Abstract
Although Hodgkin and Reed-Sternberg (HRS) cells are B lymphoid cells, they are unlike any normal cells of that lineage. Moreover, the limited proliferative potential of HRS cells belies the clinical aggressiveness of Hodgkin lymphoma (HL). More than 20 years ago, the L428 HL cell line was reported to contain a small population of phenotypic B cells that appeared responsible for the continued generation of HRS cells. This observation, however, has never been corroborated, and such clonotypic B cells have never been documented in HL patients. We found that both the L428 and KM-H2 HL cell lines contained rare B-cell subpopulations responsible for the generation and maintenance of the predominant HRS cell population. The B cells within the HL cell lines expressed immunoglobulin light chain, the memory B-cell antigen CD27, and the stem cell marker aldehyde dehydrogenase (ALDH). Clonal CD27+ALDHhigh B cells, sharing immunoglobulin gene rearrangements with lymph node HRS cells, were also detected in the blood of most newly diagnosed HL patients regardless of stage. Although the clinical significance of circulating clonotypic B cells in HL remains unclear, these data suggest they may be the initiating cells for HL.
Introduction
Hodgkin and Reed-Sternberg (HRS) cells, the hallmark of classical Hodgkin lymphoma (HL), were first recognized more than 100 years ago; nevertheless, their cellular origin has remained obscure.1 Not only do they have an unusual appearance that is unlike any normal blood cell, but their scarcity has limited biologic studies. Microdissection studies demonstrating clonal immunoglobulin gene rearrangements established their B-cell ancestry.2 In most cases, the immunoglobulin gene rearrangements demonstrated somatic hypermutation, suggesting the HRS cells arose from germinal or postgerminal center B cells. Unique among B-cell malignancies, however, HRS cells usually do not express immunoglobulin or any other B-cell markers.3-5 Initial studies suggested that the defective immunoglobulin expression in HRS was the result of crippling mutations of immunoglobulin genes, disrupting their coding capacity.6 Subsequent studies demonstrated that such crippling mutations occurred only in a minority of HL cases, and usually only in those that were Epstein-Barr Virus (EBV)–positive.4,5 In most cases, functional immunoglobulin (and other B cell-specific) genes were not transcribed, apparently as a result of epigenetic silencing of the immunoglobulin heavy chain and/or master transcription factors.7,8
Somewhat in contrast to the clinical aggressiveness of HL, especially advanced stage disease, HRS cells demonstrate limited proliferative capacity in vivo.9 More than 20 years ago, Newcom et al reported that the L428 HL cell line, while consisting predominantly of HRS cells, also contained a small population of phenotypic B cells; these phenotypic B cells appeared to be responsible for the generation of the HRS cells and the continued growth of the cell line.10 This observation, however, has never been corroborated, and such clonotypic B cells have never been documented in HL patients. Other B-cell malignancies have also been shown to contain small populations of proliferative cells that are phenotypically distinct from the predominant tumor cell population.11-15 Given this background, we explored the existence of clonotypic B cells in HL. Here we demonstrate that clonotypic B cells not only are responsible for the generation and maintenance of HL cell lines, but also circulate in most patients with newly diagnosed HL.
Methods
Human samples
Blood (10-50 mL) was obtained from 31 patients with newly diagnosed HL seen at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins (28 patients) or St Agnes Hospital, Baltimore, MD (3 patients), between November 2005 and July 2008. Blood was also obtained from 6 healthy controls. When available, a portion of the diagnostic lymph nodes was obtained for cell isolations. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki, as approved by the Johns Hopkins Medical Institutes Institutional Review Board. The diagnosis of HL was established, and in situ hybridization for Epstein-Barr virus RNA was performed, by Hematopathology at Johns Hopkins. CD19+ B cells were isolated after density centrifugation (density < 1.078; Ficoll-Paque; Pharmacia, Piscataway, NJ) using the B-cell isolation kit and the VarioMACS Separator (Miltenyi Biotec, Auburn, CA). Lymph nodes were cut into small pieces, suspended in RPMI 1640 medium (GIBCO Invitrogen, Carlsbad, CA), and filtered through a 500-μm–pore wire mesh. After continued passage through a 25-gauge needle, CD19+ and CD30+ cells (using CD30 microbeads)16 were selected with the VarioMacs Separator (Miltenyi Biotec, Bergisch Gladbach, Germany). DNA was extracted from HRS cells and circulating B cells (1-10 × 103 cells), using the QIAamp micro DNA isolation kit (QIAGEN, Valencia, CA).
HRS cell microdissection
Frozen sections from biopsied lymph nodes were cut at 5- to 10-μm thickness onto PEN-Membrane slides (PALM Microlaser Technologies, Munich, Germany) and fixed in 95% ethanol. Antigen retrieval was performed with EDTA buffer (pH 8) for 20 minutes in a steamer at 95°C; the tissue was then stained with an antibody to CD30 (clone BerH2; Ventana Medical Systems, Tucson, AZ), developed with diaminobenzidine, and dried overnight at 50°C. For each biopsy, approximately 400 to 500 CD30-positive HRS cells were microdissected using the PALM MicroBeam system (PALM Microlaser Technologies). Cells were captured into mineral oil in microtube caps, pelleted, extracted with 0.5% Tween 20 at 99°C, mixed with 2.5 volumes of 80% ethanol, and pelleted at 4°C for 10 minutes at 10 000g. Cells were resuspended in 100 μL digestion buffer (50 mM Tris pH 8, 0.5% SDS, 10 mM EDTA, 100 mM glycine, and 500 μg/mL proteinase K) and incubated overnight at 60°C. One microgram salmon sperm DNA was added as carrier. The digest was extracted with an equal volume of phenol:chloroform and then chloroform. DNA was precipitated with sodium acetate and absolute ethanol, washed with ethanol, air-dried, and resuspended in polymerase chain reaction (PCR) quality water.
Cell lines and cell culture
The L428 and KM-H2 lines were obtained from DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). The cell lines were maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS; Harlan, Indianapolis, IN), 50 units/mL penicillin (GIBCO), 50 μg/mL streptomycin (GIBCO), and 2 μM l-glutamine (GIBCO) in a humidified atmosphere at 37°C and 5% CO2. To assess the clonogenic potential of HL cell lines, 500 cells were placed into 1 mL 1.2% methylcellulose containing 30% FBS, 1% bovine serum albumin (GIBCO), 10−4 M 2-mercaptoethanol (GIBCO), and 2 μM l-glutamine. Colonies consisting of more than 40 cells were scored using an inverted microscope 14 days after plating as previously described.14,17 The HL cell lines (2 × 107cells/mL) were treated with 0.5 μM decitabine (Sigma-Aldrich, St Louis, MO) for 72 hours as previously described,7 to assess the drug's effects on immunoglobulin expression.
Flow cytometric analyses and fluorescence-activated cell sorting
The following monoclonal antibodies were used: mouse anti–human CD30-phycoerthrin (PE), CD19–fluourescein isothyocyanate (FITC), CD20-allophycocyanin (APC), CD20 APC-cyanine 7 (Cy7), CD27-APC, kappa immunoglobulin light chain-phycoerythrin (PE), and biotin-lambda immunoglobulin light chain-streptavidin-PerCP (all antibodies from BD PharMingen, San Diego, CA). Staining for aldehyde dehydrogenase (ALDH) activity used the Aldefluor reagent (StemCell Technologies, Vancouver, BC) according to the manufacturer's instructions. After the addition of 2 μg/mL propidium iodide (BD PharMingen) to discriminate dead cells, cells were analyzed and/or sorted with a FACSAria (BD Biosciences, San Jose, CA) as previously described.14 To study the proliferative potential of the cell subpopulations, varying numbers of the cell types from both cell lines were directly sorted into 24-well microtiter plates containing supplemented RPMI 1640 as described above. ALDH activity in the cell subsets is presented as the mean fluorescence intensity (MFI) difference between Aldefluor-stained cells with and without diethylaminobenzaldehyde (DEAB) as previously described.14,18
Immunoglobulin gene rearrangement detection
For the immunoglobulin heavy chain, DNA aliquots were amplified by PCR using hemi-nested degenerate primers targeting framework 3a (Fr3a). Reactions containing 5 μL of the extracted DNA underwent a first round of PCR amplification for 30 cycles in a 50-μL volume. A 5-μL aliquot of the first round reaction was reamplified for 30 cycles in a 50-μL reaction using hemi-nested primers. The primers for first- and second-round PCR were: 5′-ACACGGCYSTGTATTACTGTG-3′ (Fr3a) and 5′-ACCTGAGGAGACGGTGACC (JH, antisense); and Fr3a with 5′-GTGACCAGGGTNCCTTGGCCCCAG-3′ (VLJH, antisense); respectively, where S = G/C, Y = C/T, and N = A/G/C/T. The VLJH primer was labeled with the fluorescent molecule FAM. PCR amplification of the immunoglobulin kappa light chain gene used IgK Tube A primers (InVivoScribe Technologies, San Diego, CA) according to the manufacturer's instructions. Distilled water or control DNA encoding a known monoclonal immunoglobulin heavy or kappa light chain gene rearrangement was used as negative or positive control, respectively. PCR-amplified products were subjected to capillary electrophoresis on an ABI PRISM 3100 genetic analyzer and evaluated using the GeneScan 2.1 software package (Applied Biosystems, Foster City, CA). For sequence analysis, PCR products were reamplified using unlabeled primers and directly sequenced using standard dye terminator chemistry (Applied Biosystems).
Statistical considerations
Data are expressed as the mean plus or minus SEM. The means of 2 experimental groups were compared using a 2-tailed paired Student t test. The means of more than 2 experimental groups were compared using analysis of variance (ANOVA).
Results
HL cell lines contain small B-cell subpopulations
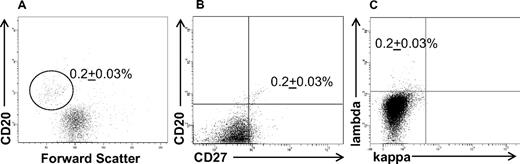
The well-characterized L428 and KM-H2 HL cell lines, consisting of cells that express the classic CD30+CD15+ HRS phenotype,19 were assessed by flow cytometry for the presence of CD30−CD15− B cells. Although the large majority of cells within the HL cell lines expressed no B-cell markers, a small population of CD30− phenotypic B cells was detected in the KM-H2 (Figure 1), as well as the L428 (Figure 2), HL cell lines. In both HL cell lines, the phenotypic B cells were smaller in size than the HRS cells (Figure 1A), and expressed CD20 (Figures 1,2), the memory B-cell marker CD27 (Figure 1B), and lambda light chain (Figures 1C,2A). The B-cell subpopulation represented 0.2% (± 0.03%) and 0.3% (± 0.08%) of the total KM-H2 (Figure 1) and L428 (Figure 2) cells, respectively.
KM-H2 cells contain a small population of phenotypic B cells. A percentage (0.2% ± 0.03%) of the total cells were (A) small-sized and expressed CD20, (B) CD27, and (C) lambda light chain. The data represent the mean ± SEM of 10 separate experiments.
KM-H2 cells contain a small population of phenotypic B cells. A percentage (0.2% ± 0.03%) of the total cells were (A) small-sized and expressed CD20, (B) CD27, and (C) lambda light chain. The data represent the mean ± SEM of 10 separate experiments.
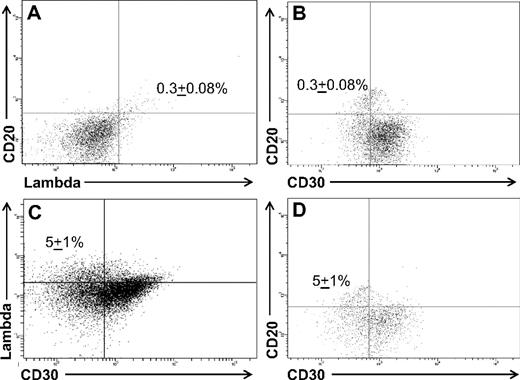
L428 cells contain a small population of phenotypic B cells. (A) A percentage (0.3% ± 0.08%) of the cells coexpressed CD20 and lambda light chain, and (B) were CD30−. Treatment of the cell line with 0.5 μM decitabine for 72 hours increased the fraction of cells expressing (C) lambda light chain and (D) CD20 to 5% ± 1% of the total cells (P = .04). The data represent the mean ± SEM of 10 separate experiments.
L428 cells contain a small population of phenotypic B cells. (A) A percentage (0.3% ± 0.08%) of the cells coexpressed CD20 and lambda light chain, and (B) were CD30−. Treatment of the cell line with 0.5 μM decitabine for 72 hours increased the fraction of cells expressing (C) lambda light chain and (D) CD20 to 5% ± 1% of the total cells (P = .04). The data represent the mean ± SEM of 10 separate experiments.
Decitabine increases the number of immunoglobulin expressing cells
Decitabine (5-aza-2′-deoxycytidine) has been reported to induce low-level expression of immunoglobulin in HL cell lines by reverse transcription (RT)–PCR, presumably by inhibition of DNA methylation.7 To determine whether a specific cell population within the cell lines was responsible for the immunoglobulin expression, both cell lines were incubated with 0.5 μM decitabine for 72 hours7 and analyzed by flow cytometry. We confirmed that immunoglobulin expression increased in both cell lines after decitabine treatment (Figure 2C). The increased expression of immunoglobulin, however, primarily reflected a higher fraction of lambda light chain-restricted CD20+ B cells in the cell lines after decitabine treatment, rather than up-regulation of immunoglobulin expression by the CD30+ HRS cells (Figure 2C,D). The mean percentage of B cells in the L428 cell line increased to 5% (± 1%; P = .04) after decitabine treatment (Figure 2C,D), and to 2% (± 0.2%; P < .001) in the KM-H2 cell line (data not shown).
The B-cell subpopulation within the HL cell lines generates HRS cells
To study the proliferative potential of the B- and HRS-cell subpopulations, varying numbers of the 2 cell types from both cell lines were directly sorted into microtiter wells (Figure S1; 3 separate experiments [available on the Blood website; see the Supplemental Materials link at the top of the online article]). Most of the wells seeded with a single sorted CD20+ L428 cell, and all wells seeded with 10 CD20+ L428 or KM-H2 cells, reproducibly expanded to more than 103 cells over 7 days; the resulting expanded cells contained both the B-cell and RS subpopulations. After 1 week of expansion, replating as few as 10 L428 and KM-H2 cells from the wells originally seeded with 10 CD20+ cells generated cellular expansion for least 4 serial replatings. No wells seeded with 1 or 10 sorted larger-sized CD30+ HRS cells from either cell line grew. Seeding 100 HRS cells from both cell lines resulted in cellular expansion over 7 days, but the expanded cells never contained the B-cell population by flow cytometry. Moreover, no further cellular expansion was seen with the second replating of 100 HRS cells from either cell line (Figure S1).
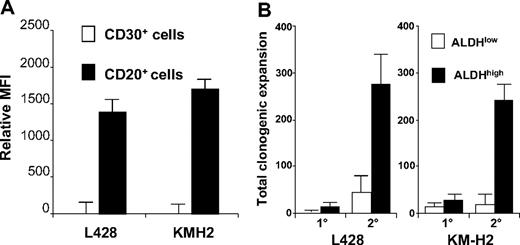
ALDH, specifically the ALDH1A1 isoenzyme, mediates the biosynthesis of all-trans-retinoic acid as well as the detoxification of a variety of compounds such as ethanol and active metabolites of cyclophosphamide.20 Adult stem cells typically exhibit higher relative levels of ALDH activity than their differentiated progeny, and the fluorescently labeled ALDH1A1 substrate Aldefluor allows isolation of viable normal stem cells,18 as well as putative cancer stem cells,14 from several tissues. The B cells within both the L428 and KM-H2 lines expressed significantly higher levels of ALDH activity than the HRS cells; in fact, essentially no ALDH activity could be detected in the HRS cells from either cell line (Figure 3A). The ALDHhigh cells from both HL cell lines exhibited higher clonogenic capacity than the ALDHlow cells, especially with secondary replating (Figure 3B).
L428 and KM-H2 CD20+ cell populations exhibit high ALDH activity and clonogenic potential. (A) ALDH activity for the CD30+ and CD20+ subsets. The data represent the mean ± SEM of the MFI difference between Aldefluor-stained cells with and without DEAB from 8 separate experiments. (B) The initial plating of 500 ALDHhigh L428 cells produced 19.7 (± 9.5) colonies compared with 5.7 ± 11 colonies from 500 ALDHlow L428 cells (P = .3), and 500 ALDHhigh KM-H2 cells generated 33.3 ± 5.4 colonies compared with 20.7 ± 6.4 from 500 ALDHlow KM-H2 cells (P = .3). Upon secondary replating, the original 500 ALDHhigh L428 generated 251 ± 25.9 colonies compared with 53.6 + 12.9 from the ALDHlow L428 cells (P = .01), and the original 500 ALDHhigh KM-H2 cells produced 243.7 ± 32.3 colonies compared with 27.7 ± 16 ALDHlow KM-H2 cells (P = .03). The data represent the mean ± SEM of 3 separate experiments.
L428 and KM-H2 CD20+ cell populations exhibit high ALDH activity and clonogenic potential. (A) ALDH activity for the CD30+ and CD20+ subsets. The data represent the mean ± SEM of the MFI difference between Aldefluor-stained cells with and without DEAB from 8 separate experiments. (B) The initial plating of 500 ALDHhigh L428 cells produced 19.7 (± 9.5) colonies compared with 5.7 ± 11 colonies from 500 ALDHlow L428 cells (P = .3), and 500 ALDHhigh KM-H2 cells generated 33.3 ± 5.4 colonies compared with 20.7 ± 6.4 from 500 ALDHlow KM-H2 cells (P = .3). Upon secondary replating, the original 500 ALDHhigh L428 generated 251 ± 25.9 colonies compared with 53.6 + 12.9 from the ALDHlow L428 cells (P = .01), and the original 500 ALDHhigh KM-H2 cells produced 243.7 ± 32.3 colonies compared with 27.7 ± 16 ALDHlow KM-H2 cells (P = .03). The data represent the mean ± SEM of 3 separate experiments.
Clonotypic B cells circulate in HL patients
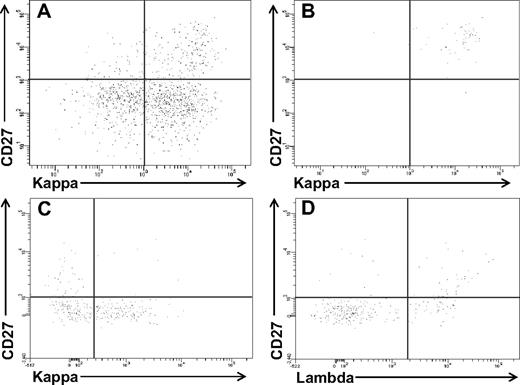
The small population of B cells within the HL cell lines had the phenotype of memory B cells, expressed the stem cell marker ALDH, and generated and maintained the growth of the HRS cells. To determine whether such cells were also present in clinical specimens, CD19+ cells were isolated from the blood of 31 patients with newly diagnosed HL (Table 1); the CD19+ cells were stained with B-cell monoclonal antibodies and Aldefluor to assess ALDH activity, as we previously described.14 No light chain-restricted populations were seen in any of the patients when the unseparated CD19+CD20+ cells or the smaller CD27+ fraction was assessed (Figure 4A). In 3 patients, high ALDH expression alone identified an immunoglobulin light chain-restricted population within the circulating CD19+ cells; this clonal population was always CD27+ (Figure 4B). In 23 patients, although the total ALDHhigh B-cell population was not clonal, there was immunoglobulin light chain-restriction within the CD27+ALDHhigh B-cell population (4C,D). Thus, all but 5 (see Figure S2) of the 31 patients with newly diagnosed HL had detectable circulating light-chain restricted CD27+ALDHhigh B cells. The circulating CD27+ALDHhigh B cells represented a median of 0.2% (range 0%-2.4%) of the total circulating CD19+ cells and expressed kappa light chain in 13 patients and lambda in 13 (Table 1). Light chain-restricted CD27+ALDHhigh B cells could not be detected in the blood of the 6 normal controls tested (data not shown).
Circulating clonotypic B cells in Hodgkin lymphoma patients
| Patient . | Sex . | Age . | Hist . | Clinical stage . | EBV . | Flow . | Percentage of CD19+ cells . |
|---|---|---|---|---|---|---|---|
| 1 | M | 21 | NS | 2A | ND | Neg | 0 |
| 2 | F | 37 | NS | 4A (marrow) | Neg | Neg | 0 |
| 3 | F | 20 | NS | 2A | ND | λ | 0.9 |
| 4 | F | 38 | NS | 3B | ND | κ | 0.2 |
| 5 | M | 23 | NS | 2A | ND | κ | 0.2 |
| 6 | M | 46 | MC | 4B (marrow) | Pos | Neg | 0 |
| 7 | F | 60 | NS | 2A | ND | λ | 0.3 |
| 8 | F | 20 | NS | 2B | Neg | λ | 0.9 |
| 9 | F | 55 | NS | 3A | Pos | κ | 0.5 |
| 10 | F | 26 | MC | 4A (pericardial) | Pos | Neg | 0 |
| 11 | F | 29 | NS | 4B (bone) | Neg | λ | 0.1 |
| 12 | F | 30 | MC | 3B | Neg | κ | 0.1 |
| 13 | M | 37 | NS | 3A | Neg | λ | 0.1 |
| 14 | M | 26 | MC | 1A | Pos | λ | 0.1 |
| 15 | F | 34 | NS | 2A | ND | λ | 0.7 |
| 16 | F | 39 | NS | 2A | Neg | κ | 0.2 |
| 17 | M | 66 | NS | 2A | Neg | κ | 0.9 |
| 18 | F | 35 | NS | 2A | Neg | κ | 0.2 |
| 19 | F | 27 | NS | 3B | Neg | Neg | 0 |
| 20 | M | 24 | NS | 2B | Pos | κ | 0.2 |
| 21 | M | 48 | NS | 3B | Neg | λ | 0.3 |
| 22 | F | 23 | NS | 2A | Neg | κ | 0.3 |
| 23 | M | 35 | NS | 2A | Neg | λ | 0.2 |
| 24 | M | 43 | NS | 4B (bone) | Neg | κ | 1.8 |
| 25 | F | 36 | NS | 3A | Neg | κ | 0.7 |
| 26 | M | 18 | NS | 4B (lung) | Neg | λ | 0.2 |
| 27 | M | 29 | NS | 2A | Neg | κ | 0.4 |
| 28 | F | 25 | NS | 2A | Neg | κ | 0.2 |
| 29 | M | 33 | NS | 4B (bone, liver) | Neg | λ | 0.4 |
| 30 | M | 20 | NS | 3A | Neg | λ | 0.2 |
| 31 | M | 37 | NS | 4B (lung) | Neg | λ | 2.4 |
| Patient . | Sex . | Age . | Hist . | Clinical stage . | EBV . | Flow . | Percentage of CD19+ cells . |
|---|---|---|---|---|---|---|---|
| 1 | M | 21 | NS | 2A | ND | Neg | 0 |
| 2 | F | 37 | NS | 4A (marrow) | Neg | Neg | 0 |
| 3 | F | 20 | NS | 2A | ND | λ | 0.9 |
| 4 | F | 38 | NS | 3B | ND | κ | 0.2 |
| 5 | M | 23 | NS | 2A | ND | κ | 0.2 |
| 6 | M | 46 | MC | 4B (marrow) | Pos | Neg | 0 |
| 7 | F | 60 | NS | 2A | ND | λ | 0.3 |
| 8 | F | 20 | NS | 2B | Neg | λ | 0.9 |
| 9 | F | 55 | NS | 3A | Pos | κ | 0.5 |
| 10 | F | 26 | MC | 4A (pericardial) | Pos | Neg | 0 |
| 11 | F | 29 | NS | 4B (bone) | Neg | λ | 0.1 |
| 12 | F | 30 | MC | 3B | Neg | κ | 0.1 |
| 13 | M | 37 | NS | 3A | Neg | λ | 0.1 |
| 14 | M | 26 | MC | 1A | Pos | λ | 0.1 |
| 15 | F | 34 | NS | 2A | ND | λ | 0.7 |
| 16 | F | 39 | NS | 2A | Neg | κ | 0.2 |
| 17 | M | 66 | NS | 2A | Neg | κ | 0.9 |
| 18 | F | 35 | NS | 2A | Neg | κ | 0.2 |
| 19 | F | 27 | NS | 3B | Neg | Neg | 0 |
| 20 | M | 24 | NS | 2B | Pos | κ | 0.2 |
| 21 | M | 48 | NS | 3B | Neg | λ | 0.3 |
| 22 | F | 23 | NS | 2A | Neg | κ | 0.3 |
| 23 | M | 35 | NS | 2A | Neg | λ | 0.2 |
| 24 | M | 43 | NS | 4B (bone) | Neg | κ | 1.8 |
| 25 | F | 36 | NS | 3A | Neg | κ | 0.7 |
| 26 | M | 18 | NS | 4B (lung) | Neg | λ | 0.2 |
| 27 | M | 29 | NS | 2A | Neg | κ | 0.4 |
| 28 | F | 25 | NS | 2A | Neg | κ | 0.2 |
| 29 | M | 33 | NS | 4B (bone, liver) | Neg | λ | 0.4 |
| 30 | M | 20 | NS | 3A | Neg | λ | 0.2 |
| 31 | M | 37 | NS | 4B (lung) | Neg | λ | 2.4 |
Hist indicates histologic subtype; Flow, light chain restriction of circulating clonotypic B cells by flow cytometry; M, male; F, female; NS, nodular sclerosis; MC, mixed cellularity; ND, not done; Pos, positive; and Neg, negative.
Immunoglobulin light chain–restricted CD27+ALDHhigh B cells circulate in newly diagnosed HL patients. CD19+ cells were isolated from newly diagnosed HL patients and stained with Aldefluor and monoclonal antibodies against CD27 and kappa and lambda light chains. Expression of CD27 and kappa light chain in the (A) CD19+ and (B) CD19+ALDHhigh peripheral blood cells from a HL patient. Expression of CD27 and (C) kappa or (D) lambda light chain in the CD19+ALDHhigh peripheral blood cells from another HL patient.
Immunoglobulin light chain–restricted CD27+ALDHhigh B cells circulate in newly diagnosed HL patients. CD19+ cells were isolated from newly diagnosed HL patients and stained with Aldefluor and monoclonal antibodies against CD27 and kappa and lambda light chains. Expression of CD27 and kappa light chain in the (A) CD19+ and (B) CD19+ALDHhigh peripheral blood cells from a HL patient. Expression of CD27 and (C) kappa or (D) lambda light chain in the CD19+ALDHhigh peripheral blood cells from another HL patient.
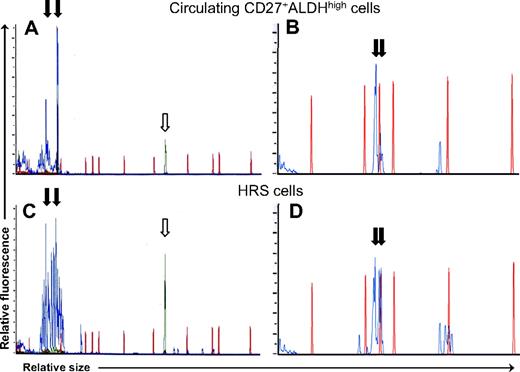
The circulating light chain-restricted B cells were clonal by immunoglobulin complementary determining region 3 (CDR3) length restriction (Figure 5A,B) and DNA sequencing (Figure S3). Using a magnetic cell separation column (Miltenyi Biotec) as previously described,16 HRS cells were isolated from fresh pathology specimens in 3 patients. In addition, HRS cells were microdissected from frozen sections of lymph node biopsies from 2 other patients. PCR demonstrated the same clonal immunoglobulin CDR3 length restriction present in the patients' circulating CD27+ALDHhigh B cells (Figure 5C,D); this was confirmed by direct sequencing of the PCR products (Figure S3). Clonotypic B cells were also found in these diagnostic lymph nodes, representing a similar percentage of CD19+ B cells as in the peripheral blood (data not shown).
Circulating light chain–restricted CD27+ALDHhigh B cells show identical immunoglobulin rearrangements to HRS cells. Capillary electrophoretic profiles of (A) immunoglobuin heavy chain CDR3 amplification products and (B) immunoglobulin kappa light chain CDR3 amplification products obtained by PCR of circulating light chain–restricted CD27+ALDHhigh B cells from 2 respective patients. Capillary electrophoretic profiles of HRS cells isolated from a diagnostic lymph node (C) from the first patient by CD30 microbeads and (D) from the second patient by microdissection. PCR products are shown in blue lines, and red lines are size markers. ↓show identical-sized PCR products in the circulating clonotypic B cells and HRS cells from (A,C) patient 1 (at 76 and 94 bp) and (B,D) patient 2 (at 146 and 150 bp).  indicates control PCR reaction product.
indicates control PCR reaction product.
Circulating light chain–restricted CD27+ALDHhigh B cells show identical immunoglobulin rearrangements to HRS cells. Capillary electrophoretic profiles of (A) immunoglobuin heavy chain CDR3 amplification products and (B) immunoglobulin kappa light chain CDR3 amplification products obtained by PCR of circulating light chain–restricted CD27+ALDHhigh B cells from 2 respective patients. Capillary electrophoretic profiles of HRS cells isolated from a diagnostic lymph node (C) from the first patient by CD30 microbeads and (D) from the second patient by microdissection. PCR products are shown in blue lines, and red lines are size markers. ↓show identical-sized PCR products in the circulating clonotypic B cells and HRS cells from (A,C) patient 1 (at 76 and 94 bp) and (B,D) patient 2 (at 146 and 150 bp).  indicates control PCR reaction product.
indicates control PCR reaction product.
Disease stage did not appear to influence the ability to detect circulating clonotypic B cells (Table 1). These cells were present in the only patient in the series with clinical stage I disease, 13 of 14 patients with stage II disease, and 7 of 8 patients with stage III disease. In fact, 3 of the 5 patients without detectable circulating clonotypic B cells (Figure S2) were among the 8 patients with stage IV disease, although 2 of these patients had EBV+ disease, which may have crippling mutations of the immunoglobulin heavy chain genes. HRS cells from approximately 40% of EBV+ HL have been reported to contain such crippling mutations,4,6 and 2 of the 5 patients with EBV+ HL in this series had no detectable circulating light chain–restricted B cells (Table 1). There was a trend toward the circulating clonotypic cell frequency to increase with increasing stage; the clone represented 0.1% of the circulating CD19+ B cells in the one patient with stage I disease, 0.4% (± 0.1%) of the circulating CD19+ B cells in the stage II patients, 0.3% (± 0.1%) in the stage III patients, and 1.0% (± 0.4%) in the stage IV patients (P = .08, ANOVA).
Discussion
Several groups of investigators have called attention to the shared features of HRS cells and plasma cells.3,21-23 Like plasma cells, HRS cells not only typically lack expression of B-cell surface markers, but they also are the only other blood cells that occasionally express CD138 (syndecan-1).3,21,22,24 Moreover, a fraction of HRS cells also express B lymphocyte–induced maturation protein 1 (Blimp-1), a transcription factor required for plasma cell differentiation.23 Recently, multiple myeloma (MM) plasma cells have been shown to be terminally differentiated like their normal counterparts, with circulating clonotypic B cells14,25-27 apparently responsible for initiation of MM.13-15 Thus, we hypothesized that HRS cells are also terminally differentiated, and as such may not represent the cells of origin of HL. We confirmed the findings of Newcom et al that the L428 HL cell line contains a rare population of small-sized, phenotypic memory B cells responsible for the generation and maintenance of the predominant population of HRS cells.10 These clonotypic B cells were also present in the KM-H2 HL cell line and the blood of most newly diagnosed HL patients, even in early stage disease. Although this is the first report describing circulating clonotypic B cells in HL, there have also been rare reports of HL arising in patients with chronic lymphocytic leukemia (CLL) and sharing identical immunoglobulin rearrangements with the CLL B cells.28-30
These data suggest that the cells of origin in MM and HL appear to be phenotypically similar, even though the biologic and clinical behaviors of the diseases are quite distinct. Similarly, several distinct malignancies, chronic myeloid leukemia, acute myeloid leukemia, myelodysplastic syndrome, and acute lymphoblastic leukemia, all appear to arise from phenotypic hematopoietic stem cells.31 Thus, the biology of a particular malignancy may be a function of both the cell of origin and the degree or type of differentiation allowed by the disease's oncogenic changes.31
We recently showed that the circulating clonotypic B cells from MM patients will generate MM when transplanted into immunodeficient mice.14 The low frequency of the circulating clonotypic B cells in the HL patients precluded such studies. Because these cells represent a median of only 0.2% of the peripheral blood CD19+ cells in HL patients (or 5- to 10-fold less than was present in MM patients14 ), less than 104 clonotypic B cells were recovered from most patients. In most patients, the small number of cells limited studies to the examination of immunoglobulin gene rearrangements to assess clonality; however, enough clonotypic B cells to study their proliferative capacity in vitro were isolated from 2 patients. Using the same conditions we used for culturing the phenotypically similar myeloma clonotypic B cells in vitro,14 no proliferation of the clonotypic B cells from the HL patients was observed. The limited expansion of the malignant clone in the face of the unique microenvironment of HL in vivo,9 however, makes finding the correct experimental culture conditions for growing HL quite problematic.
The rarity of HRS cells has also limited biologic studies with these cells. Attempts to isolate HRS cells by fluorescence-activated cell sorting have been largely unsuccessful, because of the fragility of these very large cells.16 Microdissection of lymph nodes can successfully isolate HRS cells, but cell yields are small.5,29 We successfully isolated HRS cells using a magnetic cell separation column (Miltenyi Biotec).16 The HRS cells, however, remained highly contaminated with lymphocytes and other blood cells (Figure 5C) as others have described,16 presumably because of their tendency to adhere to other cells.
It is now clear that most HRS cells have functional immunoglobulin gene rearrangements, with the suppression of immunoglobulin transcription apparently the result of epigenetic silencing.4,5,7,8 We confirmed that decitabine can increase immunoglobulin expression in HL cell lines,7 but found that this predominately reflected an increase in the fraction of light chain-expressing B cells rather than increased immunoglobulin expression in HRS-like cells.7 Possibly the B cells are more resistant to the cytotoxic properties of decitabine than the HRS cells.14
The clinical significance of circulating cancer cells is still unclear, but their presence correlates with poor risk disease in most studies.32-35 Moreover, circulating cancer cells have been reported to be quiescent and drug-resistant, possibly representing cancer stem cells.33-35 There was a trend toward stage-IV HL patients having higher levels of circulating clonotypic B cells. Perhaps a surprising finding with the propensity of HL to spread to contiguous lymph node groups, however, was that the cells could also be detected in most patients in early stage disease. Circulating occult cancer cells have also been observed in early stage malignancies, including breast cancer where local lymph nodes are also usually the initial sites of tumor spread.32-38 Recent findings have implicated a role for the microenvironment, the so-called “premetastatic niche,” in determining the pattern of metastatic spread.39,40 Such work suggests that a unique local microenvironment effect might be responsible for the homing of circulating HL B cells to contiguous lymph nodes early in the course of the disease. We did not study any patients with nodular lymphocyte predominant HL, as circulating lymphoma cells might already be expected from existing data; the malignant B cells in this disease are CD20+ and in many ways quite similar to low-grade non-Hodgkin lymphoma cells that have already been reported to circulate even in early stage disease.36,37
Although B cells appear responsible for the generation and maintenance of HRS cells in HL cell lines, the biologic relevance of the circulating clonotypic B cells in HL patients remains to be established. Recently, rituximab has shown activity in classical HL, regardless of the expression of CD20 by the HRS cells.41 Rituximab's activity was suggested to be the result of the depletion from the HL microenvironment of normal B lymphocytes required for the growth of HRS cells.41 Obviously, the existence of clonotypic CD20+ B cells potentially responsible for the generation of HRS cells provides another possible mechanism for rituximab's activity in HL. Future studies correlating circulating clonotypic B-cell numbers with progression-free survival of HL patients after treatment should help determine the clinical significance of these cells.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH; Bethesda, MD) grant PO1 CA15396.
National Institutes of Health
Authorship
Contribution: R.J.J., C.D.G., R.F.A., and W.M. designed research; R.J.J., C.D.G., R.F.A., W.M., B.P., J.P.B., M.S.V., J.M.G., L.L.G., M.S., M.V.L., and S.B. performed research; R.J.J., Y.L.K., C.B.M., and R.F.A. contributed patients; R.J.J., C.D.G., B.P., R.F.A., and W.M. analyzed data; and R.J.J., C.D.G., R.F.A., and W.M. wrote the paper.
Conflict-of-interest disclosure: R.J.J. holds the patent for Aldefluor and under a licensing agreement between Aldagen and the Johns Hopkins University is entitled to a share of royalties received by the university. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict-of-interest policies. The other authors declare no competing financial interests.
Correspondence: Richard J. Jones, Room 244, Bunting-Blaustein Cancer Research Bldg, 1650 Orleans St, Baltimore, MD 21231; e-mail: rjjones@jhmi.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal