Abstract
5-aza-2′-deoxycytidine (DAC) is approved for the treatment of myelodysplastic syndromes, but resistance to this agent is common. In search for mechanisms of resistance, we measured the half maximal (50%) inhibitory concentration (IC50) of DAC and found it differed 1000-fold among a panel of cancer cell lines. The IC50 was correlated with the doses of DAC that induced the most hypomethylation of long interspersed nuclear elements (LINE; R = 0.94, P < .001), but not with LINE methylation or DNA methyltransferase 1 (DNMT1), 3a, and 3b expression at baseline. Sensitivity to DAC showed a low correlation (R = 0.44, P = .11) to that of 5-azacytidine (AZA), but a good correlation to that of cytarabine (Ara-C; R = 0.89, P < .001). The 5 cell lines most resistant to DAC had a combination of low dCK, hENT1, and 2 transporters, and high cytosine deaminase. In an HL60 clone, resistance to DAC could be rapidly induced by drug exposure and was related to a switch from heterozygous to homozygous mutation of DCK. Transfection of wild-type DCK restored DAC sensitivity. DAC induced DNA breaks as evidenced by H2AX phosphorylation and increased homologous recombination rates by 7- to 10-fold. These results suggest that in vitro resistance to DAC can be explained by insufficient incorporation into DNA.
Introduction
Epigenetic changes have been increasingly recognized as a driving force in human leukemia.1-3 Abnormal methylation, for example, appears to accumulate over time at various sites in the genome and to promote tumorigenesis by increasing genomic instability or by silencing tumor suppressor genes. Silencing of tumor suppressor genes is closely associated with hypermethylation; methylated tumor suppressor genes can be reactivated by DNA methyltransferase (DNMT) inhibitors. These observations have led to a revival of interest in DNA methylation inhibitors as antineoplastic agents in clinical trials.4 The prototypical DNMT inhibitors 5-aza-2′-deoxycytidine (DAC) and 5-azacytidine (AZA) have recently been approved by the US Food and Drug Administration as antitumor agents for the treatment of myelodysplastic syndrome.5,6 One potential problem with both agents is that resistance can develop during treatment
Like other cytosine nucleoside analogs (NAs), DAC enters cells using the equilibrative nucleoside transporters hENT1 and hENT2. Once inside the cell, DAC is phosphorylated by deoxycytidine kinase (dCK) into the monophosphorylated derivative 5-aza-dCMP. Subsequently, 5-aza-dCMP is phosphorylated to its active form, 5-aza-dCTP, which is incorporated into DNA, where it induces demethylation. DAC metabolites might also be substrates for catabolizing enzymes such as cytidine deaminase (CDA), which catalyze the inactivation of cytidine and deoxycytidine to uridine and deoxyuridine, thereby decreasing the amount of 5-aza-dCTP that can be formed. The nucleoside analog Ara-C also relies on dCK as the initial rate-limiting step for incorporation. AZA, however, does not need dCK. Its incorporation is dependent on uridine-cytidine kinase (UCK).
dCK deficiency is the major known mechanism of resistance to cytidine NAs in vitro and was reported to be related to in vivo resistance in some patients.7,8 For example, in vitro–induced resistance to the deoxycytidine analogs cytarabine (Ara-C) and DAC in a rat model for acute myeloid leukemia was mediated by mutations in the DCK gene.9 However, there is little information regarding the origin of this kind of resistance. Resistance to treatment with anticancer drugs results from a variety of factors, including spontaneous genetic instability in tumors and drug induction mechanisms that probably play an important role in acquired anticancer drug resistance.10-16 Defects in DNA methylation might contribute to genomic instability, leading to elevated mutation rates.17-20 Therefore, we hypothesized that both spontaneous genetic instability and DAC-induced genetic instability contribute to the origin of resistance to DAC in human cancer cell lines and that resistance to DAC is due to the insufficient intracellular triphosphate of DAC. To test these hypotheses, we examined in vitro models of naturally occurring resistance to DAC in different cancer cell lines and further investigated the mechanisms and origin of resistance in the myeloid leukemia cell line HL60.
Methods
Cell culture and treatment protocols
The human leukemia and lymphoma cell lines HL60, ML-1, HEL, Raji, Jurkat, TF-1, U937, K562, and MOLT4; prostate cancer cell lines PC3 and DU145; colon cancer cell lines RKO and SW48; and breast cancer cell line Cama-1 were obtained from ATCC (Manassas, VA). The cells were grown in RPMI 1640 plus 10% heat-inactivated fetal calf serum (FCS) in plastic tissue-culture plates in a humidified atmosphere containing 5% CO2 at 37°C. For the growth inhibition assay, cells were placed at a density of 2.5 × 105/mL in 5 mL medium 24 hours before treatment. Graded concentrations of DAC were added to the medium. To measure half maximal (50%) inhibitory concentration (IC50), fresh DAC was added every 24 hours without changing medium. The doses that inhibited proliferation to 50% (IC50) were analyzed by the median-effect method with CalcuSyn software (Biosoft, Cambridge, United Kingdom).21 The proportion of live cells in treated plates was measured by trypan blue exclusion.
Pyrosequencing
We used the DNA repetitive long interspersed nuclear element (LINE) as a marker and pyrosequencing-based methylation analysis to study global genomic DNA methylation, as previously described.5 Genomic DNA was prepared from cells, and bisulfite conversion of genomic DNA was carried out. LINE was amplified by PCR using a forward primer 5′-TTTTTTGAGTTAGGTGTGGG-3′ and a 5′-biotinylated reverse primer 5′-TCTCACTAAAAAATACCAAACAA-3′. After PCR, the biotinylated reverse strand was captured on streptavidin Sepharose beads (Amersham Biosciences, Uppsala, Sweden) and annealed with the sequencing primer 5′-GGGTGGGAGTGAT-3′. To measure loss of heterozygosity (LOH) from a heterozygous mutation in exon 3 of DCK, we used a pyrosequencing-based analysis of a point mutation at nucleotide 454 of DCK mRNA (NM_000788) and an adjacent single nucleotide polymorphism (SNP) at nucleotide 459 (rs11544786). DCK exon 3 was amplified from genomic DNA and cDNA by PCR using a forward primer 5′-GGTGGGAATGTTCTTCAGA-3′ and a reverse primer 5′-AGCCATTTATACATACCTGTCAC-3′. The PCR product was used as a template and then amplified by a forward primer 5′-GGTGGGAATGTTCTTCAGA-3′, a reverse primer 5′-GGGACACCGCTGATCGTTTATTTAGCCATTTATACATACCTGTCAC-3′, and a 5′-biotinylated universal primer 5′-GGGACACCGCTGATCGTTTA-3′. The biotinylated strand was annealed with the sequencing primer 5′-TGGTCTTTTACCTTCCA-3′. Pyrosequencing was performed using PSQ HS 96 Gold SNP Reagents and the PSQ HS 96 pyrosequencing machine (Biotage, Uppsala, Sweden).
Measurement of resistance frequency
HL60 cells were plated on 6-well plates at 105 cells per well in the Iscove medium supplemented with 1% methylcellulose, 10% fetal bovine serum, 2 mM l-glutamine, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 10 μM DAC for selecting resistant colonies. The colonies were counted at 14 days. Resistance frequency was estimated as the percentage of resistant colonies divided by the plating efficiency in the medium without DAC.
Measurement of Ara-C triphosphate production
Ara-C triphosphate (Ara-CTP) production was measured as described previously.22 Briefly, HL60 cells were exposed to 10 mM [3H] Ara-CTP for 4 hours and centrifuged at 1000g for 10 minutes. Pellets were washed and extracted with trichloroacetic acid to remove proteins. The acidic extract was neutralized, and the aqueous layer was used for high-performance liquid chromatography (HPLC) separation. Ara-CTP was quantified at 262 nm by electronic integration with reference to external standards.
Measurement of dCK activity
dCK activity was measured as previously described.23 HL60 cells were centrifugated at 1000g for 10 minutes, resuspended in buffer A (50 mM potassium phosphate, pH 7.5, and 10 mM 2-mercaptoethanol), and sonicated. The suspension was centrifugated at 145 000g for 20 minutes. The supernatant was used as a crude extract after dialysis against 100 volumes of buffer A. We added 0.1 mg of the total protein to 100 μL of the reaction mixture (50 mM Tris-HCl, pH 7.8; 5 mM MgCl2, 8 mM UTP, and 25 μM [3H]Ara-C [4 Ci/mmol]) and then incubated the mixture at 37°C. At 10-minute intervals, 20-μL samples were removed (4 times) and spotted onto diethylaminoethyl (DEAE)–coated discs (DE-81; Whatman, Maidstone, United Kingdom), which were then washed 3 times for 10 to 15 minutes in 1 mM ammonium formate, twice with deionized H2O2 and once with 95% ethanol, and then dried. Radioactivity was counted in 10 mL Aquasol. Specific activities were expressed as disintegrations per minute (DPM) per milligram of protein.
Western blot analysis
For Western blot analysis, cell lysates were mixed with the same volume of 2× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA), boiled, and loaded onto 10% polyacrylamide gels containing sodium dodecyl sulfate. Proteins were transferred to polyvinylidene fluoride membranes. We used a rabbit anti–phosphohistone H2AX antibody (Sigma-Aldrich, St Louis, MO) and DNMT1, 3a, and 3b antibodies (Abcam, Cambridge, MA) and rabbit polyclonal to dCK (a generous gift from Dr Keszler Gergely at Semmelweis University, Budapest, Hungary).
DCK cDNA cloning and transfection
We cloned the full coding region of wild-type DCK into the BglII and EcoR1 restriction enzyme sites using a pEGFP-N1 vector (Clontech, Mountain View, CA). The full-length DCK cDNA was amplified by reverse transcription–polymerase chain reaction (RT-PCR) using the forward primer 5′-TATCTCAGATCTTTGCCGGACGAGCTCTG-3′ and the reverse primer 5′-ATTGAATTCTGGAACCATTTGGCTGCCTG-3′. We next transfected this vector into dCK-deficient HL60 cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. The antibiotic G418 at 0.5 mg/mL (American Bioanalytical, Natick, MA) was added to the cell culture medium to select for cells with stable integration of vector containing DCK. Mock transfection with empty pEGFP-N1 vector was used as a control.
Measurement of homologous recombination repair
To measure whether DAC can induce homologous recombination repair (HRR), we transfected pLNCX-GZ, pHit60, and pVSV-G vectors into 293T human embryonic kidney cells as described previously.24 Three days after transfection, the retrovirus-containing medium was collected. HL60 cells were incubated with dilutions of retroviral vector in RPMI 1640. The antibiotic G418 (0.5 mg/mL) was added to the cell culture medium to select for cells with stable integration of LNCX-GZ. We then treated the cells with 0.02, 0.2, 2, or 20 μM DAC for 4 days and maintained them in DAC-free medium containing G418 for 10 days. A total of 3 × 105 cells were added to 3 mL methylcellulose with G418 plus the antibiotic zeocin 200 μg/mL (Invitrogen, Carlsbad, CA) for selection. Cells were gently vortex-mixed to suspend them evenly, then plated onto 6-well plates. Colonies were counted at 14 days. The recombination frequency was estimated by taking the number of G418- and Zeocin-resistant colonies, dividing it by the total number loaded, and dividing again by the plating efficiency. Primers flanking the 2 green fluorescent protein (GFP)–zeocin cassettes were used to identify the expected recombination events. The forward primer was 5′-GCTAGCTTGCCAAACCTACAG-3′ and the reverse primer was 5′-GTGAACCGTCAGATCCGCTAG-3′. For a nonrecombined vector, the PCR fragment was 2.2 kb, whereas a 1.1-kb fragment was generated by a recombined vector.
Results
In this study, we investigated mechanisms of intrinsic resistance to DAC in a panel of cancer cell lines and acquired resistance in an HL60 cell line. First, we selected several leukemia cell lines because DAC is most active in leukemia, and also included colon, breast, and prostate cancer cell lines to represent solid tumors. We measured the IC50 of DAC, Ara-C, and AZA in the different cell lines. The IC50 of DAC was less than 0.05 μM in TF-1, U937, Raji, and HEL; between 0.05 and 0.4 μM in ML-1, HL-60, K562, SW48, and Cama-1; and greater than 2 μM in Jurkat, MOLT4, PC3, RKO, and DU145 that were defined as resistant cell lines (Table 1). The IC50 of DAC correlated with that of Ara-C (R = 0.89, P < .001), but not significantly with sensitivity to AZA (R = 0.44, P = .11; Figure 1A).
IC50 of DAC, AZA, and Ara-C in human cancer cell lines
| Cell line . | Tissue . | IC50, nM . | LINE methylation density % . | ||
|---|---|---|---|---|---|
| DAC . | AZA . | Ara-C . | |||
| TF-1 | Leukemia | 10 ± 2.1 | 76 ± 22 | 0.45 ± 0.15 | 28 |
| U93 7 | Lymphoma | 10 ± 1.8 | 281 ± 34 | 0.99 ± 0.24 | 62 |
| HEL | Leukemia | 40 ± 2.5 | 65 ± 12 | 0.65 ± 0.12 | 54 |
| Raji | Lymphoma | 54 ± 21 | 2332 ± 264 | 1.6 ± 0.32 | 75 |
| Cama-1 | Breast | 65 ± 7.0 | 280 ± 33 | 16 ± 1.5 | 30 |
| SW48 | Colon | 100 ± 9.1 | 101 ± 12 | 2.1 ± 0.21 | 62 |
| ML-1 | Leukemia | 98 ± 27 | 612 ± 55 | 1.7 ± 0.19 | 58 |
| HL60 | Leukemia | 200 ± 54 | 96 ± 37 | 3.7 ± 0.61 | 75 |
| K562 | Leukemia | 400 ± 34 | 102 ± 9.8 | 1.8 ± 0.25 | 12 |
| MOLT4 | Leukemia | 1802 ± 125 | 680 ± 112 | 10 ± 0.31 | 80 |
| Jurkat | Leukemia | 2000 ± 209 | 65 ± 15 | 17.2 ± 0.91 | 75 |
| PC3 | Prostate | 7501 ± 550 | 5600 ± 259 | 210 ± 22 | 44 |
| RKO | Colon | 9909 ± 980 | 2100 ± 147 | 20 ± 24 | 54 |
| DU145 | Prostate | 10 000 ± 660 | 2822 ± 330 | 300 ± 26 | 66 |
| Cell line . | Tissue . | IC50, nM . | LINE methylation density % . | ||
|---|---|---|---|---|---|
| DAC . | AZA . | Ara-C . | |||
| TF-1 | Leukemia | 10 ± 2.1 | 76 ± 22 | 0.45 ± 0.15 | 28 |
| U93 7 | Lymphoma | 10 ± 1.8 | 281 ± 34 | 0.99 ± 0.24 | 62 |
| HEL | Leukemia | 40 ± 2.5 | 65 ± 12 | 0.65 ± 0.12 | 54 |
| Raji | Lymphoma | 54 ± 21 | 2332 ± 264 | 1.6 ± 0.32 | 75 |
| Cama-1 | Breast | 65 ± 7.0 | 280 ± 33 | 16 ± 1.5 | 30 |
| SW48 | Colon | 100 ± 9.1 | 101 ± 12 | 2.1 ± 0.21 | 62 |
| ML-1 | Leukemia | 98 ± 27 | 612 ± 55 | 1.7 ± 0.19 | 58 |
| HL60 | Leukemia | 200 ± 54 | 96 ± 37 | 3.7 ± 0.61 | 75 |
| K562 | Leukemia | 400 ± 34 | 102 ± 9.8 | 1.8 ± 0.25 | 12 |
| MOLT4 | Leukemia | 1802 ± 125 | 680 ± 112 | 10 ± 0.31 | 80 |
| Jurkat | Leukemia | 2000 ± 209 | 65 ± 15 | 17.2 ± 0.91 | 75 |
| PC3 | Prostate | 7501 ± 550 | 5600 ± 259 | 210 ± 22 | 44 |
| RKO | Colon | 9909 ± 980 | 2100 ± 147 | 20 ± 24 | 54 |
| DU145 | Prostate | 10 000 ± 660 | 2822 ± 330 | 300 ± 26 | 66 |
Values are presented as the mean plus or minus SEM of 3 independent experiments.
Dose-dependent hypomethylation induction by DAC in different cell lines. (A) IC50 of DAC, AZA, and Ara-C in human cancer cell lines. We measured IC50 of DAC, AZA, and Ara-C in a panel of human cancer cell lines, and correlated IC50 of DAC versus IC50 of Ara-C, IC50 of DAC versus IC50 of AZA, respectively. (B) Dose-dependent hypomethylation induction by DAC in different cell lines. After treatment with DAC for 4 days, cells were collected, and DNA was extracted. LINE methylation was measured by bisulfite pyrosequencing analysis. In each cell line, except the most resistant cells (bottom graph), the dose-dependent curve was U-shaped. (B) Absence of correlation of the IC50 of DAC with LINE methylation at baseline (R = 0.05, P = .97). (C) Correlation between the IC50 of DAC with the doses of DAC required for the maximum hypomethylation of LINE (R = 0.94, P < .001).
Dose-dependent hypomethylation induction by DAC in different cell lines. (A) IC50 of DAC, AZA, and Ara-C in human cancer cell lines. We measured IC50 of DAC, AZA, and Ara-C in a panel of human cancer cell lines, and correlated IC50 of DAC versus IC50 of Ara-C, IC50 of DAC versus IC50 of AZA, respectively. (B) Dose-dependent hypomethylation induction by DAC in different cell lines. After treatment with DAC for 4 days, cells were collected, and DNA was extracted. LINE methylation was measured by bisulfite pyrosequencing analysis. In each cell line, except the most resistant cells (bottom graph), the dose-dependent curve was U-shaped. (B) Absence of correlation of the IC50 of DAC with LINE methylation at baseline (R = 0.05, P = .97). (C) Correlation between the IC50 of DAC with the doses of DAC required for the maximum hypomethylation of LINE (R = 0.94, P < .001).
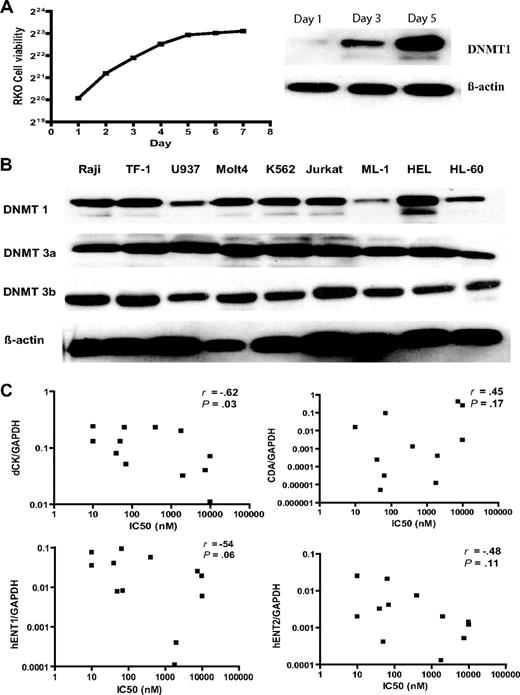
In searching for mechanisms of resistance to DAC, we first asked whether it correlated with DNA methylation. Global methylation using the DNA repetitive element LINE as a marker was measured by pyrosequencing-based analysis. There was a great variation in LINE methylation at baseline (from 12% in K562 to 79% in the MOLT4 cell line) as previously reported25 (Table 1). We next treated all cell lines with DAC at 0.03, 0.1, 0.3, 1, 3, 10, and 30 μM daily for 4 days and measured hypomethylation induction by the LINE assay. In each cell line, the hypomethylation dose response was U shaped, as previously reported,5 presumably because high doses of DAC inhibit proliferation, which is essential for hypomethylation. The doses of DAC that induced peak hypomethylation varied from 0.03 μM in U937 and TF-1, the most sensitive cell lines; to 0.3 μM in HEL, Raji, ML-1, HL60, SW48, and Cama-1, sensitive cell lines; to 1 and 3 μM in relatively resistant cell lines Jurkat and MOLT4, respectively; and to 30 μM in the most resistant cell lines, RKO and PC3 (Figure 1A). The IC50 of DAC was closely correlated with the doses that induced peak hypomethylation of LINE (R = 0.94, P < .001) but not with LINE methylation at baseline (R = 0.05, P = .97; Figure 1B,C). We next measured expression of DNMT1, 3a, and 3b proteins by Western blot analysis. DNMT1 protein is cell replication–dependent. Cells in the rapid growth phase expressed more DNMT1 protein than those in the slow growth phase (Figure 2A). For example, in RKO, DNMT1 level was high on day 1 and gradually decreased on days 3 and 5. We found that DNMT1, 3a, and 3b protein expression in log-phase growing cells was not correlated with LINE methylation at baseline (Figure 2B) and was not correlated with the IC50 of DAC.
Pharmacologic mechanisms of resistance to DAC. (A) DNMT1 protein expression was cell replication–dependent. We measured RKO cell growth curve, and DNMT1 protein expression on days 1, 3, and 5 by Western blot analysis. β-Actin was used as a control. (B) DNMT1, 3a, and 3b protein expression was independent of sensitivity to DAC and LINE methylation in different cancer cell lines. We collected exponentially growing cancer cells, extracted protein, and performed Western blot analysis of DNMT1, 3a, and 3b. β-Actin was used as a control. (C) dCK protein expression in several cell lines. dCK protein expression was measured by Western blot analysis. (D) Correlation of different nucleoside metabolic gene expression with the IC50 of DAC. DCK, CDA, hENT1, and hENT2 expressions were measured by real-time PCR using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a control. R and P values reflect Spearman correlation analysis of the IC50 of DAC with the relative gene expression.
Pharmacologic mechanisms of resistance to DAC. (A) DNMT1 protein expression was cell replication–dependent. We measured RKO cell growth curve, and DNMT1 protein expression on days 1, 3, and 5 by Western blot analysis. β-Actin was used as a control. (B) DNMT1, 3a, and 3b protein expression was independent of sensitivity to DAC and LINE methylation in different cancer cell lines. We collected exponentially growing cancer cells, extracted protein, and performed Western blot analysis of DNMT1, 3a, and 3b. β-Actin was used as a control. (C) dCK protein expression in several cell lines. dCK protein expression was measured by Western blot analysis. (D) Correlation of different nucleoside metabolic gene expression with the IC50 of DAC. DCK, CDA, hENT1, and hENT2 expressions were measured by real-time PCR using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a control. R and P values reflect Spearman correlation analysis of the IC50 of DAC with the relative gene expression.
Because of the observed correlation between the IC50 of DAC and Ara-C, which share intracellular metabolic pathways, we investigated the expression of several genes involved in nucleoside metabolism. dCK protein expression was very low in the DAC-resistant DU145 and Jurkat cell lines (Figure 2C), which was due to low dCK mRNA expression by real-time PCR (Figure 2D). The IC50 of DAC was inversely correlated with expression of dCK mRNA expression (R = −0.63, P = .038) and also showed a tendency to be inversely correlated with the nucleoside transporter hENT1 mRNA (R = −0.54, P = .068; Figure 2D). hENT1 mRNA was the lowest in the DAC-resistant MOLT4 cell line. The DAC-inactivating enzyme CDA mRNA was the highest in PC3 and DU145, respectively. Thus, 4 of the 5 cell lines most resistant to DAC have measurable alterations in one of these genes, while none of the 9 most sensitive cell lines had any such alterations. In contrast, the IC50 of AZA was not correlated with expression of DCK, CDA, hENT1, and hENT2, respectively (data not shown). Interestingly, the Raji cell line was resistant to AZA, but sensitive to DAC and Ara-C. DCK promoter is a CpG-rich region, which can be silenced by DNA hypermethylation. We next measured DCK methylation level in all these cell lines by pyrosequencing analysis, but could not detect any aberrant hypermethylation even in DU145 and Jurkat with the lowest gene expression (data not shown). In addition, alternative splicing of DCK was not not observed in resistant clones (data not shown). It is likely that reduced DCK mRNA expression in several cell lines, such as DU145 or Jurkat, might be caused by other mechanisms such as histone deacetylation or microRNAs targeting DCK.
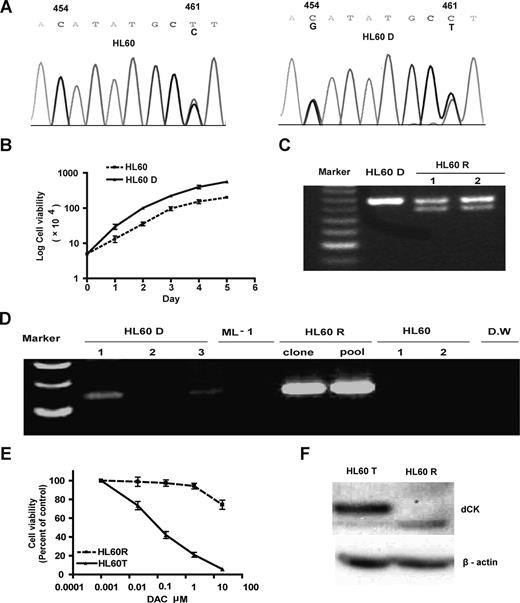
The data described above relate to natural resistance to DAC (and AZA). To study this problem in a model of induced resistance, we treated HL60 with DAC, selected resistant colonies (HL60R) in methylcellulose medium, and found a frequency of resistance of 1.2 × 10−4. We replated 11 DAC-resistant colonies and found that resistance was stable and could last for more than 2 years even when cultured in the absence of DAC. To investigate mechanisms of induced resistance to DAC in these clones, we measured DAC hypomethylation induction of LINE-repetitive elements. The hypomethylation induced by low doses of DAC (0.2 and 2 μM) in HL60D cells was almost lost in the DAC-resistant cells (Figure 3A). To study whether this resistance affects gene expression induction by DAC, we measured mRNA for RIL, a tumor suppressor gene that is silenced in HL60 by DNA hypermethylation.5 RIL reactivation by DAC was also inhibited in HL60R (Figure 3B). In contrast, AZA inhibited cell growth (Figure 3C) and induced LINE demethylation (Figure 3D) in HL60D and HL60R similarly.
dCK deficiency in resistant HL60R cells. (A) DAC hypomethylation induction in HL60D and DAC-resistant HL60R. We treated the cells with DAC (0.2-50 μM) and measured LINE methylation by bisulfite pyrosequencing analysis. (B) RILgene expression. (C) Inhibition of AZA of cell growth. Cells were treated with AZA (0.25-25 μM), and cell viability was measured by trypan blue exclusion. (D) LINE hypomethylation after AZA treatment. (E) Ara-CTP production, as measured by HPLC analysis using [3H] Ara-C as a substrate, was lost in HL60R-resistant cells. (F) dCK activity, as measured by phosphorylation of [3H]Ara-C in cell extracts, was also lost in resistant cells. (G) dCK protein expression was measured by Western blot analysis. β-Actin served as a control. (H) DCK mRNA expression was measured by quantitative PCR.
dCK deficiency in resistant HL60R cells. (A) DAC hypomethylation induction in HL60D and DAC-resistant HL60R. We treated the cells with DAC (0.2-50 μM) and measured LINE methylation by bisulfite pyrosequencing analysis. (B) RILgene expression. (C) Inhibition of AZA of cell growth. Cells were treated with AZA (0.25-25 μM), and cell viability was measured by trypan blue exclusion. (D) LINE hypomethylation after AZA treatment. (E) Ara-CTP production, as measured by HPLC analysis using [3H] Ara-C as a substrate, was lost in HL60R-resistant cells. (F) dCK activity, as measured by phosphorylation of [3H]Ara-C in cell extracts, was also lost in resistant cells. (G) dCK protein expression was measured by Western blot analysis. β-Actin served as a control. (H) DCK mRNA expression was measured by quantitative PCR.
In the resistant cells, DAC induced the most hypomethylation at the high dose of 50 μM, suggesting that resistance could be due to decreased incorporation into DNA. Consistent with this, we found these resistant cells to be also resistant to Ara-C (data not shown) and speculated that resistance was due to deficient NA incorporation into DNA. Therefore, we next measured Ara-CTP concentration by HPLC-based analysis using [3H]Ara-C as a substrate and found that Ara-CTP production in DAC-derived resistant clones was 50-fold lower than that in wild-type cells (Figure 3E). dCK activity was 10-fold lower than that in wild-type cells using [3H]Ara-C as a substrate (Figure 3F). In searching for causes of this loss of activity, we found that dCK protein expression was markedly reduced in HL60R (Figure 3G), but DCK mRNA expression in HL60R was similar to that in wild-type cell lines (Figure 3H). Thus, induced resistance to DAC in HL60 was correlated with dCK loss.
In search of a mechanism for dCK loss in HL60-resistant cells, we first sequenced DCK from cDNA and genomic DNA from the parental HL60 cells that were used to generate resistant clones. We found a heterozygous point mutation in codon 98 (ACA to AGA), resulting in a Thr/Arg change. This sequence in the parental HL60 cells designated as HL60D was absent in another batch of HL60 cells obtained from ATCC, designated as HL60 (Figure 4A). In addition, both HL60D and HL60 cells had a heterozygous synonymous SNP in codon 100 (GCC to GCT). We found that HL60D had a growth advantage over HL60, suggesting that this point mutation occurred in a subpopulation of HL60 cells (Figure 4B). Importantly, neither the point mutation nor the adjacent SNP were seen in more than 100 alleles in other investigated cell lines and normal cells. We next sequenced dCK cDNA and genomic DNA in the resistant cells HL60R (derived from HL60D), looking for additional mutations. Among the 11 resistant clones, 3 sequenced HL60R clones had exon 3 LOH of the wild-type allele, leaving behind exclusively the allele containing the AGA point mutation at codon 98 and GCC at codon 100. Eight clones had a heterozygous deletion in exon 1 between nucleotides 154 and 197 of DCK mRNA that abolished the ATG site (Table 2 and Figure 4). To determine whether this heterozygous deletion lead to biallelic mutation, we performed TA cloning (Invitrogen) and sequencing of DCK cDNA from exon 1-4. We found 2 mutant DCK alleles: the allele with a point mutation at codon 98 and another allele with a 42-bp deletion in exon 1. To determine whether the mutations were spontaneous and preexisting, we designed PCR primers spanning the deleted region of DCK exon 1 and successfully amplified that region from parental HL60D cells (Figure 4D), thus demonstrating that resistance was preexisting and spontaneous.
Spontaneous origin of resistance to DAC. (A) Two subclones of the HL60 cell line. HL60D developed a heterozygous 454C>G point mutation of DCK. This mutation was absent in another batch of HL60 obtained from ATCC. (B) The HL60D cell line had a growth advantage over the HL60 cell line. Cell number was counted from day 1 to 7. (C) Heterozygous deletion in exon 1 of DCK in HL60R cells. (D) Deletion was preexisting in HL60D cells and absent in HL60 cells. We designed a set of primers that spanned the deleted region and amplified that region from parental HL60D cells. (E) HL60R cells transfected with DCK restored sensitivity to DAC. We transfected wild-type DCK into HL60R and selected stably transfected cells by G418. HL60R and DCK-transfected (HL60T) cells were treated with DAC at 0.02, 0.2, 2, and 20 μM, respectively. Cell viability was counted. (F) Transfection of DCK cDNA restored dCK protein expression. dCK protein expression was measured in HL60R and HL60T cells by Western blot analysis.
Spontaneous origin of resistance to DAC. (A) Two subclones of the HL60 cell line. HL60D developed a heterozygous 454C>G point mutation of DCK. This mutation was absent in another batch of HL60 obtained from ATCC. (B) The HL60D cell line had a growth advantage over the HL60 cell line. Cell number was counted from day 1 to 7. (C) Heterozygous deletion in exon 1 of DCK in HL60R cells. (D) Deletion was preexisting in HL60D cells and absent in HL60 cells. We designed a set of primers that spanned the deleted region and amplified that region from parental HL60D cells. (E) HL60R cells transfected with DCK restored sensitivity to DAC. We transfected wild-type DCK into HL60R and selected stably transfected cells by G418. HL60R and DCK-transfected (HL60T) cells were treated with DAC at 0.02, 0.2, 2, and 20 μM, respectively. Cell viability was counted. (F) Transfection of DCK cDNA restored dCK protein expression. dCK protein expression was measured in HL60R and HL60T cells by Western blot analysis.
Summary of resistance to DAC in HL60D cells
| Group . | Selected resistant clones . | DAC resistance frequency . | Mutation . | ||
|---|---|---|---|---|---|
| LOH . | Deletion . | Frameshift . | |||
| HL6O | 0 | Undetected | 0 | 0 | 0 |
| HL6OD (pool) | 11 | 1.2 × 10−4 | 3 | 8 | 0 |
| HL6OD (clone) | 9 | 1.2 × 10−5 | 7 | 0 | 2 |
| Group . | Selected resistant clones . | DAC resistance frequency . | Mutation . | ||
|---|---|---|---|---|---|
| LOH . | Deletion . | Frameshift . | |||
| HL6O | 0 | Undetected | 0 | 0 | 0 |
| HL6OD (pool) | 11 | 1.2 × 10−4 | 3 | 8 | 0 |
| HL6OD (clone) | 9 | 1.2 × 10−5 | 7 | 0 | 2 |
To eliminate preexisting DAC-resistant clones, we used limited dilution to isolate individual cell-derived colonies of HL60D. After each colony expanded to approximately 106 cells, we selected for DAC-resistant clones in methylcellulose medium and obtained 9 resistant clones. The average frequency of resistance in these cells was 1.2 × 10−5 (Table 2). We measured allelic status of DCK by pyrosequencing analysis using genomic DNA and found that of 9 single resistant colonies, 7 had LOH of DCK, resulting in a single mutant allele with Arg in codon 98 in exon 3. Two clones had a single base insertion causing a frameshift (TA/TAA) in exon 6 of DCK (Table 2). cDNA cloning and sequencing confirmed that in each case, the frameshift mutation was in the allele unmutated at exon 3. LOH of DCK is therefore the most common mechanism that gives rise to induced resistance to DAC in HL60. To confirm that resistance is caused by DCK loss, we cloned wild-type DCK into a pEGFP-N1 vectors and transfected the vector into HL60R. We selected stably G418-resistant cells, designated as HL60T. Transfection of DCK restored dCK protein expression and sensitivity to DAC (Figure 4E,F), suggesting that DAC resistance was due to functional loss of dCK, which was due to LOH of the DCK locus.
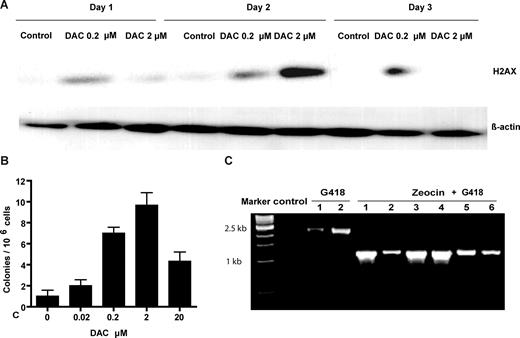
LOH can be spontaneous or can be induced by DNA damage. We reasoned that LOH in HL60R cells might be related to DNA breaks induced by DAC. We therefore measured the induction of phosphorylation of histone H2AX after DAC treatment of HL60D cells. Western blot analysis showed that treatment with DAC at 0.2 and 2 μM induced H2AX phosphorylation (Figure 5A). The phosphorylation induced by the higher concentration disappeared on day 3, possibly due to cell death, whereas the lower concentration maintained this phosphorylation on day 3. LOH at an autosomal locus is one outcome of the repair of DNA double-stranded breaks by HRR. To investigate whether DAC treatment increases the rate of HRR, we conducted retroviral transfection of a PLNCX-GZ vector24 into HL60D cells. In this assay, recombination between the tandem repeats regenerates a functional GFP-ZeoR marker that can be easily scored. The HRR rate of HL60D cells was 1.4 × 10−6. Treatment with 0.02, 0.2, 2, and 20 μM DAC increased HRR rates approximately 1.9-, 6.8-, 9.5-, and 4.1-fold, respectively (Figure 5B). To confirm that resistant colonies were recombinant, we isolated high molecular weight DNA from expanded G418 plus zeocin-resistant colonies and performed PCR with primers flanking the 2 GFP-zeocin cassettes that can identify the expected recombination events. We randomly picked 6 colonies after zeocin plus G418 selection and noted that all the colonies had a 1.1-kb fragment, confirming recombination. On the other hand, colonies after G418 selection only had an unrecombined 2.2-kb fragment (Figure 5C).
DAC-induced origin of DAC resistance. (A) Induction of the phosphorylation of the histone H2AX by DAC treatment. HL60D cells were treated with DAC (0.2 and 2 μM) for 3 days, and H2AX phosphorylation was measured by Western blot analysis. β-Actin was used as a control. (B) DAC treatment increased HRR rates. Cells with stable PLNCX-GZ transfection were treated with DAC for 4 days and maintained in drug-free medium for 1 week. Colonies resistant to Zeocin were selected and maintained in methylcellulose medium for 2 weeks. HRR rates were calculated as described in “Measurement of homologous recombination repair.” (C) Verification of intrachromosomal recombination in zeocin-resistant colonies by PCR with recombination-specific primers. PCR-amplified DNA from G418-resistant colonies produced a 2.2-kb fragment diagnostic for the nonrecombined tandem repeats. PCR-amplified DNA from G418- and zeocin-resistant colonies generated a diagnostic 1.1-kb fragment.
DAC-induced origin of DAC resistance. (A) Induction of the phosphorylation of the histone H2AX by DAC treatment. HL60D cells were treated with DAC (0.2 and 2 μM) for 3 days, and H2AX phosphorylation was measured by Western blot analysis. β-Actin was used as a control. (B) DAC treatment increased HRR rates. Cells with stable PLNCX-GZ transfection were treated with DAC for 4 days and maintained in drug-free medium for 1 week. Colonies resistant to Zeocin were selected and maintained in methylcellulose medium for 2 weeks. HRR rates were calculated as described in “Measurement of homologous recombination repair.” (C) Verification of intrachromosomal recombination in zeocin-resistant colonies by PCR with recombination-specific primers. PCR-amplified DNA from G418-resistant colonies produced a 2.2-kb fragment diagnostic for the nonrecombined tandem repeats. PCR-amplified DNA from G418- and zeocin-resistant colonies generated a diagnostic 1.1-kb fragment.
Finally, we investigated whether rapid induction of DAC resistance was common or whether it was a specific feature of HL60D cells heterozygous for DCK mutation. DAC treatment and methycellulose selection yielded no stable resistant clones in HL60 cells with 2 wild-type alleles of DCK or in ML-1, a cell line with normal dCK.
Discussion
In this study, we investigated naturally occurring resistance to DAC in vitro in a panel of cancer cell lines and found 1000-fold difference in IC50 values. These differences are likely due to pharmacologic mechanisms resulting in lower incorporation of DAC into DNA in resistant cells. Therefore, higher doses of DAC were required to induce maximum hypomethylation in resistant cells. Furthermore, we found that both spontaneous genetic instability and DAC-induced genetic instability contributed to the origin of resistance to DAC in an HL60D cells, harboring a missense point mutation in exon 3 of DCK.
Most in vitro resistance to NA results from lack of incorporation into DNA.26 Consistent with this, we found that resistance to DAC was correlated with multiple pharmacologic mechanisms of aberrant nucleoside metabolism, which result in less incorporation of DAC into DNA, but this was not directly proven by measuring DAC incorporation. It remains experimentally very difficult to answer this because DAC incorporation is at low levels. DAC has a short half-life (minutes) and is quickly deaminated by CDA once inside cells. It is also chemically unstable upon DNA extraction and HPLC analysis. The 5,6 bond in cytosine is very labile.27,28 Nevertheless, in HL60, we demonstrated that DAC resistance is caused by loss of DCK, and DAC sensitivity is highly correlated with Ara-C sensitivity, which shares NA metabolism. The AraCTP experiments are indirect, but they are a very good approximation of dCK activity. Thus, the data in favor of incorporation as a mechanism of resistance are strong. Clearly, however, multiple mechanisms may be active in different cells.
DAC exerts its effect through induction of hypomethylation at low doses and cytotoxicity at high doses. DAC induced a U-shaped curve of hypomethylation in a panel of cancer cell lines. This might be explained by the fact that low-dose DAC can covalently trap DNA methytransferases without cell-cycle arrest, whereas high dose DAC inhibits DNA synthesis and induces cell-cycle arrest, leading to less hypomethylation induction. Clinical usage of the agent is mostly at low doses, however.4 Interestingly, resistance to DAC was unrelated to DNMT levels and LINE methylation, a marker of global DNA methylation. Cancers have gene-specific hypermethylation and global hypomethylation.29 Our data suggest that differences in the methylation levels of repetitive DNA elements (reflecting global methylation) are not simply explained by DNMT levels and do not impact on sensitivity to DAC. Rather, DAC hypomethylation induction and gene reactivation are impaired in resistant cells by insufficient incorporation of DAC into DNA. A potential question is whether downstream pathways that led to gene reactivation are related to resistance to DAC, providing adequate drug incorporation. The fact that DAC treatment activates thousands of genes, not all of which are silenced by promoter hypermethylation,30 makes this kind of study difficult. However, in this panel of cell lines, the cross-resistance between DAC and Ara-C and the lack of cross-resistance between DAC and AZA suggests that resistance to gene reactivation perhaps is less important than DAC incorporation. Nevertheless, resistance to DAC is likely to be multifactorial in a specific cell line. For example, DU145 demonstrated low levels of DCK, hENT1, and high levels of CDA. The doses needed for the maximum hypomethylation are the best marker for determining the sensitivity or resistance to DAC based on our results.
In our studies, we were able to ascribe resistance to 2 potential mechanisms: (1) preexisting genetic instability and selection for resistant clones and (2) DAC-induced genetic instability. Most malignant cell populations are characterized by “genetic instability” that can be shown to be directly involved in the generation of phenotypic drug resistance. Using fluctuation analysis, researchers have demonstrated random and spontaneous origins of resistance phenotypes for some antineoplastic agents, such as topoisomerase II inhibitor, folic acid antagonists, and antibiotic agents.10-16 In this study, we amplified mutations preexisting in rare resistant cells from a pool of parental cells by PCR, providing the most direct evidence of a spontaneous origin of resistance to DAC and other NAs. On the other hand, NAs affect the structural integrity of DNA, leading to stalled replication forks and chain termination. The DNA damage sensors ATM, ATR, and DNA-PK recognize these events and signal for DNA repair.31 We observed that DAC treatment induced histone H2AX phosphorylation, a marker of double-strand DNA breaks, and increased the rate of HRR, which might give rise to LOH of DCK and resistance to DAC. Several cytostatic drugs, including aphidicolin, Ara-C, hydroxyurea, and methotrexate, induce HRR through inhibition of DNA synthesis.32
Sensitivity to DAC in our studies was closely related to sensitivity to Ara-C, but not to AZA except in the PC3 cell line, which had high CDA levels capable of deaminating all these drugs into inactive forms. DAC and Ara-C need the same dCK enzyme for initial phosphorylation. Instead, AZA uses UCK for initial phosphorylation. Therefore, DAC resistance related to dCK loss does not lead to AZA resistance, and this lack of cross-resistance could be exploited therapeutically. Indeed, a recent study has shown that some patients can respond to DAC after showing clinical resistance to AZA.33
The relevance of our findings to in vivo resistance need to be examined. Low levels of DCK gene expression or low dCK activity were correlated with a poor response to Ara-C and cladribine (2-chlorodeoxyadenosine) in childhood acute lymphoblastic leukemias (ALLs) and lymphoproliferative disorders,34,35 and to gemcitabine (a cytosine analog) in advanced pancreatic cancer.36 However, not all studies were consistent in this regard.37,38 Our combined data suggest that the best approach to examining in vivo resistance to DAC would be to measure DAC incorporation into DNA, an experiment that is not technically feasible at present because of the low doses of DAC used clinically. An alternate approach may be to measure all parameters of DAC metabolism (eg, dCK, hENT, CDA) in treated patients and correlate them with sensitivity to the drug.
In conclusion, this study provides important in vitro models for understanding the mechanisms and origin of resistance to DAC, which may lead to strategies to overcome DAC resistance in vivo. The lack of cross-resistance between DAC and AZA in some cells could also have important therapeutic implications.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr. William Plunkett and Billie Nowak for general advice and help in the measurement of Ara-CTP and dCK activity, Drs Lu Xiongbin and Lawrence A. Donehower (Baylor College of Medicine, Houston, TX) for providing the PLNCX-GZ, pVSV-G, and pHit60 vectors for studying homologous recombination, and Dr Gergely Keszler at Semmelweis University (Budapest, Hungary) for providing a dCK antibody.
This work was supported by National Institutes of Health grants CA100632 and CA108631 and by the Rosalie B. Hite Graduate Fellowship at University of Texas Health Science Center at Houston.
National Institutes of Health
Authorship
Contribution: T.Q. performed research; J.J. and J.-P.J.I. designed research; and J. Si and J. Shu analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Pierre J. Issa, Department of Leukemia, The University of Texas M. D. Anderson Cancer Center, Houston, TX; e-mail: Jpissa@mdanderson.org.


![Figure 3. dCK deficiency in resistant HL60R cells. (A) DAC hypomethylation induction in HL60D and DAC-resistant HL60R. We treated the cells with DAC (0.2-50 μM) and measured LINE methylation by bisulfite pyrosequencing analysis. (B) RILgene expression. (C) Inhibition of AZA of cell growth. Cells were treated with AZA (0.25-25 μM), and cell viability was measured by trypan blue exclusion. (D) LINE hypomethylation after AZA treatment. (E) Ara-CTP production, as measured by HPLC analysis using [3H] Ara-C as a substrate, was lost in HL60R-resistant cells. (F) dCK activity, as measured by phosphorylation of [3H]Ara-C in cell extracts, was also lost in resistant cells. (G) dCK protein expression was measured by Western blot analysis. β-Actin served as a control. (H) DCK mRNA expression was measured by quantitative PCR.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/3/10.1182_blood-2008-02-140038/5/m_zh80020929070003.jpeg?Expires=1765892143&Signature=pzzTBv0gstgObIRBMQUWROSQhHWEgXqUfS4BvNzjQ2X8t2CiWx3oY1EXiP65SAhI9oFWVjMmUr9FcIjSNH77vg6NeEF9eLx10HhtCleqxk01jWC3ln9bZ8qYf~NtXFeXYqp8WWj4n4vQaZUgP7LzTR4X720S-Y2vAOjPuYphP6m4j47uGeO4hEK7VhOj78iyD0d35vD1K9s9j4RCaytUgmkeWHpbpjj6xEjqny3xd5B1WyKWG2~dOkOv0pnoknCEUCtRFRpdSYsHCZ4qhc8QgBsXLRUdDa5ax6f~P0Asxb2PdR-aYSTRTyNIWpXVqkeAG4MOw-kDRdSZzBPbh8HTEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal