Abstract
Peripheral tolerance induction is critical for the maintenance of self-tolerance and can be mediated by immunoregulatory T cells or by direct induction of T-cell anergy or deletion. Although the molecular processes underlying anergy have been extensively studied, little is known about the molecular basis for peripheral T-cell deletion. Here, we determined the gene expression signature of peripheral CD8+ T cells undergoing deletional tolerance, relative to those undergoing immunogenic priming or lymphopenia-induced proliferation. From these data, we report the first detailed molecular signature of cells undergoing deletion. Consistent with defective cytolysis, these cells exhibited deficiencies in granzyme up-regulation. Furthermore, they showed antigen-driven Bcl-2 down-regulation and early up-regulation of the proapoptotic protein Bim, consistent with the requirement of this BH3-only protein for peripheral T-cell deletion. Bim up-regulation was paralleled by defective interleukin-7 receptor α (IL-7Rα) chain reexpression, suggesting that Bim-dependent death may be triggered by loss of IL-7/IL-7R signaling. Finally, we observed parallels in molecular signatures between deletion and anergy, suggesting that these tolerance pathways may not be as molecularly distinct as previously surmised.
Introduction
Antigen encounter alone is not sufficient to provoke priming of CD8+ T cells. Instead, for productive cytotoxic T lymphocyte (CTL)–mediated immunity, antigen recognition must occur in the context of costimulation and proinflammatory cytokines.1 In contrast, antigen encounter in the absence of any coactivators (ie, in the steady state) results in abortive tolerogenic responses,1 which are critical for maintaining peripheral T-cell tolerance.
Peripheral tolerance can be maintained by either T-cell deletion or anergy. During deletional tolerance, T cells initially proliferate in response to presented antigen but are ultimately lost by apoptotic death.2 In contrast, anergy occurs when antigen-activated T cells persist after antigen encounter in a hyporesponsive, nonproliferative (or anergic) state.3 The factors determining whether an autoreactive T cell undergoes anergy or deletion are still poorly understood, but high antigen levels have been suggested to favor anergy, whereas lower levels are thought to precipitate deletion.4,5 It has therefore been proposed that high and low levels of T-cell receptor (TCR) engagement provoke distinct signaling pathways that influence the mode of tolerance adopted.1
Although the biochemistry of anergy is well characterized,3 relatively little is known about the molecular basis of peripheral T-cell deletion. CD8+ T cells undergoing deletion fail to develop effector functions, with cells exhibiting specific deficiencies in cytolytic capacity and interferon-γ (IFN-γ) production.6 Furthermore, cell death occurs via an apoptotic pathway dependent on the proapoptotic BH3-only Bcl-2 family member Bim.2 However, the molecular basis for defective effector functions and the pathway responsible for Bim activation remain unclear.
To resolve these issues, we have examined the molecular signature of CD8+ T cells undergoing deletion. The gene expression profiles of CD8+ T cells during immunity, deletion, and lymphopenia-induced proliferation were compared to identify any deletion-specific gene expression changes. Here, we demonstrate that deletional tolerance possesses a unique molecular signature that distinguishes it from immunity and lymphopenia-induced proliferation. This gene expression signature provides a molecular basis for defective cytolysis and Bim-dependent death during deletion. Furthermore, many genes reported to be associated with anergy were also found up-regulated in deletional tolerance, suggesting a closer relationship between these 2 tolerance mechanisms than previously anticipated.
Methods
Mice
All mice were bred and maintained on a C57BL/6 background at the Walter and Eliza Hall Institute (WEHI). Transgenic OT-I7 and RIP-OVAhi8 and Rag-1−/−9 mice have been described. C57BL/6, B6.SJL-PtprcaPep3b/BoyJ (Ly5.1+), and B6.C-H2bm1/By (bm1) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal experimentation was performed in accordance with institutional guidelines and the Melbourne Health Animal Ethics Committee, which granted permission for this study.
Carboxyfluorescein succinimidyl ester labeling, adoptive transfer, and FACS analysis
For detailed methods on CD8+ T-cell bead enrichment, carboxyfluorescein succinimidyl ester (CFSE) labeling, and ppERK1/2 staining, see “Supplemental Materials and Methods” in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Instruments used for flow cytometry and sorting were a FACSCalibur, BD-LSR II (BD Biosciences, San Jose, CA), and MoFlo cell sorter (Dako North America, Carpinteria, CA). Antibodies used for flow cytometry were directed against CD8, Ly5.1, Bcl-2 (BD Biosciences), granzyme B (Invitrogen, Carlsbad, CA), PD-1, interleukin-7Rα (IL-7Rα) chain (eBioscience, San Diego, CA), ppERK1/2 (Cell Signaling Technology, Danvers, MA), Ly6C (provided by K. Shortman, WEHI), and Bim (provided by L. O'Reilly, WEHI). OVA-coated splenocytes were prepared as described previously10 and injected intravenously with 1 μg lipopolysaccharide (LPS).
Microarray sample preparation
For cell sorts, 2 × 106 enriched, naive (CD44lo) CFSE-labeled Ly5.1+ OT-I cells were injected intravenously into antigen-free C57BL/6 (B6), RIP-OVAhi, OCS/LPS-treated B6 (OCS/LPS) or Rag−/− mice. CD8+ T cells were then enriched by bead depletion from the spleen (B6, OCS/LPS, and Rag−/−) or sacral and pancreatic lymph nodes (RIP-OVAhi) at the appropriate time points after injection. Cells were stained for CD8 and Ly5.1 and the viable, CD8+Ly5.1+ cells of the appropriate CFSE level were sorted by fluorescence-activated cell sorting (FACS). RNA was isolated from sorted cells using TRI Reagent (Sigma-Aldrich, St Louis, MO) according to the manufacturer's instructions, and RNA was quantitated using an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). The appropriate amounts of RNA were then amplified and prepared for hybridization using the Affymetrix GeneChip Two-Cycle Target Labeling and Control Reagents kit (Affymetrix, Santa Clara, CA). Array hybridization was performed in the Australian Genome Research Facility using Affymetrix Mouse Expression set 430, version 2.0 microarrays. For details on real-time polymerase chain reaction (PCR) validation, see Document S1. The microarray data have been submitted to GEO under accession number GSE14699 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14699).
Microarray data analysis
Statistical analysis and quality assessment were undertaken using Bioconductor software for R.11 Expression data were background-corrected and normalized using the gcRMA algorithm,12 yielding log2-expression summaries for each probe set. Differential expression analysis used the Limma software package.13 The 4 cell types were compared by fitting a linear model and extracting contrasts for the 6 possible pairwise comparisons.14 Probe sets were flagged as differentially expressed by a “nestedF” analysis controlling the probewise false discovery rate at 0.05. This procedure has good power for detecting genes responding in more than one cell type. For the genome-wide analyses, the multiple testing adjustment was applied over all probes on the arrays. When analyzing prespecified sets of genes (Tables 3 and S3Table S4. Changes in genes up-regulated by Nur77 over-expression (PDF, 53.4 KB)Table S5. Changes in genes up-regulated in exhausted CD8+ T cells (PDF, 133 KB)–S6), the adjustment was done separately for each restricted set of genes.
Probe sets were flagged as uniquely regulated in Tol if significant for Tol.Naive but not for Imm.Naive or Rag.Naive. A shorter and more stringent list of uniquely Tol-regulated genes was also constructed (Tables 1,2). For the stringent list, probe sets were also required to be significantly different for Tol.Imm and Tol.Rag, the Tol.Naive fold change was required to be 50% or greater and probe sets with average log2 expression (A values) less than 4 were ignored. Microarray data were managed using a custom-built database that allowed genes to be organized and cross-compared on both ontologic classifications and experimental results. GOStat15 was used to calculate overrepresentation of gene ontology groups.
Results
A unique molecular signature for deletion
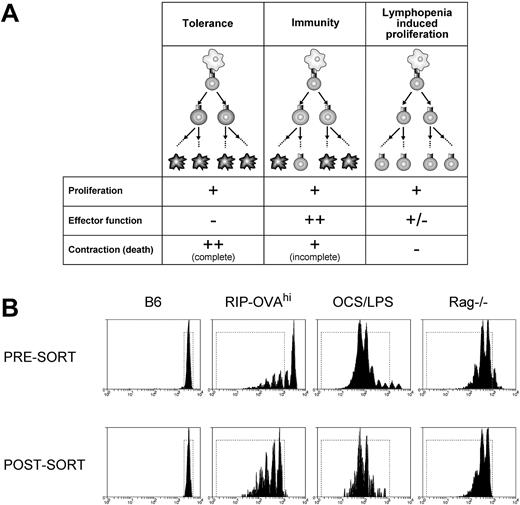
We used microarray profiling of CD8+ T cells undergoing different fates to identify deletion-specific transcriptional changes. Three cell fates were chosen for profiling: deletional tolerance, immunity, and lymphopenia-induced proliferation. Each of these conditions was compared with resting T cells to determine transcriptional changes on initiation of proliferation, and comparisons between the fates could then be used to identify deletion-specific alterations. We reasoned that deletional tolerance, immunogenic priming, and lymphopenia-induced proliferation all trigger proliferation yet differ in both the extent of effector function generated and the amount of cell death (Figure 1A). Thus, by comparing all 3 conditions, distinct components of deletional tolerance may be revealed.
A microarray approach to the analysis of peripheral deletion of CD8+ T cells. (A) Three forms of CD8+ T-cell fate were chosen for microarray comparison of gene expression profiles: deletional tolerance, immunity, and lymphopenia-induced proliferation. In deletional tolerance (left panels), naive T cells (top, round cell) proliferate in response to antigen presented by DCs (top, irregular cell). However, the responding T cells fail to acquire effector functions, and all eventually die by apoptosis (ie, the population contracts). In immunity (middle panels), naive T cells proliferate in response to antigen presented by DCs. In contrast to deletional tolerance, these cells acquire effector functions and, although most cells contract/die at the completion of the CTL immune response, some persist as memory cells. During lymphopenia-induced proliferation (right panels), T cells proliferate in response to homeostatic cytokines and low-avidity TCR/self-major histocompatibility complex interactions with DCs and the responding cells acquire a “memory” phenotype. There is no net loss of cells during this response and hence no contraction and relatively little cell death. (B) Presort and postsort profiles. A total of 2 × 106 CFSE-labeled Ly5.1+ OT-I cells were injected intravenously into antigen-free C57BL/6 (B6), RIP-OVAhi, OCS/LPS-treated C57BL/6 (OCS/LPS), or Rag−/− mice. The cells were then stained for CD8 and Ly5.1, and the viable, CD8+Ly5.1+ cells that had either not divided (B6) or had undergone 2 or more divisions (RIP-OVAhi, OCS/LPS, and Rag−/−) were sorted by FACS. (Top panels) The CFSE profiles of viable, CD8+Ly5.1+ cells presort. (Bottom panels) The same cells postsort. The dotted rectangle represents the sort gate. Representative profiles from 1 of 2 sorts are shown.
A microarray approach to the analysis of peripheral deletion of CD8+ T cells. (A) Three forms of CD8+ T-cell fate were chosen for microarray comparison of gene expression profiles: deletional tolerance, immunity, and lymphopenia-induced proliferation. In deletional tolerance (left panels), naive T cells (top, round cell) proliferate in response to antigen presented by DCs (top, irregular cell). However, the responding T cells fail to acquire effector functions, and all eventually die by apoptosis (ie, the population contracts). In immunity (middle panels), naive T cells proliferate in response to antigen presented by DCs. In contrast to deletional tolerance, these cells acquire effector functions and, although most cells contract/die at the completion of the CTL immune response, some persist as memory cells. During lymphopenia-induced proliferation (right panels), T cells proliferate in response to homeostatic cytokines and low-avidity TCR/self-major histocompatibility complex interactions with DCs and the responding cells acquire a “memory” phenotype. There is no net loss of cells during this response and hence no contraction and relatively little cell death. (B) Presort and postsort profiles. A total of 2 × 106 CFSE-labeled Ly5.1+ OT-I cells were injected intravenously into antigen-free C57BL/6 (B6), RIP-OVAhi, OCS/LPS-treated C57BL/6 (OCS/LPS), or Rag−/− mice. The cells were then stained for CD8 and Ly5.1, and the viable, CD8+Ly5.1+ cells that had either not divided (B6) or had undergone 2 or more divisions (RIP-OVAhi, OCS/LPS, and Rag−/−) were sorted by FACS. (Top panels) The CFSE profiles of viable, CD8+Ly5.1+ cells presort. (Bottom panels) The same cells postsort. The dotted rectangle represents the sort gate. Representative profiles from 1 of 2 sorts are shown.
To isolate cells differentiating down different fates, naive (ie, CD44lo) CFSE-labeled TCR transgenic ovalbumin (OVA)–specific CD8+ T cells (OT-I cells) were transferred into (1) RIP-OVAhi mice, (2) B6 mice primed with OVA coated, irradiated splenocytes mixed with LPS (OCS/LPS), or (3) Rag-1−/− mice. The RIP-OVAhi mouse model is an established transgenic deletion model in which OT-I cells proliferate8 and die2,16 in response to cross-presented, pancreatic islet β cell–derived OVA. In contrast, priming with OCS alone has been shown to induce OVA-specific CTL responses that persist into memory10,17 and is more robust in the presence of LPS (G.M.D. and W.R.H., unpublished data, 2000). Finally, the lymphopenic Rag1−/− mouse was used to provoke lymphopenia-induced proliferation.
CFSE dilution was then used to identify those cells that had responded and proliferated, and OT-I cells that had progressed through more than one division were sorted from either the draining pancreatic and sacral lymph nodes (RIP-OVAhi mice) or spleen (other conditions; Figure 1B). As a control for resting T cells, naive C57BL/6 mice were given CFSE-labeled OT-I cells, and the undivided T cells were sorted from these mice for array analysis. Sorts were performed in duplicate 60 hours after transfer, except for homeostatic proliferation, which was examined on day 5 from a single sort. Cell sorting yielded approximately 1 to 1.5 × 105 cells from each condition, and these cells were found to be of 95% to 98% purity on reanalysis (Figure 1B; data not shown). Total RNA isolated from each condition was subjected to 2 cycles of amplification before array analysis.
After array hybridization, array data were analyzed to identify differentially expressed genes within each of the 6 possible 2-way comparisons. A substantial number of genes exhibited statistically significant differential expression relative to naive cells after initial data analysis (Figure 2). The most evident trend was the relatively small overlap between lymphopenia-induced responses and the 2 other conditions (Figure 2). Although there was some overlap between all 3 conditions, cells responding in tolerogenic and immunogenic environments showed more changes in common than those responding in lymphopenic environments. This highlights that lymphopenia-induced responses are molecularly distinct from antigen-driven cell fates.
Differential expression of probe sets during deletional tolerance, immunity, and lymphopenia-induced proliferation. The Venn diagram depicts the differentially expressed probe sets detected on the array using the statistical analysis described in “Microarray data analysis” in “Methods.” The expression changes are split into probe sets up-regulated (+, left) and down-regulated (−, right) within each comparison. Tol.Naive represents probe sets differentially expressed within OT-I cells isolated from RIP-OVAhi mice relative to the undivided cells recovered from unchallenged (naive) C57BL/6 mice. Imm.Naive and Rag.Naive show the same comparison for cells recovered from OCS/LPS and Rag−/− mice, respectively.
Differential expression of probe sets during deletional tolerance, immunity, and lymphopenia-induced proliferation. The Venn diagram depicts the differentially expressed probe sets detected on the array using the statistical analysis described in “Microarray data analysis” in “Methods.” The expression changes are split into probe sets up-regulated (+, left) and down-regulated (−, right) within each comparison. Tol.Naive represents probe sets differentially expressed within OT-I cells isolated from RIP-OVAhi mice relative to the undivided cells recovered from unchallenged (naive) C57BL/6 mice. Imm.Naive and Rag.Naive show the same comparison for cells recovered from OCS/LPS and Rag−/− mice, respectively.
When the gene groupings were analyzed for overrepresentation of gene ontology groups (GOs), the most striking overrepresentation was seen within cell cycle–related GOs. Not surprisingly, cell cycle–related GOs were highly overrepresented (eg, cell-cycle GO P = 4.9 × 10−27) within genes up-regulated in common between all 3 conditions. This gene set likely defines the minimal changes required for CD8+ T-cell proliferation. Nevertheless, cell cycle–related GOs were also significantly overrepresented within the probe sets up-regulated in both deletional tolerance and immunity but not in lymphopenia-induced proliferation (eg, cell-cycle GO P = 1.8 × 10−34). This suggests that antigen-driven proliferation has its own unique cell-cycle program, which is consistent with the slower kinetics of lymphopenia-induced proliferation. There was also a moderate enrichment of the cell-cycle GO in genes selectively up-regulated in immunity (P = .02), suggesting that an immunity-specific cell-cycle program may exist. Cell cycle–related genes were not overrepresented in any other up- or down-regulated gene group.
It was also clear from the data obtained that there were a large number of probe sets exhibiting differential expression selectively within OT-I cells undergoing deletional tolerance (Figure 2). Collectively, these data indicate that peripheral deletion has a unique molecular signature and is not simply characterized by a failure to up- or down-regulate immunity-associated genes. The probe sets uniquely up- or down-regulated within deletion were stringently filtered (“Microarray data analysis” in “Methods”) to yield the gene lists in Tables 1 and 2, respectively, with unfiltered data in Tables S1 and S2. A number of these genes have established or putative associations with T-cell tolerance.
Probe sets uniquely up-regulated in deletional tolerance
| Affymetrix probe ID . | Gene symbol . | Gene name . | Log fold change . | Association with T-cell tolerance? . |
|---|---|---|---|---|
| 1419083_at | Tnfsf11 | Tumor necrosis factor (ligand) superfamily, member 11 | 6.49 | Yes19 |
| 1455265_a_at | Rgs16 | Regulator of G-protein signaling 16 | 4.8 | No |
| 1428393_at | Nrn1 | Neuritin 1 | 4.43 | No |
| 1456956_at | Ikzf2 | IKAROS family zinc finger 2 | 4.04 | Yes47 |
| 1435292_at | Tbc1d4 | TBC1 domain family, member 4 | 3.96 | No |
| 1455034_at | — | — | 3.46 | No |
| 1423091_a_at | Gpm6b | Glycoprotein m6b | 3.08 | No |
| 1426542_at | Endod1 | Endonuclease domain containing 1 | 2.93 | No |
| 1425062_at | Fcrl1 | Fc receptor-like 1 | 2.71 | No |
| 1449893_a_at | Lrig1 | Leucine-rich repeats and immunoglobulin-like domains 1 | 2.66 | Putative |
| 1434210_s_at | Lrig1 | Leucine-rich repeats and immunoglobulin-like domains 1 | 2.59 | Putative |
| 1415973_at | Marcks | Myristoylated alanine rich protein kinase C substrate | 2.46 | No |
| 1438796_at | Nr4a3 | Nuclear receptor subfamily 4, group A, member 3 | 2.38 | Yes35,61 |
| 1420353_at | Lta | Lymphotoxin A | 2.21 | No |
| 1455418_at | — | — | 2.14 | No |
| 1456028_x_at | Marcks | Myristoylated alanine-rich protein kinase C substrate | 1.81 | No |
| 1452344_at | Synj2 | Synaptojanin 2 | 1.79 | No |
| 1416505_at | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 1.74 | Yes61 |
| 1449010_at | Hspa4l | Heat shock protein 4-like | 1.73 | No |
| 1460121_at | Ptprs | Protein tyrosine phosphatase, receptor type, S | 1.65 | Yes46 |
| 1427683_at | Egr2 | Early growth response 2 | 1.63 | Yes35 |
| 1418758_a_at | Pscd3 | Pleckstrin homology, Sec7 and coiled-coil domains 3 | 1.56 | Yes48 |
| 1438331_at | Ypel2 | Yippee-like 2 (Drosophila) | 1.39 | No |
| 1427682_a_at | Egr2 | Early growth response 2 | 1.32 | Yes |
| 1422054_a_at | Skil | SKI-like | 1.26 | No |
| 1417930_at | Nab2 | Ngfi-A binding protein 2 | 1.17 | No |
| 1417695_a_at | Soat1 | Sterol O-acyltransferase 1 | 1.17 | No |
| 1452583_s_at | Galm | Galactose mutarotase | 1.15 | No |
| 1445612_at | Tox | Thymocyte selection-associated HMG box gene | 1.11 | Putative |
| 1437918_at | 4930539E08Rik | RIKEN cDNA 4930539E08 gene | 1.08 | No |
| 1424778_at | Reep3 | Receptor accessory protein 3 | 1.05 | No |
| 1438338_at | Mdh1 | Malate dehydrogenase 1, NAD (soluble) | 1.04 | No |
| 1429472_at | 4921525O09Rik | RIKEN cDNA 4921525O09 gene | 1.01 | No |
| 1422438_at | Ephx1 | Epoxide hydrolase 1, microsomal | 0.96 | No |
| 1455854_a_at | Ssh1 | Slingshot homolog 1 (Drosophila) | 0.96 | No |
| 1449037_at | Crem | cAMP-responsive element modulator | 0.95 | Yes41,45 |
| 1449381_a_at | Pacsin1 | Protein kinase C and casein kinase substrate in neurons 1 | 0.95 | No |
| 1424460_s_at | Lpcat1 | Acyltransferase-like 2 | 0.87 | No |
| 1416844_at | Prmt2 | Protein arginine N-methyltransferase 2 | 0.84 | No |
| 1439703_at | Cd200r1 | CD200 receptor 1 | 0.8 | No |
| 1434384_at | Nrip1 | Nuclear receptor interacting protein 1 | 0.76 | No |
| 1452169_a_at | Dgkz | Diacylglycerol kinase zeta | 0.76 | Yes38,39 |
| 1426476_at | Rasa1 | RAS p21 protein activator 1 | 0.74 | Yes40 |
| 1430357_at | H3f3b | H3 histone, family 3B | 0.73 | No |
| 1434273_at | A830073O21Rik | RIKEN cDNA A830073O21 gene | 0.66 | No |
| Affymetrix probe ID . | Gene symbol . | Gene name . | Log fold change . | Association with T-cell tolerance? . |
|---|---|---|---|---|
| 1419083_at | Tnfsf11 | Tumor necrosis factor (ligand) superfamily, member 11 | 6.49 | Yes19 |
| 1455265_a_at | Rgs16 | Regulator of G-protein signaling 16 | 4.8 | No |
| 1428393_at | Nrn1 | Neuritin 1 | 4.43 | No |
| 1456956_at | Ikzf2 | IKAROS family zinc finger 2 | 4.04 | Yes47 |
| 1435292_at | Tbc1d4 | TBC1 domain family, member 4 | 3.96 | No |
| 1455034_at | — | — | 3.46 | No |
| 1423091_a_at | Gpm6b | Glycoprotein m6b | 3.08 | No |
| 1426542_at | Endod1 | Endonuclease domain containing 1 | 2.93 | No |
| 1425062_at | Fcrl1 | Fc receptor-like 1 | 2.71 | No |
| 1449893_a_at | Lrig1 | Leucine-rich repeats and immunoglobulin-like domains 1 | 2.66 | Putative |
| 1434210_s_at | Lrig1 | Leucine-rich repeats and immunoglobulin-like domains 1 | 2.59 | Putative |
| 1415973_at | Marcks | Myristoylated alanine rich protein kinase C substrate | 2.46 | No |
| 1438796_at | Nr4a3 | Nuclear receptor subfamily 4, group A, member 3 | 2.38 | Yes35,61 |
| 1420353_at | Lta | Lymphotoxin A | 2.21 | No |
| 1455418_at | — | — | 2.14 | No |
| 1456028_x_at | Marcks | Myristoylated alanine-rich protein kinase C substrate | 1.81 | No |
| 1452344_at | Synj2 | Synaptojanin 2 | 1.79 | No |
| 1416505_at | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 1.74 | Yes61 |
| 1449010_at | Hspa4l | Heat shock protein 4-like | 1.73 | No |
| 1460121_at | Ptprs | Protein tyrosine phosphatase, receptor type, S | 1.65 | Yes46 |
| 1427683_at | Egr2 | Early growth response 2 | 1.63 | Yes35 |
| 1418758_a_at | Pscd3 | Pleckstrin homology, Sec7 and coiled-coil domains 3 | 1.56 | Yes48 |
| 1438331_at | Ypel2 | Yippee-like 2 (Drosophila) | 1.39 | No |
| 1427682_a_at | Egr2 | Early growth response 2 | 1.32 | Yes |
| 1422054_a_at | Skil | SKI-like | 1.26 | No |
| 1417930_at | Nab2 | Ngfi-A binding protein 2 | 1.17 | No |
| 1417695_a_at | Soat1 | Sterol O-acyltransferase 1 | 1.17 | No |
| 1452583_s_at | Galm | Galactose mutarotase | 1.15 | No |
| 1445612_at | Tox | Thymocyte selection-associated HMG box gene | 1.11 | Putative |
| 1437918_at | 4930539E08Rik | RIKEN cDNA 4930539E08 gene | 1.08 | No |
| 1424778_at | Reep3 | Receptor accessory protein 3 | 1.05 | No |
| 1438338_at | Mdh1 | Malate dehydrogenase 1, NAD (soluble) | 1.04 | No |
| 1429472_at | 4921525O09Rik | RIKEN cDNA 4921525O09 gene | 1.01 | No |
| 1422438_at | Ephx1 | Epoxide hydrolase 1, microsomal | 0.96 | No |
| 1455854_a_at | Ssh1 | Slingshot homolog 1 (Drosophila) | 0.96 | No |
| 1449037_at | Crem | cAMP-responsive element modulator | 0.95 | Yes41,45 |
| 1449381_a_at | Pacsin1 | Protein kinase C and casein kinase substrate in neurons 1 | 0.95 | No |
| 1424460_s_at | Lpcat1 | Acyltransferase-like 2 | 0.87 | No |
| 1416844_at | Prmt2 | Protein arginine N-methyltransferase 2 | 0.84 | No |
| 1439703_at | Cd200r1 | CD200 receptor 1 | 0.8 | No |
| 1434384_at | Nrip1 | Nuclear receptor interacting protein 1 | 0.76 | No |
| 1452169_a_at | Dgkz | Diacylglycerol kinase zeta | 0.76 | Yes38,39 |
| 1426476_at | Rasa1 | RAS p21 protein activator 1 | 0.74 | Yes40 |
| 1430357_at | H3f3b | H3 histone, family 3B | 0.73 | No |
| 1434273_at | A830073O21Rik | RIKEN cDNA A830073O21 gene | 0.66 | No |
Log fold change represents log (base 2) fold change. — indicates probe set has no gene annotation.
Probe sets uniquely down-regulated in deletional tolerance
| Affymetrix probe ID . | Gene symbol . | Gene name . | Log fold change . | Association with T-cell tolerance? . |
|---|---|---|---|---|
| 1417184_s_at | Hbb-bh1 | Hemoglobin, beta adult major chain | −10.2 | No |
| 1436996_x_at | Lzp-s | Lysozyme | −5.15 | No |
| 1436853_a_at | Snca | Synuclein, alpha | −5.09 | No |
| 1417898_a_at | Gzma | Granzyme A | −4.55 | Yes6 |
| 1435290_x_at | H2-Aa | Histocompatibility 2, class II antigen A, alpha | −4.34 | No |
| 1439426_x_at | Lyzs | Lysozyme | −4.34 | No |
| 1438858_x_at | H2-Aa | Histocompatibility 2, class II antigen A, alpha | −3.05 | No |
| 1459641_at | Psmb2 | Proteasome (prosome, macropain) subunit, beta type 2 | −2.35 | No |
| 1425822_a_at | Dtx1 | Deltex 1 homolog (Drosophila) | −2.08 | No |
| 1417185_at | Ly6a | Lymphocyte antigen 6 complex, locus A | −1.81 | No |
| 1444283_at | Gimap7 | GTPase, IMAP family member 7 | −1.67 | No |
| 1436845_at | Axin2 | Axin2 | −1.66 | No |
| 1421571_a_at | Ly6c | Lymphocyte antigen 6 complex, locus C | −1.64 | Yes6,20 |
| 1417398_at | Rras2 | Related RAS viral (r-ras) oncogene homolog 2 | −1.63 | No |
| 1454666_at | Klf3 | Kruppel-like factor 3 (basic) | −1.54 | No |
| 1449181_at | Fech | Ferrochelatase | −1.47 | No |
| 1438442_at | AI450236 | Expressed sequence AI450236 | −1.38 | No |
| 1450431_a_at | Nedd4 | Neural precursor cell expressed, developmentally down-regulated gene 4 | −1.34 | No |
| 1450240_a_at | Sytl1 | Synaptotagmin-like 1 | −1.13 | No |
| 1456545_at | Il18rap | Interleukin-18 receptor accessory protein | −1.12 | Putative |
| 1429723_at | 6330409N04Rik | RIKEN cDNA 6330409N04 gene | −1.1 | No |
| Affymetrix probe ID . | Gene symbol . | Gene name . | Log fold change . | Association with T-cell tolerance? . |
|---|---|---|---|---|
| 1417184_s_at | Hbb-bh1 | Hemoglobin, beta adult major chain | −10.2 | No |
| 1436996_x_at | Lzp-s | Lysozyme | −5.15 | No |
| 1436853_a_at | Snca | Synuclein, alpha | −5.09 | No |
| 1417898_a_at | Gzma | Granzyme A | −4.55 | Yes6 |
| 1435290_x_at | H2-Aa | Histocompatibility 2, class II antigen A, alpha | −4.34 | No |
| 1439426_x_at | Lyzs | Lysozyme | −4.34 | No |
| 1438858_x_at | H2-Aa | Histocompatibility 2, class II antigen A, alpha | −3.05 | No |
| 1459641_at | Psmb2 | Proteasome (prosome, macropain) subunit, beta type 2 | −2.35 | No |
| 1425822_a_at | Dtx1 | Deltex 1 homolog (Drosophila) | −2.08 | No |
| 1417185_at | Ly6a | Lymphocyte antigen 6 complex, locus A | −1.81 | No |
| 1444283_at | Gimap7 | GTPase, IMAP family member 7 | −1.67 | No |
| 1436845_at | Axin2 | Axin2 | −1.66 | No |
| 1421571_a_at | Ly6c | Lymphocyte antigen 6 complex, locus C | −1.64 | Yes6,20 |
| 1417398_at | Rras2 | Related RAS viral (r-ras) oncogene homolog 2 | −1.63 | No |
| 1454666_at | Klf3 | Kruppel-like factor 3 (basic) | −1.54 | No |
| 1449181_at | Fech | Ferrochelatase | −1.47 | No |
| 1438442_at | AI450236 | Expressed sequence AI450236 | −1.38 | No |
| 1450431_a_at | Nedd4 | Neural precursor cell expressed, developmentally down-regulated gene 4 | −1.34 | No |
| 1450240_a_at | Sytl1 | Synaptotagmin-like 1 | −1.13 | No |
| 1456545_at | Il18rap | Interleukin-18 receptor accessory protein | −1.12 | Putative |
| 1429723_at | 6330409N04Rik | RIKEN cDNA 6330409N04 gene | −1.1 | No |
Log fold change represents log (base 2) fold change.
The accuracy of the array analysis was confirmed by validating a number of the deletional tolerance-specific changes in gene expression by quantitative real-time PCR of the cRNA used for array hybridization (Figure S1). Furthermore, consistent with previous observations,6,18 the microarray analysis detected CD44 up-regulation in immunity, deletional tolerance, and lymphopenia-induced proliferation. Furthermore, in agreement with published data, the tumor necrosis factor (ligand) superfamily, member 11 (RANKL) gene was selectively up-regulated during peripheral T-cell deletion.19 Finally, there was internal consistency within the dataset. For example, the tolerance-specific up-regulation of the transcription factor Egr2 was accompanied by the concomitant, tolerance-specific up-regulation of the Egr2 target gene Cbl-b.
Deletion is associated with deficiencies in GzmB and Ly6C expression
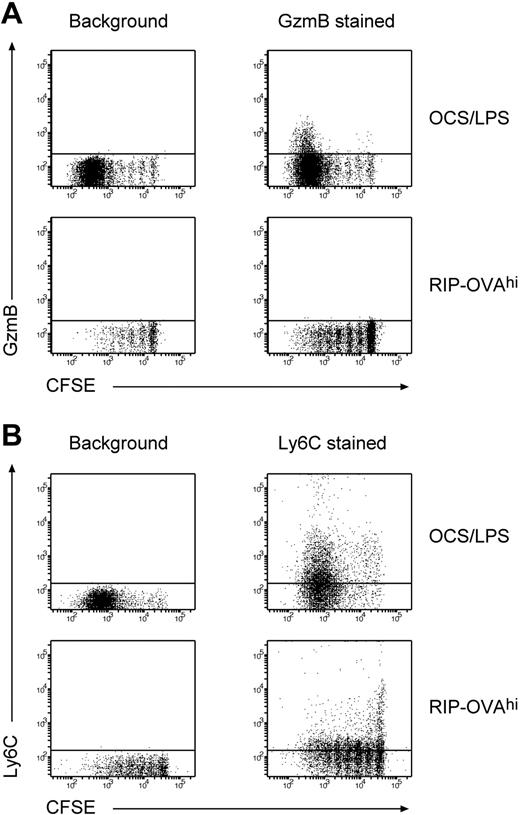
One of the hallmarks of CD8+ T cells undergoing deletion is an inability to produce IFN-γ on restimulation in culture and impaired cytolytic capacity.6 To determine the molecular basis of this phenomenon, we examined the array data for any genes that may have relevance to effector functions. Deletional tolerance was associated with a failure to up-regulate the cytolytic molecules granzyme A (GzmA) and granzyme B (GzmB). Although GzmA and GzmB were up-regulated in immunity, the expression of these genes was either unchanged (in the case of GzmB) or even down-regulated (in the case of GzmA) during deletion. The failure of OT-I cells to up-regulate GzmB during deletion was subsequently confirmed by intracellular staining for GzmB (Figure 3A). Given that granzymes participate in the process of cytolysis, these changes probably impair cytolytic capacity. Cells undergoing deletion also down-regulated expression of Ly6C, a molecule that potentiates CTL cytolytic activity and cytokine production.20 This shift in Ly6C expression was confirmed by flow cytometry (Figure 3B) and may potentiate the cytolytic defect. The only other deletion-specific change with relevance to T-cell effector functions was down-regulation of IL-18 receptor–associated protein, a molecule required for IL-18 receptor signaling and responsiveness to the IL-18 effector cytokine.21 Given that IL-18 can potentiate CTL development22 and can reverse CTL anergy in vitro,23 a defect in IL-18 signaling probably further impairs the development of effector functions during deletion.
OT-I cells undergoing deletion fail to up-regulate GzmB and Ly6C. A total of 2 × 106 Ly5.1+ CFSE labeled OT-I cells were injected intravenously into either RIP-OVAhi mice or OCS/LPS primed C57BL/6 mice (OCS/LPS). Sixty hours after transfer, the proliferating cells within the sacral and pancreatic lymph nodes (RIP-OVAhi) or spleen (OCS/LPS) were stained and analyzed by flow cytometry. (A) Dot plots of intracellular GzmB staining versus CFSE staining showing unstained (background; left panels) and GzmB-stained (right panels) cells. The plots are gated on CD8+Ly5.1+ T cells. Representative plots from 5 independent experiments (each performed with 3 mice per group) are shown. (B) Data for Ly6C staining plotted similarly to panel A. Representative plots from 3 independent experiments (each performed with 3 mice per group) are shown.
OT-I cells undergoing deletion fail to up-regulate GzmB and Ly6C. A total of 2 × 106 Ly5.1+ CFSE labeled OT-I cells were injected intravenously into either RIP-OVAhi mice or OCS/LPS primed C57BL/6 mice (OCS/LPS). Sixty hours after transfer, the proliferating cells within the sacral and pancreatic lymph nodes (RIP-OVAhi) or spleen (OCS/LPS) were stained and analyzed by flow cytometry. (A) Dot plots of intracellular GzmB staining versus CFSE staining showing unstained (background; left panels) and GzmB-stained (right panels) cells. The plots are gated on CD8+Ly5.1+ T cells. Representative plots from 5 independent experiments (each performed with 3 mice per group) are shown. (B) Data for Ly6C staining plotted similarly to panel A. Representative plots from 3 independent experiments (each performed with 3 mice per group) are shown.
In direct contrast to GzmB and Ly6C, IFN-γ transcript levels were significantly up-regulated (relative to naive cells) to a similar extent in both immunity and deletional tolerance (21- vs 16-fold, respectively; Tol vs Imm P = .49). Thus, unlike the block in cytolytic gene expression, the block in IFN-γ production that has previously been observed in deletional tolerance6 does not appear to be caused by impaired transcriptional induction.
A deletional tolerance-specific apoptotic pathway
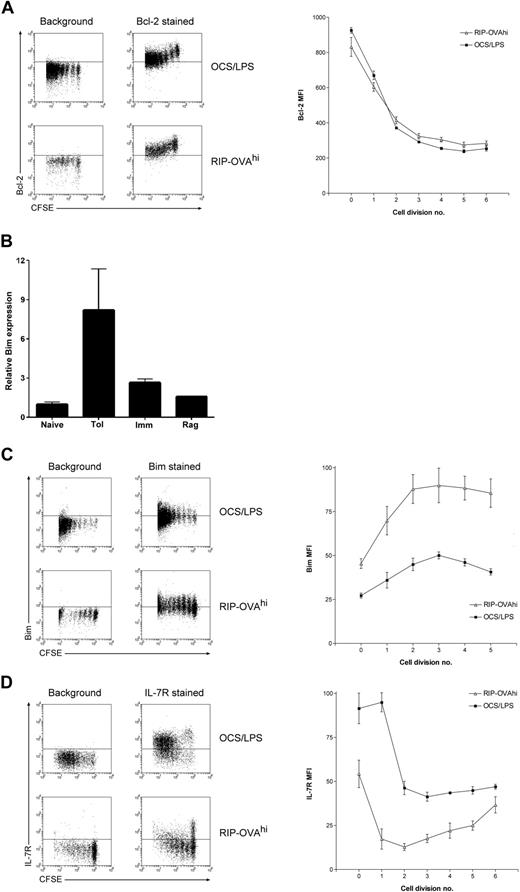
One major aim of this study was to identify the mechanism by which Bim-dependent death is activated during peripheral deletion and to determine whether this mechanism was deletion specific. To this end, the differential expression of apoptosis-related molecules was examined in detail, with P values recalculated independently for this gene group (Table S3). Although there was some differential expression of apoptosis-related genes, most of these changes were not tolerance-specific. Interestingly, the changes in apoptosis-related gene expression within cells undergoing lymphopenia-induced proliferation differed from the other 2 cell fates, consistent with the absence of a bulk contraction event after expansion within a lymphopenic environment18 (Figure 1A). In contrast, in both deletional tolerance and immunity several common expression changes in apoptosis-related genes were observed. One of the most striking of these common changes was the down-regulation of the prosurvival factor Bcl-2 in both deletional tolerance and immunity. Because OT-I deletion within RIP-OVAhi mice is Bim dependent2 and Bim induces apoptosis through direct binding to antiapoptotic Bcl-2 family members (such as Bcl-2),24 a greater degree of Bcl-2 down-regulation during deletion could sensitize cells to Bim-mediated death. We thus more rigorously examined whether there was any disparity in the magnitude of Bcl-2 down-regulation between deletional tolerance and immunity by intracellular Bcl-2 staining. A profound division-linked down-regulation of Bcl-2 protein was observed during deletional tolerance, confirming the expression change seen in the array studies (Figure 4A). However, as predicted by the array data, this drop in Bcl-2 levels was not tolerance-specific with an identical degree of Bcl-2 down-regulation observed after priming of OT-I cells (Figure 4A). Thus, Bcl-2 down-regulation was unlikely to drive deletion-specific activation of Bim.
Bcl-2, IL-7Rα chain, and Bim levels during deletion and immunity. A total of 2 × 106 CFSE-labeled OT-I cells were injected intravenously into either RIP-OVAhi mice or OCS/LPS primed C57BL/6 mice (OCS/LPS). Sixty hours after transfer, the proliferating cells within the sacral and pancreatic lymph nodes (RIP-OVAhi) or spleen (OCS/LPS) were stained and analyzed by flow cytometry. (A) Intracellular Bcl-2 staining showing dot plots (left) and quantitated mean fluorescence intensity (MFI; right). MFI graph shows Bcl-2 MFI values minus background, which was determined independently for each cell division. Error bars represent SEM. Representative data are shown from 1 of 3 independent experiments, with 3 mice per group per experiment. (B) Bim real-time PCR performed on array cRNA. The bar graphs show changes in gene expression relative to naive and represent mean data ± SEM from the duplicate array samples (with the exception of the Rag sample, for which only a single replicate was analyzed). Naive indicates naive cells; Tol, cells undergoing deletion; Imm, primed cells; and Rag, cells undergoing lymphopenia-induced proliferation. (C) Same as in panel A, except that cells were stained intracellularly for Bim. Representative data are shown from 1 of 2 independent experiments, with 3 mice per group per experiment. (D) Same as in panel A, except that cells were surface stained for IL-7Rα chain. Representative data are shown from 1 of 3 independent experiments, with 3 mice per group per experiment.
Bcl-2, IL-7Rα chain, and Bim levels during deletion and immunity. A total of 2 × 106 CFSE-labeled OT-I cells were injected intravenously into either RIP-OVAhi mice or OCS/LPS primed C57BL/6 mice (OCS/LPS). Sixty hours after transfer, the proliferating cells within the sacral and pancreatic lymph nodes (RIP-OVAhi) or spleen (OCS/LPS) were stained and analyzed by flow cytometry. (A) Intracellular Bcl-2 staining showing dot plots (left) and quantitated mean fluorescence intensity (MFI; right). MFI graph shows Bcl-2 MFI values minus background, which was determined independently for each cell division. Error bars represent SEM. Representative data are shown from 1 of 3 independent experiments, with 3 mice per group per experiment. (B) Bim real-time PCR performed on array cRNA. The bar graphs show changes in gene expression relative to naive and represent mean data ± SEM from the duplicate array samples (with the exception of the Rag sample, for which only a single replicate was analyzed). Naive indicates naive cells; Tol, cells undergoing deletion; Imm, primed cells; and Rag, cells undergoing lymphopenia-induced proliferation. (C) Same as in panel A, except that cells were stained intracellularly for Bim. Representative data are shown from 1 of 2 independent experiments, with 3 mice per group per experiment. (D) Same as in panel A, except that cells were surface stained for IL-7Rα chain. Representative data are shown from 1 of 3 independent experiments, with 3 mice per group per experiment.
Nevertheless, some deletion-specific changes in apoptosis-related genes were observed. Most striking was the deletion-specific up-regulation of Bim mRNA (Table S3). To validate this change, Bim transcript levels were examined by quantitative real-time PCR of the cRNA used for array hybridization. PCR analysis confirmed that there were high levels of Bim transcript within cells undergoing deletion relative to the other T-cell fates (Figure 4B). To further examine the kinetics of Bim up-regulation, Bim protein levels were measured by intracellular staining using an established protocol.25 Although the levels of Bim detected by this method are relatively low, there was still a clear and consistent up-regulation of Bim protein detected within OT-I cells undergoing deletion (Figure 4C). A small up-regulation of Bim levels was observed in OT-I cells undergoing priming, but it was not of the same magnitude as that observed during peripheral deletion. Thus, the molecular basis of Bim-mediated death is probably the selective up-regulation of Bim mRNA and protein during deletion.
We next examined the array data for any potential stimuli of Bim transcript up-regulation. Bim mediates death in response to IL-7 deprivation,26 and such growth factor deprivation is known to trigger Bim transcript induction.27 The array data indicated that IL-7Rα expression was significantly down-regulated in both deletional tolerance and immunity, but the data suggested that this down-regulation was more profound in deletional tolerance. When examined by flow cytometry, it was confirmed that a significant proportion of OT-I cells reexpressed IL-7Rα subsequent to stimulation under immunogenic conditions, as has been reported previously.28 In striking contrast, IL-7Rα reexpression was largely absent in T cells undergoing tolerance induction (Figure 4D). This suggests that transcriptional induction of Bim during deletion may be caused by the loss of IL-7/IL-7R survival signaling.
Other deletion-specific changes in apoptosis-related gene expression were also evident. Strikingly, all 3 members of the Nur77 transcription factor family that cause TCR activation-induced apoptosis in thymocytes, namely, Nur77 (also Nr4a1; Nuclear receptor subfamily 4, group A, member 1), Nurr1 (also Nr4a2), and Nor1 (also Nr4a3), were selectively up-regulated during deletional tolerance (Tables 1, S3; Figure S1). Thus, this family of transcription factors could also be involved in apoptosis during peripheral T-cell deletion. In addition, there was deletion-specific up-regulation of the proapoptotic p53 target gene Perp (Table S3). This was surprising because peripheral deletion is p53 independent29 and, as such, the relevance of this expression change to cell death is unclear.
Nur77 can trigger apoptosis by transcriptional30 and nontranscriptional31 mechanisms. To assess the transcriptional contribution that this transcription factor was making to the gene signature of peripheral T-cell deletion, we compared our peripheral OT-I deletion gene signature with a published list of genes that are up-regulated in thymocytes after Nur77 overexpression.30 Of the 79 genes up-regulated by Nur77 overexpression, 5 (6%) were selectively up-regulated during peripheral deletion (Table S4; P = .02), and none of the 4 genes down-regulated by Nur77 were down-regulated during peripheral deletion (data not shown). Collectively, these data suggest that Nur77 only makes a modest contribution to the peripheral deletion gene signature.
Molecular parallels between deletion and anergy
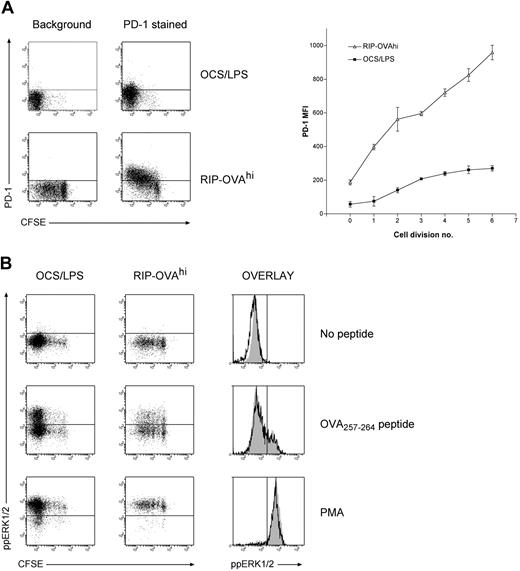
When the genes differentially expressed during deletional tolerance were examined in detail, a large proportion of these genes were found to have published or putative associations with T-cell tolerance (Tables 1,2). The most striking trend within the data was that many of the genes that were up-regulated during deletion either have functional roles in anergy induction or are associated with the anergic state. To confirm this association more rigorously, the expression changes in several genes demonstrated to be required for anergy was examined (Table 3). We chose this approach instead of direct comparison to any one dataset because there is significant disparity in array datasets between different models of anergy. Of the 14 genes required for anergy induction (as determined by knockout, knockdown, or pharmacologic inhibition studies), 5 genes (36% of the entire anergy gene list) were differentially expressed to a greater degree within OT-I cells undergoing deletion than any other cell fate. Importantly, the 5 genes were all differentially expressed in a manner expected to promote anergy. The probability of a concordant overlap is less than 10−6 by chance. One of these genes, PD-1, is essential for the induction of both anergy32 and deletion.33,34 Consistent with these studies, PD-1 was up-regulated more rapidly and to a greater extent in deletion than immunity (Figure 5A). Coupled with previous observations in CD4+ T cells,32,33 we could thus confirm that premature and excessive PD-1 up-regulation represents a universal indicator of tolerance induction.
Changes in genes required for anergy induction
| Gene symbol (no. of probe sets) . | Affymetrix probe set ID . | Tol.Naive . | Imm.Naive . | Rag.Naive . | Anergy correlation . |
|---|---|---|---|---|---|
| Cbl-b (3) | 1437304_at | 1.29* | 0.67 | 0.24 | Up |
| 1455082_at | 0.67*† | 0.33* | 0.15 | Up | |
| 1458469_at | 1.08*† | 0.39* | 0.40 | Up | |
| Cd5 (1) | No shifts | Up | |||
| Cdkn1b (p27Kip1) (2) | No shifts | Up | |||
| Ctla4 (1) | 1419334_at | 7.67* | 7.62* | −0.21 | Up |
| Dgka (1) | No shifts | Up | |||
| Dgkz (3) | 1426738_at | 1.35*† | 0.69* | 0.26 | Up |
| 1452169_a_at | 0.76*† | 0.17 | −0.07 | Up | |
| Egr2 (2) | 1427683_at | 1.63*† | −0.01 | −1.55* | Up |
| 1427682_a_at | 1.32*† | −0.07 | −1.61* | Up | |
| Egr3 (2) | 1436329_at | 0.61† | −1.38* | −1.58* | Up |
| Itch (4) | 1459332_at | −0.28* | −0.32* | −0.17 | Up |
| Nfatc2 (NFAT1) (6) | 1439205_at | 0.33* | 0.65* | 0.45* | — |
| Pag1 (4) | No shifts | — | |||
| Pdcd1 (PD-1)(1) | 1449835_at | 4.20*† | 3.28* | −0.09 | Up |
| Rnf128 (GRAIL) (2) | No shifts | Up | |||
| Tob1 (2) | 1423176_at | −0.75 | −0.92* | 0.52 | Down |
| Gene symbol (no. of probe sets) . | Affymetrix probe set ID . | Tol.Naive . | Imm.Naive . | Rag.Naive . | Anergy correlation . |
|---|---|---|---|---|---|
| Cbl-b (3) | 1437304_at | 1.29* | 0.67 | 0.24 | Up |
| 1455082_at | 0.67*† | 0.33* | 0.15 | Up | |
| 1458469_at | 1.08*† | 0.39* | 0.40 | Up | |
| Cd5 (1) | No shifts | Up | |||
| Cdkn1b (p27Kip1) (2) | No shifts | Up | |||
| Ctla4 (1) | 1419334_at | 7.67* | 7.62* | −0.21 | Up |
| Dgka (1) | No shifts | Up | |||
| Dgkz (3) | 1426738_at | 1.35*† | 0.69* | 0.26 | Up |
| 1452169_a_at | 0.76*† | 0.17 | −0.07 | Up | |
| Egr2 (2) | 1427683_at | 1.63*† | −0.01 | −1.55* | Up |
| 1427682_a_at | 1.32*† | −0.07 | −1.61* | Up | |
| Egr3 (2) | 1436329_at | 0.61† | −1.38* | −1.58* | Up |
| Itch (4) | 1459332_at | −0.28* | −0.32* | −0.17 | Up |
| Nfatc2 (NFAT1) (6) | 1439205_at | 0.33* | 0.65* | 0.45* | — |
| Pag1 (4) | No shifts | — | |||
| Pdcd1 (PD-1)(1) | 1449835_at | 4.20*† | 3.28* | −0.09 | Up |
| Rnf128 (GRAIL) (2) | No shifts | Up | |||
| Tob1 (2) | 1423176_at | −0.75 | −0.92* | 0.52 | Down |
The numbers represent log (base 2) fold change. P values were calculated independently for this gene group as described in “Microarray data analysis” in “Methods.” “No shifts” means that no statistically significant shifts were observed within any of the comparisons for the probe set(s) in question. The comparison abbreviations Tol.Naive, Imm.Naive, and Rag.Naive have the same meaning as in Figure 2. “No. of probe sets” indicates how many probe sets were present on the array for each gene listed. “Anergy correlation” indicates the shift in gene expression that has been observed in anergic cells. — indicates the gene has not been reported to shift in expression within anergic cells despite its requirement for the anergic phenotype.
Statistically significant change in gene expression.
The shift was significantly greater in Tol.Naive relative to either Imm.Naive or Rag.Naive (ie, it exhibited statistically significant up-regulation in both the Tol.Imm and Tol.Rag comparisons).
The genes listed have been shown to be required for anergy induction in various models, with findings established through knockout, knockdown, or pharmacologic inhibition studies. Most of the genes are also sufficient to induce anergy (ie, their overexpression promotes an anergic phenotype). It should be noted that most of these studies were done in CD4+ T cells and many used in vitro anergy models.
PD-1 levels and ERK1/2 activation during deletion and immunity. A total of 2 × 106 CFSE-labeled OT-I cells were injected intravenously into either RIP-OVAhi mice or OCS/LPS primed B6 mice (OCS/LPS). Sixty hours after transfer, the proliferating cells within the sacral and pancreatic lymph nodes (RIP-OVAhi) or spleen (OCS/LPS) were stained and analyzed by flow cytometry. (A) PD-1 staining showing dot plots (left) and quantitated mean fluorescence intensity (MFI; right). MFI graph shows PD-1 MFI values minus background, which was determined independently for each cell division. Error bars represent SEM. Representative data are shown from 1 of 3 independent experiments, with 3 mice per group per experiment. (B) Levels of activated (ie, phosphorylated) ERK (ppERK1/2) on in vitro peptide restimulation of T cells during immunity and deletion. Cell suspensions were either left unstimulated or restimulated with OVA257-264 peptide or PMA. Cells were then fixed, permeabilized, and stained intracellularly for ppERK1/2. Dot plots are shown for all conditions (left panels), and histograms showing ppERK1/2 levels on divided OT-I cells during both immunity (open histograms) and deletion (gray shaded histograms) are included (right panels). Representative data are shown from 1 of 2 independent experiments, with 3 mice per group per experiment.
PD-1 levels and ERK1/2 activation during deletion and immunity. A total of 2 × 106 CFSE-labeled OT-I cells were injected intravenously into either RIP-OVAhi mice or OCS/LPS primed B6 mice (OCS/LPS). Sixty hours after transfer, the proliferating cells within the sacral and pancreatic lymph nodes (RIP-OVAhi) or spleen (OCS/LPS) were stained and analyzed by flow cytometry. (A) PD-1 staining showing dot plots (left) and quantitated mean fluorescence intensity (MFI; right). MFI graph shows PD-1 MFI values minus background, which was determined independently for each cell division. Error bars represent SEM. Representative data are shown from 1 of 3 independent experiments, with 3 mice per group per experiment. (B) Levels of activated (ie, phosphorylated) ERK (ppERK1/2) on in vitro peptide restimulation of T cells during immunity and deletion. Cell suspensions were either left unstimulated or restimulated with OVA257-264 peptide or PMA. Cells were then fixed, permeabilized, and stained intracellularly for ppERK1/2. Dot plots are shown for all conditions (left panels), and histograms showing ppERK1/2 levels on divided OT-I cells during both immunity (open histograms) and deletion (gray shaded histograms) are included (right panels). Representative data are shown from 1 of 2 independent experiments, with 3 mice per group per experiment.
In addition to PD-1, a striking and selective up-regulation of the proanergy transcription factor Egr2/Krox2035 was observed in T cells undergoing deletion (Tables 1, 3; Figure S1). Egr2 expression causes up-regulation of another anergy-inducing gene, the E3 ubiquitin ligase Cbl-b.35 Consistent with elevated Egr2 transcriptional activity, Cbl-b up-regulation was detected selectively during deletion (Table 3; Figure S1). Furthermore, the related proanergy transcription factor Egr335 was significantly down-regulated in all conditions except deletion (Table 3), and Egr3 is a known inducer of Egr2 expression.36 Thus, the array data provide evidence for elevated Egr2 activity during deletion.
Anergic T cells have a selective defect in their capacity to signal via the Ras pathway on TCR triggering.37 Two negative regulators of Ras/ERK signaling associated with anergy (namely, DGKζ38,39 and Ras p21 protein activator 140 ) were transcriptionally induced within OT-I cells undergoing deletion. In addition, other negative regulators of either TCR signaling (cAMP-responsive element modulator41 and thymocyte selection-associated HMG box gene [Tox]42 ) or cell-surface receptor signaling (LIG143 and PTPσ44 ) were also up-regulated during deletion. Furthermore, several genes whose up-regulation (cAMP-responsive element modulator,45 PTPσ,46 IKAROS family zinc finger 2 [Helios],47 Pleckstrin homology, Sec7 and coiled-coil domains3 [GRP1],48 and Nr4a3 [Nor1]35 ) or down-regulation (GzmB49 ) is associated with the anergic state, were found to also be up- or down-regulated, respectively, during deletion. Thus, overall the array data suggest that similar molecular pathways are triggered in anergy and deletion.
We next examined whether there were also molecular parallels between CD8+ T-cell deletion and CD8+ T-cell exhaustion during chronic viral infection. CD8+ T-cell exhaustion represents another form of CD8+ T-cell tolerance (designed to limit immunopathology) and therefore may share features with T cells undergoing deletion. Indeed, when we compared our array dataset with the only published array data on CD8+ T-cell exhaustion,50 there was some overlap. Of the 107 genes up-regulated in exhausted cells that we examined, 17 genes (16%) displayed evidence of selective up-regulation during deletional tolerance (Table S5; P = 10−8). Furthermore, 5 of the 51 genes (10%) known to be selectively down-regulated during exhaustion were also selectively down-regulated during deletion (Table S6; P = .02). Thus, chronically stimulated cells and cells undergoing deletion exhibit some molecular parallels, but the similarities are not as extensive as those observed with anergy.
Given the overlap between the molecular signatures of deletion and anergy, we next tested whether OT-I cells undergoing deletion exhibited similar signaling defects to those seen in anergic T cells. As defective ERK1/2 phosphorylation (ie, activation) on TCR ligation is a hallmark of anergy,37 we examined the ability of OT-I cells undergoing deletion to activate (phosphorylate) ERK1/2 in response to peptide restimulation. Surprisingly, OT-I cells undergoing deletion exhibited comparable ERK1/2 phosphorylation to OT-I cells responding during immunity (Figure 5B). This was not simply the result of the high peptide dose overriding any signaling deficiency, as titrating the peptide to lower concentrations also failed to demonstrate any tolerance-specific defect (data not shown). Deficiencies in calcium signaling have also been observed in some in vivo models of anergy,51 but cells undergoing deletion fluxed calcium comparable to those being primed (data not shown). Nevertheless, it was recently demonstrated that cells acquire an anergic phenotype in deletion models if death is blocked.52 Our data thus suggest that the process of anergy induction is initiated before death, but cells do not survive long enough during deletion to develop a demonstrable anergic phenotype.
Discussion
In this study, we have provided the first detailed molecular signature of deletional tolerance through microarray analysis. Although the modest number of replicates (particularly for lymphopenia-induced proliferation) may limit the extent of our conclusions without further experimental validation, we were able to independently verify many important changes. Collectively, these data provide several novel insights into the deletion process. First, our data indicate that, although defective cytolytic gene expression may underlie the cytolytic deficiencies observed during deletion, a posttranscriptional mechanism must be responsible for impaired IFN-γ production. Second, we are able to provide a putative mechanism for Bim induction during T-cell death. Transcriptional up-regulation of Bim that is probably initiated (at least in part) by loss of IL-7/IL-7R signaling is probably sufficient to initiate apoptosis. Finally, we demonstrate that, instead of being distinct processes, the molecular pathways used in deletion and anergy overlap to a large extent. The notion of shared pathways represents an important paradigm as it implies that certain universal mechanisms, such as the PD-1 pathway, are used in disparate tolerance fates.
The deficiencies in GzmA and GzmB transcriptional induction in T cells programmed for deletion is an important observation as it provides a mechanism for the defective cytolysis observed in these cells.6 The array data did not provide evidence for differential expression of perforin between deletional tolerance and immunity, suggesting that defective expression of cytolytic effector molecules is restricted to granzymes. Nevertheless, contrary to previous observations, we did not observe perforin up-regulation relative to naive cells during priming,50 suggesting that the time point examined in these experiments may have been too early during differentiation to observe perforin up-regulation. As such, defects in perforin expression during deletion cannot be ruled out.
The potential explanation for the deficiency in granzyme (and Ly6C) induction is that expression of these factors is cell division linked and that the defect during peripheral deletion is the result of a lack of division. Indeed, during priming, maximal GzmB expression was not seen until later cell divisions and OT-I cells undergoing deletion in RIP-OVAhi mice fail to reach such late cell divisions. Nevertheless, it should be noted that the acquisition of CTL effector functions cannot be precipitated in a model of deletion by merely increasing the proliferative capacity of the responding cells.53 It therefore remains possible that active transcriptional repression of Ly6C and granzyme expression occurs during deletion. Indeed, defective GzmB expression appears to be a hallmark of CD8+ T-cell tolerance as anergic CD8+ T cells exhibit a similar defect.49 There was no differential expression of the major factors that drive granzyme up-regulation and CTL effector function between peripheral deletion and immunity (namely, Eomesodermin,54 Notch2, RBP-J, and CREB155 ; data not shown). However, there was a nonsignificant trend within the array data toward increased expression of the GzmB repressor Bcl-656 during deletion, which may have relevance to impaired granzyme expression. Overall, the observed granzyme deficiency suggests that studying GzmA and GzmB transcriptional activation will provide insights into the transcriptional regulators of tolerance. In contrast to GzmB, our array data indicated that IFN-γ transcription is up-regulated to a similar extent in deletional tolerance and immunity. This was surprising, as an inability to produce IFN-γ on peptide restimulation in culture is generally regarded as a hallmark of T cells undergoing deletion,6 an observation that we have also confirmed in the RIP-OVAhi system (I.A.P. and W.R.H., unpublished observations, 2006). These data imply that the critical point of regulation for IFN-γ production is posttranscriptional. Indeed, similar observations were recently made in exhausted CD8+ T cells.50
The array data also provided evidence for a deletional tolerance-specific death pathway. Although death during both the contraction of an immune response and peripheral deletion requires the BH3-only protein Bim,2,57 we show that the kinetics with which this pathway is activated are distinct. Rapid and early Bim transcript up-regulation was evident in T cells undergoing deletion but not in primed T cells. Such Bim up-regulation probably plays a major role in apoptosis induction as Bim overexpression alone is sufficient to trigger cell death.58 Coupled with the down-regulation of Bcl-2 that occurs on T-cell activation, Bim up-regulation could supply a potent death stimulus.
Although many factors could trigger early Bim induction, deficiencies in IL-7Rα chain expression during deletion probably contribute to this process. Interestingly, T cells undergoing deletion failed to reexpress IL-7Rα, whereas a significant fraction of primed T cells did reexpress this receptor, implying that T cells being deleted are deprived of IL-7/IL-7R survival signaling. The IL-7–dependent survival signal is predominantly transduced via the Akt signaling pathway59 and shutdown of the Akt signaling pathway can trigger Bim transcriptional up-regulation via the FOXO3A transcription factor.27 Based on this and the observation that IL-7 deprivation triggers Bim-dependent death,26 we hypothesize that defects in IL-7Rα chain expression contribute to Bim transcriptional induction during peripheral deletion.
The proapoptotic transcription factor Nur77 and its close relatives Nor1 and Nurr1 were also found to be up-regulated during T-cell deletion. Although these transcription factors have been postulated to induce transcriptional up-regulation of Bim,60 this appears doubtful as a detailed analysis of the Bim promoter failed to identify functional Nur77 binding sites (P. Bouillet and A.S., unpublished observations, 2000). Furthermore, as we failed to find evidence of a strong transcriptional contribution of Nur77 to the peripheral deletion gene expression signature, it is possible that Nur77 may be exerting its effect by nontranscriptional induction of apoptosis, as has been reported.31 Nevertheless, it is interesting that Nur77 and Bim, factors both important in T cell–negative selection within the thymus,61,62 are up-regulated during peripheral T-cell deletion. It is tempting to speculate that a conserved death pathway is used in both thymic and peripheral T-cell deletion.
The most striking pattern within the array data was the differential expression of several anergy-associated genes during deletion. This observation suggests that cells are differentiating along a pathway of anergy during deletion and that these cell fates are not as molecularly distinct as previously proposed.1 A similar conclusion was recently reached when it was demonstrated that blocking cell death during deletion by Bim ablation leads to anergy within the cells prevented from dying.52 In this study, it was argued that anergy is sufficient to maintain peripheral tolerance as cells that escape deletion will become anergic. Rather than acquiring the molecular features of anergy subsequent to escaping deletion, we can conclude that this process begins before T-cell death. Furthermore, the up-regulation of anergy-associated genes before death raises the possibility that anergy-related genes participate in the T-cell deletion process. For example, impaired TCR signaling may contribute to the deletion process by depriving T cells of the self-peptide/major histocompatibility complex signal that they require to survive.63 Coupled with the loss of the IL-7/IL-7R survival signal, CD8+ T cells being deleted could thus be deprived of the 2 main extracellular signals they require for survival, thereby triggering apoptosis. The observation that PD-1 signaling, a pathway reported to induce anergy in T cells,32 is required for peripheral T-cell deletion33,34 lends support to the idea that the anergy gene program plays a role in the T-cell deletion process.
In addition to the theoretical implications, our findings have a practical application. Thus far, relatively few definitive markers of T cells undergoing deletion have been identified, despite studies involving extensive screening.6,19 Defective IFN-γ production is regarded as a definitive indicator of cells undergoing tolerance,6 although cells may pass through a cytokine-producing effector phase en route to deletion.64 Early RANKL19 and PD-1 up-regulation33 were also identified as markers of T-cell deletion, changes that were evident in our array data. However, we were able to identify several novel markers that can be easily analyzed by flow cytometry, namely, the absence (or low levels) of GzmB, Ly6C, and IL-7Rα chain expression as well as excessive Bim up-regulation. Using this more definitive phenotypic description, it should be possible to better determine the fate of CD8+ T lymphocytes early during cell fate determination.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank J. Langley, M. Camilleri, L. Mackiewicz, and M. Dayton for their technical assistance and K. Shortman and L. O'Reilly for kindly supplying reagents.
This work was supported by the National Health and Medical Research Council (Australia), Howard Hughes Medical Institute (W.R.H.), and the Australian government's Co-operative Research Center Program (I.A.P.).
Authorship
Contribution: I.A.P., S.R., and W.R.H. designed the project; I.A.P. performed most experiments with contributions from T.J. and G.M.D.; G.K.S. designed and performed statistical analyses; G.S.D. generated important array data analysis tools; A.S. provided key concepts, supervision, and suggestions; and W.R.H. and I.A.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ian A. Parish, Immunobiology Department, Yale School of Medicine, 333 Cedar St, New Haven, CT 06520; e-mail: ian.parish@yale.edu; or William R. Heath, Department of Microbiology and Immunology, University of Melbourne, Parkville, Victoria 3010, Australia; e-mail: wrheath@unimelb.edu.au.