Abstract
Somatic rearrangements of transcription factors are common abnormalities in the acute leukemias. With rare exception, however, the resultant protein products have remained largely intractable as pharmacologic targets. One example is AML1-ETO, the most common translocation reported in acute myeloid leukemia (AML). To identify AML1-ETO modulators, we screened a small molecule library using a chemical genomic approach. Gene expression signatures were used as surrogates for the expression versus loss of the translocation in AML1-ETO–expressing cells. The top classes of compounds that scored in this screen were corticosteroids and dihydrofolate reductase (DHFR) inhibitors. In addition to modulating the AML1-ETO signature, both classes induced evidence of differentiation, dramatically inhibited cell viability, and ultimately induced apoptosis via on-target activity. Furthermore, AML1-ETO–expressing cell lines were exquisitely sensitive to the effects of corticosteroids on cellular viability compared with nonexpressers. The corticosteroids diminished AML1-ETO protein in AML cells in a proteasome- and glucocorticoid receptor–dependent manner. Moreover, these molecule classes demonstrated synergy in combination with standard AML chemotherapy agents and activity in an orthotopic model of AML1-ETO–positive AML. This work suggests a role for DHFR inhibitors and corticosteroids in treating patients with AML1-ETO–positive disease.
Introduction
The availability of new genomic tools has enabled a marked increase in the identification of potential disease targets. Despite these discoveries, however, effective targeted therapy largely remains an elusive goal. Two major obstacles to targeted drug discovery are poor functional characterization of target oncoproteins and limited screening modalities. These challenges have been particularly vexing for acute myeloid leukemia (AML) therapy, where many of the defining oncogenic events involve abnormalities of transcription factors, a class of proteins historically considered undruggable. To overcome this impasse to small molecule discovery, we developed gene expression–based high-throughput screening (GE-HTS).1,2 GE-HTS offers a powerful, generic screening approach that relies on gene signatures as surrogates for biologic phenotypes. In principle, this method can be used to identify small-molecule modulators of any oncogenic transcription factor. Here we apply this approach to discover modulators of AML1-ETO, the most common fusion protein identified in AML.
The conserved t(8;21) translocation encodes the AML1-ETO fusion protein and is identified in up to 12% of AML cases.3 The translocation creates an in-frame fusion between the N-terminal DNA-binding domain of AML1 (RUNX1, CBFA2) and almost the entire ETO (MTG8) gene.4-7 The AML1 gene encodes a subunit of the core-binding factor heterodimer, which plays an important role in transcriptional regulation during hematopoiesis. ETO recruits a nuclear receptor corepressor-histone deacetylase complex (NCoR) and the mSin3 corepressor.8-10 AML1-ETO is thus believed to act in part by repressing the transcription of AML1-driven genes associated with myeloid differentiation. A second hypothesis suggests that AML1-ETO promotes enhanced self-renewal of stem cell populations allowing for the accumulation of secondary mutations.11-15
Evidence that AML1-ETO inhibits differentiation is supported by studies in which AML1-ETO expression was blocked in cell lines. Transfection of AML1-ETO antisense oligonucleotides into t(8;21)–positive Kasumi-1 and SKNO-1 cells results in cellular differentiation and growth inhibition.16 Furthermore, AML1-ETO knockdown with transient transfection of small interfering RNAs (siRNAs) sensitizes both Kasumi-1 and SKNO-1 cell lines to transforming growth factor β1 (TGFβ1)/vitamin D3–induced cellular differentiation,17 diminishes clonogenicity, inhibits proliferation, and induces senescence.18 Blocking AML1-ETO activity in vivo has the potential to replicate these prodifferentiation and antiproliferative effects and thus presents an attractive target for therapeutic intervention.
Targeting AML1-ETO presents a formidable challenge, because the precise mechanism by which the fusion protein exerts its effects remains to be elucidated. An ideal initial screening approach would thus be broadly sensitive to multiple mechanisms that could modulate AML1-ETO function. The fusion protein could theoretically be inhibited on many discrete levels, potentially by reducing fusion protein expression, directly blocking DNA binding, inhibiting ETO-mediated histone deacetylation, or even increasing activation of alternative differentiation pathways. The GE-HTS approach enables discovery of inhibitors at any level. Here, we describe the application of GE-HTS to the identification of small molecules that inhibit an AML1-ETO expression signature.
Methods
Full details of the materials and methods are available on the Blood website (Children's Hospital Boston, Harvard Medical School, Botson, MA); see the Supplemental Materials link at the top of the online article.
Cell culture
The Kasumi-1, HL-60, U937, KG-1, and KG-1A cell lines were purchased from ATCC (Manassas, VA). MOLM-14, THP-1, and MV411 were provided by Scott Armstrong (Children's Hospital Boston, Harvard Medical School, Boston, MA). Cells were cultured in RPMI 1640 (Cellgro, Manassas, VA) with 10% fetal calf serum (FCS; Sigma-Aldrich, St Louis, MO) and 1% penicillin-streptomycin. SKNO-1 cells were obtained from Jonathan Licht (Northwestern University, Chicago, IL) and cultured in RPMI 1640 with 20% FCS, 1% penicillin-streptomycin, and 1 ng/mL granulocyte macrophage colony-stimulating factor (GM-CSF; BD Biosciences, San Jose, CA). ME-1 cells were provided by Lucio Castilla (University of Massachusetts Medical School, Worcester, MA) and grown in RPMI 1640 (ATCC) with 20% heat-inactivated FCS (Thermo Scientific Hyclone, South Logan, UT), 1% penicillin-streptomycin, and 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES). Three primary AML1-ETO–positive cryopreserved AML bone marrow samples were collected under a Dana-Farber Cancer Institute Institutional Review Board (IRB)–approved protocol, and patients' informed consent was obtained in accordance with the Declaration of Helsinki. Cells were thawed and maintained in liquid culture in RPMI 1640 with 10% FCS and 1% penicillin-streptomycin.
RNA interference (RNAi)
The siRNA constructs against AML1-ETO and firefly luciferase were designed as previously described17 and synthesized by Dharmacon (Thermo Scientific, South Logan, UT). Cells were transfected either by Amaxa nucleofection (Amaxa buffer V and program P-19; Lonza, Cologne, Germany) or by siLentFect lipid-based reagent (Bio-Rad Laboratories, Hercules, CA).
Immunoblotting
Cells were lysed in RIPA buffer with protease inhibitor (Complete Mini EDTA-free protease inhibitor tablets; Roche Diagnostics, Basel, Switzerland), resolved by electrophoresis on 10% Tris-HCl precast Ready Gels (Bio-Rad Laboratories), and transferred to polyvinylidene fluoride (PVDF; Millipore, Billerica, MA) or nitrocellulose (Bio-Rad Laboratories) membranes. Blots were probed with antibodies for AML1 (PC285; Calbiochem, San Diego, CA), ETO (PC283; Calbiochem), Bax (SC-493; Santa Cruz Biotechnology, Santa Cruz, CA), BCL-2 (551098; BD Biosciences, San Jose, CA), BIM (22-40, Calbiochem), or pan-actin antibody (ACTN05; NeoMarkers; Thermo Scientific). Blots were subsequently probed with anti–rabbit horseradish peroxidase (HRP; NA9340V; Amersham, Pittsburgh, PA) or anti–mouse-HRP (NA9310V; Amersham). Bound antibody was detected by chemiluminescence.
Real-time RT-PCR
Total RNA was isolated in a time course using TRIzol reagent (Invitrogen, Carlsbad, CA) from Kasumi-1 cells transfected with AML1-ETO–directed siRNA or the control siGL siRNA. Total RNA was isolated at 48 hours for the compound-treated samples. cDNA was synthesized from total RNA using SuperScript III Reverse Transcriptase (Invitrogen) and oligo-d(T)16 primers. cDNA was analyzed by real-time quantitative polymerase chain reactions (q-PCR) prepared with TaqMan Universal Master Mix (Applied Biosystems, Foster City, CA). Each sample was assessed in a minimum of 3 replicates to ensure reproducibility of the quantitative measurements. GAPDH, ABL1, or RPL13A expression was evaluated for each sample as a control for total RNA. Primers and probes for real-time reverse transcription PCR (qRT-PCR) were obtained from Applied Biosystems (GAPDH no. 402869, ABL1 no. Hs99999002_mH, RPL13A no. Hs01926559_g1, and AML1-ETO assay by design no. 191284).
Expression profiling
Kasumi-1 were transfected in triplicate with AML1-ETO or luciferase siRNA constructs by either Amaxa nucleofection or Bio-Rad siLentFect and incubated for 96 hours. RNA was extracted with TRIzol, and 10 μg were used to create microarray target samples for hybridization to Affymetrix U133A arrays as previously described (Affymetrix, Santa Clara, CA).19 Raw microarray data are available at http://www.broad.mit.edu/cancer/pub/AML1-ETO_GE-HTS and Cel File annotation in Tables S1 and S2. The data has also been deposited into the GEO public database under accession number GSE15648.
Marker gene selection
GeneChip MAS5 software was used for preprocessing of the raw expression data, and all scans within an experiment were scaled to the array with the median overall microarray intensity. Thresholds were set to a minimum of 10 and a maximum of 16 000, and a variation filter of 3-fold minimum and 100 unit minimum absolute difference applied. We used the signal-to-noise ratio (SNR) to rank order the genes that distinguish “AML1-ETO positive” from “AML1-ETO knockdown” and then permutation testing (1000 permutations) to identify genes distinguishing these states with P less than .05. Next, genes were identified with at least a 3-fold difference between the 2 classes. The expression of these genes was evaluated in an AML1-ETO–inducible dataset obtained from U937 cells with conditional expression of an AML1-ETO construct under the control of the tetracycline promoter20 profiled on Affymetrix U95Av2 oligonucleotide microarrays. Through a comparison of the stable knockdown and inducible rescue datasets, we selected 25 genes that were well-correlated with AML1-ETO expression in both datasets with P less than .05 in at least one of the datasets. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta-actin (ACTB), and ribosomal protein large, P0 (RPLP0) were included as controls for well-to-well variability. Probes for wild-type AML1 were also included as an additional control but were not used in the final analysis. Probes are listed in Table S3.
Small molecule library screening methods
Kasumi-1 cells were plated in 384-well tissue culture plates in 50 μL medium at 25 000 cells/well using a Multidrop (Thermo Scientific). The following controls were included on each screen plate: medium only (16 wells), Kasumi-1 cells (16 wells), AML1-ETO knockdown RNA (8 wells), and Kasumi-1 control RNA (8 wells). Compounds were added at a final approximate concentration of 20 μM in dimethyl sulfoxide (DMSO) by pin transfer and incubated for 72 hours at 37°C with 5% CO2. We screened a total of 2480 compounds in triplicate, including the National Institute of Neurological Disorders and Stroke (NINDS, Bethesda, MD) the SpecPlus, and the BIOMOL ICCB known bioactives collections. The GE-HTS assay was performed as previously described.2
Hit identification
The median fluorescence intensity for each gene (represented by an individual bead color) from the Luminex detector was used as a measure of the gene's expression. To maximize consistency between and within plates, we normalized genes using the expression ratio between each gene and an average of 3 control genes (GAPDH, ACTB, and RPLP0). Next, filtering was performed to eliminate poor performing wells from further analysis. The average across the screen of the GAPDH background intensity in the medium-only wells was used as the filtering threshold. After filtering, each plate was median scaled to reduce plate-to-plate variation. Compounds that induce the AML1-ETO abrogation signature were identified using 3 discrete analytic metrics: weighted summed score, K-nearest neighbor analysis, and naive Bayes classification. Initial chemical hits were defined as compounds that induced the AML1-ETO signature by at least 2 of the 3 analytic metrics at P less than .05.
Morphologic evaluation
Cytospin preparations were evaluated by May-Grunwald Giemsa staining (Sigma-Aldrich), and images were acquired with an Olympus BX41 microscope (Olympus, Center Valley, PA) under 1000× magnification using oil by QCapture software (QImaging, Surrey, BC).
Flow cytometry
For differentiation studies cells were plated in either duplicate or triplicate and incubated with compound for 72 hours. Cells were stained with 1:25 CD11b-FITC (Beckman Coulter IOTest IM0530U; Beckman Coulter, Fullerton, CA) and 1:25 CD14-PE (Beckman Coulter IOTest IM0650U) for 30 minutes. Cells were detected by flow cytometry (Beckman Cytomics FC500; Beckman Coulter) and data analyzed using the FlowJo software package (TreeStar, Ashland, OR). Apoptosis studies were performed in triplicate with cells treated with compounds for 3 days. Annexin V FITC/PI staining was performed with the Annexin V: FITC Apoptosis Detection kit I (BD Pharmingen, BD Biosciences). Cells were analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson, BD Biosciences) and CellQuest analytical software (BD Biosciences).
Myeloid differentiation signature
Marker genes for myeloid differentiation were chosen using previously published Affymetrix datasets containing primary AML cells, normal human neutrophils and monocytes, and the HL-60 AML cell line differentiated with either ATRA, phorbol 12-myristate 13-acetate (PMA), or 1,25-dihydroxyvitamin D3 (vitD).1 We selected 32 genes that distinguished primary AML from normal myeloid cells and undifferentiated HL-60 from HL-60 treated with at least 1 of 3 differentiation agents. These genes distinguish AML from either neutrophil or monocyte with P less than .05 by Student t test and distinguish undifferentiated versus differentiated HL-60 with either ATRA, PMA, or vitD with P less than .05 by Student t test. Probes are shown in Table S4.
Viability assay
Viability experiments were performed in a minimum of triplicate using the Promega Cell-Titer Glo ATP-based assay (Promega, Madison, WI). AML cells were evaluated at 3, 5, or 6 days with hit compounds in a 2-fold dilution to establish the drug concentrations that reduced cell viability to 50% of the vehicle controls (IC50). Values for IC50 were calculated by interpolating a natural cubic spline fit to the measured viability data in R (using the spline function). The natural spline requires that the second derivative of the interpolated curve equal 0 at the end points of the interval of interpolation. The IC50 value was found by evaluating in R the interpolated spline at 0.5 using the approx function. Synergy was assessed by analyzing the IC50 of one drug over a range of concentrations of the other drug and vice versa. The resulting concentration pairs were visualized by isobologram.21
Methylcellulose colony forming assay
Kasumi-1 cells were incubated for 24 hours with methotrexate (20-60 nM) or methylprednisolone (20-160 nM) or DMSO 0.08%. After incubation, 3200 cells were resuspended in 1.5 mL methylcellulose containing growth factors interleukin-3 (IL-3), GM-CSF, erythropoietin, stem cell factor (Methocult GF-H4434; StemCell Technologies, Vancouver, BC), and appropriate drug treatments and plated on grid culture dishes (Integrated BioDiagnostics, Munich, Germany). Colony number was counted on day 14 using an inverted microscope (Nikon Eclipse TE200; Nikon, Melville, NY) at 400× magnification. Error bars represent the SD of duplicate replicates.
Xenograft studies
SKNO-1 cells were engineered for imaging by transduction with a lentivirus encoding firefly luciferase, mCherry, and puromycin phosphotransferase.22 6-week-old male NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (NOG; The Jackson Laboratory, Bar Harbor, ME) were sublethally irradiated with 200 cGy and injected 3 hours later with 1 million SKNO-1-Luc-mCherry cells. Total body bioluminescence was determined by in vivo imaging (IVIS Spectrum; Caliper Life Sciences, Hopkinton, MA) and quantitated as previously described.23 Mice with established leukemia defined by logarithmically increasing bioluminescence signal were divided into groups and treated with vehicle, methotrexate 5 mg/kg intraperitoneally, or dexamethasone 15 mg/kg intraperitoneally for either 4 or 8 daily doses. Tumor burden was serially quantified by in vivo bioluminescence imaging. All animal studies were performed under a Dana-Farber Cancer Institute Institutional Animal Care and Use Committee (IACUC)–approved protocol.
Results
Discovery of AML1-ETO abrogation signature
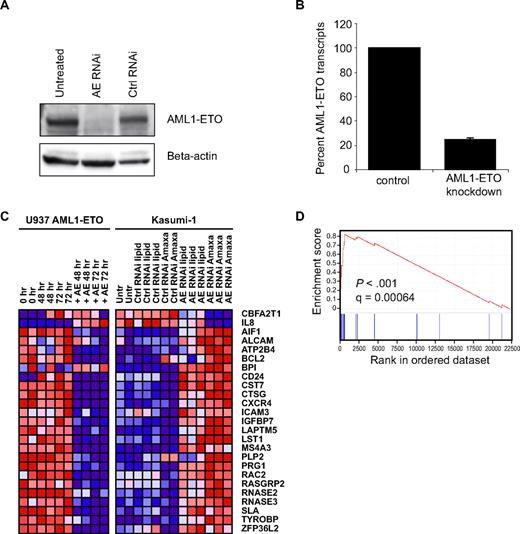
To perform GE-HTS for inhibitors of AML1-ETO, it was first necessary to define a signature for AML1-ETO abrogation. To identify the most robust signature, we used 3 approaches: RNAi-mediated knockdown of AML1-ETO, induction of AML1-ETO expression, and evaluation of primary AML expression datasets. siRNA constructs against AML1-ETO have previously been reported to deliver efficacious knockdown.17 The RNAi sequence was designed against the AML1-ETO breakpoint to minimize off-target effects on wild-type AML1 and ETO (Figure S1). After commercial siRNA synthesis, Amaxa nucleofection was used to transfect t(8;21)-containing Kasumi-1 cells. Posttransfection, near-complete AML1-ETO protein knockdown was observed by immunoblot at 96 hours (Figure 1A). In addition, knockdown of the RNA transcript was verified by real-time PCR, with only 20% of the RNA transcript remaining at 24 hours (Figures 1B, S1). A second set of Kasumi-1 cells was transfected using a lipid-based reagent to control for the effects of electroporation and achieved partial knockdown of AML1-ETO protein by immunoblot (data not shown).
Discovery of AML1-ETO abrogation signature. (A) Kasumi-1 immunoblot with AML1 antibody demonstrates loss of AML1-ETO protein expression 96 hours after transient transfection with siRNA by Amaxa. (B) Real-time PCR with AML1-ETO–specific primers reveals loss of AML1-ETO RNA at 24 hours. (C) Transcriptional profiling of Kasumi-1 cells on Affymetrix U133A microarrays reveals AML1-ETO abrogation signature in Kasumi-1 cells with both lipid and Amaxa transfection. Dark red indicates high gene expression and dark blue indicates low gene expression. The corresponding gene expression in AML1-ETO–inducible U937 cells is indicated. (D) Gene set enrichment analysis demonstrates enrichment of the AML1-ETO abrogation signature in primary AML patient cells expressing t(8;21) versus other molecular subtypes of AML.
Discovery of AML1-ETO abrogation signature. (A) Kasumi-1 immunoblot with AML1 antibody demonstrates loss of AML1-ETO protein expression 96 hours after transient transfection with siRNA by Amaxa. (B) Real-time PCR with AML1-ETO–specific primers reveals loss of AML1-ETO RNA at 24 hours. (C) Transcriptional profiling of Kasumi-1 cells on Affymetrix U133A microarrays reveals AML1-ETO abrogation signature in Kasumi-1 cells with both lipid and Amaxa transfection. Dark red indicates high gene expression and dark blue indicates low gene expression. The corresponding gene expression in AML1-ETO–inducible U937 cells is indicated. (D) Gene set enrichment analysis demonstrates enrichment of the AML1-ETO abrogation signature in primary AML patient cells expressing t(8;21) versus other molecular subtypes of AML.
Genome-wide transcriptional profiling was performed using Affymetrix U133A oligonucleotide microarrays to identify differential gene expression between Kasumi-1 cells with and without AML1-ETO knockdown. Probe sets differentially expressed upon AML1-ETO knockdown (P < .05) were identified using permutation testing. To guard against false-positive marker genes from off-target effects of RNAi, we analyzed the expression of potential marker genes in an AML1-ETO-inducible U937 cell line. Genes that were up-regulated in the knockdown data were generally down-regulated in the inducible model and vice versa. The intersection of the 2 datasets was used to select 25 marker genes. GAPDH, ACTB, and RPLP0 were included as controls for well-to-well variability in the screen because of their stable expression across the dataset. The final signature constituted 28 genes (Figure 1C).
Transcriptional profiling data from primary AML patients was analyzed to confirm physiologic relevance of our AML1-ETO abrogation signature. Gene set enrichment analysis (GSEA) was used to query a published dataset of 285 primary patient AML samples, of which 22 had cytogenetic evidence of t(8;21).24 GSEA is a nonparametric analytical approach used to determine whether the members of a gene set of interest (eg, genes associated with loss or induction of AML1-ETO) tend to occur toward the top of a ranked list of genes corresponding to class distinction (AML1-ETO-positive versus AML1-ETO–negative primary patient AML blasts).25 GSEA revealed statistically significant enrichment of the 23 unique up-regulated genes in the AML1-ETO signature, with an enrichment score of 0.84 and an FDR q-value of 6.35 × 10−4. Figure 1D displays the graphical distribution of signature genes within the AML1-ETO samples. As expected, most genes that had high expression with AML1-ETO knockdown display low expression in the presence of AML1-ETO in patient cells.
Gene expression-based screen identifies 2 drug classes
To perform a high-throughput screen, the 28-gene AML1-ETO abrogation signature was converted to the low-cost ligation-mediated amplification (LMA) format. This technique uses directly opposed gene-specific probes coupled to universal primers for PCR amplification. Final PCR amplicons are captured by complementary barcode sequences and detected by bead-based fluorescence using dual-colored flow cytometry.2 Probes were designed as described in supplemental methods and are listed in Table S3.
We sought to test FDA-approved drugs and known bioactive compounds to facilitate future clinical translation as well as inform mechanistic discovery from hit compounds. We selected 3 compound collections available at the Broad Institute: NINDS, SpecPlus, and Biomol. The 3 libraries, containing a total of 2480 compounds, were screened in triplicate. After 72 hours of incubation, cells were lysed, and cDNA synthesis was performed. Each screen plate included RNA from Kasumi-1 cells transfected with AML1-ETO siRNA as a positive control and Kasumi-1 cells treated with DMSO as a negative control. Next, LMA was carried out, and the expression level for each of the 28 genes in the signature was detected by a bead-based fluorescence system.
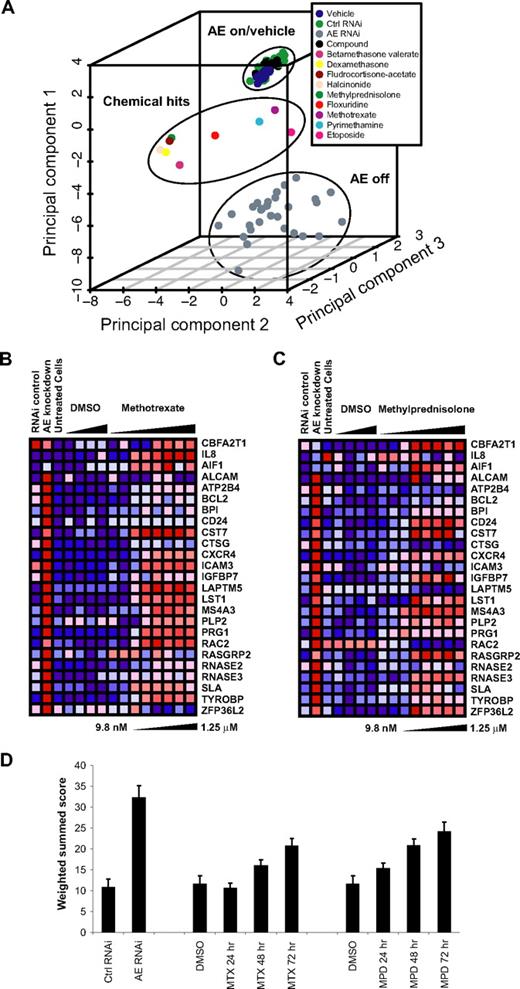
Compounds that induced the AML1-ETO abrogation signature by at least 2 discrete statistical methods (as described in detail in supplemental methods) were flagged as hits, and confirmatory testing was performed to determine signature dose and time response for a subset of these compounds. Compounds that were grossly cytotoxic at concentrations required to induce the AML1-ETO signature were excluded from further analysis. Confirmed hits are depicted graphically by principal component analysis (PCA) in Figure 2A. Within the 3-dimensional space of PCA, the chemical hits fall between full AML1-ETO knockdown and the control cells. Confirmed chemical hits primarily represented one of 2 drug classes: corticosteroids and dihydrofolate reductase (DHFR) inhibitors.
AML1-ETO GE-HTS screen identifies 2 top classes of hits. (A) Principal component analysis performed with compounds that were statistically significant by at least 2 discrete metrics for the AML1-ETO abrogation signature. The position of hits relative to controls with and without AML1-ETO expression is indicated. The points labeled “compound” represent the average of all chemicals on each individual screening plate. The hit compound datapoints represent the mean of 3 replicates. All other data points represent the mean of samples per individual screen plate: vehicle (16), Ctrl RNA (8), and AE RNAi (8). Validated hits included 2 primary drug classes: corticosteroids and DHFR antagonists. (B) Induction of AML1-ETO abrogation signature in Kasumi-1 cells treated with indicated dose of methotrexate for 72 hours. Each RNAi control or cell line control data point represents the mean of 16 samples, while the vehicle- and compound-treated points represent the mean of 3 samples. (C) Induction of abrogation signature in Kasumi-1 cells treated with methylprednisolone for 72 hours. Each RNAi control or cell line control data point represents the mean of 16 samples, while the vehicle- and compound-treated samples represent the mean of 3 samples. (D) Weighted summed score for the AML-ETO signature upon treatment of Kasumi-1 cells with 80 nM methotrexate (MTX) or 0.5 μM methylprednisolone (MPD) for 24 to 72 hours. There were 16 replicates for drug treatment and RNAi controls and 32 for DMSO controls.
AML1-ETO GE-HTS screen identifies 2 top classes of hits. (A) Principal component analysis performed with compounds that were statistically significant by at least 2 discrete metrics for the AML1-ETO abrogation signature. The position of hits relative to controls with and without AML1-ETO expression is indicated. The points labeled “compound” represent the average of all chemicals on each individual screening plate. The hit compound datapoints represent the mean of 3 replicates. All other data points represent the mean of samples per individual screen plate: vehicle (16), Ctrl RNA (8), and AE RNAi (8). Validated hits included 2 primary drug classes: corticosteroids and DHFR antagonists. (B) Induction of AML1-ETO abrogation signature in Kasumi-1 cells treated with indicated dose of methotrexate for 72 hours. Each RNAi control or cell line control data point represents the mean of 16 samples, while the vehicle- and compound-treated points represent the mean of 3 samples. (C) Induction of abrogation signature in Kasumi-1 cells treated with methylprednisolone for 72 hours. Each RNAi control or cell line control data point represents the mean of 16 samples, while the vehicle- and compound-treated samples represent the mean of 3 samples. (D) Weighted summed score for the AML-ETO signature upon treatment of Kasumi-1 cells with 80 nM methotrexate (MTX) or 0.5 μM methylprednisolone (MPD) for 24 to 72 hours. There were 16 replicates for drug treatment and RNAi controls and 32 for DMSO controls.
Hits confirmed by secondary screening
It was necessary to retest individual hit compounds to confirm signature induction in a reproducible manner. For each candidate hit compound, a 2-fold dose dilution series was performed. Cells were treated, lysed at 72 hours, and analyzed as in the initial screen. The secondary screen verified signature induction for the compounds is shown in Figure 2A. We chose to focus on methotrexate and methylprednisolone as representative of the 2 drug classes. The threshold dose required for partial signature induction by methylprednisolone was 40 nM; 78 nM or greater successfully induced expression of the majority of signature genes (Figure 2B). For methotrexate, the threshold dose for signature induction was between 40 and 80 nM (Figure 2C). In addition, a time course experiment was performed to establish the timing of signature induction after compound addition. The weighted summed score provides one metric by which to compare the degree of signature induction and was calculated for each hit compound. The weighted summed score combines expression ratios by summing them with a weight and sign determined by the SNR of each expression ratio for the positive control (AML1-ETO knockdown) and negative control (Kasumi-1 RNA) samples. Statistically significant signature induction by weighted summed score was seen at 48 and 72 hours for methylprednisolone and methotrexate (Figure 2D).
Chemical hits induce myeloid differentiation
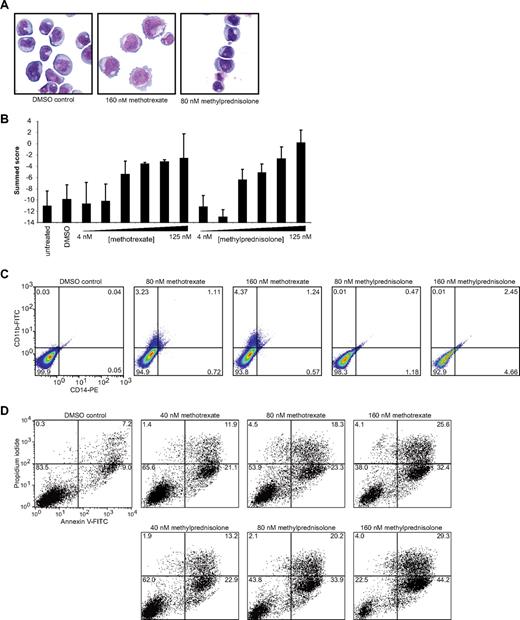
Because RNAi directed against AML1-ETO enhances differentiation, we sought to evaluate the effects of confirmed hits on differentiation by cell morphology and cell surface marker expression. May-Grunwald Giemsa staining was performed on Kasumi-1 cells, and cellular morphology was evaluated under light microscopy. Both methotrexate and methylprednisolone induced visual changes consistent with myeloid maturation. Methotrexate induced increased cell size, vacuole formation, and cytoplasmic extension. Methylprednisolone, on the other hand, induced nuclear condensation and a subtle decrease in cell size (Figure 3A). To confirm that the morphologic changes observed were indeed representative of myeloid differentiation, we determined the effects of methotrexate and methylprednisolone on a 32-gene myeloid differentiation signature measured with the LMA/fluorescent bead–based assay. Both compounds induced a dose-dependent increase in the signature compared with vehicle-treated cells at 72 hours (Figure 3B). We next measured compound effects on cell surface markers associated with myeloid maturation, CD11b and CD14. Flow cytometry revealed that methotrexate-treated cells had slightly increased expression of CD11b and, to a lesser extent, CD14 (Figure 3C). Methylprednisolone induced a subtle increase in single staining for CD14 and double staining for CD14 and CD11b. Similarly, other representative members of the class corticosteroids (dexamethasone) and DHFR inhibitors (pyrimethamine) confirmed to modulate the AML1-ETO signature induced evidence of differentiation (Figures S2, S3).
Hit compounds induce myeloid differentiation and apoptosis. (A) May-Grunwald Giemsa staining of Kasumi-1 cells treated with DMSO, 160 nM methotrexate, or 80 nM methylprednisolone for 72 hours viewed under 1000× magnification. Both methotrexate and methylprednisolone induced visual evidence of myeloid differentiation. (B) Induction of myeloid differentiation signature in Kasumi-1 cells after 72 hours of treatment with indicated concentrations of methotrexate or methylprednisolone. Each condition was evaluated in replicates of 7 for hits and 14 for DMSO and untreated cells. (C) CD11b/CD14 cell-surface staining of Kasumi-1 cells at 72 hours. Treatment with methotrexate induced CD11b up-regulation and a small population of CD11b/CD14 double-positive cells. Methylprednisolone (160 nM) induced minimal up-regulation of CD14 and double-positive cells. (D) Annexin V/propidium iodide staining of Kasumi-1 cells after 72 hours of treatment with 40 to 160 nM methotrexate or methylprednisolone. Both compounds increased annexin V cell-surface staining in a dose-responsive manner.
Hit compounds induce myeloid differentiation and apoptosis. (A) May-Grunwald Giemsa staining of Kasumi-1 cells treated with DMSO, 160 nM methotrexate, or 80 nM methylprednisolone for 72 hours viewed under 1000× magnification. Both methotrexate and methylprednisolone induced visual evidence of myeloid differentiation. (B) Induction of myeloid differentiation signature in Kasumi-1 cells after 72 hours of treatment with indicated concentrations of methotrexate or methylprednisolone. Each condition was evaluated in replicates of 7 for hits and 14 for DMSO and untreated cells. (C) CD11b/CD14 cell-surface staining of Kasumi-1 cells at 72 hours. Treatment with methotrexate induced CD11b up-regulation and a small population of CD11b/CD14 double-positive cells. Methylprednisolone (160 nM) induced minimal up-regulation of CD14 and double-positive cells. (D) Annexin V/propidium iodide staining of Kasumi-1 cells after 72 hours of treatment with 40 to 160 nM methotrexate or methylprednisolone. Both compounds increased annexin V cell-surface staining in a dose-responsive manner.
Chemical hits reduce cellular viability and induce apoptosis
Kasumi-1 cells proved to be extraordinarily sensitive to corticosteroids as measured by cellular viability. On repeat testing, the IC50 value of methylprednisolone was 37 nM at 3 days. Testing of SKNO-1 cells, a second t(8;21)-positive AML cell line, revealed a methylprednisolone IC50 of 250 nM. This result contrasts with IC50 values of greater than 100 μM for 8 cell lines that lack AML1-ETO expression, including the cell line ME-1 expressing the CBFβ-MYH11 rearrangement (Table S5). In addition, the IC50 for methotrexate was 14 nM in Kasumi-1 cells at 3 days (with an essentially static growth-inhibitory effect from 30-1000 nM) although this effect was not specific to AML1-ETO–expressing cell lines (Table S5). Furthermore, treatment with both compounds induced a dose-dependent increase in apoptosis determined by an annexin V–positive fluorescence-activated cell sorting (FACS) profile (Figure 3D) as did treatment with other class members (pyrimethamine and dexamethasone; Figure S4). Although the precise mechanism of apoptosis for these 2 molecules in AML1-ETO–positive cells has not yet been elucidated, there is evidence of decreased BCL2 protein levels with methylprednisolone treatment at 24 and 48 hours (Figure S5).
Conditional expression of AML1-ETO in U937 cells sensitizes to the effects of methotrexate and corticosteroids on viability
U937 cells conditionally expressing AML1-ETO under the control of the tetracycline promoter were grown in the presence (AML1-ETO off) or absence of tetracycline (AML1-ETO on) for 3 days (Figure S6A). They were then treated with methotrexate and methylprednisolone in a 2-fold dilution series for 3 additional days, and viability was measured by an ATP-based assay. Cells grown in the absence of tetracycline, with high expression of AML1-ETO, were more sensitive to the effects of these 2 molecules on viability than were non-AML1-ETO expressors, suggesting that expression of AML1-ETO may enhance the inhibition of cell viability with these 2 compounds (Figure S6B). Enhancement of a differentiation response was not seen in this model with the expression of AML1-ETO (Figure S6C).
DHFR inhibitor and corticosteroid activity are on-target
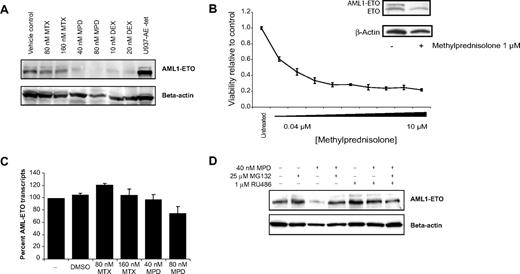
To test whether the effects of methotrexate were due to on-target DHFR inhibition, cotreatment experiments were performed with leucovorin (folinic acid), the clinical antidote to methotrexate. Cotreatment of Kasumi-1 cells with leucovorin blocked the cytotoxic effect of methotrexate and pyrimethamine by cellular viability assay (Figures 4A, S7). Importantly, leucovorin did not rescue the effects of methylprednisolone on viability (Figure 4A). RNA isolated from cotreated cells also lacked induction of the AML1-ETO abrogation signature (Figure 4B). Thus, leucovorin reverses the inhibitory effects and gene expression changes associated with DHFR inhibitor treatment.
Rescue of cellular viability and AML1-ETO gene signature with chemical antagonists. (A) Kasumi-1 cells treated with methotrexate or methylprednisolone. Rescue was performed with either 1 μM leucovorin or 1 μM RU486. Cellular viability was assessed at 72 hours with an ATP-based assay and plotted as a ratio relative to control cells. Samples were evaluated in replicates of 7. Leucovorin rescued only methotrexate, while RU486 rescued only methylprednisolone. (B) Weighted summed score for the AML1-ETO signature upon treatment of Kasumi-1 cells with 160 nM methotrexate (MTX) or 160 nM methylprednisolone (MPD) for 72 hours. Chemical rescue was performed with cotreatment of 1 μM leucovorin or 1 μM RU486 as indicated.
Rescue of cellular viability and AML1-ETO gene signature with chemical antagonists. (A) Kasumi-1 cells treated with methotrexate or methylprednisolone. Rescue was performed with either 1 μM leucovorin or 1 μM RU486. Cellular viability was assessed at 72 hours with an ATP-based assay and plotted as a ratio relative to control cells. Samples were evaluated in replicates of 7. Leucovorin rescued only methotrexate, while RU486 rescued only methylprednisolone. (B) Weighted summed score for the AML1-ETO signature upon treatment of Kasumi-1 cells with 160 nM methotrexate (MTX) or 160 nM methylprednisolone (MPD) for 72 hours. Chemical rescue was performed with cotreatment of 1 μM leucovorin or 1 μM RU486 as indicated.
To test whether the effects of methylprednisolone were due to on-target glucocorticoid receptor activation, cotreatment experiments were performed with RU486, a glucocorticoid receptor antagonist. Cotreatment of Kasumi-1 cells with RU486 blocked the cytotoxic effects of methylprednisolone and dexamethasone on cellular viability assays (Figures 4A, S8). RU486 did not rescue the effects of methotrexate (Figure 4A). RNA isolated from cotreated cells also lacked induction of the AML1-ETO abrogation signature (Figure 4B). Thus, RU486 reversed the inhibitory effects and gene expression changes associated with corticosteroid treatment.
Corticosteroids reduce AML1-ETO protein expression
To further evaluate the mechanism of signature induction, AML1-ETO protein and RNA expression were measured after incubation with hit compounds. AML1 immunoblot revealed loss of AML1-ETO protein expression in a dose- and time-responsive manner over 48 hours with both methylprednisolone and dexamethasone (Figures 5A, S9). AML1-ETO protein was almost undetectable after 80 nM methylprednisolone treatment for 48 hours. The corresponding change in protein expression level in methotrexate-treated cells was minimal. Moreover, in 3 primary patient AML samples, AML1-ETO protein was reduced with in vitro methylprednisolone treatment. In 2 samples with adequate cells for additional testing, methylprednisolone inhibited cellular viability (Figures 5B, S10). Real-time PCR testing for AML1-ETO RNA was used to evaluate whether the decrease in protein level was due to changes in gene transcription. As shown in Figure 5C, there was minimal change in AML1-ETO RNA expression upon treatment with methylprednisolone or methotrexate in Kasumi-1 cells.
Decreased AML1-ETO protein abundance upon corticosteroid treatment. (A) Kasumi-1 immunoblot with anti-AML1 antibody. AML1-ETO protein loss was seen upon treatment with methylprednisolone (MPD) or dexamethasone (DEX) for 48 hours. AML1-ETO protein expression was only minimally decreased in the methotrexate (MTX)–treated samples. (B) Methylprednisolone reduces cellular viability and AML1-ETO abundance in primary patient t(8;21) cells. Primary patients' cells were treated with methylprednisolone, and cellular viability was assessed at 3 days with an ATP-based assay and plotted as a ratio relative to control cells. Error bars denote SD across 5 replicates. At 48 hours, expression of AML1-ETO was assessed with immunoblot using an anti-ETO antibody. AML1-ETO loss was seen upon treatment with methylprednisolone. (C) Minimal change in AML1-ETO RNA expression is seen upon MTX and MPD treatment for 48 hours. Results are normalized to control ABL1 gene expression, and conditions were tested in quadruplicate. (D) Methylpredisolone-induced AML1-ETO protein loss was rescued with both MG132 and RU486. Cells were treated with methylprednisolone ± RU486 for 48 hours. MG132 was added for the final 30 minutes of incubation.
Decreased AML1-ETO protein abundance upon corticosteroid treatment. (A) Kasumi-1 immunoblot with anti-AML1 antibody. AML1-ETO protein loss was seen upon treatment with methylprednisolone (MPD) or dexamethasone (DEX) for 48 hours. AML1-ETO protein expression was only minimally decreased in the methotrexate (MTX)–treated samples. (B) Methylprednisolone reduces cellular viability and AML1-ETO abundance in primary patient t(8;21) cells. Primary patients' cells were treated with methylprednisolone, and cellular viability was assessed at 3 days with an ATP-based assay and plotted as a ratio relative to control cells. Error bars denote SD across 5 replicates. At 48 hours, expression of AML1-ETO was assessed with immunoblot using an anti-ETO antibody. AML1-ETO loss was seen upon treatment with methylprednisolone. (C) Minimal change in AML1-ETO RNA expression is seen upon MTX and MPD treatment for 48 hours. Results are normalized to control ABL1 gene expression, and conditions were tested in quadruplicate. (D) Methylpredisolone-induced AML1-ETO protein loss was rescued with both MG132 and RU486. Cells were treated with methylprednisolone ± RU486 for 48 hours. MG132 was added for the final 30 minutes of incubation.
Corticosteroid-induced AML1-ETO degradation requires proteasome activity
Given that AML1-ETO protein degradation upon corticosteroid treatment occurs in the setting of continued AML1-ETO RNA expression, one possible mechanism is increased activity of the ubiquitin-proteasome pathway. This pathway has recently been implicated in AML1-ETO degradation in experiments using the proteasome inhibitor MG132.26 Cotreatment of Kasumi-1 cells with methylprednisolone and MG132 results in increased accumulation of AML1-ETO relative to methylprednisolone treatment alone (Figure 5D). Moreover, RU486 cotreatment blocked the effects of methylprednisolone on AML1-ETO protein.
In vivo activity of DHFR antagonists and corticosteroids
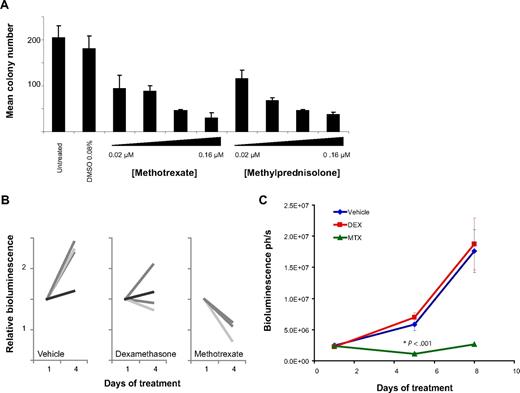
To address whether DHFR inhibitors or corticosteroids have in vivo activity, we first assessed methylcellulose colony formation with methotrexate and methylprednisolone treatment. Both compounds dramatically reduced colony formation in Kasumi-1 cells (Figures 6A, S11). We next attempted to establish an orthotopic cell line model of AML1-ETO–positive AML by tail vein injection and succeeded in establishing a luciferase positive SKNO-1 orthotopic model in NOG mice. Tumor burden was quantified using in vivo bioluminescence imaging as previously described.23 Mice were treated with placebo, dexamethasone, or methotrexate. In a pilot 4-day study, response was suggested in both the dexamethasone-treated and methotrexate-treated animals with disease stabilization with dexamethasone treatment and tumor regression with methotrexate treatment (Figure 6B). In a larger study over 8 days, only the methotrexate-treated animals demonstrated a statistically significant response (Figure 6C).
In vivo activity of DHFR antagonists and corticosteroids. (A) The ability of Kasumi-1 cells to form colonies in methylcellulose with methotrexate or methylprednisolone was assessed. Both compounds inhibited colony formation compared with a vehicle control. Error bars represent SD across duplicate measurements. (B) A luciferase positive SKNO-1 orthotopic model was established in NOG mice. Mice were treated with vehicle (n = 4), dexamethasone 15 mg/kg per day intraperitoneally (n = 4), or methotrexate 5 mg/kg per day intraperitoneally (n = 3) for 4 daily doses, and tumor burden was monitored by in vivo imaging. (C) In the luciferase-positive SKNO-1 orthotopic model as above, mice were treated with vehicle (n = 7), dexamethasone (DEX) 15 mg/kg per day intraperitoneally (n = 7), or methotrexate (MTX) 5 mg/kg per day intraperitoneally (n = 7) for 8 doses, and tumor burden was monitored by in vivo imaging. Data are represented as mean ± SEM (n = 7 in each treatment arm). *P < .001 by 1-way analysis of variance (ANOVA) with Tukey posttest.
In vivo activity of DHFR antagonists and corticosteroids. (A) The ability of Kasumi-1 cells to form colonies in methylcellulose with methotrexate or methylprednisolone was assessed. Both compounds inhibited colony formation compared with a vehicle control. Error bars represent SD across duplicate measurements. (B) A luciferase positive SKNO-1 orthotopic model was established in NOG mice. Mice were treated with vehicle (n = 4), dexamethasone 15 mg/kg per day intraperitoneally (n = 4), or methotrexate 5 mg/kg per day intraperitoneally (n = 3) for 4 daily doses, and tumor burden was monitored by in vivo imaging. (C) In the luciferase-positive SKNO-1 orthotopic model as above, mice were treated with vehicle (n = 7), dexamethasone (DEX) 15 mg/kg per day intraperitoneally (n = 7), or methotrexate (MTX) 5 mg/kg per day intraperitoneally (n = 7) for 8 doses, and tumor burden was monitored by in vivo imaging. Data are represented as mean ± SEM (n = 7 in each treatment arm). *P < .001 by 1-way analysis of variance (ANOVA) with Tukey posttest.
Corticosteroids act synergistically with standard AML chemotherapy
Although single agent efficacy in vitro and in vivo mouse models is important, combination therapy is required, with rare exception, to cure human malignancies. We thus extended testing of these compounds to combination studies. Both methotrexate and methylprednisolone were tested in combination with the standard cytotoxic AML therapies cytosine arabinoside (ARA-C) and daunorubicin in Kasumi-1 and SKNO-1 cells. In both cell lines, there was a synergistic interaction with methylprednisolone and either ARA-C or daunorubicin (Figures 7A,B, S12). Methotrexate did not have a synergistic interaction with either of these compounds (data not shown).
Methylprednisolone shows a synergistic effect on viability with either daunorubicin or cytosine arabinoside. The combined effects of cytosine arabinoside and methylprednisolone and daunorubicin and methylprednisolone on cell viability in (A) Kasumi-1 cells at 3 days for daunorubicin and 5 days for cytosine arabinoside and in (B) SKNO-1 cells at 5 days, as determined by ATP level, is shown by isobologram. Synergy appears as points below the line of additivity. Experiments were performed in duplicate.
Methylprednisolone shows a synergistic effect on viability with either daunorubicin or cytosine arabinoside. The combined effects of cytosine arabinoside and methylprednisolone and daunorubicin and methylprednisolone on cell viability in (A) Kasumi-1 cells at 3 days for daunorubicin and 5 days for cytosine arabinoside and in (B) SKNO-1 cells at 5 days, as determined by ATP level, is shown by isobologram. Synergy appears as points below the line of additivity. Experiments were performed in duplicate.
Discussion
Despite advances in our understanding of the molecular basis of AML, the mainstay of treatment remains cytotoxic chemotherapy that has changed little over the past 2 decades. While molecular diagnostics are useful for predicting response to therapy, there are few treatments tailored for specific somatic mutations other than ATRA for PML-RARα–containing leukemias. Discovery of targeted modulators has been challenging because many of the described AML mutations involve transcription factors. GE-HTS meets this challenge by providing a generic screening platform by which to identify small molecules that induce a final phenotype of interest. In this series of experiments, we used GE-HTS to identify compounds that reverse the effects of AML1-ETO in AML.
For the initial screen, we focused on libraries of FDA-approved drugs and known bioactive compounds for 2 primary reasons. First, using FDA-approved compounds enables rapid clinical translation, because their safety profiles and efficacy for other conditions are already well established. Second, knowing at least one mechanism of action facilitates initial experiments directed at mechanistic discovery once screen hits are identified. The 2 top-scoring classes of compounds were the corticosteroids and DHFR antagonists, each appearing to induce a change in the AML1-ETO signature by an on-target activity. Leucovorin rescued the activity of DHFR antagonists, and RU486 rescued the activity of corticosteroids. Interestingly, corticosteroids dramatically reduced AML1-ETO protein by Western blot analysis. AML1-ETO RNA expression, however, was maintained, thus suggesting posttranslational degradation or impaired gene translation as possible mechanisms of action. Methotrexate induced only a slight decrease in AML1-ETO protein expression, and the direct link between AML1-ETO function and DHFR inhibition remains unclear. Substantial insight into possible mechanisms driving AML1-ETO degradation by corticosteroids is provided by recent discoveries. First, corticosteroids have been shown to up-regulate genes required for activity of the ubiquitin-proteasome pathway. In particular, corticosteroids induce ubiquitin C expression and proteasomal degradation of c-maf in multiple myeloma cell lines.27 A second study revealed that AML1-ETO itself is degraded by the ubiquitin-proteasome pathway and is critically dependent on 2 genes, E2-conjugase UbcH8 and E3-ligase SIAH-1.26 Furthermore, recent studies have demonstrated degradation of AML1-ETO protein with histone deacetylase inhibitors by a proteasome-dependent mechanism in vitro, although the clinical relevance of this finding has not yet been determined.28 Taken together with our observations, corticosteroid treatment of Kasumi-1 cells may be stimulating proteasomal degradation of AML1-ETO.
Despite initial response rates in t(8;21)–positive AML patients below age 56 that approach 98%, up to a third of these patients will relapse within 5 years.29 Furthermore, in another study among older patients (median age 66), the relapse risk at 5 years in the t(8;21)–containing subgroup was 84%,30 and older patients frequently do not tolerate high-dose cytotoxic chemotherapy. New approaches to treating these patients are needed. Although corticosteroids and DHFR antagonists are frequently used in the treatment of lymphoid malignancies, they are not commonly considered beneficial for myeloid leukemia. However, clinical trials have not addressed their specific application to AML1-ETO–positive AML. The AML1-ETO GE-HTS screen independently identified 5 corticosteroids from the chemical libraries that were all confirmed on repeat testing. Particularly striking were the very low doses at which the corticosteroids inhibited cell viability in comparison to 8 other AML cell lines not bearing the translocation. This result suggests that the t(8;21)–containing cell lines bear increased sensitivity to corticosteroids. Indeed, a study from 1997 also reported growth inhibition of Kasumi-1 and SKNO-1 cells with low-nanomolar doses of dexamethasone but resistance in 6 other AML1-ETO–negative myeloid cell lines.31 The precise mechanism of apoptosis induction by corticosteroids in AML1-ETO–expressing cells remains unclear, although our results suggest down-regulation of BCL2 as a contributing factor. Additional reports show that AML1-ETO increases AP-1–dependent transcription32 and that corticosteroids regulate AP-1 activity by a posttranslational mechanism,33 thus providing another hypothesis regarding the activity of corticosteroids on AML1-ETO–positive leukemia.
A few reports in the literature suggest efficacy of corticosteroids in some patients with AML. One case report described a patient with severe pneumonia at diagnosis of AML1-ETO–positive leukemia who received a short course of high-dose methylprednisolone. Leukemia cells were cleared, and fluorescence in situ hybridization (FISH) for AML1 split signal was negative after single agent corticosteroid.34 Although this report is suggestive of a complete response with steroids alone, the contribution of an endogenous immune response to severe pneumonia cannot be excluded. Spontaneous AML remission with sepsis has very rarely been reported.35 In a second case report, 2 pediatric patients with M2 AML and myeloid tumor had a significant reduction in peripheral blast percentage with short-course oral methylprednisolone.36 Interestingly, although molecular information was not reported for these 2 patients, the presence of granulocytic sarcoma is associated with the AML1-ETO translocation.37
Similarly, AML is generally considered to be a methotrexate-resistant disease. There is growing evidence, however, that methotrexate may remain active against certain AML subtypes. For instance, acute monocytic leukemia, Down syndrome–associated AML, and acute megakaryoblastic leukemia have all been shown to bear increased sensitivity to methotrexate relative to overall AML.38-40 Follow-up experiments compared the concentrations of methotrexate required to inhibit thymidylate synthetase (TS) activity in M5 AML cells versus non-M5 AML and pre-B acute lymphoblastic leukemia (pre-B ALL) patient cells. The authors found that the 50% of TS activity was inhibited at 390 nM (pre-B ALL), 730 nM (M5 AML), and 2.16 μM (non-M5 AML).41 Our own findings indicate that the methotrexate IC50 for cellular viability at 72 hours in Kasumi-1 cells is 14 nM, although the majority of AML cell lines that we tested were also sensitive to the effects of methotrexate on viability. Testing of primary t(8;21)–containing AML cells versus nonexpressers will be required to establish specificity in actual patient cells.
Our pilot in vivo testing of DHFR antagonists and corticosteroids demonstrates efficacy for both compound classes in a methylcellulose colony-forming assay and in an orthotopic xenograft model of AML1-ETO–positive AML. A longer-term response was demonstrated with methotrexate in vivo. More extensive pharmacokinetic and pharmacodynamic studies will be needed to optimize these 2 drugs in vivo and to determine the possible contributions of tumor-host microenvironmental interactions to response. Our studies also demonstrate in vitro synergy between methylprednisolone in combination with either cytarabine or daunorubicin in AML1-ETO–positive AML cell lines. Testing of these combinations in animal models of AML1-ETO disease will also be necessary to establish the in vivo relevance of these findings.
There remains a critical need for improved cancer therapy. Once solely the domain of hematopoietic and pediatric malignancies, transcription factor mutations and effector pathway dysregulation are now broadly implicated in a wide range of malignancies.42,43 Innovative strategies are required to discover new means of targeting such disease-specific abnormalities, with the ultimate goal of providing less toxic and more effective cancer treatment. Using the most common AML-associated chromosomal translocation as a model, we have demonstrated the application of GE-HTS to small molecule discovery for previously intractable cancer-associated proteins.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Nicola Tolliday, Paul Clemons, Stephanie Norton, and Stuart Schreiber for chemistry/screening-related guidance; David Peck for GE-HTS platform support; Erica Esrick, Tina Davis, and Koichi Sasaki for technical support; Lucio Castilla, Dmitri Madera, Scott Armstrong, and Jonathan Licht for providing cell lines; Dong-Er Zhang for the U937 conditional AML1-ETO cell line; and Jacob Berchuck for assistance with figure design.
This work was funded by the American Society of Hematology (Washington, DC; K.S.), Sidney Kimmel Foundation for Cancer Research (Baltimore, MD; K.S.), the National Cancer Institute (Bethesda, MD; K.S.), and the Howard Hughes Medical Institute (K.S., T.R.G., and S.M.C.).
National Institutes of Health
Howard Hughes Funding
Authorship
Contribution: S.M.C., G.R., and K.S. designed and performed experiments and wrote the manuscript; K.N.R. performed statistical analyses; K.T.C. and A.L.K. designed and conducted experiments; I.G., D.J.D., and R.M.S. facilitated access to primary patient samples; and T.R.G. designed experiments and wrote the manuscript. All authors read the manuscript and contributed to its final presentation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kimberly Stegmaier, Department of Pediatric Oncology, Dana 322A, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: kimberly_stegmaier@dfci.harvard.edu.
References
Author notes
*S.M.C. and G.R. contributed equally to this work.