Abstract
The Lmo2 gene encodes a transcriptional cofactor critical for the development of hematopoietic stem cells. Ectopic LMO2 expression causes leukemia in T-cell acute lymphoblastic leukemia (T-ALL) patients and severe combined immunodeficiency patients undergoing retroviral gene therapy. Tightly controlled Lmo2 expression is therefore essential, yet no comprehensive analysis of Lmo2 regulation has been published so far. By comparative genomics, we identified 17 highly conserved noncoding elements, 9 of which revealed specific acetylation marks in chromatin-immunoprecipitation and microarray (ChIP-chip) assays performed across 250 kb of the Lmo2 locus in 11 cell types covering different stages of hematopoietic differentiation. All candidate regulatory regions were tested in transgenic mice. An extended LMO2 proximal promoter fragment displayed strong endothelial activity, while the distal promoter showed weak forebrain activity. Eight of the 15 distal candidate elements functioned as enhancers, which together recapitulated the full expression pattern of Lmo2, directing expression to endothelium, hematopoietic cells, tail, and forebrain. Interestingly, distinct combinations of specific distal regulatory elements were required to extend endothelial activity of the LMO2 promoter to yolk sac or fetal liver hematopoietic cells. Finally, Sfpi1/Pu.1, Fli1, Gata2, Tal1/Scl, and Lmo2 were shown to bind to and transactivate Lmo2 hematopoietic enhancers, thus identifying key upstream regulators and positioning Lmo2 within hematopoietic regulatory networks.
Introduction
The identification and functional characterization of transcriptional regulatory elements remain principal challenges of the postgenome era. Comparative genomic analysis across vertebrates ranging from fish to mammals has enabled the discovery of highly conserved noncoding evolutionary conserved regions, yet many known distal regulatory elements are not conserved across this large evolutionary distance. By contrast, comparisons across smaller evolutionary distances, such as human/mouse, often lack sufficient discriminative power, presumably due to relatively short evolutionary distances not being sufficient to specifically highlight all regions under purifying selection.1,2 The recent development of large-scale techniques for the mapping of histone modification status or transcription factor binding therefore hold great promise as a complementary strategy to improve our ability to predict functionality of noncoding sequences. For example, studies using chromatin immunoprecipitation and microarrays (ChIP-chip) or ChIP and sequencing (ChIP-Seq), have shown that specific histone modifications are associated with either transcriptionally active or inactive chromatin.3-7 However, none of the above studies has performed in vivo validation of predicted regulatory elements, and therefore it is still unclear to what extent the combination of computational approaches and ChIP-chip/ChIP-Seq will be useful for the identification of regulatory elements.
The Lim Domain Only 2 gene (Lmo2) encodes a transcriptional cofactor originally identified through its involvement in recurrent chromosomal translocations in T-cell acute lymphoblastic leukemia (T-ALL).8,9 Mice lacking Lmo2 die around embryonic day 10.5 because of a complete failure of erythropoiesis.10 Studies of chimeric mice produced from Lmo2−/− embryonic stem (ES) cells showed that Lmo2 is also required for the formation of adult hematopoietic cells11 as well as for vascular endothelial remodeling.12 After differentiation of hematopoietic stem cells (HSC), Lmo2 expression is down-regulated in T lymphocytes, where aberrant expression of LMO2 results in T-cell leukemias.9,13-15 Transcriptional activation, as a consequence of retroviral vector integration into the LMO2 locus, has also been implicated in the development of clonal T-cell proliferation in patients undergoing gene therapy for X-linked severe combined immunodeficiency.16-18 Together, these data indicate that appropriate transcriptional control of Lmo2 is crucial for the formation and subsequent behavior of blood cells.
A stringent search for homology between evolutionarily distant species demonstrated that, apart from the coding exons, high levels of identity between mammalian, amphibian, and fish Lmo2 sequences were restricted to the proximal promoter (pP) region.19 The pP was functional in hematopoietic progenitor and endothelial cell lines, where its activity was dependent on conserved Ets sites bound by Fli1, Ets1, and Elf1. Although transgenic analysis demonstrated that the Lmo2 pP was sufficient for expression in endothelial cells in vivo, expression levels were weak, and no expression in any other Lmo2-expressing tissues was observed,19 indicating that additional as yet uncharacterized regulatory elements are present within the Lmo2 locus.
Here we have used a combination of comparative genomics, locus-wide ChIP-chip and transgenic mouse assays, which led to the identification of 8 distinct regulatory elements spread over more than 100 kb and sufficient to target expression to all embryonic tissues expressing endogenous Lmo2. Modular combinations of specific distal elements were required to extend endothelial activity of the pP to hematopoietic cells, suggesting that hematopoietic expression of Lmo2 is established on top of a preexisting endothelial regulatory framework. Moreover, identification of key hematopoietic transcription factors acting through these elements allowed us to position Lmo2 within the transcriptional networks that control blood and endothelial development.
Methods
Design and fabrication of custom array
Primers to generate the Lmo2 polymerase chain reaction (PCR) tiling array were designed using Primer320 on repeat masked sequence spanning Lmo2 and flanking genes (chr2:103636099-103886024 in build mm7). Resulting PCR fragments (median size 532 bp) were spotted in triplicate using a BioRobotics MicroGrid II Total Array System (Digilab Genomic Solutions, Ann Arbor, MI). Array design files have been submitted to ArrayExpress (accession nos. A-MEXP-1020 and A-MEXP-1021).
ChIP-chip assays
ChIP assays were performed as previously described.21 Briefly, cells were treated with formaldehyde, and cross-linked chromatin was sonicated to 300 bp averaged size. Immunoprecipations were performed using anti-acetyl histone H3 antibody (06-599; Upstate Biotechnology, Lake Placid, NY), anti-Tal (provided by C. Porcher, MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, Oxford, United Kingdom), anti-Lmo2 (AF2726; R&D Systems, Minneapolis, MN), anti-Gata2 (SC-9008X; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Fli1 (SC-356X; Santa Cruz Biotechnology), and anti-Sfpi1 (SC-352X; Santa Cruz Biotechnology). ChIP material was labeled with Cy3 and Cy5 fluorochromes and hybridized as described.22 Microarrays were scanned using an Agilent scanner (Agilent, Santa Clara, CA), and median spot intensities were quantified using GenePix Pro version 6.0 (Molecular Devices, Sunnyvale, CA) with background subtraction. A Perl script was developed to normalize the resulting data and calculate mean ratios of normalized ChIP signals over input, using the triplicate values on the array. Resulting data were plotted using the Variable Width Bar Graph Drawer (http://hscl.cimr.cam.ac.uk/genomic_tools.html). All experiments have been deposited in ArrayExpress under accession number E-TABM-431.
Sequence analysis
Genomic LMO2 sequences were downloaded from Ensembl, aligned using multi-Lagan,23 and displayed using mVista24 or Genedoc (http://www.psc.edu/biomed/genedoc). Candidate transcription factor binding sites were identified using TFBSsearch.25
Reporter constructs and transgenic analysis
LMO2 LacZ and luciferase reporter constructs were amplified from human genome using primers listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) and confirmed by sequencing. Their selection was based on the combined results of comparative genomics and ChIP-chip experiments. Detailed information on reporter constructs is available on request. Plasmids were linearized and founder transgenic embryos produced by pronuclear injection, which were subsequently harvested between E11.5 and E12.5 and analyzed as described.26 A total of 27 reporter constructs were screened using transient transgenic mouse assay. Selected embryos were cleared as described.27 Whole-mount images were acquired using a Nikon Digital Sight DS-FL1 camera attached to a Nikon SM7800 microscope (Nikon, Kingston upon Thames, United Kingdom). Images of sections were acquired with the Zeiss AxioCam MRc5 camera attached to a Zeiss Axioscope2plus microscope (Carl Zeiss, Welwyn Garden City, United Kingdom) using Olympus UPlanApo 40×/0.85 numeric aperture (NA) and 100×/1.35 NA objectives (Olympus, Tokyo, Japan). Axio Vision Rel version 4.3.1.0 software (Carl Zeiss) was used for acquisition of digital images, which were processed using Adobe Photoshop and Adobe Illustrator (Adobe Systems, San Jose, CA). All animal experiments were performed in accordance with United Kingdom Home Office rules and were approved by Home Office inspectors.
Cell culture, flow cytometry, and cell sorting
ES cells were maintained and differentiated as previously described.28 Briefly, embryoid bodies (EB) from an ES cell line with green fluorescent protein (GFP) targeted to the Brachyury gene were harvested and trypsinized, and single-cell suspensions were sorted on a MoFlo cell sorter (Cytomation Systems, Fort Collins, CO). Staining with monoclonal antibody (mAb) Flk1 bio (BD Pharmingen, San Diego, CA) was performed as previously described.29,30 HPC7 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 1.5 × 10−4 M monothioglycerol (MTG), and Steel factor as described.31 The myeloid progenitor cell line 416B, murine erythroleukemia cell line F4N (MEL), endothelial cell line MS1, and the T-lymphoid cell line BW5147 (BW) were maintained as described.32,33 Fetal liver (FL) and adult thymus cell suspensions were obtained by direct pipetting of freshly dissected tissues from mice.
Transfection assays
416B cells were stably transfected by electroporation as described.33 G418 was added 24 hours posttransfection, and cells were assayed 7 to 10 days later. For transactivation assays, 293T cells were transfected with luciferase constructs alone or in combination with the following expression constructs: pEFBOSMycTLMO2, pEFBOSMycTGATA1 or GATA2, pEFBOSFlagTal1 or Ldb1, and pcDNA3MycE47. An equivalent quantity of DNA was transfected using the empty vectors pcDNA3 and pEFBOS as controls when necessary. Each transfection and transactivation was performed on at least 2 different days in triplicate.
Results
Locus-wide comparative genomic analysis identifies 17 noncoding conserved regions representing candidate Lmo2 distal regulatory elements
Past studies have shown that highly conserved noncoding elements are often associated with genes encoding important developmental regulators, such as Lmo2.34-36 We have previously demonstrated that pan-vertebrate noncoding sequence conservation of the Lmo2 locus was restricted to a small region containing the pP.19 This region was sufficient to drive expression in endothelial cells in vivo. However, expression levels were weak, and no expression in any other Lmo2-expressing tissues was observed, suggesting the presence of additional as yet uncharacterized elements elsewhere in the Lmo2 locus. To explore whether an “intermediate” evolutionary distance would be more informative to reveal these additional elements, we took advantage of the publication of the opossum genome and compared the human, mouse, dog, and rat LMO2 loci to the opossum locus. The resulting multiple sequence alignment (see Figure 1) revealed 15 conserved regions in addition to the pP and distal promoters (dP), thus suggesting that selection of an adequate evolutionary distance may be critical for identifying candidate regulatory elements.
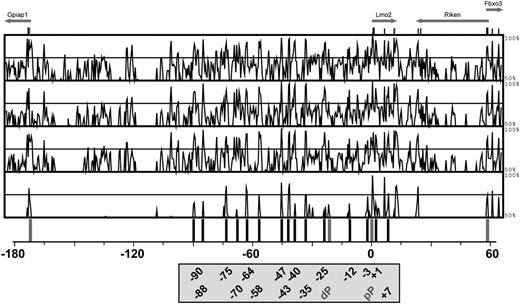
Sequence conservation between Lmo2 loci from eutherian mammals and opossum identifies 15 distal candidate regulatory elements. MVista representation of sequence conservation across 250 kb of the mouse Lmo2 locus. The conservation panels correspond to, from top to bottom, mouse/human, mouse/dog, mouse/cow, and mouse/opossum alignments. The conservation plots show regions with at least 50% of conservation (y-axis) across the 250-kb tiling path spanning the mouse Lmo2 locus, where the translation initiation (ATG) is marked as position 0 (x-axis). Shown at the top of the figure are exons with arrows pointing in the direction of transcription. The gray lines indicate promoters, and the black lines highlight the 15 noncoding conserved regions between opossum and eutherian sequences. The positions of the conserved regions are named relative to the ATG of Lmo2 and depicted in the box at the bottom of the figure (black), together with the distal (dp) and proximal (pP) Lmo2 promoters (gray).
Sequence conservation between Lmo2 loci from eutherian mammals and opossum identifies 15 distal candidate regulatory elements. MVista representation of sequence conservation across 250 kb of the mouse Lmo2 locus. The conservation panels correspond to, from top to bottom, mouse/human, mouse/dog, mouse/cow, and mouse/opossum alignments. The conservation plots show regions with at least 50% of conservation (y-axis) across the 250-kb tiling path spanning the mouse Lmo2 locus, where the translation initiation (ATG) is marked as position 0 (x-axis). Shown at the top of the figure are exons with arrows pointing in the direction of transcription. The gray lines indicate promoters, and the black lines highlight the 15 noncoding conserved regions between opossum and eutherian sequences. The positions of the conserved regions are named relative to the ATG of Lmo2 and depicted in the box at the bottom of the figure (black), together with the distal (dp) and proximal (pP) Lmo2 promoters (gray).
Locus-wide ChIP-chip analysis identifies 9 Lmo2 candidate distal regulatory elements
Driven by the previously highlighted limitations in sensitivity and specificity of comparative genomic approaches, we decided to explore experimental validation using locus-wide functional assays. To this end, we performed histone acetylation ChIP-chip analysis (H3K9ac) in 11 cell types covering different stages of hematopoiesis. We used a 250-kb tiling array, spanning the Lmo2 locus and flanking genes, to explore possible enrichment of active histone marks at regions highlighted by comparative genomic analysis (Figure 2). The cell types included non-Lmo2–expressing ES cells as well as their in vitro differentiated mesodermal and hemangioblast progeny, thus covering the earliest time point during ontogeny where Lmo2 expression is induced. Additional cell types included Lmo2-expressing murine cell lines (endothelial, hematopoietic progenitor, erythroid) and primary cells (FL) as well as a T-lymphoid cell line and whole adult thymus, cell types in which Lmo2 expression would have been extinguished. As shown in Figure 2, enrichments of H3K9ac were present at the promoters of the 2 Lmo2 flanking genes (Gpiap1 and Fbxo3) in all cell types tested. In Lmo2-expressing cells (MS1, HPC7, 416B, MEL, and FL), the pP of Lmo2 was highly acetylated with generally much lower enrichment present at the dP. Small peaks of enrichment for H3K9ac were also found at the pP of Lmo2 in nonexpressing ES and in vitro differentiated ES cells.
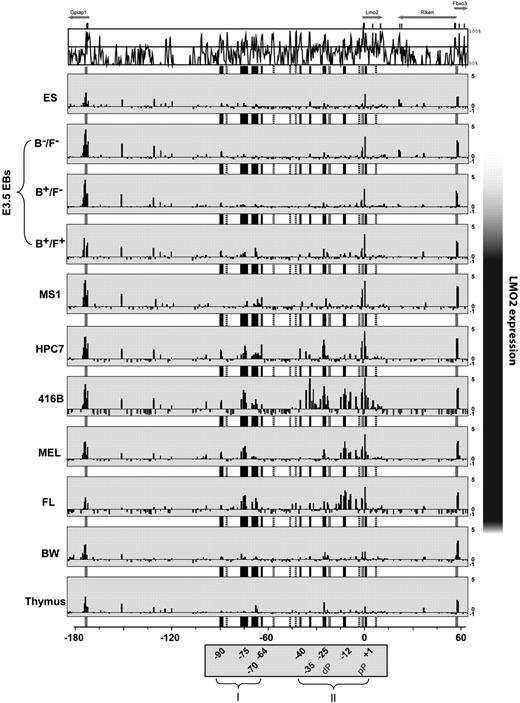
ChIP-chip assays across 250 kb of the mouse Lmo2 locus reveal candidate distal regulatory elements. ChIP-chip analysis of histone H3 acetylation in 11 hematopoietic and endothelial cell types. MVista representation of mouse/human sequence conservation is shown on the top and annotations of promoters (gray), and all candidate regulatory elements (black) are as in Figure 1, except that regions not enriched (−88, −58, −47, −43, −3, and +7) are marked by dotted lines. Enrichment cluster I comprises −90, −75, −70, −64; cluster II covers −40, −35, −25, −12, +1. The y-axis represents the log 2 enrichment of ChIPed DNA over input DNA ranging from −1 to 5, whereas the x-axis depicts the 250 kb spanning the mouse Lmo2 locus, where the translation initiation codon (ATG) is marked as position 0. The black bar on the right hand side indicates relative levels of Lmo2 expression in the 11 different cell types. The cells surveyed included non-Lmo2–expressing ES cells, as well their in vitro–differentiated EB sorted for Brachyury−/Flk1− (B−/F−: premesoderm), Brachyury+/Flk1− (B+/F−: prehemangioblast mesoderm), and Brachyury+/Flk1+ (B+/ F+: hemangioblast mesoderm). Additional cell types included Lmo2-expressing cell lines, representing endothelial progenitor MS1, multipotential hematopoietic progenitor HPC7, myeloid progenitors 416B, erythroid progenitor MEL, and primary cells derived from day 11.5 FL. In addition, cell types were used, in which Lmo2 expression is supposed to be extinguished, such as a T-lymphoid cell line (BW) and whole adult murine thymus.
ChIP-chip assays across 250 kb of the mouse Lmo2 locus reveal candidate distal regulatory elements. ChIP-chip analysis of histone H3 acetylation in 11 hematopoietic and endothelial cell types. MVista representation of mouse/human sequence conservation is shown on the top and annotations of promoters (gray), and all candidate regulatory elements (black) are as in Figure 1, except that regions not enriched (−88, −58, −47, −43, −3, and +7) are marked by dotted lines. Enrichment cluster I comprises −90, −75, −70, −64; cluster II covers −40, −35, −25, −12, +1. The y-axis represents the log 2 enrichment of ChIPed DNA over input DNA ranging from −1 to 5, whereas the x-axis depicts the 250 kb spanning the mouse Lmo2 locus, where the translation initiation codon (ATG) is marked as position 0. The black bar on the right hand side indicates relative levels of Lmo2 expression in the 11 different cell types. The cells surveyed included non-Lmo2–expressing ES cells, as well their in vitro–differentiated EB sorted for Brachyury−/Flk1− (B−/F−: premesoderm), Brachyury+/Flk1− (B+/F−: prehemangioblast mesoderm), and Brachyury+/Flk1+ (B+/ F+: hemangioblast mesoderm). Additional cell types included Lmo2-expressing cell lines, representing endothelial progenitor MS1, multipotential hematopoietic progenitor HPC7, myeloid progenitors 416B, erythroid progenitor MEL, and primary cells derived from day 11.5 FL. In addition, cell types were used, in which Lmo2 expression is supposed to be extinguished, such as a T-lymphoid cell line (BW) and whole adult murine thymus.
As our 250-kb custom array contained the entire Lmo2 locus, we were in a position to look beyond the acetylation status of promoter elements and explore the remaining noncoding section of the Lmo2 locus. Significant levels of enrichment were defined by an empirical threshold of 1 on a log 2 scale, identified on at least 2 adjacent tiles or 2 different cell-types. Interestingly, a region 1 kb downstream of the Lmo2 pP (+1 region) showed substantial levels of acetylation even in nonexpressing ES cells, which was further enhanced in all Lmo2-expressing cell types. Additional prominent peaks of enrichment found in hematopoietic cell types fell into 2 clusters: −90 to −64 and −40 to +1 (distances in kb relative to the ATG start codon). No enrichments were found on −88, −58, −47, −43, −3, or +7. The acetylation pattern of the endothelial cell line MS1 was similar to hemangioblasts (Brachyury/Flk1 double-positive cells), with prominent peaks on the pP and only minor peaks on the 2 clusters. By contrast, −90 and −75 were enriched in all Lmo2-expressing cells of hematopoietic, but not endothelial origin. In HPC7 hematopoietic progenitor cells, an additional robust peak was found at −25 and a minor peak at −40. The myeloid progenitor cell line 416B displayed extended enrichment on all elements of both clusters, with specific enrichment at −35. Consistent with its predominant erythroid nature, the pattern of FL was most similar to erythroid MEL cells, showing robust enrichments on −75 and −12, and minor enrichment on −70 and −25. T-lymphoid cells (BW, thymus) showed only very minor peaks of enrichment consistent with the fact that they represent cell types that would have turned off Lmo2 expression during their differentiation from a hematopoietic stem/progenitor cell. Peaks of acetylated histones in blood/endothelial cells were conserved between mouse and opossum and accounted for approximately two-thirds (9/15, or 12/17 if promoters included) of the regions with more than 60% sequence identity between mouse and opossum. In summary, the ChIP-chip survey allowed us to delimit 9 candidate distal regulatory elements in addition to the 2 Lmo2 promoters (Table S4).
Extension of the LMO2 pP dramatically increases activity in transgenic assays
Aside from its hematopoietic expression, Lmo2 is expressed in endothelium, specific regions of the developing brain, somites, and limbs12 (Figure 3A). We had previously shown that a 349-bp fragment of the LMO2 pP displayed weak yet reproducible endothelial-specific activity when tested in transgenic mice.19 Our new comparative genomic analysis highlighted the fact that mouse/opossum conservation was much broader than this small region of pan-vertebrate conservation. We therefore generated a new extended LMO2 pP construct (pPex) that contains 1.3-kb sequence upstream of the ATG start codon in exon 4 and compared its activity to the original smaller promoter in transgenic analysis. Our investigation focused on representative hematopoietic and endothelial tissues from the FL, dorsal aorta (DA), heart (H), yolk sac (YS) and peripheral vessels (V) with the Lmo2 LacZ knock-in serving as reference point (Figure 3A). To complete transgenic analysis of LMO2 promoters, a transgenic reporter construct for the dP, which also showed conservation across all mammals, was included.
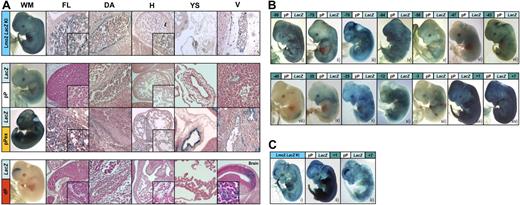
Transgenic analysis of Lmo2 candidate regulatory elements at midgestation. (A) Transgenic mouse embryos at E12.5 showing X-Gal reporter expression in a Lmo2 LacZ knock-in (Lmo2 LacZ KI) and X-Gal reporter expression driven by 3 different LMO2 promoter constructs (pPLacZ, pPexLacZ, and dpLacZ). Whole-mount staining (WM) and representative histologic sections of FL, dorsal aorta (DA), heart (H), yolk sac (YS), and peripheral vessels (V) are depicted. The Lmo2 LacZ KI embryo is macroscopically characterized by staining of vessels, brain, eyes, somites, apical ridges of the limb buds, and tail. Histologically, X-Gal is expressed in peripheral and main vessels, FL, DA, and circulating erythrocytes (see inset at higher magnification of representative areas in FL and H). Compared with the LMO2 349-bp minimal pP (pPLacZ) a 1.3-kb extended version of the pP (pPexLacZ) increases dramatically the expression in endothelial cells. In contrast, the LMO2 dP (dPLacZ) directs expression to the forebrain only (note that the brain section is placed instead of a vessel section). (B) Transgenic mouse embryos at E12.5 showing whole-mount X-Gal staining of the LMO2 minimal pP in collaboration with 1 of 14 putative enhancers. The numbers in the green box of each panel correspond to the distance in kilobases of the putative regulatory elements with respect to the mouse Lmo2 ATG as shown in Figures 1 and 2. Eight enhancer elements −90, −75, −70, −64, −25, −12, +1, and +7 significantly augment the endothelial staining of pPLacZ and/or induced LacZ expression in several additional tissues, such as tail, apical ridges of the limb buds, brain, and potentially FL. In contrast, the elements −58, −47, −43, −40, −35, and −3 show similar or less LacZ expression compared with pP. (C) Comparison of cleared, X-Gal-stained transgenic mouse embryos at E12.5 bearing the Lmo2 LacZ KI (i) and pP combined with the 2 strongest enhancers +1 (ii) and +7 (iii). Besides the strong endothelial enhancement the pP +1 construct shows strong expression in the apical ridges of the limb buds and the tail (ii), whereas pP +7 confers additional brain staining (iii).
Transgenic analysis of Lmo2 candidate regulatory elements at midgestation. (A) Transgenic mouse embryos at E12.5 showing X-Gal reporter expression in a Lmo2 LacZ knock-in (Lmo2 LacZ KI) and X-Gal reporter expression driven by 3 different LMO2 promoter constructs (pPLacZ, pPexLacZ, and dpLacZ). Whole-mount staining (WM) and representative histologic sections of FL, dorsal aorta (DA), heart (H), yolk sac (YS), and peripheral vessels (V) are depicted. The Lmo2 LacZ KI embryo is macroscopically characterized by staining of vessels, brain, eyes, somites, apical ridges of the limb buds, and tail. Histologically, X-Gal is expressed in peripheral and main vessels, FL, DA, and circulating erythrocytes (see inset at higher magnification of representative areas in FL and H). Compared with the LMO2 349-bp minimal pP (pPLacZ) a 1.3-kb extended version of the pP (pPexLacZ) increases dramatically the expression in endothelial cells. In contrast, the LMO2 dP (dPLacZ) directs expression to the forebrain only (note that the brain section is placed instead of a vessel section). (B) Transgenic mouse embryos at E12.5 showing whole-mount X-Gal staining of the LMO2 minimal pP in collaboration with 1 of 14 putative enhancers. The numbers in the green box of each panel correspond to the distance in kilobases of the putative regulatory elements with respect to the mouse Lmo2 ATG as shown in Figures 1 and 2. Eight enhancer elements −90, −75, −70, −64, −25, −12, +1, and +7 significantly augment the endothelial staining of pPLacZ and/or induced LacZ expression in several additional tissues, such as tail, apical ridges of the limb buds, brain, and potentially FL. In contrast, the elements −58, −47, −43, −40, −35, and −3 show similar or less LacZ expression compared with pP. (C) Comparison of cleared, X-Gal-stained transgenic mouse embryos at E12.5 bearing the Lmo2 LacZ KI (i) and pP combined with the 2 strongest enhancers +1 (ii) and +7 (iii). Besides the strong endothelial enhancement the pP +1 construct shows strong expression in the apical ridges of the limb buds and the tail (ii), whereas pP +7 confers additional brain staining (iii).
The results of the whole-mount transgenic analysis and histologic sections of the 3 promoter constructs (pP, pPex, and dP) are summarized in Tables S2A and S3A. Only 3 of 10 pPLacZ transgenic embryos showed any LacZ expression, which in all cases was weak and restricted to endothelial specific expression in small vessels (Figure 3A). By contrast, expression was dramatically increased in pPexLacZ transgenic embryos with strong staining of endothelium (9/10 embryos; Figure 3A). Only 1 of 8 transgenic embryos carrying the dP construct (dPLacZ) showed transgene expression, which was restricted to neuronal cells within the posterior part of the forebrain (Figure 3A). Taken together, the transgenic analysis was consistent with our ChIP-chip survey, which suggested that the pP was the predominant promoter used in endothelial and hematopoietic mouse tissues. Moreover, extension of the pP to 1.3 kb resulted in a dramatic increase of endothelial activity.
Transgenic analysis identifies 8 enhancer elements that recapitulate the whole-mount expression pattern of Lmo2 at midgestation
To test in vivo function of candidate distal regulatory elements identified by comparative genomics and ChIP-chip, we generated transgenic embryos with 14 of the candidate regions driving LacZ expression from the minimal pP construct (pPlacZ). We chose this minimal promoter construct because its activity was weaker than the extended pPex construct, which would aid the identification of possible enhancer activities. Candidate regulatory elements were assayed by transgenic analysis of E12.5 embryos (Figure 3B; whole-mount staining patterns of pP enhancer constructs are summarized in Tables S2B and S4). Eight regions (−90, −75, −70, −64, −25, −12, +1, and +7) significantly augmented the endothelial staining of pPLacZ and/or induced LacZ expression in several additional tissues, such as tail, apical ridges of the limbs, brain, and potentially FL. Constructs containing elements −58, −47, −43, −40, −35, and −3 showed similar LacZ expression as the parental pP minimal promoter suggesting that these regions may not function as classical enhancers (Figure 3B).
Because of the very strong activity of the +1 and +7 elements, it was not possible to assess the staining of internal structures. Whole-mount staining was therefore reassessed after clearing of embryos and compared with age-matched cleared Lmo2 LacZ knock-in embryos12 (Figure 3C). In addition to strong endothelial staining, limb and tail staining was present in embryos with the +1 construct, while +7 conferred brain staining (Figure 3Cii,iii). Interestingly, the Lmo2 LacZ knockin embryo displayed the same staining features, expressing the transgene in the tail, limb, brain, FL, and blood vessels (Figure 3Ci). Apart from blood expression, which is difficult to ascertain from whole-mount analysis, the above survey had therefore allowed us to identify 8 enhancer elements, which together could mediate the full pattern of endogenous Lmo2 expression in midgestation embryos.
Robust hematopoietic expression of Lmo2 requires combinatorial interaction of multiple elements
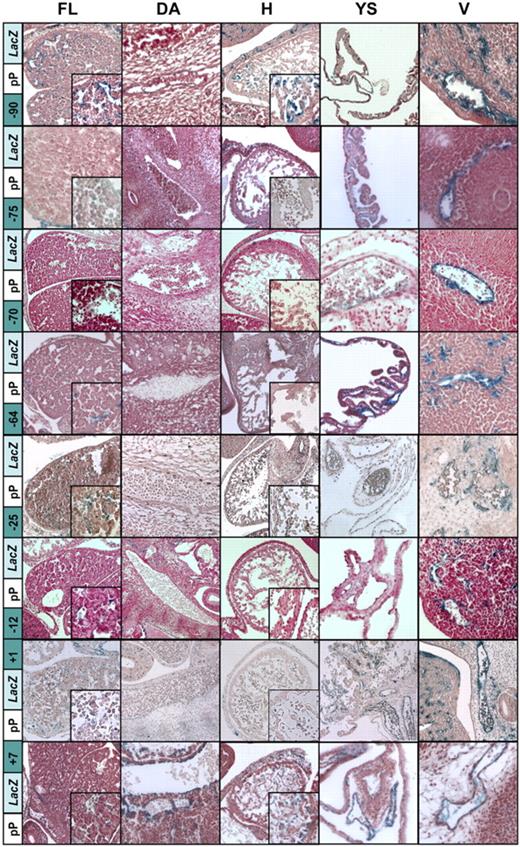
To further investigate possible hematopoietic specific activity of candidate regulatory elements, representative embryos from all 8 constructs conferring enhancer activity by whole-mount analysis were sectioned for histologic analysis (Figure 4; results are summarized in Tables S3B and S4). Consistent with the whole-mount pattern, the pPLacZ alone construct could direct only weak endothelial specific expression in small vessels in transgenic mice (Figure 3A). This weak endothelial staining pattern could be significantly enhanced by adding any of the 8 enhancer elements. Of note, the −90 and +7 elements were able to extend endothelial expression to endocardium and large vessels, including the DA. Most interestingly, the elements −90, −75, −64, −25, −12, and +1 displayed weak yet consistent expression in a minority of FL cells, whereas element −75 mediated weak staining of circulating blood cells, although overall hematopoietic staining was much less intense compared with the Lmo2 LacZ knock-in. To further quantify the hematopoietic activity of the Lmo2 enhancers, all regions tested in transgenic constructs were subcloned into luciferase reporter plasmids and stably transfected in 416B cells. The −75, −70, −64, and −25 enhancers increased the activity of the pP between 4- and 10-fold (Figure S1 and Table S4). Of note, these elements included the hematopoietic elements identified by transgenic analysis.
Histologic analysis of single enhancer constructs reveals multiple elements conferring weak hematopoietic expression. Histologic sections of FL, dorsal aorta (DA), heart (H), yolk sac (YS), and peripheral vessels (V) from transgenic mouse embryos at E12.5. X-Gal reporter expression driven by the LMO2 349-bp minimal pP (pPLacZ) combined with each of the 8 enhancers (−90, −75, −70, −64, −25, −12, +1, +7). As previously shown, the pPLacZ construct can mediate only a weak endothelial-specific activity (Figure 3B). The endothelial expression is enhanced by collaboration of pP with any of the presumed enhancer elements under investigation. The strongest endothelial enhancer activity is conferred by the elements −90 and +7, which are able to direct expression to the DA and endocardium. Most interestingly, the elements −90, −75, −64, −25, −12, and +1 confer weak and focal FL expression, whereas the element −75 mediates weak staining to circulating erythrocytes (see inset in FL, H, and V). However, overall hematopoietic staining is still less pronounced compared with the Lmo2 LacZ KI (Figure 3A).
Histologic analysis of single enhancer constructs reveals multiple elements conferring weak hematopoietic expression. Histologic sections of FL, dorsal aorta (DA), heart (H), yolk sac (YS), and peripheral vessels (V) from transgenic mouse embryos at E12.5. X-Gal reporter expression driven by the LMO2 349-bp minimal pP (pPLacZ) combined with each of the 8 enhancers (−90, −75, −70, −64, −25, −12, +1, +7). As previously shown, the pPLacZ construct can mediate only a weak endothelial-specific activity (Figure 3B). The endothelial expression is enhanced by collaboration of pP with any of the presumed enhancer elements under investigation. The strongest endothelial enhancer activity is conferred by the elements −90 and +7, which are able to direct expression to the DA and endocardium. Most interestingly, the elements −90, −75, −64, −25, −12, and +1 confer weak and focal FL expression, whereas the element −75 mediates weak staining to circulating erythrocytes (see inset in FL, H, and V). However, overall hematopoietic staining is still less pronounced compared with the Lmo2 LacZ KI (Figure 3A).
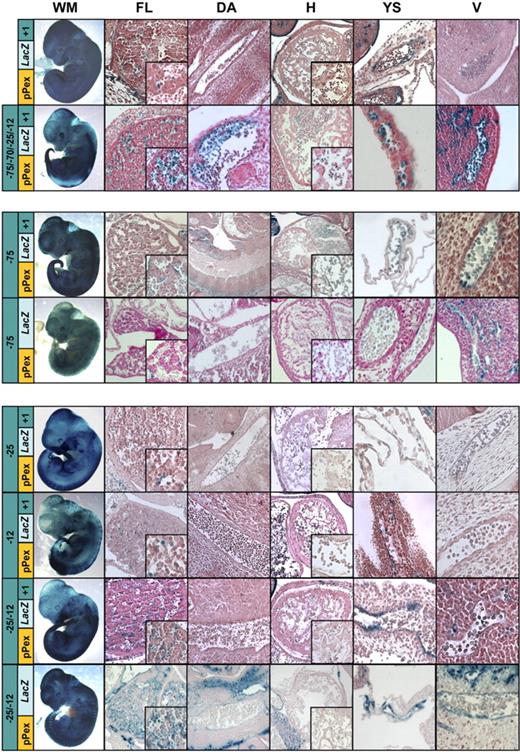
Because the extended pP, pPex, was much stronger than the minimal pP fragment, we reasoned that interactions between the extended pP and distal fragments with weak hematopoietic activity might be required to achieve more robust expression in hematopoietic tissues. We therefore generated 10 pPex multienhancer constructs and repeated the transgenic analysis. The selection of distal regulatory elements for multienhancer constructs was based on the presence of acetylation marks in ChIP-chip experiments, performance in stable transfection and hematopoietic activity in transgenic assays (summarized in Table S4). The results of the whole-mount staining and the sectioning of pPex multienhancer transgenic embryos are summarized in Tables S2C and S3C. A construct containing a combination of 5 distinct enhancer regions (−75, −70, −25, −12, and +1) showed strong staining of circulating erythrocytes and FL (Figure 5). Subsequent analysis of constructs with smaller combinations of elements demonstrated that a combination of the −75 and +1 elements (−75pPexLacZ+1 construct) was sufficient to mediate highly specific and strong staining of circulating erythrocytes, whereas −75 alone showed weaker, but still erythroid-specific activity (Figure 5). On the other hand, we found that elements −25 and −12 were able to direct consistent staining to FL cells, but not to circulating erythrocytes (Figure 5). Combinations of −25/−12, with and without +1, demonstrated that −25/−12 was sufficient to direct strong reporter gene expression to FL cells (Figure 5). Stable transfection of pPex multienhancer constructs confirmed the cell-type specific activity of the erythroid −75 element and the hematopoietic progenitor cell elements −25/−12, respectively (Figure S2). In summary, our transgenic analysis suggests that robust hematopoietic Lmo2 expression requires a combination of cell-type specific distal enhancers, which are deployed on top of a largely endothelial pP
Transgenic analysis of multienhancer constructs reveals distinct combinations able to drive expression to circulating erythroid or FL cells. Whole-mount staining (WM) and histologic sections of FL, dorsal aorta (DA), heart (H), yolk sac (YS), and peripheral vessels (V) from transgenic embryos harvested between E11.5 and E12.5. X-Gal reporter expression driven by the LMO2 1.3-kb extended pP (pPexLacZ) combined with candidate hematopoietic enhancers. The +1 construct is characterized by reduced endothelial and hematopoietic activity compared with pPex alone (Figure 3A). Addition of 5 putative hematopoietic enhancer elements (−75/−70/−25/−12/+1) induce specific expression in circulating erythrocytes and enhanced staining of FL. Systematic exclusion of elements −70, −25, and −12 reveals that the erythroid-specific expression can be attributed to the element −75, possibly in combination with element +1 (see −75 pPexLacZ+1 and −75 pPexLacZ). Robust FL expression, on the other hand, is conferred by collaboration of elements −25 and −12 (see −25 pPexLacZ+1, −12 pPexLacZ+1, −25/−12 pPexLacZ+1, and −25/−12 pPexLacZ).
Transgenic analysis of multienhancer constructs reveals distinct combinations able to drive expression to circulating erythroid or FL cells. Whole-mount staining (WM) and histologic sections of FL, dorsal aorta (DA), heart (H), yolk sac (YS), and peripheral vessels (V) from transgenic embryos harvested between E11.5 and E12.5. X-Gal reporter expression driven by the LMO2 1.3-kb extended pP (pPexLacZ) combined with candidate hematopoietic enhancers. The +1 construct is characterized by reduced endothelial and hematopoietic activity compared with pPex alone (Figure 3A). Addition of 5 putative hematopoietic enhancer elements (−75/−70/−25/−12/+1) induce specific expression in circulating erythrocytes and enhanced staining of FL. Systematic exclusion of elements −70, −25, and −12 reveals that the erythroid-specific expression can be attributed to the element −75, possibly in combination with element +1 (see −75 pPexLacZ+1 and −75 pPexLacZ). Robust FL expression, on the other hand, is conferred by collaboration of elements −25 and −12 (see −25 pPexLacZ+1, −12 pPexLacZ+1, −25/−12 pPexLacZ+1, and −25/−12 pPexLacZ).
Lmo2/Tal1 and Gata factors occupy hematopoietic elements in vivo but do not bind to the pP
Given the critical function of Lmo2 in hematopoietic cells and having identified hematopoietic cell-type specific regulatory elements, we next set out to identify upstream factors to establish the hierarchies within which Lmo2 functions in hematopoietic cells. The Lmo2 protein lacks direct DNA binding capacity, but instead functions as a bridging molecule serving to assemble multiprotein DNA-binding complexes, with the best known complex composed of the bHLH factor Tal1 and Gata factors Gata1 or Gata2.37,38 In addition to the E-box and GATA motifs bound by Tal1 and Gata factors, respectively, we have previously shown that binding sites for the Ets family of transcription factors characterize functional hematopoietic enhancers.32,39-42 We therefore surveyed the entire Lmo2 locus for the occurrence of evolutionarily conserved GATA sites, E-boxes (CANNTG) and Ets (GGAW) sites revealing the presence of such motifs in the −90, −75, −70, and −25 elements (Figure S3A-E; Table S5).
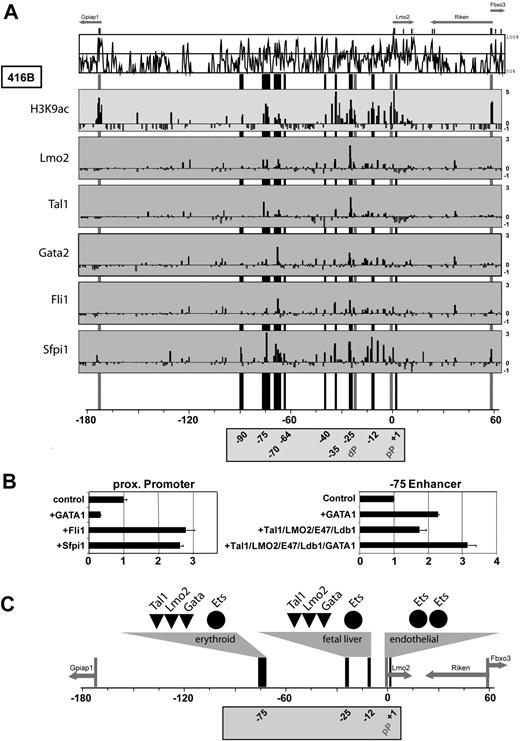
To verify, if these sites were bound in vivo, we performed ChIP assays with antibodies against Lmo2, Tal1, Gata2, Fli1, and Sfpi1. We had previously shown that Elf1, Fli1, and Ets1 bind the conserved noncoding region of the pP. For the new series of ChIP assays, we used our Lmo2 ChIP-chip platform allowing us to survey the entire 250 kb for binding events of candidate upstream factors. These experiments, shown in Figure 6A, validated our earlier ChIP–quantitative PCR results on Ets factor binding to the Lmo2 pP.19 In addition, all regulatory regions in the 2 5′ clusters were bound by Sfpi1, whereas Fli1 binding was found in the pP and the −25, −35, and −70 regions. The −75 and −25 enhancers, and to a lesser level the −70, −35, and −12 elements, were bound by Tal1 and Lmo2, but both factors were absent on the pP, +1 and +7 elements. The most prominent binding for Gata2 was seen at the −25 and −70 elements. Taken together, the combination of in silico comparative genomics and in vivo ChIP-chip revealed that Lmo2/Tal1 and Gata-factors are binding to the upstream hematopoietic elements, while Ets factors bind to distal elements as well as the pP.
Lmo2/Tal1 and Gata factors act through hematopoietic elements, while Ets factors control the pP. (A) ChIP-chip analysis in myeloid progenitor cell line 416B performed with antibodies against histone H3 acetylation (H3K9ac) or transcription factors Lmo2, Tal1, Gata2, Fli1, and Sfpi1. MVista representation of mouse/human sequence conservation is shown on the top and annotations of promoters (gray), and the 9 candidate regulatory elements (black) are as in Figure 1. The y-axis represents the log 2 enrichment of ChIPed DNA over input DNA ranging from −1 to 5 or 3, respectively. The H3K9ac panel is derived from Figure 2, to highlight accessible areas in the Lmo2 locus of 416B cells. Specific binding of Tal1/Lmo2/Gata2 can be found to hematopoietic elements but not to the endothelial pP region. This is in contrast with Ets factors, which do bind the pP region. (B) Transactivation assays in 293T cells using the LMO2 pP and hematopoietic enhancer constructs. The pP can be transactivated by Ets factors Fli1 and Sfpi1, whereas the −75 element is transactivated by a multiprotein complex containing Tal1, LMO2, E47, Ldb1, and GATA1. Transactivation assays were performed in at least 2 biologic replicates and assayed in triplicates. The values shown for the −75 enhancer were normalized using the values obtained with the pP construct without enhancer. (C) Differential regulation of Lmo2 elements. Autoregulatory complexes composed of Lmo2, Tal1, and Gata factors activate distal hematopoietic elements (erythroid and FL), whereas Ets factors are acting on the endothelial promoter. Annotations are as in Figure 1.
Lmo2/Tal1 and Gata factors act through hematopoietic elements, while Ets factors control the pP. (A) ChIP-chip analysis in myeloid progenitor cell line 416B performed with antibodies against histone H3 acetylation (H3K9ac) or transcription factors Lmo2, Tal1, Gata2, Fli1, and Sfpi1. MVista representation of mouse/human sequence conservation is shown on the top and annotations of promoters (gray), and the 9 candidate regulatory elements (black) are as in Figure 1. The y-axis represents the log 2 enrichment of ChIPed DNA over input DNA ranging from −1 to 5 or 3, respectively. The H3K9ac panel is derived from Figure 2, to highlight accessible areas in the Lmo2 locus of 416B cells. Specific binding of Tal1/Lmo2/Gata2 can be found to hematopoietic elements but not to the endothelial pP region. This is in contrast with Ets factors, which do bind the pP region. (B) Transactivation assays in 293T cells using the LMO2 pP and hematopoietic enhancer constructs. The pP can be transactivated by Ets factors Fli1 and Sfpi1, whereas the −75 element is transactivated by a multiprotein complex containing Tal1, LMO2, E47, Ldb1, and GATA1. Transactivation assays were performed in at least 2 biologic replicates and assayed in triplicates. The values shown for the −75 enhancer were normalized using the values obtained with the pP construct without enhancer. (C) Differential regulation of Lmo2 elements. Autoregulatory complexes composed of Lmo2, Tal1, and Gata factors activate distal hematopoietic elements (erythroid and FL), whereas Ets factors are acting on the endothelial promoter. Annotations are as in Figure 1.
Ets factors transactivate the pP, whereas Lmo2/Tal1 and Gata factors transactivate hematopoietic elements
The combination of ChIP-chip and transgenic assays suggested differential regulation of Lmo2 elements with Ets factors acting on the endothelial promoter, while an autoregulatory complex composed of Lmo2, Tal1, and Gata factors might activate distal hematopoietic elements. To assess whether the transcription factors identified by ChIP-chip were indeed able to activate Lmo2 regulatory elements, we performed transactivation assays. Reporter constructs containing the pP alone or the promoter combined with the −75 element were transfected in conjunction with expression vectors for Fli1, Sfpi1, Tal1, LMO2, E47, Ldb1, and GATA1 (Figure 6B). Both Fli1 and Sfpi1 were able to transactivate the pP, while addition of Gata factors reduced baseline activity. By contrast, addition of Gata factors or the Lmo2 complex (Tal1, LMO2, E2A, Ldb1) enhanced activity of the −75 enhancer constructs, which could be enhanced further by supplying Gata factors and the Lmo2 complex simultaneously. Taken together, the transactivation results are consistent with the notion that robust hematopoietic Lmo2 expression requires a positive feedback loop involving Gata/Lmo2/Tal1 complexes, which is deployed on top of preexisting and Ets factors dependent promoter activity in endothelial cells (Figure 6C).
Discussion
Lmo2 is a key regulator of hematopoietic and vascular development. Appropriate temperospatial control of Lmo2 expression is therefore vital for early endothelial and blood differentiation. Here, we have used a combination of bioinformatics, ChIP-chip and transgenic assays to explore the entire Lmo2 locus to delineate the cis elements that dictate its transcription. This study represents the most comprehensive locus-wide analysis of the regulation of any key regulator of early HSC specification and as such, many of the lessons learned from this benchmark examination will provide useful guidelines for future work.
A multipronged approach for locus-wide identification of transcriptional regulatory elements
The complexity of mammalian genomes is underlined by the fact that regulatory elements for a given gene can be spread over several hundred kilobases and are thus essentially hidden within the bulk of nonregulatory sequence. The postgenomic era has seen the development of both computational and experimental approaches for the identification of regulatory elements. Computational approaches take advantage of the observation that regulatory sequences are often more highly conserved than neighboring nonregulatory DNA and contain clusters of candidate transcription factor binding sites.2,43,44 Experimental techniques are based on the notion that distal regulatory elements are hypersensitive to DNase I and carry specific histone marks, which can be surveyed using genome-scale approaches such as ChIP-chip or ChIP-Seq.3,22,45,46
Here we have explored the potential of combining comparative genomic and ChIP-chip analyses to discover regulatory elements across the entire Lmo2 locus. Importantly, while comparative genomic analysis has been used before to interpret mammalian ChIP-chip data,47,48 previous studies lacked comprehensive in vivo functional validation of predicted elements. However, without in vivo validation in transgenic mice, studies of mammalian gene regulation can never be definitive. Our current study, therefore, moves significantly beyond these previous reports and provides several lessons likely to be of wider significance: (1) Comparative genomic analysis in vertebrates greatly depends on a somewhat arbitrary decision about the evolutionary distance used. Comparisons between eutherian and marsupial mammals proved useful for Lmo2, but this is likely to be different for other gene loci. Of note, all predicted Lmo2 regulatory elements showed increased regulatory potential (RP) scores,49 and a subset, including the Lmo2 erythroid and FL elements, also matched the criteria recently reported for the computational identification of erythroid elements50 (see Figure S4), thus underlining the potential power of computational genomics. (2) Candidate elements flagged up by elevated marks of histone acetylation in at least 1 of the 11 cell types accounted for 11 of 17 regions of noncoding sequence conservation, suggesting that a carefully chosen set of cell types for ChIP analysis will be sufficient to predict possible tissue-specific regulatory activity for a large proportion of noncoding conserved sequences, in line with recent conclusions from the Encyclopedia of DNA Elements (ENCODE) pilot project.51 (3) ChIP-chip and ChIP-Seq assays require substantial cell numbers thus precluding the use of primary cells in many instances. Cell lines may be good predictors of in vivo activity, as we saw with the hematopoietic lines used in this study. However, cell lines may also give false negative results, as seen in the current study, where the +7 region was a powerful endothelial enhancer but was marked by neither histone acetylation nor transcription factor binding in the endothelial cell line. (4) With an ever-increasing understanding of transcriptional regulatory codes, transcription factor ChIP-chip (or ChIP-Seq) may emerge as the method of choice for the identification of gene regulatory elements. For the Lmo2 locus, transcription factor ChIP-chip alone proved to be a highly effective strategy to not only identify regulatory elements but also, based on the transcription factor binding, predict in vivo activity with hematopoietic elements bound by Tal1/Lmo2 and Gata factors, whereas endothelial elements were bound largely by Ets factors. (5) In vivo validation of predicted regulatory regions remains a cornerstone for reliable assessment of the biologic function of regulatory elements. Through comprehensive transgenic analysis, we have identified 6 hematopoietic elements that, in different combinations, were able to direct expression to circulating blood cells and FL.
However, even though in vivo transgenic analysis can provide definitive answers, there are still limitations. Firstly, although transgenic assays show whether an element is sufficient for expression, they do not address the question whether an element is absolutely required in the context of the wider gene locus. Secondly, complete analysis of all potential combinatorial interactions between multiple elements is prohibitive in terms of both cost and time. Educated guesses based on ChIP results as well as activity of the individual elements can clearly be successful, as shown in the current study, but may not always be so.
Dynamic deployment of Lmo2 regulatory elements during ontogeny
Early specification of hematopoietic cells from developing mesoderm has been dissected in great detail with much evidence in support of the notion that cells with largely endothelial characteristics will give rise to both maturing endothelial and hematopoietic cells. The close biologic relatedness between endothelium and blood stem/progenitor cells has therefore been repeatedly cited as the prime reason for the extensive overlap of transcriptional control mechanisms between these tissues. For example, the Tal1 +19 stem cell enhancer is not only active in blood stem/progenitor cells but also targets expression to endothelium and hemangioblasts,39,52,53 suggesting that such elements provide an efficient strategy to control expression of genes important for both lineages.40,44,54
The Tal1 and Lmo2 knockout phenotypes in blood and endothelium are virtually identical, which has been attributed to the fact that the 2 proteins function together as key components of a multiprotein complex. It might therefore have been expected that, like Tal1, Lmo2 would contain powerful bipotential hemtoendothelial regulatory elements, as coregulation would ensure simultaneous availability of the 2 proteins. By contrast, however, our new data suggest that endothelial and hematopoietic expressions of Lmo2 are largely decoupled. Endothelial expression appears to be mainly conferred by sequences close to the pP dependent on upstream regulators of the Ets family. Transcriptional control in hematopoietic cells on the other hand seems more elaborated with modular deployment of several distal regulatory elements responsive to additional upstream inputs such as Tal1/Lmo2 and Gata2. Several distinct Lmo2- and Tal1-containing multiprotein complexes have been described suggesting that independent control of Lmo2 may provide an important means to shift the balance between these distinct complexes.
Unraveling the dynamics of differential deployment of modular regulatory elements during ontogeny will be critical to understand how genes such as Lmo2 act in concert with other key regulators by assembling the transcriptional regulatory networks that drive tissue development. In the case of Lmo2 regulation for example, Gata2 is expressed in hemangioblasts, endothelium, and blood stem/progenitor cells, yet only in the latter appears to be important for directly controlling Lmo2 expression. One can only speculate, therefore, that specific changes in the regulatory environment occur when mesodermal progenitors commit to the blood fate and that at least some of these changes trigger Gata2 occupancy of Lmo2 hematopoietic regulatory elements. Identification of the underlying mechanisms is likely to reveal fundamental aspects of early hematopoietic differentiation.
Concluding remarks
In conclusion, we have demonstrated that comparative genomics paired with ChIP-chip analysis is a powerful combination to identify tissue-specific enhancers. Our data indicate that hematopoietic expression of Lmo2 requires multiple distal regulatory elements bound by Tal1/Lmo2 and Gata factors, which are deployed during ontogeny to build on preexisting Ets factors' control of the pP already established in hemangioblasts and persisting into mature endothelial cells. This study provides the most comprehensive locus-wide analysis of the transcriptional control of a key regulator of early hematopoiesis, and many of the lessons learned will provide useful guidelines for future work. Moreover, this report lays the foundation for further locus-wide studies aiming to identify transcriptional pathways, which, when perturbed, lead to ectopic expression of Lmo2 in acute leukemias or tumor angiogenesis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Terry Rabbits for the Lmo2 LacZ knock-in mice and Richard Auburn from FlyChip for printing custom arrays.
This work was supported by grants from the Leukaemia Research Fund, Newton Trust, Leukemia & Lymphoma Society, Kay Kendall Leukaemia Fund, Cancer Research UK, and fellowships from the Canadian Institutes of Health Research (J.R.L.) and Swiss National Science Foundation (N.B.).
Authorship
Contribution: J.-R.L. designed research, performed research, analyzed data, and wrote the paper; N.B. performed research, analyzed data, and wrote the paper; S.K., K.K., N.K.W., S.H.O., M.J., S.P., M.H., J.C., T.H., I.J.D., G.L., G.F., and V.K. performed research; J.F. contributed vital new reagents; and B.G. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Berthold Göttgens, Department of Haematology, Cambridge Institute for Medical Research, Cambridge University, Hills Rd, Cambridge, CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk.
References
Author notes
*J.-R.L. and N.B. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal