Abstract
Therapy-related acute myeloid leukemia (t-AML) is a rare but fatal complication of cytotoxic therapy. Whereas sporadic cancer results from interactions between complex exposures and low-penetrance alleles, t-AML results from an acute exposure to a limited number of potent genotoxins. Consequently, we hypothesized that the effect sizes of variants associated with t-AML would be greater than in sporadic cancer, and, therefore, that these variants could be detected even in a modest-sized cohort. To test this, we undertook an association study in 80 cases and 150 controls using Affymetrix Mapping 10K arrays. Even at nominal significance thresholds, we found a significant excess of associations over chance; for example, although 6 associations were expected at P less than .001, we found 15 (Penrich = .002). To replicate our findings, we genotyped the 10 most significantly associated single nucleotide polymorphisms (SNPs) in an independent t-AML cohort (n = 70) and obtained evidence of association with t-AML for 3 SNPs in the subset of patients with loss of chromosomes 5 or 7 or both, acquired abnormalities associated with prior exposure to alkylator chemotherapy. Thus, we conclude that the effect of genetic factors contributing to cancer risk is potentiated and more readily discernable in t-AML compared with sporadic cancer.
Introduction
It is believed that the genetic contribution to cancer susceptibility is similar to that of other common diseases; that is, cancer results from the complex interplay of environmental exposures and many susceptibility alleles, each of which contributes only a small amount to overall risk. Recent large genome-wide association studies (GWASs) in breast cancer and prostate cancer support this model,1–4 as do several studies in a variety of other cancers.5 Although these studies have yielded a small number of inherited genetic variants associated with cancer risk, they remain plagued by high rates of false positivity, and their success remains dependent on large sample sizes. Furthermore, the odds ratios attributable to even those risk alleles with evidence for consistent and highly significant associations are often too low to be clinically meaningful.
Therapy-related acute myeloid leukemia (t-AML) and myelodysplastic syndrome (t-MDS; collectively referred to as t-AML) are increasingly common complications of prior cytotoxic therapy. Currently comprising 10% to 20% of all cases of AML,6,7 t-AML is typically resistant to conventional AML treatment and is associated with poor outcome8–12 ; the median life expectancy from diagnosis is 8 to 10 months.7,13 Two distinct subtypes of t-AML have been described. The more common, comprising approximately 75% of cases, occurs 3 to 10 years after exposure to alkylating agents or radiation, is often preceded by a myelodysplastic syndrome, and is frequently accompanied by clonal unbalanced cytogenetic abnormalities, such as the loss of all or part of chromosomes 5 or 7 or both.7,14,15 Mutations of the TP53 tumor suppressor gene are also common.16 Risk is related to total cumulative dose of alkylating agents. The less common subtype occurs among persons treated with topoisomerase II inhibitors such as etoposide, doxorubicin, or mitoxantrone. It is characterized by a typical latency to t-AML of only 1 to 3 years, antecedent MDS is rare, and balanced rearrangements involving MLL at 11q23 or RUNX1/AML1 at 21q22 are common. Risk is less clearly related to cumulative dose, but it is associated with dosing schedule.7,14,17 Ominously, some data suggest that 12% to 30% of patients treated with epipodophyllotoxin-type topoisomerase II inhibitors develop t-AML.18
There are data to suggest that genetic factors contribute to t-AML risk. Specifically, variants in drug-metabolizing genes, DNA repair genes, and genes that regulate hematopoietic development are associated with increased t-AML susceptibility.19–24 Those studies, however, were small in size and investigated only a small number of single nucleotide polymorphisms (SNPs) in candidate genes chosen because of a priori assumptions about their likely contribution to leukemogenesis.
One tenet of pharmacogenetic research is that well-characterized clinical outcomes that can be ascribed to a drug exposure are often associated with relatively large genetic effects (reviewed in Nelson et al25 ). Hence, we reasoned that the potent mutagenic exposure associated with t-AML would magnify the contribution of genetic factors to t-AML risk. We hypothesized that the effect sizes of risk alleles in t-AML would be greater than in sporadic cancer, and, consequently, that associations would be detectable in a much smaller patient cohort than is typically required for a GWAS.
To test this, we genotyped nonmalignant DNA from 80 well-characterized t-AML patients of European descent followed at the University of Chicago and 150 healthy controls with the use of the Affymetrix GeneChip Human Mapping 10K array platform. A validation cohort of 70 white t-AML cases and 95 cancer-free controls was assembled at a second site, Washington University School of Medicine. Given that germ line samples from patients with t-AML are extremely scarce, showing that a robust association signal can be detected in a modest-sized sample would be a particularly important first step for future genomic investigations in t-AML.
Methods
Patients and controls
Cases.
The discovery cohort was composed of patients of European descent with t-AML (n = 80) ascertained in the Section of Hematology/Oncology at the University of Chicago from 1980 to 2002 (UC cases). The criterion for inclusion was availability of an EBV-immortalized lymphoblastoid cell line for germ line DNA preparation. Sixty-two patients presented initially with t-AML and 18 with t-MDS. Among those treated with chemotherapy, most were treated with multiple agents, often including both alkylating agents and topoisomerase II inhibitors. The clinical characteristics of these patients are described in Table 1. A validation cohort composed of 70 patients of European descent with t-AML from Washington University in St Louis (WU cases) is also described in Table 1. All samples from both case cohorts were collected at the time of diagnosis before the initiation of therapy for t-AML.
Clinical characteristics of the t-AML cases in the 2 study cohorts
| Characteristic . | University of Chicago (n = 80) . | Washington University (n = 70) . |
|---|---|---|
| Sex, n (%) | ||
| Female | 44 (55) | 36 (51) |
| Male | 36 (45) | 34 (49) |
| Primary therapy, n (%) | ||
| Radiotherapy (RT) | 11 (14) | 7 (10) |
| Chemotherapy (CT) | 32 (40) | 23 (33) |
| Combined modality therapy (CMT) | 37 (46) | 40 (57) |
| Primary diagnosis, n (%) | ||
| Hematologic malignancies | 40 (50)* | 27 (39)† |
| Solid tumors | 38 (48)‡ | 36 (51)§ |
| Nonmalignant disorders | 2 (3)‖ | 7 (10)¶ |
| Therapy-related myeloid neoplasm, n (%) | ||
| t-AML | 62 (78) | 33 (47) |
| t-MDS | 18 (23) | 37 (53) |
| Cytogenetic features, n (%) | ||
| Loss of chromosome 5, 7, or both | 51 (65)# | 25 (36) |
| MLL or RUNX1 translocations | 9 (12) | 8 (11)** |
| Normal karyotype | 8 (10) | 12 (17) |
| Other karyotypes | 10 (13) | 25 (36) |
| Age at first cancer, y†† | 50 (7-85) | 51 (12-78) |
| Age at t-MDS/t-AML diagnosis, y†† | 59 (13-90) | 59 (19-87) |
| Latency, mo†† | 60.5 (11-639) | 56.4 (4-187) |
| Characteristic . | University of Chicago (n = 80) . | Washington University (n = 70) . |
|---|---|---|
| Sex, n (%) | ||
| Female | 44 (55) | 36 (51) |
| Male | 36 (45) | 34 (49) |
| Primary therapy, n (%) | ||
| Radiotherapy (RT) | 11 (14) | 7 (10) |
| Chemotherapy (CT) | 32 (40) | 23 (33) |
| Combined modality therapy (CMT) | 37 (46) | 40 (57) |
| Primary diagnosis, n (%) | ||
| Hematologic malignancies | 40 (50)* | 27 (39)† |
| Solid tumors | 38 (48)‡ | 36 (51)§ |
| Nonmalignant disorders | 2 (3)‖ | 7 (10)¶ |
| Therapy-related myeloid neoplasm, n (%) | ||
| t-AML | 62 (78) | 33 (47) |
| t-MDS | 18 (23) | 37 (53) |
| Cytogenetic features, n (%) | ||
| Loss of chromosome 5, 7, or both | 51 (65)# | 25 (36) |
| MLL or RUNX1 translocations | 9 (12) | 8 (11)** |
| Normal karyotype | 8 (10) | 12 (17) |
| Other karyotypes | 10 (13) | 25 (36) |
| Age at first cancer, y†† | 50 (7-85) | 51 (12-78) |
| Age at t-MDS/t-AML diagnosis, y†† | 59 (13-90) | 59 (19-87) |
| Latency, mo†† | 60.5 (11-639) | 56.4 (4-187) |
Non-Hodgkin lymphoma (n = 16), Hodgkin lymphoma (n = 14), nonmyeloid leukemia (n = 9), multiple myeloma (n = 1).
Non-Hodgkin lymphoma (n = 14), multiple myeloma (n = 6), Hodgkin lymphoma (n = 4), nonmyeloid leukemia (n = 3).
Includes cancers of the breast (n = 17), ovaries (n = 4), prostate (n = 3), cervix (n = 2), thyroid (n = 2), esophagus, head and neck, lung, rectum, stomach, testis, vulva, astrocytoma, sarcoma, and a primary site unknown.
Includes cancers of the breast (n = 18), rectum (n = 4), prostate (n = 2), uterus (n = 2), testis (n = 2), central nervous system (n = 2), thyroid, lung, head and neck, Ewing sarcoma, melanoma, and carcinoid.
Crohn disease (n = 2).
¶Multiple sclerosis (n = 3), scleroderma, rheumatoid arthritis, aplastic anemia, cyclic neutropenia.
Cytogenetic data available for 78 of 80 patients.
Fluorescence in situ hybridization for translocations was performed in 23 of 70 patients.
Values are median; minimum to maximum in parentheses.
Controls.
Unrelated North American persons (n = 150) of European descent with no history of cancer or cytotoxic therapies were used as a comparison group (UC controls, for details see Suresh et al26 ). As controls for the WU validation cohort, 95 persons of European descent 64 years of age or older with no personal history of cancer (excluding nonmelanoma skin cancer) were selected from the Cancer Free Control cohort accrued by the Siteman Cancer Center Hereditary Cancer Core (WU controls).
Both groups of controls were selected because the cases are drawn from a broad catchment area, and these controls represent a similarly broad sampling of Americans of European descent.
DNA methods
For the UC cases, genomic DNA from nonmalignant cells was isolated from cryopreserved EBV-immortalized lymphoblastoid cell lines (2 × 106 cells) established from peripheral blood lymphocytes. For the UC controls, DNA was isolated from 3 mL of whole blood. For the WU cases, nonmalignant DNA was extracted from 6-mm punch biopsies of skin, and, for the WU controls, DNA was isolated from peripheral blood leukocytes. DNA was isolated from all UC samples with the use of the PureGene DNA extraction kit (Gentra Systems, Minneapolis, MN). For the WU cases, DNA was isolated with the use of the Puregene Core Kit A (Gentra Systems), and, for the WU controls, DNA was isolated with the use of the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA). DNA isolated from lymphoblastoid cell lines (LCLs) is well accepted as virtually indistinguishable from the original source DNA (http://www.hapmap.org/hapmappopulations.html.en).27,28 Thus, differences in source DNA are extremely unlikely to affect genotype or association results.
Informed consent was obtained from all participants in accordance with the institutional review board requirements of the University of Chicago and Washington University and the Declaration of Helsinki.
Array methods
Control samples were genotyped on the Affymetrix GeneChip Human Mapping 10K Xba131 Array (Affymetrix, Santa Clara, CA), containing 11 560 SNPs throughout the genome. Cases were genotyped on the Affymetrix GeneChip Human Mapping 10K Xba142 Array, containing 10 204 SNPs. This array is an improved version of the 10K Xba131 array, in which noninformative SNPs were replaced by more informative SNPs. These 2 arrays share 10 136 SNPs, 9771 of which are autosomal. For each sample, 250 ng of total genomic DNA was arrayed according to the manufacturer's protocol as previously described.29 Genotypes were assigned with the use of the DM algorithm as either AA, AB, BB, or NoCall.30,31
Genotyping quality control methods
Tests of association and genotyping quality control measures were undertaken with the use of PLINK32 (http://pngu.mgh.harvard.edu/purcell/plink/). We excluded from further analysis persons with call rates less than 90%, SNPs with call rates less than 90% in either cases or controls, and SNPs with a minor allele frequency (MAF) less than 0.01 in controls or less than 0.025 in the combined case–control cohort. We tested for departures from Hardy-Weinberg equilibrium (HWE) in controls with the use of an exact test33 and excluded from further analysis SNPs with P values less than .001. Population substructure was assessed with the use of STRUCTURE 2.2.34,35
Pyrosequencing
Genotyping in the WU validation cohort was undertaken by pyrosequencing. Primers (sequences provided in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were designed with the use of Pyrosequencing Assay Design Software (version 1.0.6; Biotage, Uppsala, Sweden). Pyrosequencing was performed as previously described.36
Statistical methods
Allele frequency comparisons between case and control samples were made with an allele-based Fisher exact test.37 To determine whether there was an excess of associations in our data relative to what is expected by chance, we randomly reassigned case and control status for all samples while leaving the genotypes unperturbed, recalculated the Fisher exact test for each SNP in the permuted dataset, repeated the permutation 100 000 times, and stored the P values. Permuting case-control status disassociates any existing disease or genotype association. Thus, given any statistical threshold for association, all P values that exceed that threshold in a permuted dataset represent chance associations. If the number of SNPs exceeding a given significance threshold in the original dataset is much greater than the number observed in the permuted datasets, this provides evidence for an enrichment of true SNP–disease associations in our original dataset. We assessed the significance of this enrichment at multiple significance thresholds by determining empirical enrichment P values. For each P value threshold investigated, these enrichment P values were calculated as the proportion of permuted datasets with as many or more associations than our observed dataset. For example, if we observed 400 P values less than or equal to .05 in the original dataset, but only 50 of the 100 000 permuted datasets had 400 or more significant associations at this P value threshold, then the Penrich would be 50/100 000 or less than .001. Permutations and statistical tests of significance were conducted with the use of R version 2.7.1 (ISBN 3-900051-07-0; http://www.R-project.org).
We searched for recurrent copy number variation (CNV) in t-AML with the CNAT 4.0 software (Affymetrix). Details are available at http://www.affymetrix.com/support/technical/whitepapers/cnat_4_algorithm_whitepaper.pdf.
Results
Enrichment of associations in t-AML
Two of 80 UC cases and 2 of 150 controls were excluded from analysis because of call rates less than 90%. Autosomal SNPs (n = 9771)were genotyped in both cases and controls, of which 6218 remained for analysis after applying quality control (QC) methods to remove rare or noninformative SNPs, with a mean intermarker distance of 467 kb (range, 330 kb-1.09 Mb). These QC filters improved call rates from 94.8% to 98.0% in controls and from 92.4% to 98.3% in cases. The overall call rate for the combined case–control cohort in overlapping SNPs was 98.4%. Using the program STRUCTURE, we found no evidence of population substructure in our data.
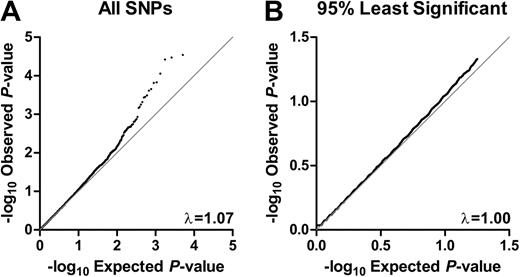
P values were individually calculated for each SNP with a Fisher exact test of allele frequency differences between cases and controls. The distribution of P values for the entire dataset was compared with that of the expected distribution (Figure 1A), and the genomic control parameter38 was found to be 1.07. When the distribution of only the bottom 95% of P values was compared with that of the expected distribution, however, the genomic control parameter was 1.00, suggesting that there was no systematic increase in false positives because of population stratification, genotyping errors, or other forms of bias in our data such as batch effect as a result of genotyping chips, platform, or DNA source (Figure 1B).
Quantile–quantile plots. Quantile-quantile plots for (A) all associations and (B) the bottom 95% of associations. Dots are the uncorrected test statistics. Under the null hypothesis of no association at any locus, the dots would be expected to follow the black line. (A) For all P values, the genomic control parameter = 1.07. (B) For the bottom 95% of P values, the genomic control parameter = 1.00.
Quantile–quantile plots. Quantile-quantile plots for (A) all associations and (B) the bottom 95% of associations. Dots are the uncorrected test statistics. Under the null hypothesis of no association at any locus, the dots would be expected to follow the black line. (A) For all P values, the genomic control parameter = 1.07. (B) For the bottom 95% of P values, the genomic control parameter = 1.00.
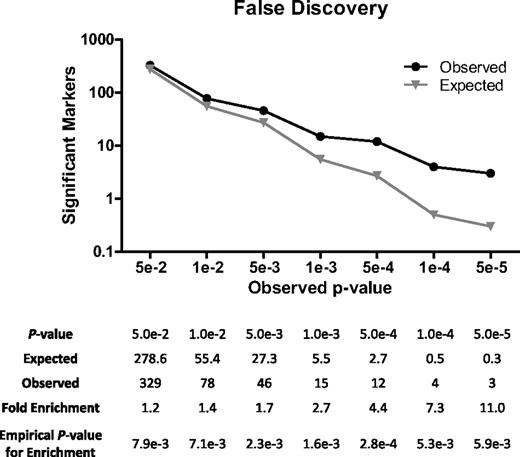
Below a P value threshold of .05, we found a significant excess of associations over what would have been expected by chance (Figure 2). At P value less than or equal to .05, for example, 329 markers were associated with t-AML compared with 279 expected by chance (a 1.2-fold enrichment, Penrich = .008). AtP values less than or equal to .005, 46 markers were associated compared with 27 expected by chance (a 1.7-fold enrichment, Penrich = .002). And at a P value less than or equal to .001, 15 markers were associated compared with only 6 expected by chance (a 2.7-fold enrichment, Penrich = .002). Thus, we conclude that SNPs truly associated with t-AML are markedly overrepresented even at these nominal significance thresholds and that this enrichment is unlikely to be the result of chance.
Assessment of false discovery. Case and control identifiers were removed, and SNP data were permuted 100 000 times between cases and controls while maintaining the relationship of each genotype to all others for each person. Association with cases was determined for each genotype in the permuted datasets and contrasted against the number of genotypes associated with cases in the real dataset at various levels of significance.
Assessment of false discovery. Case and control identifiers were removed, and SNP data were permuted 100 000 times between cases and controls while maintaining the relationship of each genotype to all others for each person. Association with cases was determined for each genotype in the permuted datasets and contrasted against the number of genotypes associated with cases in the real dataset at various levels of significance.
Association of polymorphic variants and t-AML
At a significance threshold (P = .005), 46 SNPs were associated with t-AML compared with 27 expected by chance. These SNPs are described in Table 2 along with associated genes. All SNPs are common, with MAFs in controls ranging from 0.02 to 0.54 (using MAFs in cases as the reference values) and were in HWE in both cases and controls. For most variants, the MAFs in controls were quite similar to those reported in the HapMap project for persons of European descent (CEU MAFs), suggesting that there were no systematic genotyping errors. The odds ratios (ORs) for these associated SNPs deviated from one more so than is commonly observed in sporadic cancer and other complex diseases, suggesting they are of higher penetrance. For example, the most significant SNP (rs953509) had an OR of 2.68 (CI, 1.70-4.24), and the third most significant SNP (rs1394384) had an OR of 0.29 (CI, 0.16-0.55). Twenty-five of 46 SNPs are associated with known or predicted genes; of these, 15 are in introns, and 1 results in a synonymous change in a hypothetical protein (FLJ40243 encoding LOC133558). There was no evidence to suggest that the observed associations were modified by either sex or primary cancer type (solid tumor or hematologic; data not shown).
SNP markers associated with t-AML at P < .005
| Rank . | SNP . | Chrom . | Physical position . | MAF . | Fisher exact P . | Odds ratio (95% CI) . | Gene* . | ||
|---|---|---|---|---|---|---|---|---|---|
| CEU . | Controls . | Cases . | |||||||
| 1 | rs953509 | 9q21.31 | 81560347 | 0.217 | 0.172 | 0.357 | 2.88E−05 | 2.68 (1.70-4.24) | TLE4† |
| 2 | rs719293 | 2p16.3 | 50516523 | 0.100 | 0.085 | 0.000 | 3.36E−05 | NA | NRXN1‡ |
| 3 | rs1394384 | 17q12 | 28813156 | 0.200 | 0.250 | 0.089 | 3.79E−05 | 0.29 (0.16-0.55) | ACCN1‡ |
| 4 | rs1609772 | 1q31.1 | 186820222 | 0.250 | 0.355 | 0.180 | 8.76E−05 | 0.40 (0.25-0.64) | |
| 5 | rs556831 | 18p11.31 | 4379879 | 0.050 | 0.025 | 0.118 | 1.48E−04 | 5.20 (2.12-12.76) | |
| 6 | rs1381392 | 3p24.1 | 28724318 | 0.175 | 0.128 | 0.282 | 1.54E−04 | 2.68 (1.62-4.45) | |
| 7 | rs2375990 | 4p14 | 36289759 | 0.092 | 0.075 | 0.000 | 2.22E−04 | NA | |
| 8 | rs1374284 | 2q13 | 113470054 | 0.467 | 0.395 | 0.577 | 2.44E−04 | 2.09 (1.41-3.09) | IL1F6†/IL1F9† |
| 9 | rs1335546 | 10p12.1 | 26683539 | 0.483 | 0.378 | 0.558 | 3.24E−04 | 2.07 (1.40-3.07) | GAD2† |
| 10 | rs1394605 | 5q33.3 | 155692365 | 0.150 | 0.151 | 0.040 | 3.55E−04 | 0.23 (0.10-0.56) | SGCD‡ |
| 11 | rs2133508 | 4p15.2 | 24779283 | 0.058 | 0.068 | 0.000 | 3.68E−04 | NA | SEPSECS† |
| 12 | rs957553 | 1q31.1 | 186692232 | 0.300 | 0.356 | 0.195 | 4.79E−04 | 0.44 (0.27-0.70) | |
| 13 | rs1394606 | 5q33.3 | 155692424 | 0.150 | 0.155 | 0.047 | 5.62E−04 | 0.27 (0.12-0.60) | SGCD‡ |
| 14 | rs1199098 | 10q21.1 | 59619742 | 0.192 | 0.269 | 0.128 | 6.37E−04 | 0.40 (0.23-0.69) | IPMK† |
| 15 | rs666282 | 1q31.1 | 186162828 | NA | 0.226 | 0.092 | 6.88E−04 | 0.34 (0.34-0.65) | |
| 16 | rs996725 | 1p31.1 | 82534997 | 0.392 | 0.297 | 0.455 | 1.16E−03 | 1.98 (1.32-2.97) | |
| 17 | rs2320289 | 4p15.32 | 17771202 | 0.233 | 0.223 | 0.103 | 1.31E−03 | 0.40 (0.22-0.72) | |
| 18 | rs2187987 | 11q23.2 | 114491510 | 0.250 | 0.192 | 0.333 | 1.42E−03 | 2.10 (1.34-3.29) | |
| 19 | rs1378094 | 15q12 | 25039127 | 0.050 | 0.017 | 0.083 | 1.55E−03 | 5.15 (1.80-14.72) | GABRG3‡ |
| 20 | rs722575 | 5p13.1 | 41033992 | 0.425 | 0.327 | 0.481 | 1.56E−03 | 1.91 (1.28-2.84) | FLJ40243§ |
| 21 | rs718220 | 6q15 | 88606514 | 0.300 | 0.267 | 0.136 | 1.78E−03 | 0.43 (0.26-0.73) | AY927641‡ |
| 22 | rs217190 | 10q25.3 | 116451412 | 0.058 | 0.170 | 0.066 | 1.84E−03 | 0.34 (0.17-0.70) | ABLIM1‡ |
| 23 | rs1938684 | 11q13.2 | 68986287 | 0.183 | 0.166 | 0.064 | 1.94E−03 | 0.35 (0.17-0.70) | |
| 24 | rs2255408 | 15q24.2 | 74290350 | 0.142 | 0.142 | 0.263 | 2.11E−03 | 2.16 (1.33-3.50) | C15orf27†/ETFA† |
| 25 | rs925261 | 11q23.3 | 118921537 | 0.108 | 0.075 | 0.007 | 2.12E−03 | 0.09 (0.01-0.65) | |
| 26 | rs2046733 | 11p11.2 | 45404666 | NA | 0.307 | 0.173 | 2.13E−03 | 0.47 (0.29-0.77) | |
| 27 | rs1394999 | 4q31.22 | 145544980 | 0.442 | 0.459 | 0.307 | 2.16E−03 | 0.52 (0.34-0.79) | |
| 28 | rs2416733 | 9q33.1 | 121784551 | 0.492 | 0.538 | 0.380 | 2.18E−03 | 0.53 (0.35-0.79) | |
| 29 | rs1595752 | 2p25.2 | 4863708 | 0.117 | 0.139 | 0.260 | 2.31E−03 | 2.19 (1.33-3.59) | |
| 30 | rs35000 | 5q14.1 | 80312992 | 0.375 | 0.257 | 0.401 | 2.35E−03 | 1.94 (1.28-2.94) | RASGRF2‡ |
| 31 | rs1326251 | 10q11.23 | 52918572 | 0.133 | 0.068 | 0.162 | 2.52E−03 | 2.67 (1.43-4.99) | PRKG1‡ |
| 32 | rs723147 | 4p13 | 44241768 | NA | 0.098 | 0.021 | 2.59E−03 | 0.19 (0.06-0.65) | |
| 33 | rs1017002 | 7p21.3 | 8717957 | 0.397 | 0.372 | 0.519 | 2.72E−03 | 1.83 (1.23-2.71) | NXPH1‡ |
| 34 | rs1351865 | 3p26.3 | 653347 | 0.475 | 0.500 | 0.351 | 2.73E−03 | 0.54 (0.36-0.81) | AK126307‡ |
| 35 | rs1116180 | 5p12 | 44478700 | 0.133 | 0.153 | 0.056 | 2.93E−03 | 0.33 (0.15-0.72) | |
| 36 | rs728676 | 5q23.1 | 118087433 | 0.483 | 0.429 | 0.577 | 3.01E−03 | 1.82 (1.23-2.69) | |
| 37 | rs951848 | 4q31.22 | 145544797 | 0.442 | 0.459 | 0.314 | 3.43E−03 | 0.54 (0.36-0.81) | |
| 38 | rs1878275 | 4p16.1 | 9386499 | NA | 0.051 | 0.000 | 3.56E−03 | NA | DRD5† |
| 39 | rs34999 | 5q14.1 | 80312801 | 0.375 | 0.260 | 0.397 | 3.70E−03 | 1.88 (1.24-2.83) | RASGRF2‡ |
| 40 | rs1390669 | 5p14.3 | 21667361 | 0.050 | 0.038 | 0.112 | 3.77E−03 | 3.17 (1.45-6.96) | BC038535‡ |
| 41 | rs564367 | 1p32.1 | 60891519 | 0.408 | 0.503 | 0.359 | 3.92E−03 | 0.55 (0.37-0.82) | AK097193† |
| 42 | rs1980888 | 9q22.2 | 91090376 | 0.100 | 0.080 | 0.173 | 4.35E−03 | 2.41 (1.33-4.37) | |
| 43 | rs1961495 | 13q34 | 109679374 | 0.142 | 0.153 | 0.061 | 4.77E−03 | 0.36 (0.17-0.76) | COL4A1‡ |
| 44 | rs1343700 | 3q21.1 | 125054444 | 0.308 | 0.309 | 0.449 | 4.81E−03 | 1.82 (1.21-2.73) | MYLK‡ |
| 45 | rs959100 | 2q36.1 | 224162196 | 0.242 | 0.284 | 0.160 | 4.82E−03 | 0.48 (0.29-0.80) | SCG2† |
| 46 | rs1603681 | 8q11.1 | 47400858 | 0.367 | 0.346 | 0.218 | 4.99E−03 | 0.53 (0.34-0.83) | |
| Rank . | SNP . | Chrom . | Physical position . | MAF . | Fisher exact P . | Odds ratio (95% CI) . | Gene* . | ||
|---|---|---|---|---|---|---|---|---|---|
| CEU . | Controls . | Cases . | |||||||
| 1 | rs953509 | 9q21.31 | 81560347 | 0.217 | 0.172 | 0.357 | 2.88E−05 | 2.68 (1.70-4.24) | TLE4† |
| 2 | rs719293 | 2p16.3 | 50516523 | 0.100 | 0.085 | 0.000 | 3.36E−05 | NA | NRXN1‡ |
| 3 | rs1394384 | 17q12 | 28813156 | 0.200 | 0.250 | 0.089 | 3.79E−05 | 0.29 (0.16-0.55) | ACCN1‡ |
| 4 | rs1609772 | 1q31.1 | 186820222 | 0.250 | 0.355 | 0.180 | 8.76E−05 | 0.40 (0.25-0.64) | |
| 5 | rs556831 | 18p11.31 | 4379879 | 0.050 | 0.025 | 0.118 | 1.48E−04 | 5.20 (2.12-12.76) | |
| 6 | rs1381392 | 3p24.1 | 28724318 | 0.175 | 0.128 | 0.282 | 1.54E−04 | 2.68 (1.62-4.45) | |
| 7 | rs2375990 | 4p14 | 36289759 | 0.092 | 0.075 | 0.000 | 2.22E−04 | NA | |
| 8 | rs1374284 | 2q13 | 113470054 | 0.467 | 0.395 | 0.577 | 2.44E−04 | 2.09 (1.41-3.09) | IL1F6†/IL1F9† |
| 9 | rs1335546 | 10p12.1 | 26683539 | 0.483 | 0.378 | 0.558 | 3.24E−04 | 2.07 (1.40-3.07) | GAD2† |
| 10 | rs1394605 | 5q33.3 | 155692365 | 0.150 | 0.151 | 0.040 | 3.55E−04 | 0.23 (0.10-0.56) | SGCD‡ |
| 11 | rs2133508 | 4p15.2 | 24779283 | 0.058 | 0.068 | 0.000 | 3.68E−04 | NA | SEPSECS† |
| 12 | rs957553 | 1q31.1 | 186692232 | 0.300 | 0.356 | 0.195 | 4.79E−04 | 0.44 (0.27-0.70) | |
| 13 | rs1394606 | 5q33.3 | 155692424 | 0.150 | 0.155 | 0.047 | 5.62E−04 | 0.27 (0.12-0.60) | SGCD‡ |
| 14 | rs1199098 | 10q21.1 | 59619742 | 0.192 | 0.269 | 0.128 | 6.37E−04 | 0.40 (0.23-0.69) | IPMK† |
| 15 | rs666282 | 1q31.1 | 186162828 | NA | 0.226 | 0.092 | 6.88E−04 | 0.34 (0.34-0.65) | |
| 16 | rs996725 | 1p31.1 | 82534997 | 0.392 | 0.297 | 0.455 | 1.16E−03 | 1.98 (1.32-2.97) | |
| 17 | rs2320289 | 4p15.32 | 17771202 | 0.233 | 0.223 | 0.103 | 1.31E−03 | 0.40 (0.22-0.72) | |
| 18 | rs2187987 | 11q23.2 | 114491510 | 0.250 | 0.192 | 0.333 | 1.42E−03 | 2.10 (1.34-3.29) | |
| 19 | rs1378094 | 15q12 | 25039127 | 0.050 | 0.017 | 0.083 | 1.55E−03 | 5.15 (1.80-14.72) | GABRG3‡ |
| 20 | rs722575 | 5p13.1 | 41033992 | 0.425 | 0.327 | 0.481 | 1.56E−03 | 1.91 (1.28-2.84) | FLJ40243§ |
| 21 | rs718220 | 6q15 | 88606514 | 0.300 | 0.267 | 0.136 | 1.78E−03 | 0.43 (0.26-0.73) | AY927641‡ |
| 22 | rs217190 | 10q25.3 | 116451412 | 0.058 | 0.170 | 0.066 | 1.84E−03 | 0.34 (0.17-0.70) | ABLIM1‡ |
| 23 | rs1938684 | 11q13.2 | 68986287 | 0.183 | 0.166 | 0.064 | 1.94E−03 | 0.35 (0.17-0.70) | |
| 24 | rs2255408 | 15q24.2 | 74290350 | 0.142 | 0.142 | 0.263 | 2.11E−03 | 2.16 (1.33-3.50) | C15orf27†/ETFA† |
| 25 | rs925261 | 11q23.3 | 118921537 | 0.108 | 0.075 | 0.007 | 2.12E−03 | 0.09 (0.01-0.65) | |
| 26 | rs2046733 | 11p11.2 | 45404666 | NA | 0.307 | 0.173 | 2.13E−03 | 0.47 (0.29-0.77) | |
| 27 | rs1394999 | 4q31.22 | 145544980 | 0.442 | 0.459 | 0.307 | 2.16E−03 | 0.52 (0.34-0.79) | |
| 28 | rs2416733 | 9q33.1 | 121784551 | 0.492 | 0.538 | 0.380 | 2.18E−03 | 0.53 (0.35-0.79) | |
| 29 | rs1595752 | 2p25.2 | 4863708 | 0.117 | 0.139 | 0.260 | 2.31E−03 | 2.19 (1.33-3.59) | |
| 30 | rs35000 | 5q14.1 | 80312992 | 0.375 | 0.257 | 0.401 | 2.35E−03 | 1.94 (1.28-2.94) | RASGRF2‡ |
| 31 | rs1326251 | 10q11.23 | 52918572 | 0.133 | 0.068 | 0.162 | 2.52E−03 | 2.67 (1.43-4.99) | PRKG1‡ |
| 32 | rs723147 | 4p13 | 44241768 | NA | 0.098 | 0.021 | 2.59E−03 | 0.19 (0.06-0.65) | |
| 33 | rs1017002 | 7p21.3 | 8717957 | 0.397 | 0.372 | 0.519 | 2.72E−03 | 1.83 (1.23-2.71) | NXPH1‡ |
| 34 | rs1351865 | 3p26.3 | 653347 | 0.475 | 0.500 | 0.351 | 2.73E−03 | 0.54 (0.36-0.81) | AK126307‡ |
| 35 | rs1116180 | 5p12 | 44478700 | 0.133 | 0.153 | 0.056 | 2.93E−03 | 0.33 (0.15-0.72) | |
| 36 | rs728676 | 5q23.1 | 118087433 | 0.483 | 0.429 | 0.577 | 3.01E−03 | 1.82 (1.23-2.69) | |
| 37 | rs951848 | 4q31.22 | 145544797 | 0.442 | 0.459 | 0.314 | 3.43E−03 | 0.54 (0.36-0.81) | |
| 38 | rs1878275 | 4p16.1 | 9386499 | NA | 0.051 | 0.000 | 3.56E−03 | NA | DRD5† |
| 39 | rs34999 | 5q14.1 | 80312801 | 0.375 | 0.260 | 0.397 | 3.70E−03 | 1.88 (1.24-2.83) | RASGRF2‡ |
| 40 | rs1390669 | 5p14.3 | 21667361 | 0.050 | 0.038 | 0.112 | 3.77E−03 | 3.17 (1.45-6.96) | BC038535‡ |
| 41 | rs564367 | 1p32.1 | 60891519 | 0.408 | 0.503 | 0.359 | 3.92E−03 | 0.55 (0.37-0.82) | AK097193† |
| 42 | rs1980888 | 9q22.2 | 91090376 | 0.100 | 0.080 | 0.173 | 4.35E−03 | 2.41 (1.33-4.37) | |
| 43 | rs1961495 | 13q34 | 109679374 | 0.142 | 0.153 | 0.061 | 4.77E−03 | 0.36 (0.17-0.76) | COL4A1‡ |
| 44 | rs1343700 | 3q21.1 | 125054444 | 0.308 | 0.309 | 0.449 | 4.81E−03 | 1.82 (1.21-2.73) | MYLK‡ |
| 45 | rs959100 | 2q36.1 | 224162196 | 0.242 | 0.284 | 0.160 | 4.82E−03 | 0.48 (0.29-0.80) | SCG2† |
| 46 | rs1603681 | 8q11.1 | 47400858 | 0.367 | 0.346 | 0.218 | 4.99E−03 | 0.53 (0.34-0.83) | |
SNPs are listed by decreasing significance of association, along with their chromosomal location (Chrom) and physical position along the chromosome. Genes tagged by SNPs are indicated. Data are from release 22 (HapMap Phase II), based on dbSNP build 36 (http://www.hapmap.org).
MAF indicates minor allele frequency; NA, not applicable. CEU (Centre d'Etude du Polymorphisme Humain (CEPH) European) MAFs are from the 60 unrelated Utah residents with ancestry in northern and western Europe genotyped as part of the HapMap project.
A gene is defined as its genomic sequence ± 10 kb.
SNP is in linkage disequilibrium (LD) with the gene.
SNP is intronic to the gene.
SNP is in the coding region of the gene, resulting in a synonymous amino acid substitution.
At P value of .001, the enrichment for SNPs associated with t-AML was almost 3-fold. Of the 15 SNPs surpassing this threshold, 3 were absent in cases but had MAFs ranging from 0.068 to 0.085 in controls (rs719293, rs2375990, and rs2133508). For 5 of the 15 SNPs, the minor allele was associated with increased t-AML risk, whereas for the other 10 SNPs the more common variant was associated with t-AML. Nine of these 15 SNPs are in linkage disequilibrium (LD) with known genes; of these, 4 SNPs are intronic to known genes.
Of note, 3 markers are located within 660 kb of each other at 1q31.1 (rs666282, rs957553, and rs1609772). No known or predicted gene, regulatory element, or microRNA maps within 500 kb of this region. Two of these SNPs (rs957553 and rs1609772) are in LD (r2 = 0.62), whereas the third is not in LD with the other 2. To determine whether these 2 SNPs in LD represent a single association, we conditioned on the genotype of one SNP while testing for association in the other. These tests were not significant, suggesting that they are probably tagging the same as-yet-unidentified disease-associated variant. Thus, these 3 SNPs represent 2 distinct association results within this 660-kb region. Although it remains unclear why this region of 1q is associated with t-AML, SNPs in similar so-called gene deserts have been reported and validated in other complex diseases such as type 2 diabetes39 and chronic lymphocytic leukemia.5
We also analyzed our samples for copy number alterations, but found no evidence of recurrent copy number variants in the nonmalignant DNA of patients with t-AML.
Replication of associations in a validation cohort
Replication analysis in the entire validation cohort.
We hypothesized that it would be easier to replicate associations in t-AML than in other complex diseases, first, because risk alleles for t-AML have larger effect sizes than associations in other complex diseases and sporadically occurring cancers, and, second, because the heterogeneity of environmental exposures contributing to t-AML is markedly reduced relative to other complex diseases. We attempted to replicate our most significant associations (15 SNPs with P < .001) by genotyping these SNPs in an independent validation cohort (the WU cohort) of 70 white patients with t-AML (Table 1) and 95 WU controls. Two SNPs in strong LD with other SNPs among these (rs1609772 and rs1394605) were excluded. Three SNPs with MAF less than 0.1 in cases and controls (rs719293, rs2375990, and rs2133508) were also excluded, because there was insufficient power to detect a significant difference below this threshold (power = 37%, α = 0.05) in the WU cohort. All 10 SNPs genotyped in the validation cohort were in HWE in both cases and controls.
Of the 10 SNPs, we found that 2 SNPs significantly associated with t-AML in the UC discovery cohort trended toward significance in the WU validation cohort as well (Table 3I). For rs1394384, the distribution of genotypes in the WU controls was similar to that observed for the UC controls, and the allele frequencies in cases similarly differed from the allele frequencies in controls in both cohorts with regard to direction and magnitude (UC: fcontrols = 0.25, fcases = 0.09; WU: fcontrols = 0.23, fcases = 0.16). This variant was the third most significant SNP in the UC cohort, and its P value approached significance in the WU validation cohort (P = .094). For rs1381392, again, the genotype frequencies were similar between controls in the 2 cohorts, and allele frequencies in cases differed similarly from controls in both cohorts (UC: fcontrols = 0.13, fcases = 0.28; WU: fcontrols = 0.14, fcases = 0.19). This variant was the sixth most significant SNP in the UC cohort, and again the P value approached significance in the WU cohort (P = .14).
Replication results for tests of association in the WU validation cohort as compared with the original UC cohort
| Rank . | SNP . | University of Chicago . | Washington University . | Combined . | |||||
|---|---|---|---|---|---|---|---|---|---|
| MAF . | Fisher exact P* . | MAF . | Fisher exact P† . | ||||||
| Controls . | Cases . | Controls . | Cases . | Fisher exact P* . | OR (95% CI) . | ||||
| I. Allele frequency comparisons conducted between all cases and all controls in each cohort (UC: 78 cases, 148 controls; WU: 70 cases, 95 controls) | |||||||||
| 1 | rs953509 | 0.172 | 0.357 | 2.88E−05 | 0.204 | 0.169 | .826 | 8.18E−03 | 1.62 (1.14-2.30) |
| 3 | rs1394384 | 0.250 | 0.089 | 3.79E−05 | 0.228 | 0.162 | .094 | 6.88E−05 | 0.45 (0.29-0.67) |
| 5 | rs556831 | 0.025 | 0.118 | 1.48E−04 | 0.042 | 0.051 | .455 | 2.20E−03 | 2.85 (1.48-5.50) |
| 6 | rs1381392 | 0.128 | 0.282 | 1.54E−04 | 0.142 | 0.193 | .140 | 3.22E−04 | 2.02 (1.38-2.96) |
| 8 | rs1374284 | 0.395 | 0.577 | 2.44E−04 | 0.458 | 0.448 | .615 | 9.15E−03 | 1.48 (1.11-1.98) |
| 9 | rs1335546 | 0.378 | 0.558 | 3.24E−04 | 0.420 | 0.421 | .536 | 7.39E−03 | 1.49 (1.12-2.00) |
| 12 | rs957553 | 0.356 | 0.195 | 4.79E−04 | 0.268 | 0.321 | .875 | .050 | 0.72 (0.52-0.99) |
| 13 | rs1394606 | 0.155 | 0.047 | 5.62E−04 | 0.121 | 0.121 | .574 | .016 | 0.55 (0.33-0.89) |
| 14 | rs1199098 | 0.269 | 0.128 | 6.37E−04 | 0.210 | 0.239 | .775 | .046 | 0.68 (0.47-0.98) |
| 15 | rs666282 | 0.226 | 0.092 | 6.88E−04 | 0.117 | 0.181 | .962 | .102 | 0.70 (0.46-1.07) |
| II. Allele frequency comparisons conducted only between cases with abnormalities of chromosomes 5 or 7 or both and all controls in each cohort (UC: 51 cases, 148 controls; WU: 25 cases; 95 controls) | |||||||||
| 1 | rs953509 | 0.172 | 0.296 | .015 | 0.204 | 0.130 | .917 | .176 | 1.38 (0.87-2.19) |
| 3 | rs1394384 | 0.250 | 0.083 | 7.54E−04 | 0.228 | 0.087 | .022 | 4.55E−05‡ | 0.29 (0.15-0.56) |
| 5 | rs556831 | 0.025 | 0.140 | 1.80E−04 | 0.042 | 0.060 | .411 | 1.02E−03 | 3.74 (1.78-7.87) |
| 6 | rs1381392 | 0.128 | 0.256 | 8.81E−03 | 0.142 | 0.220 | .131 | 4.19E−03‡ | 2.08 (1.29-3.35) |
| 8 | rs1374284 | 0.395 | 0.589 | 1.55E−03 | 0.458 | 0.435 | .672 | .019 | 1.60 (1.09-2.35) |
| 9 | rs1335546 | 0.378 | 0.544 | 7.05E−03 | 0.420 | 0.280 | .977 | .243 | 1.26 (0.86-1.84) |
| 12 | rs957553 | 0.356 | 0.182 | 1.71E−03 | 0.268 | 0.348 | .894 | .071 | 0.66 (0.43-1.03) |
| 13 | rs1394606 | 0.155 | 0.068 | .034 | 0.121 | 0.140 | .733 | .156 | 0.63 (0.34-1.18) |
| 14 | rs1199098 | 0.269 | 0.122 | 4.06E−03 | 0.210 | 0.146 | .218 | 3.43E−03‡ | 0.46 (0.27-0.79) |
| 15 | rs666282 | 0.226 | 0.105 | .023 | 0.117 | 0.200 | .956 | .353 | 0.75 (0.43-1.30) |
| Rank . | SNP . | University of Chicago . | Washington University . | Combined . | |||||
|---|---|---|---|---|---|---|---|---|---|
| MAF . | Fisher exact P* . | MAF . | Fisher exact P† . | ||||||
| Controls . | Cases . | Controls . | Cases . | Fisher exact P* . | OR (95% CI) . | ||||
| I. Allele frequency comparisons conducted between all cases and all controls in each cohort (UC: 78 cases, 148 controls; WU: 70 cases, 95 controls) | |||||||||
| 1 | rs953509 | 0.172 | 0.357 | 2.88E−05 | 0.204 | 0.169 | .826 | 8.18E−03 | 1.62 (1.14-2.30) |
| 3 | rs1394384 | 0.250 | 0.089 | 3.79E−05 | 0.228 | 0.162 | .094 | 6.88E−05 | 0.45 (0.29-0.67) |
| 5 | rs556831 | 0.025 | 0.118 | 1.48E−04 | 0.042 | 0.051 | .455 | 2.20E−03 | 2.85 (1.48-5.50) |
| 6 | rs1381392 | 0.128 | 0.282 | 1.54E−04 | 0.142 | 0.193 | .140 | 3.22E−04 | 2.02 (1.38-2.96) |
| 8 | rs1374284 | 0.395 | 0.577 | 2.44E−04 | 0.458 | 0.448 | .615 | 9.15E−03 | 1.48 (1.11-1.98) |
| 9 | rs1335546 | 0.378 | 0.558 | 3.24E−04 | 0.420 | 0.421 | .536 | 7.39E−03 | 1.49 (1.12-2.00) |
| 12 | rs957553 | 0.356 | 0.195 | 4.79E−04 | 0.268 | 0.321 | .875 | .050 | 0.72 (0.52-0.99) |
| 13 | rs1394606 | 0.155 | 0.047 | 5.62E−04 | 0.121 | 0.121 | .574 | .016 | 0.55 (0.33-0.89) |
| 14 | rs1199098 | 0.269 | 0.128 | 6.37E−04 | 0.210 | 0.239 | .775 | .046 | 0.68 (0.47-0.98) |
| 15 | rs666282 | 0.226 | 0.092 | 6.88E−04 | 0.117 | 0.181 | .962 | .102 | 0.70 (0.46-1.07) |
| II. Allele frequency comparisons conducted only between cases with abnormalities of chromosomes 5 or 7 or both and all controls in each cohort (UC: 51 cases, 148 controls; WU: 25 cases; 95 controls) | |||||||||
| 1 | rs953509 | 0.172 | 0.296 | .015 | 0.204 | 0.130 | .917 | .176 | 1.38 (0.87-2.19) |
| 3 | rs1394384 | 0.250 | 0.083 | 7.54E−04 | 0.228 | 0.087 | .022 | 4.55E−05‡ | 0.29 (0.15-0.56) |
| 5 | rs556831 | 0.025 | 0.140 | 1.80E−04 | 0.042 | 0.060 | .411 | 1.02E−03 | 3.74 (1.78-7.87) |
| 6 | rs1381392 | 0.128 | 0.256 | 8.81E−03 | 0.142 | 0.220 | .131 | 4.19E−03‡ | 2.08 (1.29-3.35) |
| 8 | rs1374284 | 0.395 | 0.589 | 1.55E−03 | 0.458 | 0.435 | .672 | .019 | 1.60 (1.09-2.35) |
| 9 | rs1335546 | 0.378 | 0.544 | 7.05E−03 | 0.420 | 0.280 | .977 | .243 | 1.26 (0.86-1.84) |
| 12 | rs957553 | 0.356 | 0.182 | 1.71E−03 | 0.268 | 0.348 | .894 | .071 | 0.66 (0.43-1.03) |
| 13 | rs1394606 | 0.155 | 0.068 | .034 | 0.121 | 0.140 | .733 | .156 | 0.63 (0.34-1.18) |
| 14 | rs1199098 | 0.269 | 0.122 | 4.06E−03 | 0.210 | 0.146 | .218 | 3.43E−03‡ | 0.46 (0.27-0.79) |
| 15 | rs666282 | 0.226 | 0.105 | .023 | 0.117 | 0.200 | .956 | .353 | 0.75 (0.43-1.30) |
SNP markers genotyped in the WU cohort are listed by descending order of significance along with their rank in the UC cohort. Site-adjusted odds ratios (ORs) are given for the combined cohort with 95% CIs.
MAF indicates minor allele frequency.
Denotes a 2-sided Fisher exact test.
Denotes a 1-sided Fisher exact test.
SNPs are more significantly associated with t-AML in the combined cohort than in the original UC cohort.
Replication analysis in patients with abnormalities of chromosomes 5 or 7 or both.
In other studies, it has been shown that genetic associations with t-AML can be modified, based on prior treatment.40 t-AML resulting from prior exposure to alkylator therapy is frequently associated with acquired abnormalities of chromosomes 5 or 7 or both. Although these abnormalities were found in 65% of the UC case cohort, they were found in only 36% of the WU case cohort (P < .001). We hypothesized that by analyzing cohorts composed of differing proportions of patients with causatively distinct types of t-AML, we were masking true associations; consequently, to compare homogeneous populations, we undertook a subset analysis by testing for replication only in the subset of patients in each case cohort with abnormalities of chromosomes 5 or 7 or both (Table 3II).
For rs1394384, although the allele frequencies in cases differed between the entire UC and WU case cohorts (MAF = 0.089 and 0.162, respectively), they were similar in the abnormal 5/7 subset (UC: fcontrols = 0.25, fcases = 0.083; WU: fcontrols = 0.23, fcases = 0.087). This SNP was significantly associated with t-AML in the WU cohort (P = .022) as well as in the UC cohort. For rs1381392, the allele frequencies were also more similar in the abnormal 5/7 subset of both cohorts than in the entire cohorts (UC: fcontrols = 0.13, fcases = 0.26; WU: fcontrols = 0.14, fcases = 0.22). Finally, for rs1199098, although the allele frequencies were quite dissimilar in the entire UC and WU case cohorts (UC: fcontrols = 0.27, fcases = 0.13; WU: fcontrols = 0.21, fcases = 0.24), they were similar in patients with abnormalities of chromosomes 5 or 7 or both (UC: fcontrols = 0.27, fcases = 0.12; WU: fcontrols = 0.21, fcases = 0.15). For all 3 SNPs, the associations with t-AML were more significant in the combined UC and WU cohort of patients with abnormalities of chromosomes 5 or 7 or both than in the UC cohort alone, suggesting that their associations with t-AML are indeed robust (Table 3II). That the P value for neither rs1381392 nor rs1199098 was significant in patients with abnormalities of chromosomes 5 or 7 or both in the WU cohort probably reflects the limited size of this subset of patients in this cohort.
Thus, in this analysis undertaken in a biologically homogeneous subset of patients with t-AML, 3 of 10 associations detected in only 80 cases and 150 controls were validated. These results strongly support our hypothesis that t-AML is a powerful model to detect cancer-associated genetic variation.
Discussion
t-AML results from DNA damage induced by cytotoxic therapy for a primary condition, most often a malignant disease. This damage engages response pathways in hematopoietic stem and progenitor cells, leading to DNA repair or cell death. Cells that survive with acquired mutations because of non- or misrepair are at risk for leukemic transformation. Genetic variation in pathways that mediate cellular responses to DNA damage can affect the risk of developing t-AML, presumably by influencing the survival of hematopoietic cells with proleukemogenic mutations.
Currently, it is not possible to identify the patients at greatest risk for t-AML. For common diseases, considerable theoretical and empirical data suggest that the contribution to disease risk of most associated genetic variants is modest. For example, in the Wellcome Trust Case Control Consortium (WTCCC) GWAS of 7 common diseases with 14 000 cases and 3000 shared controls, the ORs of disease-associated variants ranged from 1.2 to 1.5.41 Recent studies suggest the contribution of genetic factors to sporadic cancer risk is similar.1,3 Because these effect sizes are so small, it has been necessary to genotype large numbers of samples to achieve sufficient power for the reliable identification of disease-associated genetic variants in GWASs. As an illustration, Easton et al1 screened more than 225 000 SNPs tagging 58% of the common genetic variation in persons of European descent (at an r2 > 0.8) for association with breast cancer in almost 400 cases and 400 controls. Of these, the top 12 500 (5%) were analyzed in another 4000 cases and 4000 controls, of which the top 30 were tested in more than 21 000 cases and 21 000 controls. Ultimately, only 6 of these 225 000 SNPs were replicated as the result of this massive effort, with effect sizes ranging from 1.2 to 1.6.1
In contrast, we genotyped 6218 SNPs, tagging a much smaller proportion of the genome, in the 80 cases and 150 controls comprising the UC discovery cohort, and subsequently analyzed only 10 SNPs in the 70 cases and 95 controls comprising the WU validation cohort. We validated 3 of these 10 SNPs in the biologically homogeneous subset of patients with abnormalities of chromosomes 5 or 7 or both (rs1394384, rs1381392, and rs1199098). Although chip or hybridization-batch differences may have introduced some undetected systemic bias or error in the data, our rigorous QC analysis and the fact that our validation cohort was genotyped by a different method (pyrosequencing) strongly argue against this possibility.
These results are particularly compelling in that they suggest that exposure is a potent modifier of t-AML susceptibility. They also underscore the importance of incorporating biology into assessments of associations in complex diseases. Thus, our study suggests that conditioning on a potent exposure such as cytotoxic therapy is a novel and powerful strategy to enhance the detection of genetic variation truly associated with complex diseases in GWAS. Furthermore, it suggests that t-AML, even when limited by small sample size, is a robust model for the identification of cancer-associated genetic variation.
None of the 3 validated SNPs identified here have been studied previously in t-AML. rs1394384 is intronic to ACCN1, a gene encoding an amiloride-sensitive cation channel that is a member of the degenerin/epithelial sodium channel (DEG/ENaC) superfamily.42 It is highly conserved throughout evolution and has been associated with neurodegeneration in Caenorhabditis elegans,43 is expressed in bone marrow and hematopoietic cells, and has been associated with autism44 and multiple sclerosis.45 rs1199098 is in LD with IPMK, which encodes a multikinase that positively regulates the prosurvival AKT kinase and may modulate Wnt/beta-catenin signaling.46,47 Finally, rs1381392 is not near any known genes, miRNAs, or regulatory elements, although it lies in a region recurrently deleted in lung cancer.48
Of the other top associations identified in the UC cohort, rs953509 is in LD with TLE4, a candidate tumor suppressor commonly deleted in AML that encodes a transcriptional corepressor of PAX5-mediated transcriptional activation49 and WNT-pathway signaling.50,51 rs1374284 is linked to a cluster of 9 genes comprising the inflammatory cytokine IL-1 family. Although the function of many proteins encoded by these genes remains unknown, IL-1β has been shown to regulate cell proliferation and apoptosis resistance in AML blasts52 and also promotes tissue invasion by leukemic cells.53 Surprisingly, 2 other top associations in addition to rs1394384 in ACCN1 (rs719293 and rs1335546) are linked to genes involved in determining neuronal phenotypes or associated with neurodegenerative disorders (rs719293 and NRXN1, and rs1335546 and GAD2), as are several SNPs exceeding the significance threshold (P = .005; GABRG3, PRKG1, NXPH1, DRD5, and SCG2). Ultimately, the contribution of these variants to t-AML risk awaits the analysis of larger t-AML patient cohorts.
This study represents an important step toward the translational goal of identifying persons at risk for t-AML at the time of their original cancer diagnosis so that their initial cancer therapy can be modified to minimize this risk. Our major findings are (1) in contrast to sporadic cancer, associations are markedly enriched in t-AML even at nominally significant P values; (2) even in a small sample set, this enrichment allows for the identification and replication of likely t-AML–predisposing genetic variants, each of which may contribute significantly to overall risk; and (3) distinct subsets of patients with t-AML may have distinct inherited susceptibilities toward t-AML. Furthermore, because cytotoxic therapy is a potent surrogate for the environmental exposures that drive sporadically occurring cancers, t-AML may be a powerful model for the study of gene-exposure interactions in sporadically occurring cancers.
More broadly, despite considerable effort, the hope of personalized medicine remains largely unrealized. Major barriers continue to limit the translation of GWAS data to the clinical arena. Here, we propose a novel strategy to identify cancer-associated genetic variation by conditioning on a potent exposure, namely, cytotoxic therapy. This pharmacogenetic approach may prove to be a highly effective new paradigm for genomic studies not only in cancer but in a variety of other complex diseases as well.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Bear Necessities Pediatric Cancer Foundation (Chicago, IL; K.O.), Leukaemia Research (London, United Kingdom; J.M.A.), the Barnes Jewish Hospital Foundation (St Louis, MO; T.A.G.), and National Institutes of Health (Bethesda, MD) grants HL007088 (R.A.W.), CA40046 and CA14599 (M.M.L.B. and R.A.L.), CA101937 (T.A.G.), and HD0433871 (K.O.).
National Institutes of Health
Authorship
Contribution: J.A.K. performed research and analyzed and interpreted the data; A.D.S. designed research, designed and performed statistical analysis, and drafted the manuscript; A.S., D.H., and T.R.T. performed research; R.A.W. and J.S. performed research and statistical analysis; M.B. generated lymphoblastoid cell lines; J.M.A. designed research and analysis and drafted the manuscript; M.M.L.B. designed research and analyzed data, contributed data and vital new reagents, and drafted the manuscript; R.A.L. analyzed data and contributed vital new reagents; T.A.G. designed and performed research, analyzed data, and drafted the manuscript; N.J.C. designed analysis and drafted the manuscript; and K.O. designed the research, analyzed and interpreted the data, and wrote the paper. All authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kenan Onel, Section of Hematology/Oncology, Department of Pediatrics, University of Chicago, 5841 S Maryland Ave, C-425, MC 4060, Chicago, IL 60637; e-mail: konel@uchicago.edu.
References
Author notes
*J.A.K. and A.D.S. contributed equally to this study.