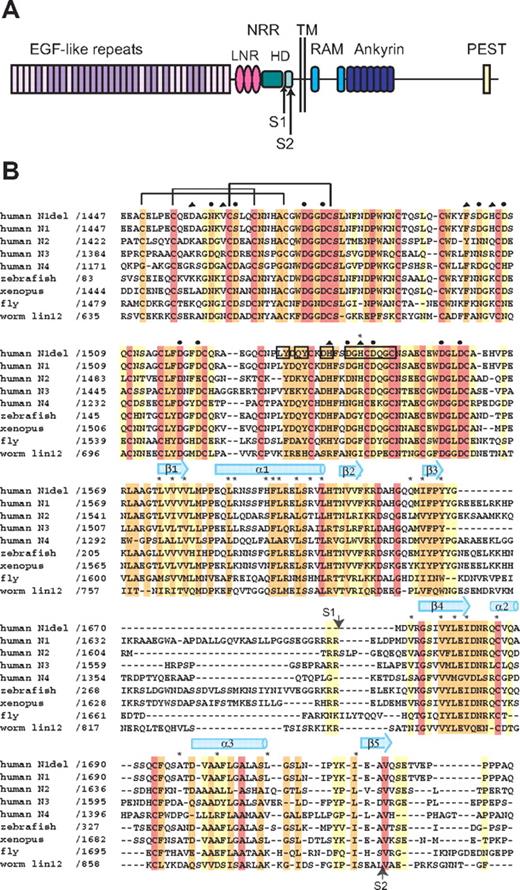
Domain organization and multiple sequence alignment. (A) Domain organization of human Notch1. The NRR consists of the LNR and HD domains. Adapted from Gordon et al.40 (B) Sequence alignment of the NRR region of various Notch receptors, colored according to sequence conservation: red indicates absolutely conserved; orange, highly conserved (defined by ClustalW41 strong conservation groups and/or > 80% sequence identity); yellow, moderately conserved (defined by ClustalW weak conservation groups or > 50% sequence identity); and white, nonconserved. Amino acid residues of special importance are denoted as follows: side-chain and main-chain Ca++-coordinating residues, circles and triangles, respectively; residues mutated in Notch1 in T-ALL, asterisks. Residues from LNR-C engaged in crystal lattice contacts are boxed. Representative disulfide connectivity is shown for LNR-A and HD secondary structural elements are represented by arrows (beta strands) and cylinders (alpha helices).
Domain organization and multiple sequence alignment. (A) Domain organization of human Notch1. The NRR consists of the LNR and HD domains. Adapted from Gordon et al.40 (B) Sequence alignment of the NRR region of various Notch receptors, colored according to sequence conservation: red indicates absolutely conserved; orange, highly conserved (defined by ClustalW41 strong conservation groups and/or > 80% sequence identity); yellow, moderately conserved (defined by ClustalW weak conservation groups or > 50% sequence identity); and white, nonconserved. Amino acid residues of special importance are denoted as follows: side-chain and main-chain Ca++-coordinating residues, circles and triangles, respectively; residues mutated in Notch1 in T-ALL, asterisks. Residues from LNR-C engaged in crystal lattice contacts are boxed. Representative disulfide connectivity is shown for LNR-A and HD secondary structural elements are represented by arrows (beta strands) and cylinders (alpha helices).
Data collection, phasing, and refinement statistics
| . | Human Notch1 NRR . |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c, Å | 169.9, 91.8, 59.8 |
| α, β, γ, ° | 90, 109, 90 |
| Resolution, Å | 50.0-2.0 (2.07-2.0)* |
| Rsym | 4.6 (20.2) |
| I / σI | 23.3 (3.2) |
| Completeness (%) | 93.1 (60.7) |
| Redundancy | 3.5 (2.4) |
| Refinement | |
| Resolution, Å | 30.0-2.0 |
| No. reflections | 52 098 |
| Rwork/Rfree | 21.3/25.2 |
| No. atoms | |
| Protein | 3580 |
| Ligand/ion | 8 |
| Water | 282 |
| Glycerol | 7 |
| Overall B-factors | 43.6 |
| R.m.s deviations | |
| Bond lengths, Å | 0.015 |
| Bond angles, ° | 1.47 |
| . | Human Notch1 NRR . |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c, Å | 169.9, 91.8, 59.8 |
| α, β, γ, ° | 90, 109, 90 |
| Resolution, Å | 50.0-2.0 (2.07-2.0)* |
| Rsym | 4.6 (20.2) |
| I / σI | 23.3 (3.2) |
| Completeness (%) | 93.1 (60.7) |
| Redundancy | 3.5 (2.4) |
| Refinement | |
| Resolution, Å | 30.0-2.0 |
| No. reflections | 52 098 |
| Rwork/Rfree | 21.3/25.2 |
| No. atoms | |
| Protein | 3580 |
| Ligand/ion | 8 |
| Water | 282 |
| Glycerol | 7 |
| Overall B-factors | 43.6 |
| R.m.s deviations | |
| Bond lengths, Å | 0.015 |
| Bond angles, ° | 1.47 |
Highest resolution shell is shown in parentheses.
Structure overview
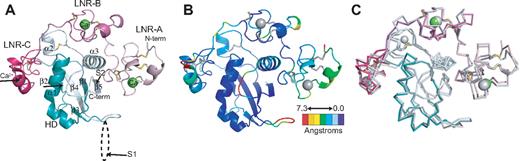
The Notch1 NRR adopts an autoinhibited conformation resembling that of the NRR from human Notch2,28 confirming the overall role of the NRR as the regulatory switch that controls Notch activation. In both the Notch1 and Notch2 structures, the 3 LNR repeats wrap around the compact HD domain to stabilize it and sterically occlude the S2 site (Figure 2). A direct comparison of the Notch1 and Notch2 structures reveals a striking similarity in overall architecture, but with several notable differences in the interactions responsible for maintaining the autoinhibited conformation (Figure 2B,C).
Overall structure of the human Notch1 NRR and comparison with the human Notch2 NRR. (A) Overall structure of the human Notch1 NRR, shown in ribbon representation. Shades of pink and purple represent LNR modules; shades of light and dark cyan, HD domain (on N- and C-terminal sides of the furin cleavage loop, respectively). Disulfide bonds are rendered as yellow sticks, and calcium ions as green spheres. Arrows denote positions of the S1 and S2 cleavage sites. (B) Ribbon representation of the Notch1 NRR colored according to the root mean square deviation (RMSD, in angstroms) between corresponding alpha carbon atoms of the Notch1 and Notch2 structures. Colors are assigned on a sliding scale from blue (RMSD = 0) to red (RMSD = 7.25 Å). Residues present in Notch1 that are absent in Notch2 were set to the maximum RMSD of the range. (C) Overlay of the backbone traces of the Notch1 (colored as in A) and Notch2 (gray) NRRs.
Overall structure of the human Notch1 NRR and comparison with the human Notch2 NRR. (A) Overall structure of the human Notch1 NRR, shown in ribbon representation. Shades of pink and purple represent LNR modules; shades of light and dark cyan, HD domain (on N- and C-terminal sides of the furin cleavage loop, respectively). Disulfide bonds are rendered as yellow sticks, and calcium ions as green spheres. Arrows denote positions of the S1 and S2 cleavage sites. (B) Ribbon representation of the Notch1 NRR colored according to the root mean square deviation (RMSD, in angstroms) between corresponding alpha carbon atoms of the Notch1 and Notch2 structures. Colors are assigned on a sliding scale from blue (RMSD = 0) to red (RMSD = 7.25 Å). Residues present in Notch1 that are absent in Notch2 were set to the maximum RMSD of the range. (C) Overlay of the backbone traces of the Notch1 (colored as in A) and Notch2 (gray) NRRs.
The 3 LNR repeats serve as the cap of the structure, whereas the HD domain constitutes the “stem.” Each LNR module is characterized by 3 disulfide bonds and a calcium coordination site, and has little secondary structure. The HD domain, which immediately follows the LNR repeats in the sequence, folds into an alpha-beta sandwich with a β1-α1-β2-β3-β4(-α2)-α3-β5 topology. The base of the HD fold is a 4-stranded beta sheet, with the helices packed against the “top” face of this sheet to create an extensive hydrophobic core. Several loops, together with a cysteine “knuckle” in the HD domain, connect the secondary structural elements that bear the side chains comprising the hydrophobic core.
Although a blast search for protein sequences homologous to the Notch1 HD yields only other Notch HD domains, a search for homologous structures using the program MSD-fold (http://www.ebi.ac.uk/msd-srv/ssm; European Bioinformatics Institute) reveals that other domains adopt similar folds with the same secondary structure topology. These homologous folds include the SEA domains of mucins,42,43 the RNA recognition motifs (RRMs) of several proteins,44 and a juxtamembrane domain in the receptor protein-tyrosine phosphatase AI-2.45 Remarkably, the composition of the hydrophobic core is conserved among the different groups of proteins that share the HD-domain fold, despite the high level of overall sequence divergence elsewhere (not shown).
Interdomain interactions
In both the Notch1 and Notch2 NRRs, the LNR domain stabilizes the overall fold of the HD domain by making extensive packing interactions with the helices, buttressing them against the central 4-stranded sheet and stabilizing the hydrophobic core. Both proteins also use side chains from the linker connecting the LNR-A and LNR-B modules to form a hydrophobic plug that straddles the scissile bond cleaved by metalloprotease. In the normal receptor, therefore, this hydrophobic plug and its flanking LNR modules must be displaced away from the HD domain to expose the S2 site for metalloprotease cleavage.
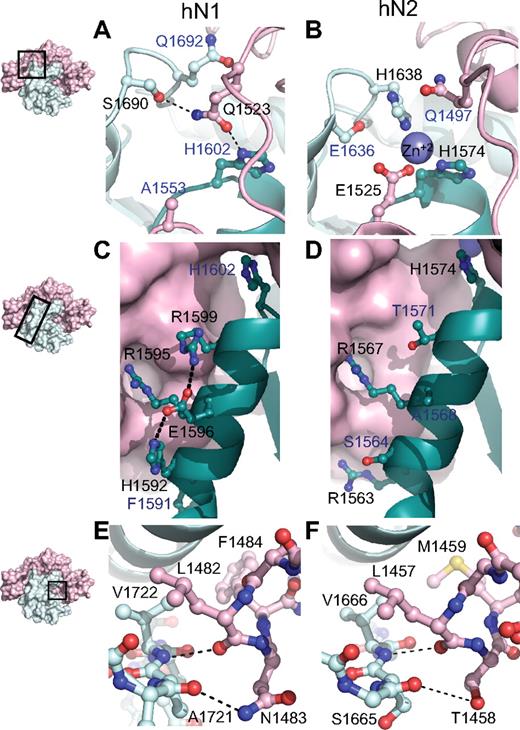
Although the overall architecture and general features of autoinhibition are preserved in both the Notch1 and Notch2 NRR structures, stabilization of the metalloprotease-resistant conformation of each protein is achieved through different packing interactions between the LNR and HD domains. At the most global level, the amount of surface area buried between the LNR and HD domains in the Notch1 NRR is 3300 Å2 compared with 3000 A2 in Notch2. The most striking difference between the 2 interfaces is at the junction between the LNR-C repeat and the HD domain, where 3 different structural elements of the protein converge (Figure 3A,B). The Notch2 NRR harbors a Zn++ coordination site that acts to fix the relative positions of LNR-C, helix 1 of the HD domain, and the cysteine knuckle immediately preceding helix 3 of the HD domain (Figure 3B). In contrast, the Notch1 NRR does not bind Zn++, because only 1 of the 3 Zn++-coordinating side chains is preserved (Figure 3A). Instead, side chains from residues in the LNR B-C linker substitute for the Zn++ ion of Notch2 by forming packing interactions and hydrogen bonds with the cysteine knuckle and the C-terminal end of helix 1. These interactions include contacts between the side chain of Q1523 and Q1692, the residue analogous to the Zn++-coordinating H1638 from the cysteine knuckle of Notch2, and a hydrogen bond to the S1690 side chain hydroxyl group of the cysteine knuckle. Thus, an intriguing feature of these 2 proteins is the coalescence of 3 different structural features at this junction, which may serve as a fulcrum for movements that accompany the switch from the autoinhibited to the activation-competent conformation. The differences between the 2 proteins may represent 2 structural solutions for achieving the same end (stabilization of the autoinhibited structure), or alternatively, may serve to generate different levels of sensitivity to activation by ligands.
Structural divergence between Notch1 and Notch2 NRRs. (A,B) Convergence of HD-N, HD-C, and LNR-C is mediated by a hydrogen bond in the Notch1 NRR (A) and a Zn2+ coordination site in the Notch2 NRR (B). (C,D) Comparison of interactions stabilizing helix 1 in the Notch1 and Notch2 NRRs. In Notch1, there is an intrahelical salt bridge and the helix is anchored to LNR-C via a single salt bridge. In Notch2, there are several electrostatic interactions between the helix and LNR-C (D). (E,F) The LNR A-B linker of the Notch1 (E) and Notch2 (F) NRRs masks the metalloprotease cleavage site. In each structure, a 3-residue sequence from the linker occludes the S2 site, even though 2 of the 3 amino acid residues comprising the protective plug are not conserved (see “Interdomain interactions” for details). Black labels identify residues participating in the interactions discussed, whereas blue labels identify residues that form analogous interactions in the other receptor.
Structural divergence between Notch1 and Notch2 NRRs. (A,B) Convergence of HD-N, HD-C, and LNR-C is mediated by a hydrogen bond in the Notch1 NRR (A) and a Zn2+ coordination site in the Notch2 NRR (B). (C,D) Comparison of interactions stabilizing helix 1 in the Notch1 and Notch2 NRRs. In Notch1, there is an intrahelical salt bridge and the helix is anchored to LNR-C via a single salt bridge. In Notch2, there are several electrostatic interactions between the helix and LNR-C (D). (E,F) The LNR A-B linker of the Notch1 (E) and Notch2 (F) NRRs masks the metalloprotease cleavage site. In each structure, a 3-residue sequence from the linker occludes the S2 site, even though 2 of the 3 amino acid residues comprising the protective plug are not conserved (see “Interdomain interactions” for details). Black labels identify residues participating in the interactions discussed, whereas blue labels identify residues that form analogous interactions in the other receptor.
Another difference between the 2 structures is the nature of the packing interactions that determine the interface between the LNR-C module and helix 1 of the HD domain (Figure 3C,D). In Notch2, the interface between helix 1 and LNR-C is primarily established by the Zn++-binding site in conjunction with 2 salt bridges to the LNR-C module, which together anchor helix 1 and position it over the beta sheet to help define the hydrophobic core. In the Notch1 structure, on the other hand, only 1 of these 2 salt bridges remains to fix the helix in position with respect to the LNR-C module. R1595, the residue of the Notch1 helix involved in this interaction (R1567 in Notch2), is highly conserved among different Notch proteins, suggesting that it plays a key role in positioning the helix against LNR-C.
A key set of interactions that maintain the Notch1 and Notch2 NRR regions in an autoinhibited conformation prior to ligand binding is the hydrophobic “plug” that straddles the scissile bond of the S2 site. This plug consists of 3 residues that envelop the amide bond, sterically precluding metalloprotease access to S2. In both Notch1 and Notch2, a leucine residue (L1482 of Notch1) extending from the LNR A-B linker packs tightly against the valine residue (V1722 of Notch1) that lies immediately terminal to the scissile bond. Although the other 2 residues of the plug differ between Notch1 and Notch2, key interactions are nevertheless preserved in both proteins (Figure 3E,F): a hydrogen bond from the main chain carbonyl group of the leucine residue to the main chain amide group protects the scissile bond, another polar interaction between the carbonyl group of the residue preceding the scissile bond (E1720 in Notch1 and V1664 in Notch2) and the side chain of either N1483 in Notch1 or T1458 in Notch2 stabilizes the orientation of the long-range interactions, and a hydrophobic cap provided by F1484 in Notch1 and M1459 in Notch2 is also buried within contact distance of both the valine residue following the S2 cleavage site and an aliphatic side chain from helix 3. Together, these shared structural features, despite the divergence in primary sequence, affirm the importance of the long-range interface between the LNR repeats and the HD domain in enforcing autoinhibition of Notch receptors in the absence of ligand stimulation, and argues strongly that this mechanism is general among different Notch family receptors.
Conserved residues at the Notch1 crystal packing interface
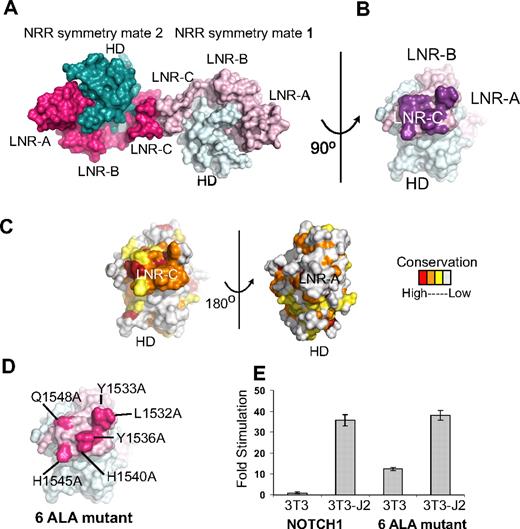
Analysis of the packing of Notch1 NRR molecules in the crystal lattice suggests the possibility that the surface of LNR-C may contain a conserved interface for intramolecular or intermolecular protein-protein interactions (Figure 4A-C). The total interface area at this crystal contact is 998 Å2 and the interface contains a total of 12 hydrogen bonds, suggesting that the surface involved at the contact interface may be biologically significant. Indeed, the surface patch engaged in the lattice contacts is highly conserved among the LNR-C modules of Notch receptors, but is not conserved in the LNR-A or LNR-B modules of Notch receptors, or among LNR repeats in general. The program Pisa, which predicts the potential biologic significance of interfaces based on several criteria including buried solvent accessible surface area and hydrogen bonding interactions, also scores the interface between LNR-C modules in the crystal lattice as “highly significant,” and predictive of an interaction site on the protein surface.
Conserved residues at a Notch1 crystal packing interface. (A) The Notch1 NRR and a symmetry mate are depicted in molecular surface representation. The LNR domains are colored in light and dark pink and the HD domains colored in light and dark cyan in molecules 1 and 2, respectively. (B) Molecule one surface after a counterclockwise 90-degree rotation about the axis shown. In this view, the surface-exposed face of LNR-C is facing outward. Amino acid residues involved in the crystal contact interface are colored purple. (C) Surface of the Notch1 NRR colored according to amino acid conservation using the scale shown (as described in the legend for Figure 1B). (D) Surface of the Notch1 NRR illustrating the 6 crystal contact residues that have been mutated to Ala (hot pink). (E) Effect of mutating the LNR-C crystal contact interface in Notch reporter gene assays. Signaling in coculture assays with NIH 3T3 cells alone or stably expressing the ligand Jagged-2 was examined using chimeric full-length receptors in which the RAM and ANK domains of NOTCH1 were replaced with the DNA-binding domain of the transcription factor GAL4 (“Luciferase reporter/urea sensitivity assays”). Firefly luciferase activity was normalized to the internal Renilla control and expressed relative to the reporter gene activity of the unmutated chimeric receptor cocultured with NIH 3T3 cells alone. Error bars represent the SE of the 3 replicate measurements made for each experimental condition.
Conserved residues at a Notch1 crystal packing interface. (A) The Notch1 NRR and a symmetry mate are depicted in molecular surface representation. The LNR domains are colored in light and dark pink and the HD domains colored in light and dark cyan in molecules 1 and 2, respectively. (B) Molecule one surface after a counterclockwise 90-degree rotation about the axis shown. In this view, the surface-exposed face of LNR-C is facing outward. Amino acid residues involved in the crystal contact interface are colored purple. (C) Surface of the Notch1 NRR colored according to amino acid conservation using the scale shown (as described in the legend for Figure 1B). (D) Surface of the Notch1 NRR illustrating the 6 crystal contact residues that have been mutated to Ala (hot pink). (E) Effect of mutating the LNR-C crystal contact interface in Notch reporter gene assays. Signaling in coculture assays with NIH 3T3 cells alone or stably expressing the ligand Jagged-2 was examined using chimeric full-length receptors in which the RAM and ANK domains of NOTCH1 were replaced with the DNA-binding domain of the transcription factor GAL4 (“Luciferase reporter/urea sensitivity assays”). Firefly luciferase activity was normalized to the internal Renilla control and expressed relative to the reporter gene activity of the unmutated chimeric receptor cocultured with NIH 3T3 cells alone. Error bars represent the SE of the 3 replicate measurements made for each experimental condition.
To examine whether this conserved surface patch on LNR-C might be functionally relevant, we constructed a mutated form of the full-length Notch1 receptor in which 6 of the residues engaged in lattice contacts were substituted with alanine (Figure 4D), and tested the effect of these mutations on signaling in coculture assays. The mutated receptor responds indistinguishably from the normal receptor when cocultured with NIH 3T3 cells stably expressing Jagged-2 (Figure 4E), suggesting that it is fully competent for ligand-induced signaling. However, when cocultured with 3T3 cells alone, the mutated receptor exhibits a significant increase in reporter gene activity over the normal receptor. This observation suggests that the mutated receptor exhibits substantial ligand-independent signaling and is consistent with the idea that the residues engaged in the lattice contacts are functionally important in the normal restraint of signaling, a possibility that can be further explored in future studies. Because functional studies of Notch1 signaling argue against an important role for homodimerization in transmission of signals,46 and Notch2 crystals do not exhibit similar contacts in their crystal lattice, it seems unlikely that the contacts constitute a biologically important homodimerization interface. Instead, the lattice contacts point to the possibility that this site is used for other intramolecular or intermolecular interactions. Interestingly, a novel T-ALL mutation, H1545P,21 maps to this region of LNR-C, and is further investigated in “Functional analysis of the H1545P mutation.”
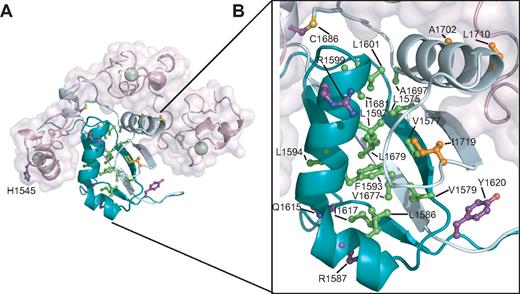
Analysis of T-ALL–associated mutations mapped to the Notch1 NRR
Approximately 40% to 50% of T-ALLs harbor mutations lying within the NRR of Notch1.18-22,24 Mapping the sites of the known T-ALL–associated mutations onto the Notch1 NRR reveals that more than half (13 of 22) of the mutations are located in the conserved hydrophobic core of the HD domain (Figure 5). This hydrophobic core includes residues from the inner faces of the 2 primary alpha helices and the top face of the 4-stranded beta sheet. All of the mutations in the hydrophobic core belong to class I, as defined by destabilization of the NRR under native conditions (class 1A) or in the presence of urea (class 1B). Several of these substitutions introduce structure-breaking proline residues, which readily explains their destabilizing effects. Other less drastic mutations still add or remove steric bulk or introduce charged or polar side chains into the hydrophobic interior of the protein, again providing an explanation for their effects on the stability of the heterodimeric receptor.
Mapping of T-ALL tumor-associated mutations onto the Notch1 NRR. (A) Structural representation highlighting T-ALL mutations. Side chains of residues mutated in T-ALL patients are shown in ball-and-stick form. Residues are colored according to mutation site: core (green), interface (orange), or partially exposed (purple). (B) Close-up view of mutations in the HD domain.
Mapping of T-ALL tumor-associated mutations onto the Notch1 NRR. (A) Structural representation highlighting T-ALL mutations. Side chains of residues mutated in T-ALL patients are shown in ball-and-stick form. Residues are colored according to mutation site: core (green), interface (orange), or partially exposed (purple). (B) Close-up view of mutations in the HD domain.
Although R1599 is not itself a residue in the hydrophobic core, the R1599P mutation strongly destabilizes the protein. It seems likely that the mutation has 2 concurrent effects: (1) it eliminates the stabilizing salt-bridge from the R1599 side-chain to E1595, and (2) it unravels the C-terminal end of the helix, propagating its disruptive effects into the hydrophobic core of the HD with further deleterious effects on the stability of the protein.
Other mutations within the HD domain outside of the hydrophobic core tend to cause less destabilization of the HD domain. Some of these mutations alter residues at the interface with the LNR domain (Figure 5). Three mutations fall into this category: A1702P, L1710P, and I1719T. The reduced stability of proteins with these mutations is consistent with the notion that they disengage the LNR domain from the HD domain, relaxing a clamp that normally stabilizes the HD domain, thereby indirectly destabilizing the HD domain and the heterodimer. A summary of published mutations and their putative structural classification is presented in Table 2.
Summary of Notch1 single point mutations found in T-ALL
| Mutation . | Reference . | Location . | % buried . | Ligand-independent reporter activity . | Urea dissociation propensity . | Putative classification . |
|---|---|---|---|---|---|---|
| L1575P | 18 | β1 | 99 | +++ | Core | |
| L1575Q | 19 | |||||
| V1577E | 19 | β1 | 100 | ++ | +++ | Core |
| V1579G | 19 | β1 | 100 | Core | ||
| L1586P | 18 | α1 | 100 | Core | ||
| L1586R | 19 | |||||
| R1587P | 19 | α1 | 50 | Partially exposed | ||
| F1593S | 18 | α1 | 100 | +++ | +++ | Core |
| F1593C | 19 | |||||
| L1594P | 18 | α1 | 93 | ++ | ++ | Core |
| L1597H | 18,27 | α1 | 100 | +++ | +++ | Core |
| R1599P | 18 | α1 | 45 | +++ | +++ | Partially exposed |
| L1601P | 18 | α1 | 99 | ++++ | Core | |
| L1601Q | 19 | |||||
| Q1615K | 19 | Loop β2, β3 | 74 | Partially exposed | ||
| I1617T | 18,27 | β3 | 95 | + | ++ | Core |
| I1617N | 18 | +++ | ||||
| Y1620N | 19 | Furin loop | 66 | Partially exposed | ||
| V1677D | 18 | β4 | 100 | + | ++++ | Core |
| L1679P | 18 | β4 | 100 | + | ++++ | Core |
| L1679Q | 24 | |||||
| I1681N | 18 | β4 | 98 | ++ | +++ | Core |
| C1686F | 19 | Cysteine-knuckle | 55 | Partially exposed | ||
| A1697D | 22 | α3 | 97 | Core | ||
| A1702P | 18 | α3 | 94 | + | + | Interface |
| L1710P | 20 | α3 | 92 | Interface | ||
| I1719T | 18,27 | Loop α3, β5 | 86 | + | ++ | Interface |
| H1545P | 21 | LNR-C | 17 | ++ | + | Partially exposed |
| Mutation . | Reference . | Location . | % buried . | Ligand-independent reporter activity . | Urea dissociation propensity . | Putative classification . |
|---|---|---|---|---|---|---|
| L1575P | 18 | β1 | 99 | +++ | Core | |
| L1575Q | 19 | |||||
| V1577E | 19 | β1 | 100 | ++ | +++ | Core |
| V1579G | 19 | β1 | 100 | Core | ||
| L1586P | 18 | α1 | 100 | Core | ||
| L1586R | 19 | |||||
| R1587P | 19 | α1 | 50 | Partially exposed | ||
| F1593S | 18 | α1 | 100 | +++ | +++ | Core |
| F1593C | 19 | |||||
| L1594P | 18 | α1 | 93 | ++ | ++ | Core |
| L1597H | 18,27 | α1 | 100 | +++ | +++ | Core |
| R1599P | 18 | α1 | 45 | +++ | +++ | Partially exposed |
| L1601P | 18 | α1 | 99 | ++++ | Core | |
| L1601Q | 19 | |||||
| Q1615K | 19 | Loop β2, β3 | 74 | Partially exposed | ||
| I1617T | 18,27 | β3 | 95 | + | ++ | Core |
| I1617N | 18 | +++ | ||||
| Y1620N | 19 | Furin loop | 66 | Partially exposed | ||
| V1677D | 18 | β4 | 100 | + | ++++ | Core |
| L1679P | 18 | β4 | 100 | + | ++++ | Core |
| L1679Q | 24 | |||||
| I1681N | 18 | β4 | 98 | ++ | +++ | Core |
| C1686F | 19 | Cysteine-knuckle | 55 | Partially exposed | ||
| A1697D | 22 | α3 | 97 | Core | ||
| A1702P | 18 | α3 | 94 | + | + | Interface |
| L1710P | 20 | α3 | 92 | Interface | ||
| I1719T | 18,27 | Loop α3, β5 | 86 | + | ++ | Interface |
| H1545P | 21 | LNR-C | 17 | ++ | + | Partially exposed |
+ represents relative strength of measured effect as reported in listed references.
Functional analysis of the H1545P mutation
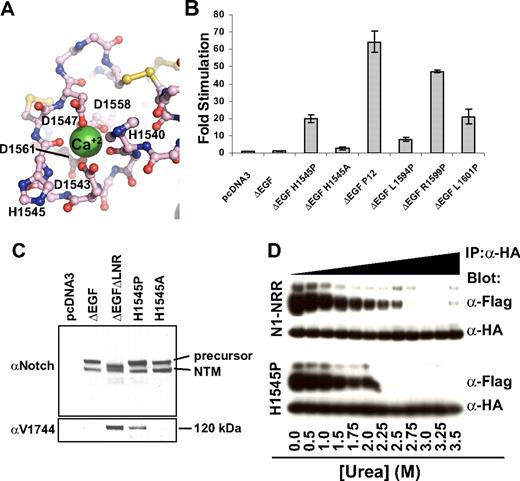
The H1545P substitution represents the first T-ALL–associated mutation of the NRR that lies outside of the HD domain.21 H1545P is located in a loop of LNR-C, and its solvent-exposed imidazole side chain, as previously noted, is located within the conserved crystal contact interface. Its backbone carbonyl oxygen also is one of the calcium-coordinating ligands within the LNR-C repeat (Figure 6A). Thus, a more complete mechanistic characterization of this mutation is of considerable interest.
Characterization of the H1545P mutation in LNR-C. (A) Structure of the region surrounding H1545. Calcium coordinating residues are shown in ball-and-stick representation and are colored by atom type (gray indicates carbon; red: oxygen; and blue: nitrogen). Disulfide bonds are yellow, and the calcium ion is a green sphere. (B) Reporter gene assay for ligand-independent Notch1 activation. H1545P and several other well-characterized T-ALL–associated mutations were tested for their ability to induce reporter gene transcription in the context of a Notch1 polypeptide lacking the EGF-like ligand binding repeats (ΔEGF). The ΔEGF forms of the receptors were transiently transfected into U2OS cells together with a plasmid encoding a luciferase reporter gene under control of 4 iterated CSL binding sites, and an internal control plasmid expressing Renilla luciferase. Firefly luciferase activities were normalized to the internal Renilla control and expressed relative to the activity produced by the unmutated ΔEGF form of the receptor, which was assigned a relative value of 1. Error bars represent the SE of the 3 replicate measurements made for each experimental condition. (C) The H1545P mutation confers ligand-independent proteolytic sensitivity. U2OS cells were transfected with plasmids encoding the indicated Notch1 receptor variants. Western blots with antibodies specific for the intracellular portion of human Notch1 (top panel) and for the S3-cleaved product (bottom panel) are shown. (D) Effect of the H1545P mutation on sensitivity to urea-induced subunit dissociation. Conditioned media from HEK 293T cells expressing epitope-tagged Notch1 NRR minireceptors were immunoprecipitated with α-HA coupled beads followed by incubation in buffer containing different concentrations of urea for 30 minutes (0 to 3.5 M). Subunit dissociation was evaluated by SDS–polyacrylamide gel electrophoresis (PAGE) followed by Western blot analysis. The N-terminal and C-terminal subunits were detected with α-FLAG and α-HA antibodies, respectively.
Characterization of the H1545P mutation in LNR-C. (A) Structure of the region surrounding H1545. Calcium coordinating residues are shown in ball-and-stick representation and are colored by atom type (gray indicates carbon; red: oxygen; and blue: nitrogen). Disulfide bonds are yellow, and the calcium ion is a green sphere. (B) Reporter gene assay for ligand-independent Notch1 activation. H1545P and several other well-characterized T-ALL–associated mutations were tested for their ability to induce reporter gene transcription in the context of a Notch1 polypeptide lacking the EGF-like ligand binding repeats (ΔEGF). The ΔEGF forms of the receptors were transiently transfected into U2OS cells together with a plasmid encoding a luciferase reporter gene under control of 4 iterated CSL binding sites, and an internal control plasmid expressing Renilla luciferase. Firefly luciferase activities were normalized to the internal Renilla control and expressed relative to the activity produced by the unmutated ΔEGF form of the receptor, which was assigned a relative value of 1. Error bars represent the SE of the 3 replicate measurements made for each experimental condition. (C) The H1545P mutation confers ligand-independent proteolytic sensitivity. U2OS cells were transfected with plasmids encoding the indicated Notch1 receptor variants. Western blots with antibodies specific for the intracellular portion of human Notch1 (top panel) and for the S3-cleaved product (bottom panel) are shown. (D) Effect of the H1545P mutation on sensitivity to urea-induced subunit dissociation. Conditioned media from HEK 293T cells expressing epitope-tagged Notch1 NRR minireceptors were immunoprecipitated with α-HA coupled beads followed by incubation in buffer containing different concentrations of urea for 30 minutes (0 to 3.5 M). Subunit dissociation was evaluated by SDS–polyacrylamide gel electrophoresis (PAGE) followed by Western blot analysis. The N-terminal and C-terminal subunits were detected with α-FLAG and α-HA antibodies, respectively.
To determine whether the H1545P mutation leads to increases in ligand-independent Notch signaling, we examined the effect of the mutation on reporter gene transcription in the context of ΔEGF, a receptor that lacks the ligand-binding EGF repeats, and compared it with other well-characterized T-ALL–associated mutations (Figure 6B). In this assay, the mutation results in a 20-fold increase in ligand-independent reporter gene transcription, a level intermediate between the weak activating mutation L1594P, and the more potent “P12” insertion, which is a strong ligand-independent activator in these assays. Unlike the H1545P mutation, the more conservative H1545A mutation does not result in substantial induction of reporter gene transcription. Western blots performed on lysates from cells transfected with H1545P confirm that the observed transcriptional induction relies on canonical Notch signaling, as blots for the activated form of Notch1 detect the proteolytic product of H1545P and a control receptor that undergoes constitutive proteolysis, but do not detect the activated form for either the inactive ΔEGF receptor or the H1545A control protein (Figure 6C).
Typical class I mutations of the HD domain lead to reduced stability of the Notch1 NRR, as judged by increased sensitivity to urea-induced subunit dissociation. The assay design relies on the construction of minireceptors with N-terminal FLAG and C-terminal HA tags for monitoring the dissociation reaction, which is induced with increasing concentrations of urea.27 To test whether the H1545P mutation reduces the stability of the Notch1 NRR, we compared the sensitivity of the normal Notch1 NRR with the H1545P mutant form of the NRR in this well-established urea-induced dissociation assay. Remarkably, the H1545P mutation results in only a mild increase in sensitivity toward urea-induced dissociation (Figure 6D). This finding is consistent with the anticipated effect of a mutation that is not in the hydrophobic core, and suggests that the observed ligand-independent signaling that results from the mutation may be an indirect consequence of increased disengagement of the LNR clamp from the HD domain to expose its S2 site, rather than direct destabilization of the HD domain itself.
One explanation for how the H1545P mutation results in disengagement of the LNR clamp is that the proline substitution directly or indirectly disrupts the calcium-binding site in LNR-C, interfering with the integrity of LNR-C and disengaging it from the HD domain. To investigate whether loss of calcium binding by LNR-C might result in ligand-independent signaling (and thus mimic the effect of the H1545P mutation), we constructed the D1561A mutation in LNR-C. This mutation removes a side-chain carboxylate group that participates in calcium coordination for LNR-C, and is likely to entirely disrupt calcium binding by this domain. Then, we tested the effect of this mutation on ligand-independent signaling in a reporter assay using a ΔEGF form of the receptor. The data show that the D1561A mutation in LNR-C leads to a ligand-independent increase in signaling activity (Figure S1), supporting the notion that disruption of the calcium-binding site can cause enough of a conformational disruption to allow inappropriate access of metalloproteases to the S2 cleavage site.
Discussion
Here we have presented the structure of the Notch1 NRR, compared it with the structure of the Notch2 NRR, and examined the effects of a novel T-ALL–associated mutation in LNR-C on receptor stability and signaling properties.
Conformational events associated with normal and pathogenic Notch activation
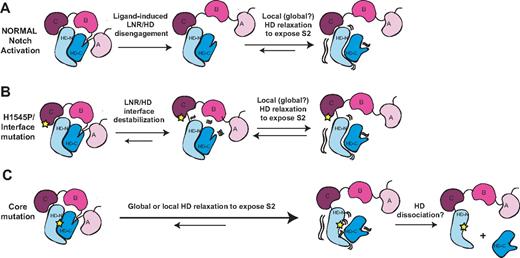
The structures of the Notch NRRs and a wealth of published biochemical and functional data suggest a model for normal signaling in which ligand binding promotes a series of conformational movements that expose the S2 site (Figure 7). Regardless of whether mechanical force28,47,48 or allosteric events produce this conformational movement, the steps leading to S2 exposure must first include the displacement of at least the first 2 LNR modules (the autoinhibitory “clamp”) away from the HD domain.28 To release the autoinhibitory clamp, ligand activation must remove the hydrophobic “plug” that directly occludes the metalloprotease site and undo the “latch” that holds the LNRs on top of helices 1 and 3 of the HD domain. However, mere peeling of the LNR modules away from the HD domain is unlikely to suffice for exposure of the S2 cleavage site, because the active site of metalloproteases such as TACE is buried in a deep cleft,49 and the strand housing the S2 site still remains in a compact conformation when retained within a fully folded HD domain. Thus, we believe that either local or global relaxation of the HD domain will be required to fully expose the S2 site during activation, an idea in accordance with studies that favor a requirement for complete dissociation of the heterodimeric Notch1 receptor prior to activation.47
Models for normal Notch activation and aberrant activation by T-ALL mutations. (A) Ligand-mediated Notch activation. Ligand binding triggers a conformational movement that first disengages the LNR and HD domains. Without the stabilizing interactions provided by the LNR domain, the HD domain then relaxes locally or globally to expose the S2 site. (B) H1545P and other interface mutations promote disengagement of the LNR/HD interface. This step then allows local or global HD relaxation to expose the S2 site in a manner analogous to ligand binding. (C) Core mutations directly destabilize the HD domain, precluding stable interaction with the LNR domain and promoting exposure of S2. Extremely destabilizing mutations such as those of class 1A may lead to complete HD dissociation.
Models for normal Notch activation and aberrant activation by T-ALL mutations. (A) Ligand-mediated Notch activation. Ligand binding triggers a conformational movement that first disengages the LNR and HD domains. Without the stabilizing interactions provided by the LNR domain, the HD domain then relaxes locally or globally to expose the S2 site. (B) H1545P and other interface mutations promote disengagement of the LNR/HD interface. This step then allows local or global HD relaxation to expose the S2 site in a manner analogous to ligand binding. (C) Core mutations directly destabilize the HD domain, precluding stable interaction with the LNR domain and promoting exposure of S2. Extremely destabilizing mutations such as those of class 1A may lead to complete HD dissociation.
Mechanism of activation by T-ALL mutations
In the pathogenic signaling associated with T-ALL, the normal autoinhibitory constraints are bypassed as a result of mutations in the Notch1 NRR (Figure 7B,C). Mutations in the hydrophobic core, which comprise all class IA and several class IB mutations, cause substantial destabilization of heterodimeric NRR molecules.27 In these mutated proteins, the ensuing local or global unraveling of the HD disfavors the clamped, autoinhibited conformation, precludes stable intramolecular binding of the LNR clamp, and directly exposes the S2 site. Other more subtle class IB mutations located outside of the hydrophobic core may increase the rate of access to an open, proteolytically sensitive conformation despite a modest effect on the global stability of the NRR.
The new H1545P mutation of LNR-C characterized here also causes ligand-independent Notch1 signaling. As with the more conservative class IB mutations, the overall effect of the mutation on the stability of the NRR is modest, with a small shift in the urea-induced dissociation curve. The most straightforward explanation for this observation is that the mutation acts to disable the calcium-binding site of LNR-C and destabilize the LNR/HD interface, loosening or releasing the autoinhibitory LNR domain clamp that normally protects the S2 site, effectively mimicking deletion of the LNR domain.5 In essence, the mutation bypasses the first step of the normal Notch activation mechanism (Figure 7A) by aberrantly disengaging the LNR from the HD domain without a requirement for ligand. The local or global relaxation of the HD inferred to occur during normal ligand-induced activation would then ensue to expose the S2 site.
Utility of the Notch1 NRR in a search for compounds to inhibit Notch1 signaling in T-ALL
Modulatory antibodies that can either activate or inhibit Notch3 signaling have recently been reported. The inhibitory antibodies are highly potent, selective for Notch3, and block signaling from either Jagged or Delta-like ligands.50 Similar antibodies or inhibitors targeting the NRR of Notch1 might find utility as targeted therapeutics in the management of T-ALL. For class I mutations that destabilize the NRR without causing total unfolding, antibodies or stabilizing small-molecular-weight compounds would shift the conformational preference of the mutated receptor back toward the autoinhibited conformation, and might prevent ligand-independent signaling. Inhibitors targeting the autoinhibited conformation of the Notch1 NRR would also directly attenuate the enhanced signaling by T-ALLs associated with PEST deletions. The structure of the Notch1 NRR can also serve as a valuable starting point for a computational search to identify compounds that directly stabilize the native conformation of the HD domain or that mimic the LNR domain in occluding the S2 site to confer resistance to proteolysis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (NIH, Bethesda, MD; P01 CA119070 [J.C.A. and S.C.B.] and R01 CA092433 [S.C.B.]), and by a SCOR award from the Leukemia & Lymphoma Society (Mamoroneck, NY; J.C.A. and S.C.B.). W.R.G. is a Leukemia & Lymphoma Society Special Fellow.
National Institutes of Health
Authorship
Contribution: W.R.G., D.V.-U., J.C.A., and S.C.B. designed research; W.R.G., M.R., D.V.-U. and M.G. performed research and collected data; M.R.M. contributed vital new reagents; all authors analyzed and interpreted data; and W.R.G. and S.C.B. wrote the paper with critical input from J.C.A. and M.R.M.
Conflict-of-interest disclosure: S.C.B. and J.C.A. have received funding previously from Merck Research Laboratories (West Point, PA) for a related project, and have filed patent applications related to the structure of the Notch1-negative regulatory region.
Correspondence: Stephen C. Blacklow, 77 Ave Louis Pasteur, Rm 652E, Boston, MA 02115; e-mail: sblacklow@rics.bwh.harvard.edu.